Abstract
Antimicrobial drug resistance can limit the ability to effectively treat patients. Numerous factors have been proposed to impact the development of antimicrobial resistance, including those specific to the drug and the dosing regimen. The field of investigation that examines the relationship between dosing regimen and outcome is termed antimicrobial pharmacokinetics and pharmacodynamics. Our prior in vivo investigations examined the relationship between fluconazole pharmacodynamics and the modulation of isogenic resistant and susceptible Candida albicans populations in a mixed-inoculum design (1). The goal of the current studies was to examine the impact of fluconazole pharmacodynamics on resistance emergence from a susceptible parent population over time using a murine systemic-candidiasis model. Both microbiologic and transcriptional endpoints were examined during the evolution of cell populations. As in our previous investigation, the more frequently administered dosing regimen prevented the emergence of a resistant cell phenotype. Conversely, dosing regimens that produced prolonged sub-MIC concentrations were associated with resistance development. The studies also demonstrated a striking relationship between fluconazole pharmacodynamic exposures and the mRNA abundance of drug resistance-associated efflux pumps. Global transcriptional profiling of cell populations during the progressive emergence of a resistance phenotype provides insight into the mechanisms underlying this complex physiologic process.
Antimicrobial resistance has become an increasingly serious public health problem in a wide range of infectious diseases (5, 15, 24, 26, 34, 42, 47, 48). Thus, it is imperative to understand the factors that lead to the evolution of resistance and to design strategies to prevent or delay the emergence of antimicrobial-resistant pathogens.
It is well accepted that antimicrobial exposure is correlated with the prevalence of antimicrobial resistance (7, 8, 13, 16, 17, 19, 21). However, the relationship in the context of use pattern and resistance development is complex and remains for the most part undefined. A recent study examined these relationships for the fungistatic triazole antifungal fluconazole in an in vivo candidiasis model (1). The experimental model utilized isogenic strain pairs of Candida albicans in which the parent population was fluconazole susceptible and the other isolate was drug resistant. The treatment studies examined modulation of the resistant and susceptible cell populations from an initial inoculum consisting of a small percentage of the less susceptible group of cells. Study of the relationship between fluconazole pharmacokinetics and amplification of each cell population was undertaken with six strain pairs. Fluconazole dosing regimens producing prolonged sub-MIC exposures or a brief period of time in which drug concentrations exceeded the MIC (%T > MIC) resulted in the selection of resistant cells in each of the experiments.
The current studies further examined the relationship between the dose level and dosing frequency with fluconazole and resistance development in an in vivo test system. The goals of the experiments used in these studies included defining the exposure conditions that prevent the emergence of a resistant cell phenotype from a previously sensitive population over time. In addition, the experiments involved examination of the transcriptional profiles of these cell populations in order to begin to understand the relationship between antimicrobial pharmacodynamics and gene expression for this drug-pathogen combination.
MATERIALS AND METHODS
Microorganisms and in vitro susceptibility testing.
The starting parent Candida albicans strain, K1, is a clinical isolate from a patient with disseminated candidiasis and endophthalmitis (3). The parent strain was chosen both because it is fluconazole susceptible and because it has been used extensively in prior triazole pharmacodynamic studies. Eighty cell populations evolved in vivo in response to various antimicrobial exposures over time were catalogued for further phenotypic and genotypic evaluation (Table 1). Susceptibility testing was performed in duplicate on two occasions using CLSI (formerly NCCLS) M-27A methods (32). The final results were expressed as the geometric mean of these results.
TABLE 1.
Fluconazole treatment regimens and population MIC for each regimen over time
| Study | MIC (μg/ml)a for treatment regimen:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| A | 1A | 2A | 3A | 4A | 5A | 6A | 7A | 8A |
| 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 1.0 | 0.5 | |
| B | 1B | 2B | 3B | 4B | 5B | 6B | 7B | 8B |
| 1.0 | 0.5 | 0.5 | 0.5 | 0.5 | 1.0 | 1.0 | 1.0 | |
| C | 1C | 2C | 3C | 4C | 5C | 6C | 7C | 8C |
| 2.0 | 0.5 | 0.5 | 1.0 | 0.5 | 2.0 | 1.0 | 1.0 | |
| D | 1D | 2D | 3D | 4D | 5D | 6D | 7D | 8D |
| 1.0 | 0.5 | 0.5 | 2.0 | 0.25 | 2.0 | 1.0 | 1.0 | |
| E | 1E | 2E | 3E | 4E | 5E | 6E | 7E | 8E |
| 2.0 | 0.5 | 0.5 | 2.0 | 0.5 | 4.0 | 1.0 | 0.5 | |
| F | 1F | 2F | 3F | 4F | 5F | 6F | 7F | 8F |
| 2.0 | 0.5 | 1.0 | 2.0 | 1.0 | 4.0 | 1.0 | 0.5 | |
| G | 1G | 2G | 3G | 4G | 5G | 6G | 7G | 8G |
| 4.0 | 0.5 | 0.5 | 2.0 | 1.0 | 4.0 | 2.0 | 0.5 | |
| H | 1H | 2H | 3H | 4H | 5H | 6H | 7H | 8H |
| 4.0 | 0.5 | 0.5 | 8.0 | 0.5 | 8.0 | 1.0 | 1.0 | |
| I | 1I | 2I | 3I | 4I | 5I | 6I | 7I | 8I |
| 4.0 | 0.5 | 0.5 | 8.0 | 0.5 | 8.0 | 2.0 | 1.0 | |
| J | 1J | 2J | 3J | 4J | 5J | 6J | 7J | 8J |
| 4.0 | 0.5 | 0.5 | 8.0 | 0.5 | 8.0 | 1.0 | 1.0 | |
Parent K1 MIC, 0.5 μg/ml. Parent K1 MIC, 0.5 μg/ml. Archived isolates and MICs are shown.
Inoculum preparation.
For the initial infection, C. albicans K1 was grown to log phase in YPD (1% yeast extract, 2% Bacto Peptone, and 2% glucose), and the inoculum was adjusted to a cell concentration of 5.5 log10 CFU/ml with a hemacytometer. Viable counts were confirmed by plate counts. The inoculum for the remaining archive treatment studies was prepared from the entire viable population (isolated on Sabouraud dextrose agar [SDA] plates) from the prior treatment period.
Animal infection model.
A neutropenic murine disseminated-candidiasis model was used for all studies. The model has been described in detail in previous publications (3, 4). The disseminated-candidiasis model was chosen for these studies to allow consistent comparison to prior fluconazole pharmacodynamic studies. Briefly, 6-week-old specific-pathogen-free female CD1 mice weighing 23 to 27 g were rendered neutropenic prior to infection by intraperitoneal injection of cyclophosphamide at 4 days (150 mg/kg of body weight) and 1 day (100 mg/kg). The cyclophosphamide protocol resulted in profound reduction in neutrophil levels (<100/mm3) throughout the period of study, allowing adequate growth of Candida albicans in the mice (Harlan Sprague-Dawley, Indianapolis, IN) (1, 2). The inoculum was injected via the lateral tail veins of neutropenic mice. Groups of two animals were used for each treatment regimen. We chose the kidney as the initial end organ of study, as it is the most extensively characterized in vivo infection model for this pathogen (1, 2). The kidneys were processed for CFU enumeration in duplicate. The lower limit of detection for both susceptible and resistant cells was 100 CFU/kidney. The results were expressed as the mean number of CFU per two kidneys from two mice.
Tracking cell populations.
To quantify all viable organisms, the kidney tissue homogenate was grown on SDA plates. To quantify the fluconazole-resistant isolate, the homogenate was grown on CHROMagar plates containing fluconazole (4 μg/ml) (1, 33). The quantity of the susceptible isolate was then calculated as the difference in growth between the two plates.
Impact of fluconazole exposure on emergence of the resistant isolate.
Following infection with the parent strain (K1) of C. albicans, groups of two animals were treated with eight different fluconazole regimens and a no-drug control. These regimens were chosen to widely vary the pharmacokinetic profiles of fluconazole in animals relative to the MIC of the parent organism. The pharmacodynamic exposures anticipated with each of the eight dosing regimens are shown in Table 2. Groups of mice were treated for 72 h for each round of therapy. At the end of each round, kidneys were harvested and homogenized, and the homogenate was serially 10-fold diluted and plated for CFU enumeration. Homogenate dilutions were plated to quantify each cell population as described above. The entire lawn of growth on SDA plates from each treatment was collected. One half of the cell growth was placed in 30% glycerol plus YPD and frozen at −80°C. The eight cell populations resulting from the first round of treatment were labeled A1 through A8. The second half of the cell growth was used to prepare an inoculum to infect a second group of neutropenic mice. These groups of animals were then treated with the same eight fluconazole treatment regimens for 72 h. At the end of therapy, homogenates were similarly processed. The eight cell populations from the second round of therapy were labeled B1 through B8. The sequence of infection, treatment, kidney harvest, CFU enumeration (total and resistant), and archiving was undertaken 10 times, or for 30 days of fluconazole in vivo exposure.
TABLE 2.
Fluconazole treatment regimens and population MIC for each regimen over time
| Regimen no. | Regimena | %T > MIC (h) | 24-h AUC/MIC (mg · h/liter) | Cmax/MIC |
|---|---|---|---|---|
| 1 | 0.2 mg/kg q6h | 0 | 9 | 0.34 |
| 2 | 0.78 mg/kg q6h | 42 | 33 | 1.34 |
| 3 | 3.13 mg/kg q6h | 100 | 133 | 5.3 |
| 4 | 0.39 mg/kg q12h | 0 | 7 | 0.66 |
| 5 | 1.56 mg/kg q12h | 73 | 27 | 2.66 |
| 6 | 0.78 mg/kg q24h | 10 | 6 | 1.34 |
| 7 | 3.13 mg/kg q24h | 40 | 26 | 5.32 |
| 8 | 12.5 mg/kg q24h | 57 | 102 | 16.3 |
q6h, every 6 h; q12h, every 12 h; q24h, every 24 h.
Fluconazole pharmacokinetics and pharmacodynamic analyses.
The serum kinetics used in the analyses were from our prior study using this model (1). The pharmacokinetic/pharmacodynamic (PK/PD) index values (percent time serum levels remained above the MIC and the ratios of the maximum concentration of drug in serum [Cmax] and 24-h area under the curve [AUC] relative to the MIC) were calculated for each of the eight dosing regimens. Calculations were performed relative to the MIC of the parent population. A sigmoid maximal effect model was used to characterize the relationship between the PK/PD indices for the treatment regimens and quantitation of the viable burdens of both the susceptible and any resistant populations that might emerge over the 10 72-h treatment periods. The coefficient of determination (R2) was used as a measure of the strength of the relationships. The various relationships were examined to define the PK/PD exposure(s) that led to and prevented the emergence of the resistant cell populations.
Phenotypic studies. (i) C. albicans strain fitness in vitro and in vivo.
To detect potential differences in fitness between the parent strains and select populations evolved in animals with various fluconazole exposures, the rate and extent of growth were measured in vitro and in vivo. Twenty-four-hour subcultures from frozen stocks were diluted to an optical density at 600 nm (OD600) of 0.02 in RPMI buffered with MOPS (morpholinepropanesulfonic acid). The cultures were grown with shaking at 35°C. At 1- to 2-h intervals, samples were withdrawn for turbidity measurements over 10 h to assess the generation rate. To similarly assess fitness in vivo, groups of mice were infected with the same inoculum size of both the parent and the final set of archived strains (J1, J2, J3, J4, J5, J6, J7, and J8). The mice were euthanized 24 h after infection, and the burden of organisms in the kidneys was used as a measure of in vivo fitness.
(ii) Stability of resistant and fitness phenotypes in vitro and in vivo.
Isolates found to exhibit a fluconazole resistance phenotype were serially cultured in fluconazole-free RPMI-MOPS. After each 24-h subculture, the percentage of the total cell population that retained a drug-resistant phenotype was determined by CFU enumeration on SDA and CHROMagar with fluconazole. Antifungal susceptibility was also measured in the 24-h subculture cell population as described above. A total of eight 24-h subcultures were performed.
In a similar manner, the stability of a drug-resistant phenotype was examined in infected animals in the absence of fluconazole exposure. Neutropenic mice were infected as described above. After 72 h, the animals were sacrificed and the percentage of the total cell population that retained a drug-resistant phenotype was determined by CFU enumeration on SDA and CHROMagar with fluconazole. The growth of organisms on SDA was used to prepare an inoculum for a subsequent round of infection in untreated mice. Eight rounds of infection or 24 days of in vivo growth were studied.
The stability of the fitness cost observed in the drug-resistant cell populations was also assessed in these animal passage studies. The burden of growth in the kidneys of untreated mice was measured for each of the resistant cell populations after 72 h and compared to the growth of the parent strain. Kidney homogenates were diluted, plated on SDA, incubated, and enumerated as described above.
Strain genetic similarity.
Randomly amplified polymorphic DNA analysis was chosen as a fingerprinting method to assess the relatedness between the parent strain and the final archived cell populations from the eight in vivo drug exposures (J1, J2, J3, J4, J5, J6, J7, and J8) (36). Genomic DNAs from C. albicans strains 98-234 and 98-17 were used as unrelated strain controls. The intent of this study was to demonstrate that the archived populations were not acquired exogenously during the study period. All amplifications were repeated at least twice. Similarity coefficients (SAB) were used to estimate the relatedness among strains. The SAB measures the proportion of bands with the same molecular weights in the patterns of two isolates by the following formula: SAB = 2E/(2E + a + b), where E is the number of bands shared by the two strains A and B, a is the number of bands unique to strain A, and b is the number of bands unique to strain B. An SAB of 1.00 was considered identically matched, an SAB of 0.0 represented no matches, and SABs from 0.01 to 0.99 represented increasing proportions of matched bands.
ERG11 sequencing.
The coding regions from the start codon at position 148 to position 1694 near the stop codon of the ERG11 genes from the C. albicans parent K1 and each of the final archived cell populations (J1, J2, J3, J4, J5, J6, J7, and J8) were sequenced to determine if the reduced susceptibility might be due to previously described mutations (4, 23, 47). A previously described dye terminator method was used for sequencing studies (38, 39). Sequences were compared to the published sequences in GenBank.
Transcriptional profile of cell populations.
Genomewide expression analysis was undertaken using cDNA microarrays (version 6.31) produced at the Biotechnology Research Institute, National Research Council, Montreal, Canada (http://ghi-igs.nrc-cnrc.gc.ca/home_e.html) (2, 11). The microarray is based on C. albicans SC5314. The arrays consisted of 6,737 PCR open reading frames covering 98% of the open reading frames. Studies were undertaken to compare the global responses of C. albicans during resistance development in vivo. The in vivo transcriptional profiles of two cell populations evolved during exposure to treatment regimen number 4 over time were examined (see Tables S2 and S3 in the supplemental material). The first cell archive (4E) examined evolved over five in vivo treatment periods (15 days) and demonstrated a fourfold-reduced susceptibility to fluconazole. The second population (4J) studied was recovered following the final (10th) round of therapy and exhibited a 16-fold reduction in susceptibility to fluconazole. The global transcriptional responses of these cell populations were compared to that of the parent cells. For each of three independent sets of cultures, an aliquot of the frozen stock from each archived population was grown overnight in YNB with shaking at 35°C. Cultures were diluted to an OD600 of 0.05 in 200 ml of YNB and incubated with shaking at 35°C until mid-log phase (OD600 = 0.6). Cells were collected by centrifugation, and the cell pellets were flash frozen in an ethanol-dry ice bath. Candida RNA was isolated by the hot-phenol method (40). The RNA was purified using the Fast Track mRNA isolation kit (Invitrogen). Each set of samples was used for separate array hybridizations with cDNA from the parent, K1, and either of the evolved populations. In the first hybridization, the parent was labeled with Cy3 and the evolved population with Cy5. In the second hybridization, the parent was labeled with Cy5 and the evolved population with Cy3. A total of 12 hybridizations were performed.
Microarrays were scanned using ScanArray 5000 (Packard BioScience, Billerica, MA) at a 10-μm resolution. The intensities of the spots were quantified with QuantArray (GSI Lumonics, Billerica, MA). QuantArray files were analyzed in EXCEL (Microsoft, Redmond, WA). To be included in the normalization and analysis, each spot had to satisfy three quality control criteria (2, 11). The signal intensity minus half of the standard deviation had to be greater than the local background plus half of the standard deviation. The signal intensity had to be within the dynamic range of the photomultiplier tube. The raw intensities of the duplicate spots for each gene had to be within 50% of each other. For spots that met these criteria, the log ratio of the intensity of the expression from yeast from the evolved cell population relative to that from the parent cell was considered. The results presented consist of the average of three independent experiments with dye swaps. When either channel value was below 100.0, the data point was considered unacceptable. A normalization factor was applied to account for systematic differences in probe label intensities. A two-sided Student's t test was used to assess the statistical significance of log2 ratios. Gene ontology identification was undertaken for differentially expressed genes using Candida (CandidaDB, http://genolist.pasteur.fr/CandidaDB/; Candida Genome Database, http://www.candidagenome.org/; Stanford, http://www-sequence.stanford.edu/group/candida/search.html) and Saccharomyces (Saccharomyces Genome Database, http://genome-www.Stanford.edu/Saccharomyces/) databases.
Confirmation of microarray expression data. (i) Northern blotting.
The transcript abundances of several genes of interest were independently confirmed using either Northern blotting or quantitative reverse transcription (RT)-PCR (38). The parent strain (K1) and the final archived cell populations (J1, J2, J3, J4, J5, J6, J7, and J8) of C. albicans from frozen stocks were grown and harvested, and RNA was isolated as described above. Probes were generated for CDR1, CDR2, MDR1, ERG11, and ACT1 by PCR amplification from Candida genomic DNA (see Table S1 in the supplemental material) (29, 35, 38, 46, 48, 49). Both actin and rRNA bands were utilized to account for potential differences in RNA loading. The normalized levels were divided by the background measurement to determine the change relative to background of each sample.
(ii) Quantitative RT-PCR.
Quantitative real-time PCR was also used to compare the mRNA abundances of the genes of interest (2, 10, 27). TaqMan MGB probe and primer sets were designed for the target genes CDR1, CDR2, MDR1, ERG11, TAT2, RTA1, SGE1, SUR2, and GFA1 and for the normalizing gene, ACT1, of C. albicans (see Table S1 in the supplemental material) using Primer Express 1.5 software (Applied Biosystems, Foster City, CA). The QIAGEN Quanti Tect Probe RT-PCR Kit (Valencia, CA) was used in an ABI PRISM 7700 Sequence Detection System v1.7 (Applied Biosystems, Foster City, CA). Reactions were performed in triplicate according to the kit manufacturer's instructions. The quantitative data analysis was completed using the delta Ct (2−DΔCt) method of Livak and Schmittgen (28). The comparative expression method generated data as change (n-fold) in gene expression normalized to a constitutive reference gene and relative to a control or baseline condition. The baseline condition in this case was mRNA isolated from C. albicans K1 cells.
RESULTS
In vitro susceptibility testing.
The in vitro susceptibilities of the C. albicans parent, K1, and each of the 80 archived cell populations isolated after serial in vivo fluconazole exposures are presented in Table 2. The MIC was unchanged throughout the 10 rounds of in vivo drug exposures for five of the eight treatment regimens. The MIC increased up to 16-fold (two cell populations) from the 0.5-μg/ml (one cell population) starting population for three of the eight archived populations (no change for five cell populations). The initial rise in MIC was not observed until the third in vivo round of exposure, or 9 days.
Differential growth on drug-containing plates.
The parent cell population, C. albicans K1, did not grow on fluconazole-containing CHROMagar plates. The drug-containing plates did support the growth of several of the evolved cell populations following in vivo drug exposure over time.
Emergence of resistance phenotype in vivo.
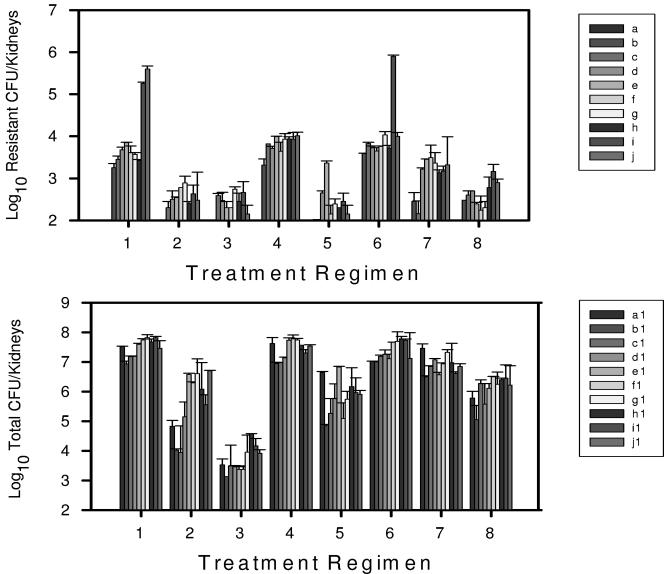
Detection of viable cells at low levels on the fluconazole-containing plates began as early as 3 days into drug exposure for three of the eight treatment regimens (Fig. 1). The burden of cells exhibiting a drug-resistant phenotype steadily increased for these three treatment regimens through day 21 of therapy. Over the final 9 days of in vivo fluconazole exposure with these three regimens, the burden of the resistant population increased markedly, and it became the predominant cell phenotype.
FIG. 1.
(Top) Burden of viable C. albicans with a resistance phenotype (based on growth on fluconazole-containing media) isolated from mouse kidneys following treatment with one of eight fluconazole treatment regimens. Each bar (A through J) indicates 72 h of therapy. Each bar represents data from two mice. (Bottom) Total burden of viable C. albicans based on growth on SDA plates. The error bars represent standard deviations.
Stability of resistant phenotype in vitro and in vivo.
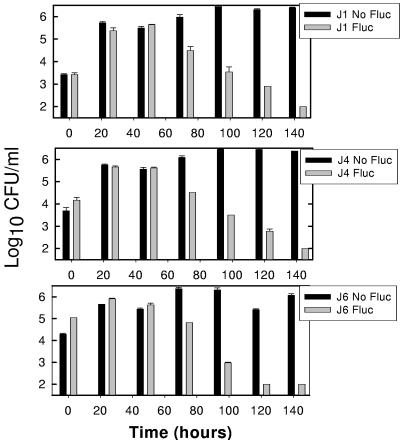
Isolates found to exhibit fluconazole resistance were serially cultured in fluconazole-free media. A less susceptible phenotype was detected throughout the estimated 120 in vitro generations. However, the percentage of the viable cell population exhibiting this phenotype began to decline after nearly 50 generations (Fig. 2). Similarly, in the in vivo studies, the MIC remained at the highest recorded level for at least 9 days (range, 9 to 18 days) of passage in untreated animals. The MIC did not return to the level of the parent cell population after 24 days of passage for any of the three resistant archived cell populations (Table 3). For the J6 population, the final MIC after eight rounds of in vivo passage declined by only a single twofold dilution.
FIG. 2.
Stability of the drug resistance phenotypes of three strains (J1, J4, and J6) following passage of the strains in vitro in the absence of fluconazole (Fluc). The black bars represent the total viable cell count. The gray bars represent viable growth of cells with a drug-resistant phenotype. The error bars represent standard deviations.
TABLE 3.
Stability of resistant phenotype in archived cell populations
| Passage no. | MIC (μg/ml) following animal passage without fluconazole
|
|||
|---|---|---|---|---|
| K1 | 1J | 4J | 6J | |
| End of archive | 0.5 | 4.0 | 8.0 | 8.0 |
| 1 | 0.5 | 4.0 | 8.0 | 8.0 |
| 2 | 0.5 | 4.0 | 8.0 | 8.0 |
| 3 | 0.5 | 4.0 | 4.0 | 4.0 |
| 4 | 0.5 | 4.0 | 4.0 | 2.0 |
| 5 | 0.5 | 4.0 | 2.0 | 4.0 |
| 6 | 0.5 | 2.0 | 2.0 | 4.0 |
| 7 | 0.5 | 2.0 | 2.0 | 4.0 |
| 8 | 0.5 | 2.0 | 2.0 | 4.0 |
Fitness in evolved populations and stability of the fitness phenotype.
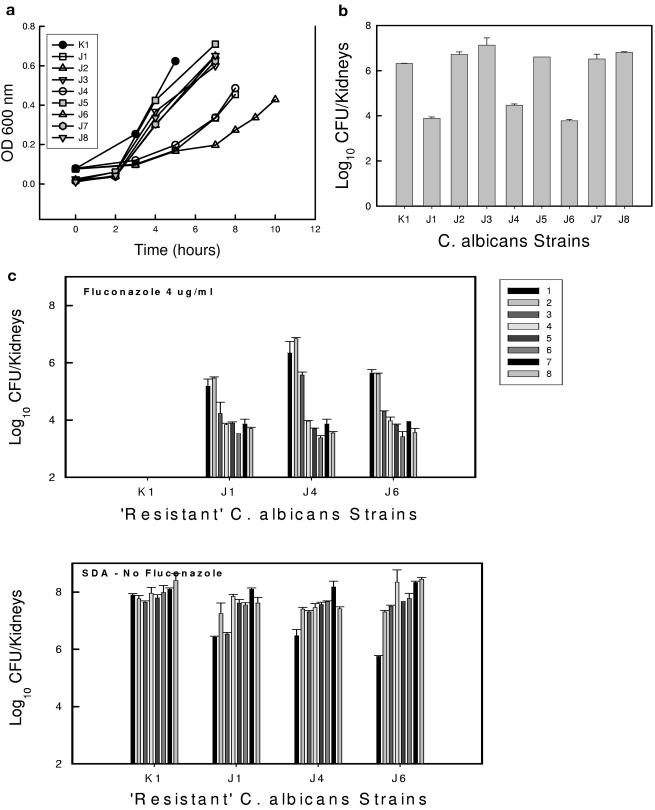
The in vitro rates of growth for the five evolved populations that did not develop a drug-resistant phenotype were similar to that of the parent (Fig. 3a). However, the in vitro generation times were lengthened in the less susceptible cell populations. Doubling times increased from near 90 min in the parent and susceptible populations to more than 2 to 5 h in the drug-resistant cells. The in vivo burden of organisms in the kidneys of mice infected with the three resistant populations was more than 2 orders of magnitude less than that of the parent over 24 h (Fig. 3b). Each of the drug-resistant reduced-fitness cell populations regained fitness to the level of the parent population over time. Within 144 h, all three of the populations were able to grow in mice to a burden similar to that of the parent population (Fig. 3c).
FIG. 3.
(a) In vitro growth of C. albicans in RPMI broth as estimated by the OD600 measurement over time of the parent, C. albicans K1, and each of eight evolved isolates following 30 days of different fluconazole exposures in vivo. Each bar represents mean data from two independent cultures. (b) In vivo burden of the parent, C. albicans K1, and each of eight evolved isolates after 24 h. Each bar represents mean data from two mice. The error bars represent standard deviations. (c) Stabilities of the drug resistance phenotypes of three strains (J1, J4, and J6) following passage of the strains in vivo in the absence of fluconazole. Each bar indicates 72 h of growth in the mice (the bars from left to right represent consecutive periods of time). The top graph represents viable growth of cells exhibiting a resistant phenotype based on growth on fluconazole-containing agar. The bottom graph represents total viable-organism recovery based on growth on SDA plates. Each bar represents mean data from two mice.
Strain genetic similarities.
Following amplification with each of the three randomly amplified polymorphic DNA primers, between three and five major bands were generated for each individual isolate, with an average of four bands for the 11 isolates analyzed (see Table S1 in the supplemental material). The range of SAB values for the parent relative to the evolved archive was 0.85 to 1.0. The SAB values for the parent relative to the negative control isolates ranged from 0 to 0.5.
ERG11 sequence comparison.
In order to determine if the reduced fluconazole susceptibility in the archived cell populations was caused by a change in ERG11, we compared sequences from strains J1, J4, and J6 to that of K1. We did not identify nucleotide changes in the isolates, suggesting that ERG11 mutations were not responsible for fluconazole resistance in the in vivo-evolved populations.
Transcriptional profiles of cell populations.
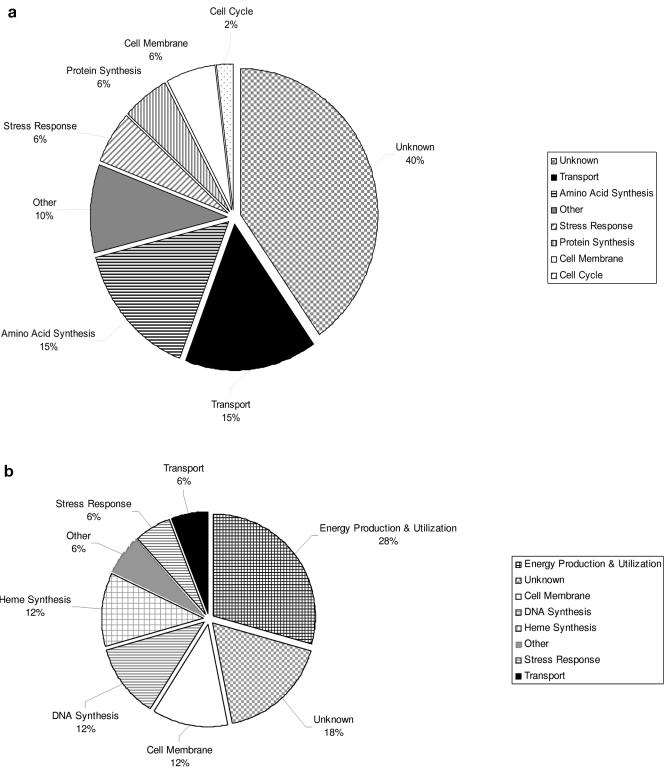
Adaptation of C. albicans to pharmacodynamically defined in vivo PK/PD fluconazole exposures resulted in a stepwise reduction in cell susceptibility to the inhibitory effect of the triazole. The transcriptional profiles of two cell populations during this evolution were compared to that of the starting drug-susceptible parent population. The first population (4E) exhibited a small reduction in susceptibility (twofold) that evolved 15 days into therapy. The second population (4J) represented the final evolved population from the fourth treatment regimen and was more drug resistant (a 16-fold rise in the MIC) to fluconazole. Among the differentially expressed genes, the majority of genes were up-regulated (80%). Sixty-six percent of the differentially expressed genes encoded a protein of known function in either C. albicans or Saccharomyces cerevisiae. The functional categories of the differentially expressed genes are shown in Tables S2 and S3 in the supplemental material and Fig. 4a and b. In the population exhibiting the most marked resistance phenotype (4J), 69 genes, or 1% of the genes, were differentially expressed. Genes responsible for transport across and maintenance of the cell membrane represented 21% of the up-regulated transcripts. Genes encoding proteins associated with amino acid synthesis and uptake were also common in the up-regulated category. The most commonly down-regulated functional category of genes was related to energy production and utilization. Early in the process of resistance development (4E), 167 genes, or 2.5% of the genes, were differentially expressed. The functions of the largest groups of genes up-regulated during early resistance evolution were associated with protein and ribosomal synthesis and processing (26%). Genes associated with energy production and utilization were the most common functional group among the suppressed genes during this period of evolution. The expression profiles of the known genes previously linked to azole drug resistance were confirmed in Northern blot and RT-PCR studies.
FIG. 4.
(a) Categories and percentages of C. albicans genes up-regulated in the evolved cell population J4 compared to the parent, K1. (b) Categories and percentages of C. albicans genes down-regulated in the evolved cell population J4 compared to the parent, K1.
Relationship between PK/PD exposure and emergence of less susceptible cell populations or transcriptional profiles.
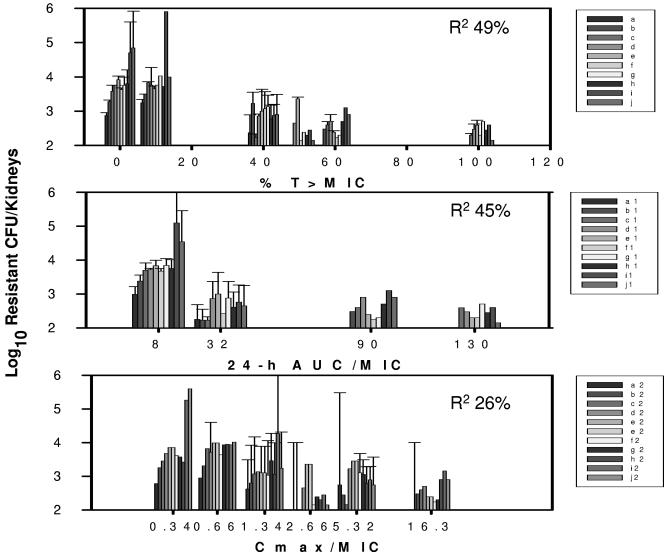
The pharmacokinetic/pharmacodynamic exposures resulting from the eight fluconazole treatment regimens included %T > MIC values ranging from 0 to 100% and a 16- and 47-fold range in 24-h AUC/MIC and Cmax/MIC values, respectively. The relationships between the range of %T > MIC (R2 = 49%) and 24-h AUC/MIC (R2 = 45%) and modulation of emergence of the less susceptible cell populations were strong (Fig. 5 and 6). The association between the fluconazole Cmax/MIC exposure and the emergence of a less susceptible cell phenotype was less evident (R2 = 26%). The %T > MIC values of less than 40% were associated with increasingly larger populations of less susceptible cells over the 30-day treatment period. Conversely, the drug exposures producing exposures with values from 40 to 100% performed similarly in regard to delaying and reducing the emergence of these populations. The 24-h AUC/MIC values of less than 32 also resulted in the appearance of increasingly larger numbers of Candida cells exhibiting reduced drug susceptibility. Among the range of Cmax/MIC values, the higher-value exposures did result in a trend toward lower concentrations of the less susceptible cell population; however, the confidence intervals were quite large. None of the drug exposures entirely suppressed the emergence of less susceptible cell populations.
FIG. 5.
Relationship between fluconazole %T > MIC, 24-h AUC/MIC, and Cmax/MIC and the burden of growth of C. albicans in kidneys of mice with a resistant phenotype based on growth on fluconazole-containing agar. Each bar indicates a 72-h in vivo drug exposure. Each bar represents mean data from two mice. The error bars represent standard deviations. R2 is the coefficient of determination.
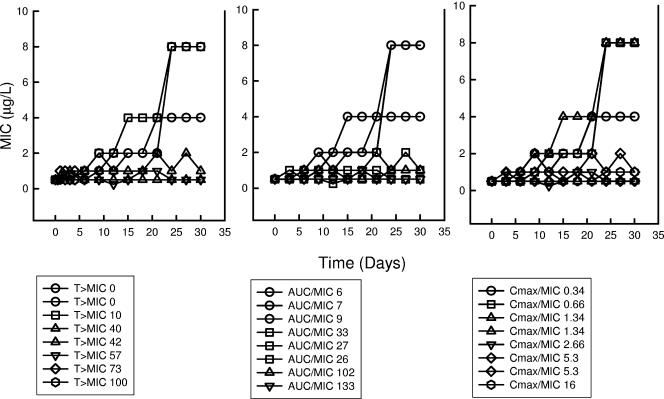
FIG. 6.
Relationship between in vivo fluconazole %T > MIC, 24-h AUC/MIC, and Cmax/MIC and the in vitro MIC of the cell population after each 72-h treatment period. Each symbol represents the MIC of the entire isolated population after each 72-h period. The MIC value is the mean from three independent experiments.
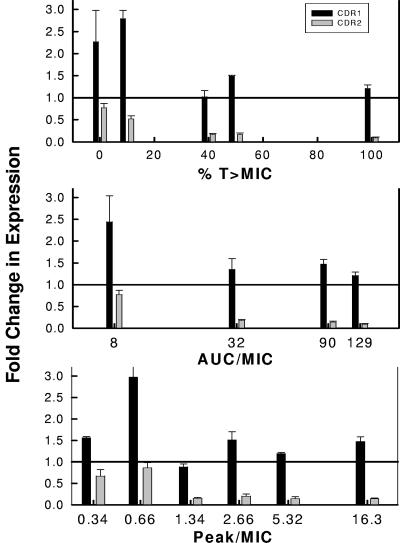
We also examined the relationship between the various PK/PD exposures and the mRNA abundances of four genes previously associated with fluconazole drug resistance. Transcript levels for these four genes were estimated from the final eight archived cell populations, J1 to J8, and the parent cells, K1. The abundances of ERG11 and MDR1 were not different in the cell populations exposed to the range of PK/PD exposures utilized (data not shown). However, the relationship between CDR1 abundance and the fluconazole PK/PD exposure was quite strong (Fig. 7). Drug exposures producing %T > MIC values of less than 40% and 24-h AUC/MIC values of less than 32 were associated with increasing CDR1 transcripts. CDR2 abundance was relatively less in drug-exposed than in parent cell populations. However, among the drug-exposed populations, we did observe a similar relationship between the %T > MIC and 24-h AUC/MIC exposures and CDR2 abundance. The association between mRNA levels of these genes and drug exposure expressed as the Cmax/MIC was poor.
FIG. 7.
Relationship between in vivo fluconazole %T > MIC, 24-h AUC/MIC, and Cmax/MIC and the expression of CDR1 and CDR2 in C. albicans from infected mice. Each bar represents the change (n-fold) in mRNA abundance of an archived cell population relative to the beginning C. albicans K1 population. The bars represent the means and standard deviations from three biological replicates using quantitative RT-PCR. The RT-PCR assay was performed in triplicate for each biological replicate.
DISCUSSION
It is generally accepted that anti-infective drug exposure is an important factor in the development and emergence of drug resistance (17, 25, 30). Recent studies have begun to investigate the relationship between drug exposure to the fungistatic antifungal drug fluconazole and resistance emergence in the fungal pathogen C. albicans (1). The prior fluconazole pharmacodynamic investigation involved tracking amplification of susceptible and resistant cell populations in a mixture design. The studies utilized well-characterized clinical isogenic strain pairs of C. albicans. For each of the six pairs examined, the parent was fluconazole susceptible and the other strain was resistant. The initial parent cell population was isolated from a patient prior to drug therapy. The subsequent isolate was collected from the patient after various periods of fluconazole therapy in association with treatment failure. In the mixture experiments, the starting inoculum used to infect animals consisted primarily of the susceptible parent cells spiked with a low concentration of the resistant isolate. These studies defined fluconazole drug-dosing regimens that both led to and suppressed the emergence of resistance.
The experimental design used in the current investigations began with a single susceptible progenitor cell. Similar to the prior study design, a wide range of antifungal drug dose levels and dosing frequencies allowed analyses to examine the relative importance of these dosing regimen variables. Regimens from the current study resulted in the emergence, as well as prevention of the emergence, of a less susceptible cell population. Cell populations evolved with three of eight dosing regimens exhibited a reduced-susceptibility phenotype over the treatment period. The duration of exposure associated with maximal changes in the susceptibility phenotype of the cell population occurred after 21 to 24 days of exposure. For each dose level examined, the emergence of the less susceptible population occurred with the more widely spaced dosing intervals. Providing brief periods of supra-MIC exposure and, conversely, more prolonged sub-MIC contact was associated with resistance emergence. Consideration of the PK/PD indices associated with each of these dosing regimens also suggested that more frequent administration that provided drug levels above the MICs of the infecting pathogens was associated with a reduction in amplification of the resistant strains. The findings from the current investigations are similar to those from the mixture design with this drug-pathogen combination (1).
Evolution of a less susceptible phenotype was also associated with a change in the ability of the organism to proliferate both in media and in animals. However, the fitness cost phenotype was short lived, resolving following generation in the absence of fluconazole both in vitro and in vivo within 24 to 72 h. The transient nature of a fitness cost has been recognized in this fungal species, as well as in other prokaryotic pathogens, most commonly associated with compensatory mutations (9, 12, 30, 41).
The present study also examined the relationship between drug exposure and resistance development utilizing genomics tools in an attempt to further understand the molecular basis underlying this evolution. Among the previously identified resistance mechanisms, modulation of the CDR1 efflux pump was the only differentially expressed transcript (47). The up-regulation of this gene was confirmed by several methodologies in each of the three less susceptible cell populations. Many of the other differentially expressed transcripts identified in the current studies have been identified in prior array analysis of fluconazole-resistant C. albicans strains (6, 11, 22, 37). Among the most highly expressed were those related to cell and plasma membrane maintenance (TAT2, YAH1, RTA1, GFA1, SGE1, and SUR2) (14, 18, 20, 31, 44, 45). The expression of these genes may implicate cell membrane changes contributing to resistance or may simply represent a response to cell-damaging conditions.
The experimental model system utilized in the current studies was able to follow resistance development over time in relation to in vivo antimicrobial exposures. The model provides insight into the relationship between drug dosing and treatment failure due to drug resistance. The emergence of resistance with the triazole antifungal was closely related to the time-above-MIC parameter. These data demonstrate that the static killing characteristics and presence of sub-MIC effects contribute to the selection of resistant mutants. The current investigations suggest that resistance to a fungistatic antimicrobial will rarely emerge in a population of drug-susceptible Candida during in vivo treatment when the times that cells are exposed to sub-MIC concentrations are limited.
Predictions from the current archive and the prior reconstruction experiments may be valuable for design of optimal dosing regimens for currently available triazole drugs, as well as those under development (43). In addition, the experimental approaches utilized in these studies provide a means to address these relationships for other drug-pathogen combinations for which resistance is an emerging problem. Lastly, these experiments demonstrate an important link between antimicrobial pharmacodynamics and gene expression. The relationship between the pharmacodynamic exposure and the pathogen transcriptional response may provide insight into drug resistance mechanisms and may identify novel drug targets.
Supplementary Material
Acknowledgments
David Andes is supported by NIH/NIAID AI01767-01A1.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Andes, D., A. Forrest, A. Lepak, J. Nett, K. Marchillo, and L. Lincoln. 2006. Impact of antimicrobial dosing regimen on evolution of drug resistance in vivo: fluconazole and Candida albicans. Antimicrob. Agents Chem. 50:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andes, D., A. Lepak, A. Pitula, K. Marchillo, and J. Clark. 2005. A simple approach for estimating gene expression of Candida albicans directly from a systemic infection site. J. Infect. Dis. 192:893-900. [DOI] [PubMed] [Google Scholar]
- 3.Andes, D., and M. Van Ogtrop. 1999. Characterization and quantitation of the pharmacodynamics of fluconazole in a neutropenic murine disseminated candidiasis model. Antimicrob. Agents Chemother. 43:2116-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andes, D. 2003. In vivo pharmacodynamics of antifungal drugs in treatment of candidiasis. Antimicrob. Agents Chemother. 47:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baddley, J. W., and S. A. Moser. 2004. Emerging fungal resistance. Clin. Lab. Med. 24:721-735. [DOI] [PubMed] [Google Scholar]
- 6.Barker, K. S., S. Crisp, R. E. Wiederhold, R. E. Lewis, B. Bareither, J. Eckstein, R. Barbuch, M. Bard, and P. D. Rogers. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J. Antimicrob. Chemother. 54:376-385. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, J., B. B. Stone, M. C. Groner, and S. H. Zinner. 1987. Comparative study with enoxacin and netilmicin in a pharmacodynamic model to determine importance of ratio of antibiotic peak concentration to MIC for bactericidal activity and emergence of resistance. Antimicrob. Agents Chemother. 31:1054-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonhoeffer, S., M. Lipsitch, and B. R. Levin. 1997. Evaluating treatment protocols to prevent antibiotic resistance. Proc. Natl. Acad. Sci. USA 94:12106-12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouma, J. E., and R. E. Lenski. 1988. Evolution of a bacteria/plasmid association. Nature 335:351-352. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 11.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowen, L. E., L. M. Kohn, and J. B. Anderson. 2001. Divergence of fitness and evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 183:2971-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drusano, G. L., D. E. Johnson, M. Rosen, and H. C. Standiford. 1993. Pharmacodynamics of a fluoroquinolone antimicrobial agent in a neutropenic rat model of Pseudomonas sepsis. Antimicrob. Agents Chemother. 37:483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrenhofer-Murray, A. E., M. U. Keller Seitz, and C. Sengstag. 1998. The Sge1 protein of Saccharomyces cerevisiae is a membrane-associated multidrug transporter. Yeast 14:49-65. [DOI] [PubMed] [Google Scholar]
- 15.Fan-Harvard, P., D. Capano, S. M. Smith, A. Mangia, and R. H. K. Eng. 1991. Development of resistance in Candida isolates from patients receiving prolonged antifungal therapy. Antimicrob. Agents Chemother. 35:2302-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firsov, A. A., S. N. Vostrov, I. Y. Lubenko, K. Drlica, Y. A. Portnoy, and S. H. Zinner. 2003. In vitro pharmacodynamic evaluation of the mutant selection window hypothesis using four fluoroquinolones against Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fish, D. N., S. C. Piscitelli, and L. H. Danziger. 1995. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy 15:279-291. [PubMed] [Google Scholar]
- 18.Gabriel, I., J. Olchowy, A. Stanislawska-Sachadyn, T. Mio, J. Kur, and S. Milewski. 2004. Phosphorylation of glucosamine-6-phosphate synthase is important but not essential for germination and mycelial growth of Candida albicans. FEMS Microbiol. Lett. 235:73-80. [DOI] [PubMed] [Google Scholar]
- 19.Gerber, A. U., A. P. Vastola, J. Brandel, and W. A. Craig. 1982. Selection of aminoglycoside-resistant variants of Pseudomonas aeruginosa in an in vivo model. J. Infect. Dis. 146:691-697. [DOI] [PubMed] [Google Scholar]
- 20.Grilley, M. M., S. D. Stock, R. C. Dickson, R. L. Lester, and J. Y. Takemoto. 1998. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J. Biol. Chem. 273:11062-11068. [DOI] [PubMed] [Google Scholar]
- 21.Jumbe, N., A. Louie, R. Leary, W. Liu, M. R. Deziel, V. H. Tam, R. Bachhawat, C. Freeman, J. B. Kahn, K. Bush, M. N. Dudley, M. H. Miller, and G. L. Drusano. 2003. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J. Clin. Investig. 112:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karababa, M., A. T. Coste, B. Rognon, J. Bille, and D. Sanglard. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb, D. C., D. E. Kelly, W. H. Schunck, A. Z. Shyadehi, M. Akhtar, D. J. Lowe, B. C. Baldwin, and S. L. Kelly. 1997. The mutation T315A in Candida albicans sterol 14 alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272:5682-5688. [DOI] [PubMed] [Google Scholar]
- 24.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Mede. 10(Suppl. 12):S122-S129. [DOI] [PubMed] [Google Scholar]
- 25.Lipsitch, M., and B. R. Levin. 1997. The population dynamics of antimicrobial chemotherapy. Antimicrob. Agents Chemother. 41:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipsitch, M., C. T. Bergstrom, and B. R. Levin. 2000. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proc. Natl. Acad. Sci. USA 97:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 28.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−DΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 29.Losberger, C., and J. E. Ernst. 1989. Sequence of Candida albicans gene encoding actin. Nucleic Acids Res. 17:9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez, J. L., and F. Baquero. 2002. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15:647-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mio, T., T. Yamada-Okabe, T. Yabe, T. Nakajima, M. Arisawa, and H. Yamada-Okabe. 1997. Isolation of the Candida albicans homologs of Saccharomyces cerevisiae KRE6 and SKN1: expression and physiological function. J. Bacteriol. 179:2363-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.Patterson, T. F., S. G. Revankar, W. R. Kirkpatrick, O. Dib, A. W. Fothergill, S. W. Redding, D. A. Sutton, and M. G. Rinaldi. 1996. A simple method for detecting fluconazole-resistant yeasts with chromogenic agar. J. Clin. Microbiol. 34:1794-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perea, S., and T. F. Patterson. 2002. Antifungal resistance in pathogenic fungi. Clin. Infect. Dis. 35:1073-1080. [DOI] [PubMed] [Google Scholar]
- 35.Prasad, R., W. P. De, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 36.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, enzyme electrophoresis, and Southern blot hybridization and the repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers, P. D., and S. K. Barker. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob. Agents Chemother. 46:3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sanglard, D., R. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 40.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14 alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrag, S. J., V. Perrot, and B. R. Levin. 1997. Adaptation to the fitness cost of antibiotic resistance in Escherichia coli Proc. R. Soc. Long B 264:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selkon, J. B., S. Devadatta, K. G. Kulkarni, D. A. Mitchison, A. S. L. Narayana, C. Narayanan Nair, and K. Ramachandran. 1964. The emergence of isoniazid-resistant cultures in patients with pulmonary tuberculosis during treatment with isoniazid alone or isoniazid plus PAS. Bull. W. H. O. 31:273-294. [PMC free article] [PubMed] [Google Scholar]
- 44.Sheehan, D. J., C. A. Hitchcock, and C. M. Sibley. 1999. Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 12:40-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soustre, I., Y. Letourneux, and F. Karst. 1996. Characterization of the Saccharomyces cerevisiae RTA1gene involved in 7-aminocholesterol resistance. Curr. Genet. 30:121-125. [DOI] [PubMed] [Google Scholar]
- 46.Umebayashi, K., and A. Nakano. 2003. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 161:1117-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White, T. C., M. A. Pfaller, R. G. Rinaldi, J. Smith, and S. W. Redding. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 3(Suppl. 1):102-109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.