Abstract
Root hairs are specialized epidermal cells that play a role in the uptake of water and nutrients from the rhizosphere and serve as a site of interaction with soil microorganisms. The process of root hair formation is well characterized in Arabidopsis (Arabidopsis thaliana); however, there is a very little information about the genetic and molecular basis of root hair development in monocots. Here, we report on isolation and cloning of the β-expansin (EXPB) gene HvEXPB1, tightly related to root hair initiation in barley (Hordeum vulgare). Using root transcriptome differentiation in the wild-type/root-hairless mutant system, a cDNA fragment present in roots of wild-type plants only was identified. After cloning of full-length cDNA and genomic sequences flanking the identified fragment, the subsequent bioinformatics analyses revealed homology of the protein coded by the identified gene to the EXPB family. Reverse transcription-PCR showed that expression of HvEXPB1 cosegregated with the root hair phenotype in F2 progeny of the cross between the hairless mutant rhl1.a and the wild-type Karat parent variety. Expression of the HvEXPB1 gene was root specific; it was expressed in roots of wild-type forms, but not in coleoptiles, leaves, tillers, and spikes. The identified gene was active in roots of two other analyzed root hair mutants: rhp1.a developing root hair primordia only and rhs1.a with very short root hairs. Contrary to this, a complete lack of HvEXPB1 expression was observed in roots of the spontaneous root-hairless mutant bald root barley. All these observations suggest a role of the HvEXPB1 gene in the process of root hair formation in barley.
Specialized root epidermis cells of higher plants produce long, tubular outgrowths, called root hairs. By contributing to the root surface area, root hairs are known to play an important role in the absorption of water and nutrients from the soil (Grabov and Bottger, 1994; Lauter et al., 1996; Gahoonia et al., 1997, 2001; White, 1998). Additionally, root hairs serve as a site of interaction for fungal and bacterial soil microorganisms, among them nitrogen-fixing bacteria, such as Rhizobium, Azorhizobium, and Bradyrhizobiumin (Mylona et al., 1995; Long, 1996).
Because of their accessibility, simplicity of observation, and rapid growth, root hairs are an important model in studies of higher plant cell differentiation. Large collections of Arabidopsis (Arabidopsis thaliana) root hair mutants made possible identification of approximately 40 loci responsible for root hair development (Schiefelbein and Somerville, 1990; Masucci and Schiefelbein, 1994; Grierson et al., 1997; Parker et al., 2000; Schiefelbein, 2000). Genetic interactions between genes controlling each step of root hair development (i.e. trichoblast specification, root hair initiation, bulge formation, transition to tip growth, and hair elongation) were determined. Many genes were also well characterized at the molecular level. Nucleotide sequences and protein products were described and characterized for at least seven genes directly related to the process of trichoblast specification (Galway et al., 1994; Masucci and Schiefelbein, 1996; Lee and Schiefelbein, 1999, 2001; Wada et al., 2002; Bernhardt et al., 2003; Kwak et al., 2005). The processes of root hair initiation and bulge formation are controlled by genes encoding nuclear proteins (Masucci and Schiefelbein, 1996; Schneider et al., 1998), genes responsible for hormone signal transduction (Kieber et al., 1993; Nagpal et al., 2000), and genes involved in cell wall metabolism (Favery et al., 2001; Vissenberg et al., 2001; Cho and Cosgrove, 2002). At least 16 other genes are involved in transition to tip growth and root hair elongation and some of their products have been characterized (Wymer et al., 1997; Rigas et al., 2001; Wang et al., 2002; Foreman et al., 2003; Hu et al., 2003; Carol et al., 2005; Hemsley et al., 2005).
In comparison to dicotyledonous species, the process of root hair formation in monocots, including major cereals, is poorly characterized. The main reason is the unavailability of the comparable mutant collections. There are only a few root hair mutants described in barley (Hordeum vulgare), rice (Oryza sativa), and maize (Zea mays), and none of them has been characterized at the molecular level. Gahoonia and coworkers (2001) isolated a root-hairless mutant bald root barley (brb) in barley, a spontaneous form found in the Pallas variety. Genetic studies revealed the monogenous recessive character of the analyzed trait. Engvild and Rasmussen (2004) described several barley mutants with various changes in root hair phenotype, but many of the presented forms show unstable or variable phenotypes, segregating after selfing. None of the mutants were analyzed genetically. In rice, there has only been one root hair mutant reported up to now, a recessive hairless form, RH2, which is able to develop very short root hairs after naphthaleneacetic acid treatment (Suzuki et al., 2003). Using the Mutator transposon system, Wen and Schnable (1994) isolated three root hair mutants designated rth1, rth2, and rth3 in maize. A single recessive gene controlled the phenotype of each mutant. Mutant rth1 produced root hair primordia that were defective in elongation growth. Similarly, root hair development in the mutant rth2 was blocked at the primordium stage, but was able to elongate to one-fifth the length of wild-type root hairs. Mutant rth3 developed only irregularly shaped root hair bulges. The chromosomal location of each gene was determined and, recently, the protein product for RTH1 was characterized (Wen et al., 2005).
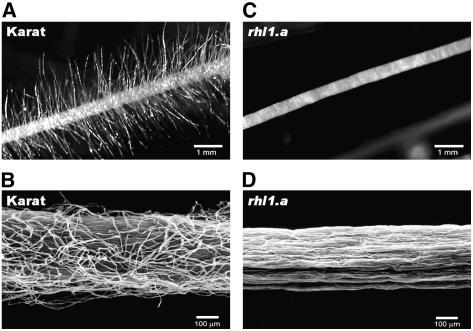
The collection of barley root hair mutants obtained by treatment with chemical mutagens N-methyl-N-nitrosourea (MNU) and sodium azide in our department (Szarejko et al., 2005) made it possible to initiate studies on genetic and molecular characterization of root hair morphogenesis in barley. The collection includes several monogenically inherited recessive mutant lines with root hairs blocked at different stages of development. The analysis described here was aimed at the identification and isolation of the gene responsible for root hair initiation with the use of the root-hairless mutant rhl1.a and its parent variety, Karat (Fig. 1).
Figure 1.
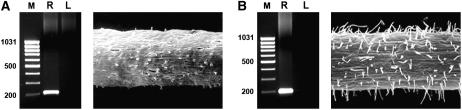
Roots of barley plants used in the study. A and B, Wild-type Karat parent variety. C and D, Root-hairless mutant rhl1.a obtained by chemical mutagenesis of Karat variety. A and C, Stereomicroscope. B and D, Scanning electron microscope.
Most of the genes responsible for root hair development in Arabidopsis were identified by insertional mutagenesis or map-based cloning methods. These approaches are very cumbersome for species with large genomes, such as barley, due to insufficient saturation of genetic maps and unavailability or poor accessibility of insertional mutagenesis techniques. In such cases, methods based on transcriptome analysis are often applied. Among them, differential display reverse transcription-PCR (DDRT-PCR; Liang and Pardee, 1992) was widely used for identification of genes differentially expressed under various environmental conditions, in specific stages of development, or in various individuals (for review, see Liang, 2002). However, besides numerous positive experiments carried out with the use of DDRT-PCR, a range of disadvantages and unsuccessful attempts to apply this method were reported (for review, see Appel et al., 1999). Our studies on the use of DDRT-PCR in the root-hairless mutant/wild-type variety system failed to identify genes differentiating the compared genotypes through qualitative profiling of their root transcriptomes (M. Kwasniewski and I. Szarejko, unpublished data). The main limitation of this technique was a high proportion of false-positive results not confirmed by RT-PCR with specific primers. We decided to use the transcriptome subtractive hybridization method, the cDNA representational difference analysis (cDNA-RDA; Hubank and Schatz, 1994), to identify genes expressed differentially during initiation of root hair formation in barley. Contrary to DDRT-PCR analysis, the cDNA-RDA method was infrequently used in plant functional genomics. Nevertheless, there are numerous positive reports from human genomics (Frohme et al., 2000; Yoon et al., 2001; Simkhovich et al., 2003) and rare examples from plants (Li et al., 1998; Zhu et al., 1998; Ling et al., 2003) that indicate this technique as an attractive tool for transcriptome differentiation in the mutant/wild-type plant system.
RESULTS
Root-Hairless Mutant rhl1.a Is Defective in Expression of the HvEXPB1 Gene Encoding β-Expansin Protein
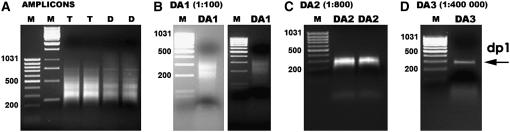
We applied the cDNA-RDA method to identify genes controlling the root-hairless phenotype of the mutant rhl1.a. F2 progeny from the rhl1.a × Karat parent variety cross was used in the analysis. To obtain the tester and driver amplicons, double-stranded (ds) cDNA was synthesized using as a template total mRNA isolated from the roots of 10 wild-type and 10 mutant phenotype F2 individuals, respectively. Such an approach made possible the elimination from the reaction of most of the cDNAs not corresponding directly to the analyzed phenotype. Three rounds of subtractive hybridization in 1:100, 1:800, and 1:400,000 tester-to-driver ratios, followed by selective amplification, were carried out. In the final PCR reaction, only one fragment, 228 bp long, was amplified as a product of tester and driver transcriptome differentiation (Fig. 2). After cloning and sequencing analyses, the identified fragment, designated dp1, proved to be homogeneous.
Figure 2.
Results of cDNA-RDA analysis after three rounds of subtractive hybridization. A, Electrophoresis of tester (T) and driver (D) amplicons. B to D, Results of differential amplification (DA) after first (B), second (C), and third (D) round of subtractive hybridization. The increased tester-to-driver ratios are indicated. dp1, Homogeneous product of Karat/rhl1.a root transcriptome differentiation.
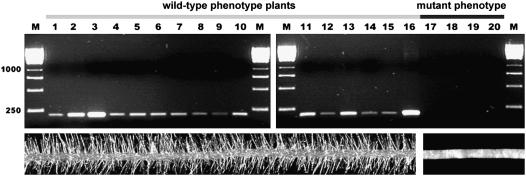
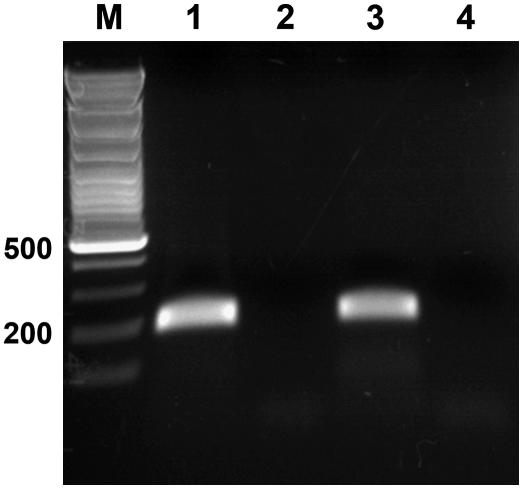
To confirm the results of cDNA-RDA analysis, a pair of primers for RT-PCR based on the dp1 sequence was designed. As the identified fragment was a product of high-stringency cDNA subtractive hybridization (1:400,000 tester-to-driver ratio), a complete lack of dp1 expression in roots of root-hairless plants was expected. RT-PCR with specific primers revealed a lack of dp1 expression in roots of the rhl1.a mutant and a presence of the expected product in roots of the Karat parent variety. Analysis of 220 F2 progeny from the rhl1.a × Karat cross showed that expression of the dp1 product cosegregated completely with the root hair phenotype. The gene represented by the identified cDNA fragment was expressed in roots of all of 164 wild-type F2 individuals, whereas it was undetected in roots of all of 56 root-hairless plants (χ3:12 = 0.02; Fig. 3). This result confirmed the data obtained from transcriptome differentiation analysis.
Figure 3.
Analysis of HvEXPB1 expression in roots of F2 progeny of rhl1.a × Karat cross. RT-PCR with 3′-UTR-specific primers: 1 to 16, Wild-type F2 plants; 17 to 20, root-hairless F2 plants.
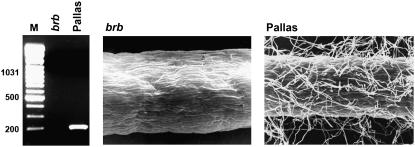
A similar analysis of expression of the identified cDNA fragment was carried out in roots of the spontaneous mutant brb, a root-hairless form of the Pallas variety (Gahoonia et al., 2001). As in rhl1.a, the identified gene was completely unexpressed in roots of this hairless form, whereas its cDNA was detectable in the wild type (Fig. 4). To analyze whether two root-hairless mutants, rhl1.a and brb, are allelic, a complementation test was performed parallel to the molecular studies. The analysis revealed that all of 11 F1 and 87 F2 individuals of the rhl1.a × brb cross were completely hairless, proving that both root-hairless mutants were allelic (B. Chmielewska, M. Kwasniewski, J. Mizeracka, and I. Szarejko, unpublished data).
Figure 4.
Analysis of HvEXPB1 expression in roots of the spontaneous root-hairless mutant brb and its wild-type parent Pallas.
The obtained results strongly suggested the connection of the identified cDNA with root hair formation in barley. To check whether the identified gene is expressed in root hair cells, an analysis of its expression in root hair outgrowth was carried out. PolyA+ mRNA isolated directly from cut root hair outgrowth served as a template in this analysis. Two independent tests clearly demonstrated that the identified cDNA is expressed in root hair cells (Fig. 5). It supports the thesis of the involvement of identified cDNA in root hair formation in barley.
Figure 5.
Analysis of HvEXPB1 expression in isolated root hair outgrowth of the Karat variety. 1 and 3, Two independent RT-PCRs with 3′-UTR-specific primers; 2 and 4, negative controls (PCR amplification using isolated polyA+ mRNA as a template).
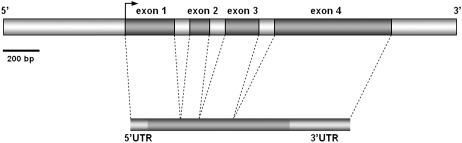
To characterize the gene represented by the identified fragment, full-length cDNA cloning and genomic sequence analysis were carried out. Using the GenomeWalking strategy, a 2,536-bp-long fragment of the genomic DNA corresponding to the dp1 fragment was cloned. Additionally, using the RNA ligase-mediated RACE method, a 1,265-bp-long cDNA fragment was identified. cDNA mapping and analysis performed with GenScan software revealed the genomic structure of the identified gene (covering 1,495 bp of the genomic sequence) and the corresponding regions of its mRNA (Fig. 6). Conceptual translation enabled determination of the sequence of a putative protein encoded by the identified gene. Similarity searching identified homology of the analyzed protein to the β-expansin (EXPB) family, with the highest similarity to the EXPB 5 of rice (Fig. 7). Comparative analysis of the deduced amino acid sequence with known rice EXPBs using the ClustalW method revealed the conserved Cys, Trp, and His-Phe-Asp motifs shared between all EXPBs examined (data not shown). Additionally, analyses carried out with the use of PSORT and SOSUI software made it possible to determinate a 25-amino acid-long N-terminal sorting signal for the vesicular pathway, characteristic for all known premature EXPB proteins (Fig. 7). As a result of these analyses, in conformity with the accepted nomenclature for expansins (Kende et al., 2004), the identified gene was called HvEXPB1. Information about the gene and mRNA sequences is deposited in the GenBank database (accession nos. AY351785 and AY351786 for the gene and mRNA sequences, respectively).
Figure 6.
Genomic and mRNA structure of the HvEXPB1 gene. A 2,536-bp-long genomic sequence with 1,495 bp of the HvEXPB1 gene (with subsequent exons) and a 1,265-bp-long mRNA with marked UTRs are shown.
Figure 7.
Alignment of the amino acid sequences of EXPB 5 of rice and putative EXPB 1 of barley obtained with ClustalW and BoxShade software. Conserved Cys, Trp, His-Phe-Asp motif, and N-terminal sorting signals are indicated.
HvEXPB1 Expression Correlates with Initiation of Root Hair Development
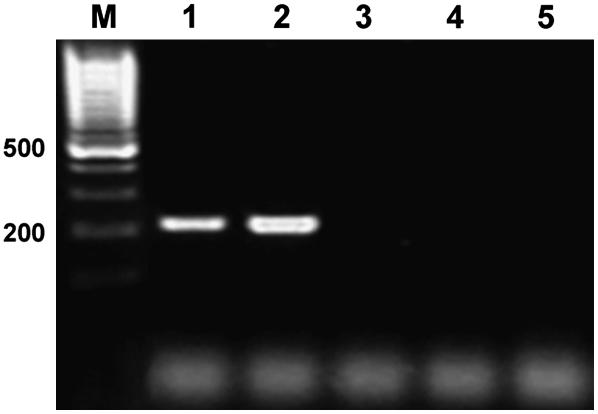
To characterize the role of the HvEXPB1 gene during root hair morphogenesis, analysis of gene expression in other root hair mutants was carried out. Two mutant lines, obtained after chemical mutagenesis in our department, representing different stages of root hair development were used in this study: mutant rhp1.a, with root hair growth blocked at the primordium stage, and mutant rhs1.a, producing very short root hairs, reaching approximately 10% of the length of root hairs in the parent variety (Szarejko et al., 2005). RT-PCR analysis with 3′-untranslated region (UTR)-specific primers carried out on mRNA isolated from roots of mutants rhp1.a and rhs1.a revealed normal, wild-type-like expression of the HvEXPB1 gene (Fig. 8). The results of analyses of HvEXPB1 expression in different root hair mutants suggest its role in the initiation of root hair formation in barley.
Figure 8.
Analysis of HvEXPB1 expression in roots (R) and leaves (L) of root hair mutants. RT-PCR with 3′-UTR-specific primers. A, Mutant rhp1.a producing root hair primordia only. B, Mutant rhs1.a producing very short root hairs.
Expression of HvEXPB1 Is Restricted to Roots Only
To check whether HvEXPB1 expression in barley is related to root hair formation only, different organs of wild-type plants were examined at the seedling and further developmental stages using RT-PCR. As shown above, the HvEXPB1 gene was expressed in roots of seedlings in all lines producing normal root hairs, short root hairs, or root hair primordia only. Detailed analysis of HvEXPB1 expression in different root fragments of 6-d-old seedlings of the wild-type Karat variety revealed its normal activity both in 1-mm-long root tips and in root fragments lacking a 1-cm-long differentiation zone (Fig. 9). This result suggests that HvEXPB1 is activated before differentiation of epidermal cells and its expression is maintained during root growth.
Figure 9.
HvEXPB1 expression profile analysis in various organs of barley. 1, 1-mm-long tip of the root; 2, root fragment without differentiation zone; 3, coleoptile; 4, last internode; 5, spike at anthesis.
Different results were obtained in analyses of HvEXPB1 expression in above-ground plant organs of wild-type plants. HvEXPB1 was not expressed in coleoptiles, seedling and tiller leaves, last tiller internodes, and immature spikes at anthesis (Fig. 9). These data clearly indicate root-specific expression of the HvEXPB1 gene in barley.
Root-Hairless Phenotype Is Not a Result of the HvEXPB1 Gene Mutation
As indicated previously, HvEXPB1 was not expressed in roots of two allelic root-hairless mutants examined in this study: rhl1.a obtained after MNU treatment of the Karat variety and brb, a spontaneous mutant from the Pallas variety. To check whether the lack of HvEXPB1 expression resulted from mutations in the gene sequence or upstream promoter regulatory elements, the genomic fragments covering HvEXPB1 and its 5′-flanking region were sequenced in both mutants and their parent varieties. No sequence difference was found between the rhl1.a mutant and the Karat parent variety in the analyzed 2,067-bp genomic fragment covering the gene and 603 bp of the promoter region (Fig. 10). The same lack of sequence differences was obtained when the HvEXPB1 gene and the promoter of the mutants brb and Pallas variety were compared (data not shown). These data suggest that the root-hairless phenotype in both mutants could result from the malfunction of a mechanism controlling HvEXPB1 expression and this malfunction is not caused by a change in the identified gene and promoter sequences.
Figure 10.
Comparison of 2,067 bp of a genomic sequence covering the HvEXB1 gene and 603 bp of its promoter in the wild-type Karat variety and the root-hairless mutant rhl1.a. Four exons of the HvEXPB1 are underlined, coding sequences are bold, and first transcribed nucleotide is indicated as +1.
DISCUSSION
The Role of Expansins in Root Hair Development
Root transcriptome differentiation carried out in the system of the root-hairless mutant/wild-type parent variety made it possible to identify the first barley EXPB gene, designated HvEXPB1, and indicated it as a critical factor in the initiation of root hair development in barley. Expansins are a large family of plant proteins that show unique cell-wall-loosening action and are involved in many biological processes related to plant growth and development (Cosgrove, 2000; Li et al., 2003b). Up to now, the detailed mechanism of expansin action has not been well established, but it appears to involve the disruption of hydrogen bonds between cellulose microfibrils and cross-linking glycans in the wall (McQueen-Mason and Cosgrove, 1994). Expansin sequences were found in di- and monocotyledonous species, pine, fern, moss, and the genome of amoeba, Dictyostelium discoidium, suggesting its ancient evolutionary origin (Li et al., 2002). On the basis of sequence alignment and phylogenetic analysis, the expansin superfamily of plants was divided into four families, designated α-expansin (EXPA), EXPB, expansin-like A, and expansin-like B (Kende et al., 2004). Analysis of the two sequenced plant genomes revealed a total of 36 expansin sequences in the Arabidopsis genome and 58 expansin-coding genes in rice (Sampedro and Cosgrove, 2005). Whereas the EXPA family is of similar size in Arabidopsis and rice, there are less EXPB-encoding genes in dicots than in monocots. The explanation of the predominance of EXPB genes in grass genomes could be that EXPA and EXPB proceed on different polysaccharide matrixes in the cell wall (Li et al., 2003a), whereas its composition in grasses differs significantly from that of dicotyledonous species (Carpita, 1996). The redundancy of expansin genes in plants indicates their specific role in various processes related to growth and development, including root elongation (Wu et al., 1996, 2001; Lee et al., 2003), leaf primordium initiation (Reinhardt et al., 1998), elongation of internodes (Lee and Kende, 2001), and fruit development and ripening (Rose et al., 1997; Brummell et al., 1999).
The report by Baluška et al. (2000) suggested the role of expansins as proteins that are involved in initiation of root hair outgrowth. Using labeled antiexpansin antibodies, the authors showed high accumulation of expansins at outgrowth bulges of maize trichoblasts. High levels of expansin accumulation were also observed in the further stages of root hair development (i.e. during primordium formation and transition to tip growth). Similar results were obtained in the studies of Cho and Cosgrove (2002), who examined expression of two EXPA genes, AtEXP7 and AtEXP18, in roots of Arabidopsis. Analyses revealed that expression of both genes was directly related to root hair formation. Using the AtEXP7 promoter:β-glucuronidase or the AtEXP7 promoter:green fluorescent protein constructs, the authors proved that AtEXP7 expression was restricted solely to root hair cell files. The gene was already active in cells before the root hair bulges appeared (indicating close temporal expression with the process of root hair initiation) and remained active in the subsequent stages of root hair growth. In the root hair mutants ttg and gl/2, which have hairs in all epidermal cells regardless of their position in relation to the underlying cortical cells, AtEXP7 was active in all cells producing hairs. Analysis of AtEXP7 expression in two other Arabidopsis mutants, axr2, which is defective in hair elongation but produces few root hair bulges, and rhd6, which develops almost no hair bulges, showed gene activity, although reduced, only in the axr2 mutant. The same expression pattern was observed for the AtEXP18 gene (Cho and Cosgrove, 2002).
In this study, the HvEXPB1 gene was not active in two completely hairless barley mutants, rhl1.a and brb, which did not develop even root hair bulges. HvEXPB1 was expressed, however, in the mutant rhp1.a, which produced root primordia shown as slightly elongated bulges, and in the mutant rhs1.a with very short root hairs, similar to the root-specific expansin genes AtEXP7 and AtEXP18 in Arabidopsis. Those results, supported by the analysis of HvEXPB1 expression in isolated root hair outgrowths, indicate that HvEXPB1 expression is probably root hair specific. Analysis of genetic relationships between barley mutants showed that the gene controlling the root-hairless phenotype was epistatic to genes responsible for inhibition of hair development at the transition to tip growth or at the hair elongation stage (Szarejko et al., 2005). Molecular and genetic data suggest that the HvEXPB1 gene is necessary for initiation of root hair development and acts upstream of the other genes involved in root hair growth.
As with AtEXP7 and AtEXP18 genes in Arabidopsis, the HvEXPB1 gene is already active in cells that do not yet show visible signs of bulge formation. In this experiment, expression of HvEXPB1 was observed in 1-mm-long root tips devoid of a differentiation zone in 6-d-old barley seedlings. The role of expansins in the initiation of root hair development was supported by the studies of Bibikova et al. (1998), who observed local acidification of the cell wall in place of subsequent root hair formation. It is a well-known phenomenon that acidification enhances activity of expansins (McQueen-Mason et al., 1992). Decreasing pH to a value of 4.0 to 4.5 strongly stimulated growth of root hairs in Lactuca sativa (Inoue et al., 2000). Accumulation of expansins in the cell wall domain in place of subsequent root hair outgrowth can promote local disassembling of cell wall components. In the next step, the locally loosened cell wall bulges out due to the high internal turgor pressure that might arise from apoplast acidification through activation of potassium channels. As a result, local depletion of cortical microtubules is observed in place of root hair bulge formation (Baluška et al., 2000). In this scenario, proper production of a specific expansin is the first step leading to the formation of root hair in trichoblasts.
In this study, we have successfully demonstrated the use of the cDNA-RDA approach for identification of genes expressed differentially in the mutant/parent variety system. The cDNA-RDA was developed on the basis of RDA of genomic sequences, described by Lisitsyn et al. (1993), and can be applied to identify genes differentially expressed in different individuals, organs, or tissues under various environmental conditions, different stages of development, etc. Combining PCR with a subtractive hybridization approach made this technique extremely sensitive and effective even in identification of differences between gene products expressed at less than one copy per cell (O'Neill and Sinclair, 1997). Additionally, what is an important advantage in the studies of poorly described genomes, such as barley, cDNA-RDA requires no prior knowledge of genome sequences and could help in identification of genes not previously described in the databases (Hubank et al., 2004). In the course of these experiments, after three series of subtractive hybridization, we have identified only one homogeneous, 228-bp-long fragment of a cDNA (973–1,200 of GenBank accession no. AY351786) with no statistically significant similarity to the GenBank expressed sequence tag genomic sequences even for barley and rice. Annotation of the gene represented by the identified cDNA fragment was possible after cloning of the full-length cDNA and flanking genomic sequences.
Although these studies clearly indicated a relation between HvEXPB1 expression and initiation of root hair formation in barley, no differences were found in the HvEXPB1 sequence, including the 603-bp promoter region, between the analyzed root-hairless mutants and their respective parent varieties. The cDNA-RDA approach used in this study is based on the differential appearance of the short cDNA fragments generated by a frequently cutting restriction enzyme in the two compared transcriptomes. It can detect a mutation in a gene responsible for a mutant phenotype only when the mutation results in lack of a gene transcript or its 3′ polyadenylated fragment. This, in turn, can be caused by deletion in the gene sequence, nucleotide substitution at the splice junction site, or any mutation at the regulatory regions. None of these were detected in the compared HvEXPB1 sequences of the analyzed mutants and their parent varieties. The other assumption explaining the lack of HvEXPB1 activity in the hairless mutants is a mutation in one of the genes encoding transcription factors responsible for HvEXPB1 regulation. Such a mutation could result in the malfunction of a mechanism controlling HvEXPB1 expression. Further studies are required to confirm this hypothesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Three root hair mutants obtained in the Department of Genetics, University of Silesia, by MNU treatment were used in the study. Mutant rhl1.a was completely hairless, mutant rhp1.a developed root hair primordia blocked at the bulge stage, and mutant rhs1.a had very short root hairs reaching about 10% of the length of hairs in the parent variety. The analyzed mutants were derived from spring barley (Hordeum vulgare) Karat, Dema, and Diva varieties, respectively. Root hair phenotypes in each mutant were controlled by a single recessive gene (Szarejko et al., 2005). In addition to the mutants produced by chemical mutagenesis, the spontaneous root-hairless mutant brb, found in the Pallas variety, was included in the experiments. Seeds of brb were kindly provided by Tara Gahoonia (Royal Veterinary and Agricultural University, Copenhagen). Mutants rhl1.a and brb proved to be allelic (B. Chmielewska, M. Kwasniewski, J. Mizeracka, and I. Szarejko, unpublished data).
In all experiments on gene cloning and root transcriptome analyses, 6-d-old seedlings were used for DNA and mRNA isolation. Seeds were surface sterilized with 20% bleach solution (v/v) and germinated in sterile aeroponic conditions using two glass tubes linked with Parafilm. For analysis of gene expression in further stages of plant development, plants were grown in pots with soil. All plants used in this study were grown in a growth room under a 16-/8-h photoperiod at 20°C.
cDNA-RDA
A cDNA-RDA experiment was carried out according to the original protocol of Hubank and Schatz (1994), with modifications of Wallrapp and Gress (2001). The tester and driver populations were prepared after Lisitsyn et al. (1994), who suggested genetically directed designing of the probes for generating markers linked to a trait of interest. As a tester population, 10 wild-type plants of the F2 generation from the rhl1.a × Karat cross were used. Ten root-hairless F2 individuals from the same cross served as a driver population. This manner of probe creation made elimination from the reaction differences not linked directly to the analyzed phenotype possible.
Total RNA from roots of each population was isolated using TRIzol reagent (Life Technologies/Invitrogen) according to the manufacturer's instructions. Poly(A)+ mRNA was isolated from 100 μg of RNA using the PolyATtract mRNA isolation system III (Promega). ds cDNA was synthesized using the cDNA synthesis system (Roche). Two micrograms of ds cDNA, used as a starting material for amplicon preparation, were digested with DpnII (New England Biolabs), purified, and ligated to RDpn adaptors (8 μg of RDpn24 oligo/4 μg of RDpn12 oligo; Table I) in a 60-μL reaction using 600 units of T4 DNA ligase (New England Biolabs) at 16°C overnight. Before addition of the enzyme, the oligonucleotides were annealed to each other and to cDNA by incubation of the reaction in 55°C on a heating block for 5 min and then cooling to 10°C over a period of 1.5 h. After ligation, probes were subjected to multiple PCR reactions using RDpn24 oligo to generate at least 120 μg of the driver and 20 μg of the tester amplicons. After amplicon preparation, RDpn adaptors were removed by DpnII restriction and further purification. A new set of JDpn adaptors was ligated to the tester amplicon, as described above. The first round of subtractive hybridization was carried out by combining 400 ng of the tester amplicon and 40 μg of the driver amplicon. After denaturation at 98°C, temperature was decreased to 67°C and probes were hybridized for 24 h. After hybridization, probes were amplified in 14 PCR cycles using JDpn24 oligos. Amplification products were purified and treated with mung bean (Vigna radiata) nuclease (New England Biolabs). After treatment, probes were reamplified, the obtained products were purified, and JDpn adaptors were replaced, as described previously, with a new set of NDpn adaptors. Two additional rounds of subtractive hybridization in the tester:driver ratios 1:800 and 1:400,000 were carried out, as previously. After the last round of hybridization, probes were amplified in 28 cycles of PCR and resolved by agarose gel electrophoresis. The differentiation product was isolated from the agarose gel using the Qiaex II gel extraction kit (Qiagen), cloned in pGEM-T Easy vector (Promega), and sequenced. The detailed protocol of cDNA-RDA methodology used in this study is available from the authors upon request.
Table I.
Primer sequences used during HvEXPB1 gene isolation, cloning, sequencing, and expression profiling (see text for details)
| Primer Name | Sequence 5′–3′ |
|---|---|
| RDpn24 | AGCACTCTCCAGCCTCTCACCGCA |
| RDpn12 | GATCTGCGGTGA |
| JDpn24 | ACCGACGTCGACTATCCATGAACA |
| JDpn12 | GATCTGTTCATG |
| NDpn24 | AGGCAACTGTGCTATCCGAGGGAA |
| NDpn12 | GATCTTCCCTCG |
| DP1GW1F | CCTTGTCAAAGGCCCGTATCCGGTAA |
| DP1GW1FN | GGCCGCGTCAGAAGAGGAAGAAAGTC |
| DP1GW1R | CATCAGCATCGATCCTTTGCTGTGAC |
| DP1GW1RN | TCGGAGAGGGAAGAGCGAAGAACACA |
| DP1GW2R | TGCCGCTCATGTCGAAATGATCCTC |
| DP1GW2RN | AATGATCCTCCGAGGCGCATATGC |
| DP1GW3R | TAGCATGCACCGCAGCCTTTGC |
| DP1GW3RN | GCCGACGGCGCTTTGGTATCCG |
| M13F | TGTAAAACGACGGCCAGT |
| M13R | CAGGAAACAGCTATGACC |
| DP1F | TACCTTGTCAAAGGCCCGTA |
| DP1R | CCTTTGCTGTGACTACAACTGAA |
| UBQF | GAGGCCCAAGAAGAAGATCA |
| UBQR | ACGATAGGAGGCTGAAAGCA |
RACE, GenomeWalking, and Sequencing
The ends of the HvEXPB1 mRNA were cloned by RACE reactions using the GeneRacer kit (Invitrogen) with total RNA isolated from roots of the wild-type Karat variety. PCR products were cloned in pGEM-T Easy vector (Promega) and sequenced.
The genomic sequence of the HvEXPB1 gene was obtained by cloning with the GenomeWalking strategy using the GenomeWalker kit (CLONTECH). The genomic libraries were created using the DNA isolated from leaves of the Karat variety with the use of DraI, EcoRV, PvuII, and StuI restriction enzymes. PCR amplification of DNA fragments from genomic libraries was carried out with the use of DyNAzym EXT polymerase mix (Finnzymes) in a touchdown reaction according to the following temperature profile: 94°C, 2 min; 80°C, 3 min (HotStart); 94°C, 30 s; 72°C, 4 min (last two steps repeated three times); 94°C, 30 s; 70°C, 4 min (repeated three times); 94°C, 30 s; 68°C, 4 min (repeated three times); 94°C, 30 s; 66°C, 4 min (repeated 31 times); 66°C, 7 min; and 4°C (pause). The whole genomic sequence containing the HvEXPB1 gene was cloned in four steps by nested PCR with primers presented in Table I. PCR products were isolated from agarose gels using the Qiaex II gel extraction kit (Qiagen), cloned in pGEM-T Easy vector (Promega), and sequenced.
All sequencing analyses were carried out in simultaneous bidirectional sequencing reactions using the SequiTherm EXCEL II DNA sequencing kit-LC (Epicentre). DNA templates were cloned in pGEM-T Easy plasmids (Promega). M13F and M13R primers, labeled with IRD-700 and IRD-800 dyes, respectively (MWG-Biotech), were used in the sequencing reactions. Sequencing products were separated on 6% polyacrylamide gel (acrylamide/Bis 29:1; Sigma) with the use of a LI-COR IR2 DNA sequencer.
Expression Analysis
All expression analyses related to this study were carried out in a qualitative manner by RT-PCR. For most experiments, total RNA was isolated using TRIzol reagent (Invitrogen). The exception was the analysis of HvEXPB1 expression in the root tips and in isolated root hair outgrowth, where polyA+ mRNA was isolated using the Dynabeads mRNA direct micro kit (Dynal Biotech). In all cases prior to RT, to prevent DNA contamination, RNA was treated with RQ1 RNase-free DNase (Promega) for 30 min at 37°C using 1 unit of enzyme for 1 μg of RNA. After DNase digestion, mRNA was reverse transcribed using the RevertAid first-strand cDNA synthesis kit (Fermentas), following the manufacturer's instructions. In HvEXPB1 expression analyses, primers DP1F and DP1R (Table I) complementary to the highly specific 3′-UTR region were used. PCR was performed according to the following temperature profile: 94°C, 3 min; 94°C, 45 s; 58°C, 30 s; 72°C, 1 min (last three steps repeated three times); 94°C, 45 s; 57°C, 30 s; 72°C, 1 min (repeated three times); 94°C, 45 s; 56°C, 30 s; 72°C, 1 min (repeated 32 times; in the case of HvEXPB1 expression analysis in isolated root hair outgrowth, this cycle was repeated 42 times); 72°C, 5 min; and 4°C (pause). As a positive control, RT-PCR amplifications of the ubiquitin/ribosomal protein CEP52 gene were carried out using UBQF and UBQR primers at the temperature profile identical to HvEXPB1 expression analysis.
Analysis of HvEXPB1 expression in isolated root hair outgrowth was carried out using 6-d-old seedlings of the Karat variety. Approximately 3-cm-long fragments with completely differentiated root hairs were excised from three roots using sterile forceps, placed on RNase-free microscopic slides on an ice-cold base of a stereomicroscope, and immediately covered by 100 μL of Lysis/Binding buffer (Dynabeads mRNA direct micro kit; Dynal Biotech). During the next 2 min, root hair outgrowths were carefully cut out directly to the buffer using a sterile scalpel blade, then sucked out from the slide using a pipette tip, and used for polyA+ mRNA isolation, cDNA synthesis, and further PCR amplification were as described.
Bioinformatics
All gene-specific primers used in this study were designed using Jellyfish 1.5 software (http://jellyfish.labvelocity.com). Analyses of a gene and its mRNA structure, as well as conceptual translation, was carried out using GENSCAN software (Burge and Karlin, 1997, 1998; http://genes.mit.edu/GENSCAN) with maize (Zea mays) weight matrixes and limiting values equal to 1. For comparative analysis of expansin amino acid sequences, ClustalW 1.8 software was used (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) with a PAM250 substitution matrix. Results of alignments were visualized using BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). Analysis of the EXPB1 sequence for recognition of signal peptides was carried out using PSORT (Nakai and Horton, 1999; http://psort.nibb.ac.jp) and SOSUI software (Mitaku et al., 2002; http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0E.html) with standard settings.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY351785 and AY351786.
Acknowledgments
The authors are grateful to Dr. Tara Gahoonia (Royal Veterinary and Agricultural University, Copenhagen) for kindly providing seeds of the mutant brb and the Pallas variety. We also thank Dr. Jagna Karcz (Scanning Electron Microscopy Laboratory, University of Silesia) for scanning electron microscope photographs, and Damian Gruszka (Department of Genetics) for technical assistance.
This work was supported by the International Atomic Energy Agency (project no. 10763) and the Polish Committee for Scientific Research (grant no. 3–P04C–090–25).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Iwona Szarejko (szarejko@us.edu.pl).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.078626.
References
- Appel M, Bellstedt DU, Gresshoff PM (1999) Differential display of eukaryotic mRNA: meeting the demands of the new millennium? J Plant Physiol 154: 561–570 [Google Scholar]
- Baluška F, Salaj J, Matur J, Braun M, Kasper F, Šamaj J, Chua N-H, Barlow PW, Volkmann D (2000) Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profiling-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev Biol 227: 618–632 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J (2003) The bHLH genes GLABRA 3 (GL3) and ENHANCER OF GLABRA 3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Jacob T, Dahse I, Gilroy S (1998) Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125: 2925–2934 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P (1999) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11: 2203–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Burge CB, Karlin S (1998) Finding the genes in genomic DNA. Curr Opin Struct Biol 8: 346–354 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Cho H-T, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14: 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Engvild KC, Rasmussen SK (2004) Root hair mutants of barley. Barley Genet Newsl 34: 13–15 [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonek K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Frohme M, Scharm B, Delius H, Knecht R, Hoheisel JD (2000) Use of representational difference analysis and cDNA arrays for transcriptional profiling of tumor tissue. Ann NY Acad Sci 910: 85–105 [DOI] [PubMed] [Google Scholar]
- Gahoonia TS, Care D, Nielsen NE (1997) Root hairs and phosphorus acquisition of wheat and barley cultivars. Plant Soil 191: 181–188 [Google Scholar]
- Gahoonia TS, Nielsen NE, Joshi PA, Jahoor A (2001) A root hairless barley mutant for elucidating genetic of root hairs and phosphorus uptake. Plant Soil 235: 211–219 [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Grabov A, Bottger M (1994) Are redox reactions involved in regulation of K+ channels in the plasma membrane of Limnobium stoloniferum root hairs? Plant Physiol 105: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson CS, Roberts K, Feldmann KA, Dolan L (1997) The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol 115: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Kemp AC, Grierson CS (2005) The TIP GROWTH DEFECTIVE1 S-acyltransferase regulates plant cell growth in Arabidopsis. Plant Cell 17: 2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhong R, Morrison WH III, Ye ZH (2003) The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217: 912–921 [DOI] [PubMed] [Google Scholar]
- Hubank M, Bryntesson F, Regan J, Schatz DG (2004) Cloning of apoptosis-related genes by representational difference analysis of cDNA. Methods Mol Biol 282: 255–273 [DOI] [PubMed] [Google Scholar]
- Hubank M, Schatz DG (1994) Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res 22: 5640–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Yamaoka K, Kimura K, Sawai K, Arai T (2000) Effects of low pH on the induction of root hair formation in young lettuce (Lactuca sativa L. cv. Grand Rapids) seedlings. J Plant Res 113: 39–44 [Google Scholar]
- Kende H, Bradford K, Brummell D, Cho HT, Cosgrove DJ, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, et al (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55: 311–314 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lauter FR, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB (1996) Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA 93: 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-K, Ahn JH, Song S-K, Choi YD, Lee JS (2003) Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol 131: 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (2001) Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128: 1539–1546 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kende H (2001) Expression of β-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol 127: 645–654 [PMC free article] [PubMed] [Google Scholar]
- Li HY, Guo ZF, Zhu YX (1998) Molecular cloning and analysis of a pea cDNA that is expressed in darkness and very rapidly induced by gibberellic acid. Mol Gen Genet 259: 393–397 [DOI] [PubMed] [Google Scholar]
- Li L-C, Bedinger PA, Volk C, Jones AD, Cosgrove DJ (2003. a) Purification and characterization of four β-expansins (Zea m 1 isoforms) from maize pollen. Plant Physiol 132: 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S (2003. b) Expansins and cell growth. Curr Opin Plant Biol 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Liang P (2002) A decade of differential display. Biotechniques 33: 338–346 [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967–971 [DOI] [PubMed] [Google Scholar]
- Ling JQ, Kojima T, Shiraiwa M, Takahara H (2003) Cloning of two cysteine proteinase genes, CysP1 and CysP2, from soybean cotyledons by cDNA representational difference analysis. Biochim Biophys Acta 1627: 129–139 [DOI] [PubMed] [Google Scholar]
- Lisitsyn N, Lisitsyn N, Wigler M (1993) Cloning the differences between two complex genomes. Science 259: 946–951 [DOI] [PubMed] [Google Scholar]
- Lisitsyn NA, Segre JA, Kusumi K, Lisitsyn NM, Nadeau JH, Frankel WN, Wigler MH, Lander ES (1994) Direct isolation of polymorphic markers linked to a trait by genetically directed representational difference analysis. Nat Genet 6: 57–63 [DOI] [PubMed] [Google Scholar]
- Long SR (1996) Rhizobium symbiosis: nod factors in perspective. Plant Cell 8: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1994) The rhd6 mutant of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall expansion in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitaku S, Hirokawa T, Tsuji T (2002) Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18: 608–616 [DOI] [PubMed] [Google Scholar]
- Mylona P, Pawlowski K, Bisseling T (1995) Symbiotic nitrogen fixation. Plant Cell 7: 869–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–35 [DOI] [PubMed] [Google Scholar]
- O'Neill MJ, Sinclair AH (1997) Isolation of rare transcripts by representational difference analysis. Nucleic Acids Res 25: 2681–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C (1998) Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 10: 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit specific and ripening-regulated. Proc Natl Acad Sci USA 94: 5955–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW (2000) Constructing a plant cell: the genetic control of root hair development. Plant Physiol 124: 1525–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K (1998) The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev 12: 2013–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkhovich BZ, Kloner RA, Poizat C, Marjoram P, Kedes LH (2003) Gene expression profiling—a new approach in the study of myocardial ischemia. Cardiovasc Pathol 12: 180–185 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Taketa S, Ichii M (2003) Morphological and physiological characteristics of a root-hairless mutant in rice (Oryza sativa L.). Plant Soil 255: 9–17 [Google Scholar]
- Szarejko I, Janiak A, Chmielewska B, Nawrot M (2005) Genetic analysis of several root hair mutants of barley. Barley Genet Newsl 35: 36–38 [Google Scholar]
- Vissenberg K, Fry SC, Verbelen J-P (2001) Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol 127: 1125–1135 [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K (2002) Role of positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419 [DOI] [PubMed] [Google Scholar]
- Wallrapp C, Gress TM (2001) Isolation of differentially expressed genes by representational difference analysis. Methods Mol Biol 175: 279–294 [DOI] [PubMed] [Google Scholar]
- Wang H, Lee MM, Schiefelbein JW (2002) Regulation of the cell expansion gene RHD3 during Arabidopsis development. Plant Physiol 129: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T-J, Hochholdinger F, Sauer M, Bruce W, Schnable PS (2005) The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol 138: 1637–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T-J, Schnable PS (1994) Analyses of mutants of three genes that are influence root hair development in Zea mays (Gramineae) suggest that root hairs are dispensable. Am J Bot 81: 833–842 [Google Scholar]
- White PJ (1998) Calcium channels in the plasma membrane of root cells. Ann Bot (Lond) 81: 173–183 [Google Scholar]
- Wu Y, Meeley RB, Cosgrove DJ (2001) Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiol 126: 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer CL, Bibikova TN, Gilroy S (1997) Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J 12: 427–439 [DOI] [PubMed] [Google Scholar]
- Yoon DY, Buchler P, Saarikoski ST, Hines OJ, Reber HA, Hankinson O (2001) Identification of genes differentially induced by hypoxia in pancreatic cancer cells. Biochem Biophys Res Commun 288: 882–886 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zhang Y, Luo J, Davies PJ, Ho DT (1998) PPF-1, a post-floral-specific gene expressed in short-day-grown G2 pea, may be important for its never-senescing phenotype. Gene 208: 1–6 [DOI] [PubMed] [Google Scholar]