Abstract
In many plant species, a subset of the genes of the chloroplast genome is transcribed by RpoTp, a nuclear-encoded plastid-targeted RNA polymerase. Here, we describe the positional cloning of the SCABRA3 (SCA3) gene, which was found to encode RpoTp in Arabidopsis (Arabidopsis thaliana). We studied one weak (sca3-1) and two strong (sca3-2 and sca3-3) alleles of the SCA3 gene, the latter two showing severely impaired plant growth and reduced pigmentation of the cotyledons, leaves, stem, and sepals, all of which were pale green. The leaf surface was extremely crumpled in the sca3 mutants, although epidermal cell size and morphology were not perturbed, whereas the mesophyll cells were less densely packed and more irregular in shape than in the wild type. A significant reduction in the size, morphology, and number of chloroplasts was observed in homozygous sca3-2 individuals whose photoautotrophic growth was consequently perturbed. Microarray analysis showed that several hundred nuclear genes were differentially expressed in sca3-2 and the wild type, about one-fourth of which encoded chloroplast-targeted proteins. Quantitative reverse transcription-PCR analyses showed that the sca3-2 mutation alters the expression of the rpoB, rpoC1, clpP, and accD plastid genes and the SCA3 paralogs RpoTm and RpoTmp, which respectively encode nuclear-encoded mitochondrion or dually targeted RNA polymerases. Double-mutant analysis indicated that RpoTmp and SCA3 play redundant functions in plant development. Our findings support a role for plastids in leaf morphogenesis and indicate that RpoTp is required for mesophyll cell proliferation.
Leaf development is regulated by environmental signals and endogenous cues, together with their cross talk, by means of genetic networks that modulate cell division, expansion, and differentiation. Light is one of the most important signals controlling leaf development because it triggers differentiation of nonphotosynthetic proplastids into fully functional photosynthetic chloroplasts (López-Juez and Pyke, 2005; Waters and Pyke, 2005). The identification and characterization in different plant species of mutants with aberrant leaf anatomy, affected in nuclear genes encoding proteins with chloroplast-related functions, highlights the important role of chloroplast biogenesis in leaf morphogenesis and overall plant development. Thus, the Antirrhinum majus differentiation and greening (dag) mutant (Sommer et al., 1985; Chatterjee et al., 1996), the Arabidopsis (Arabidopsis thaliana) mutants pale cress (pac; Reiter et al., 1994; Grevelding et al., 1996), DAG-like1 (dal1; Babiychuk et al., 1997; despite the sequence similarity between DAL1 and DAG proteins, the corresponding genes are not orthologs [Bisanz et al., 2003]), cloroplastos alterados1 (cla1; Mandel et al., 1996), yellow variegated1 and 2 (var1 and var2; Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002), immutans (im; Wetzel et al., 1994; Carol et al., 1999; Wu et al., 1999), CAB underexpressed1 (cue1; Li et al., 1995; Streatfield et al., 1999), and phosphatidylglycerolphosphate synthetase1 (pgp1; Hagio et al., 2002) and the tomato (Lycopersicon esculentum) defective chloroplast and leaves-mutable (dcl; Keddie et al., 1996) mutant are albinos or display pale or whitish sectors as a consequence of damaged nuclear genes encoding chloroplast proteins. The fact that the structure of the internal leaf tissues is altered in all of these mutants suggests a connection between chloroplast and mesophyll development (Chatterjee et al., 1996; Keddie et al., 1996) and supports the hypothesis that leaf morphogenesis is controlled by plastid-to-nucleus signaling (Pyke et al., 2000; Rodermel, 2001).
The effect on leaf and whole-plant development of mutations in genes encoding proteins of the transcriptional machinery of plastids is beginning to emerge. The transcription of chloroplast genes is carried out by two types of plastid RNA polymerases (RNAPs), which are plastid encoded (PEP) or nuclear encoded (NEP; for review, see Hess and Börner, 1999). PEPs are multisubunit polymerases that require σ-like factors encoded by the nucleus, similar to the RNAPs found in eubacteria such as Escherichia coli (Igloi and Kössel, 1992; Allison et al., 1996; Allison, 2000), whereas NEPs belong to the family of bacteriophage-type T3 and T7 RNAPs (RpoTs; Lerbs-Mache, 1993). It has been proposed that transcription of the photosynthetic genes harbored by the plastid genome is preferentially driven by PEP, whereas NEP transcribes genes encoding components of the plastid genetic system, such as those of the translational apparatus and PEP core subunits (Allison et al., 1996; Hajdukiewicz et al., 1997; Magee and Kavanagh, 2002). However, other studies indicate an overlap of PEP and NEP activities (Krause et al., 2000; Legen et al., 2002).
Two dually targeted organelle NEPs have been identified in the moss Physcomitrella patens (Richter et al., 2002). Monocotyledonous plants possess one NEP specific to mitochondria and another specific to chloroplasts (Ikeda and Gray, 1999; Emanuel et al., 2004; Kusumi et al., 2004). Three organelle NEPs have been identified in Arabidopsis and other dicotyledonous species: RpoTm is targeted to the mitochondria, RpoTp to the chloroplasts, and RpoTmp to both of these organelles (Hedtke et al., 1997, 1999, 2000). Orthologs of the RpoT genes of Arabidopsis have been cloned in P. patens (Richter et al., 2002), Chenopodium album (Weihe et al., 1997), Hordeum vulgare (Emanuel et al., 2004), tobacco (Nicotiana tabacum), Nicotiana sylvestris, Nicotiana tomentosiformis (Kobayashi et al., 2001; Hedtke et al., 2002), Oryza sativa (Kusumi et al., 2004), Triticum aestivum (Ikeda and Gray, 1999), and Zea mays (Chang et al., 1999). It is assumed that plastid-type RpoT genes derive from an original mitochondrion-type RpoT gene by gene duplication (Hess and Börner, 1999; Kabeya et al., 2002). Mutational analyses on the role of RpoT genes in plant development are scarce, one of the few exceptions being the recent isolation and characterization of the rpoT;2 mutant of Arabidopsis (Baba et al., 2004), which exhibits altered leaf morphology and a general reduction in growth, consistent with the assumed relationship between chloroplast and leaf development.
To gain insight into the genetic and molecular mechanisms involved in the control of leaf ontogeny, we performed a large-scale screening for ethyl methanesulfonate (EMS)-induced mutants with abnormal leaves in Arabidopsis (Berná et al., 1999). We report here our studies of one of these EMS-induced mutations, scabra3-1 (sca3-1), together with that of two additional insertional alleles of the SCA3 gene, which was positionally cloned and found to encode the RpoTp protein. Loss of function of SCA3 affected plastid and nuclear gene expression in Arabidopsis, severely perturbing chloroplast development and leaf morphogenesis.
RESULTS
Positional Cloning of the SCA3 Gene
In a large-scale screening for EMS-induced mutants with altered leaf morphology, we isolated seven mutants of Arabidopsis displaying rounded and pale-green leaves, protruding leaf laminae, and irregular leaf margins (Fig. 1). The corresponding mutations were found to be recessive and completely penetrant, with only small variations in expressivity. They were grouped in a phenotypic class that we named Scabra (Sca) and found to fall into five complementation groups (SCA1 to SCA5; Berná et al., 1999). The SCA genes have been low-resolution mapped (Robles and Micol, 2001).
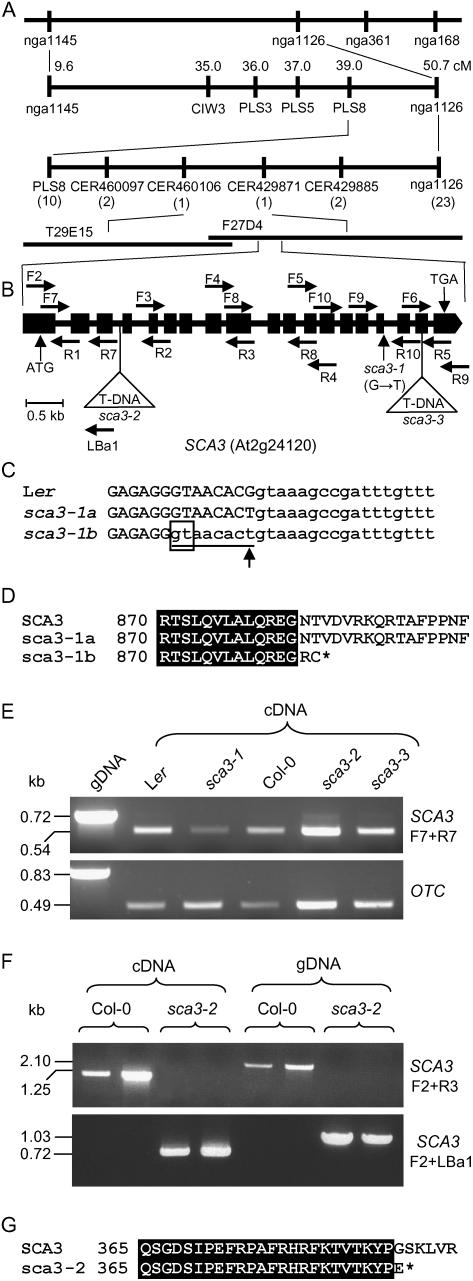
Figure 1.
Some morphological traits of the phenotype of sca3 mutants. Images correspond to cotyledons of 7-d-old seedlings (A–D), 21-d-old rosettes (E–H), and third node vegetative leaves (I–L) of the sca3-1 and sca3-2 mutants and their corresponding wild types (Ler and Col-0, respectively). Six-week-old plants (M) grown on soil and details of their siliques (N and O) and inflorescences (P and Q). R, Fifteen-day-old plants vertically grown on agar medium. All plants were homozygous for the mutations shown. Scale bars indicate 1 mm (A–D, H–L, and N–Q), 5 mm (E–G), 10 mm (R), and 25 mm (M).
To positionally clone the SCA3 gene, we performed outcrosses of sca3-1/sca3-1 plants in a Landsberg erecta (Ler) genetic background to Columbia-0 (Col-0). Linkage analysis of the F2-mapping populations obtained allowed us to delimit a candidate region of 70 kb, containing 16 annotated genes (Fig. 2A). We then searched for publicly available lines bearing T-DNA insertions within the candidate interval and found two (N593884 and N567191) that displayed a recessive phenotype more extreme than that of the sca3-1 mutant (Fig. 1). T-DNA insertions were confirmed at nucleotide positions 1,894 (numbering from the predicted translation initiation codon) in N593884 and 5,194 in N567191, disrupting the third intron and eighteenth exon, respectively, of the At2g24120 transcription unit (Fig. 2B), as annotated at the SIGnAL Web site (http://signal.salk.edu). Allelism tests were performed by crossing sca3-1/sca3-1 plants to phenotypically mutant N593884 and N567191 individuals, and the F1 progeny were phenotypically mutant. Because these results indicated that the gene perturbed in the EMS-derived sca3-1 mutant is At2g24120, we consequently named the T-DNA alleles carried by the N593884 and N567191 lines as sca3-2 and sca3-3, respectively. Given that the phenotypes of the latter two mutants were indistinguishable, the sca3-2 allele was chosen for further study.
Figure 2.
Positional cloning and structural analysis of the SCA3 gene. A, Map-based strategy followed to identify the SCA3 gene. After studying 882 chromosomes, a total of 33 recombinant events (in parentheses) were identified in a region of 11.7 cm on chromosome 2, flanked by the PLS8 and nga1126 markers. A candidate interval of 70 kb was finally delimited, flanked by the CER460106 and CER429871 markers and encompassing the T29E15 and F27D4 bacterial artificial chromosome clones, which included the At2g24060 to At2g24200 annotated genes. B, Structure of the SCA3 gene, with indication of the position and nature of the sca3 mutations. Exons and introns are indicated by black boxes and lines, respectively. Triangles indicate T-DNA insertions. Horizontal arrows indicate the oligonucleotides used to characterize the structure and expression of SCA3, which are not drawn to scale. C, Alignment of the sixteenth exon-intron junction region of the SCA3 gene in the wild-type Ler and the sca3-1 mutant. The sequences shown correspond to the single splicing pattern found in the wild-type SCA3 allele (Ler) and the two detected in the sca3-1 mutant (here named sca3-1a and sca3-1b). Upper- and lowercase letters indicate exonic and intronic sequences, respectively. The eight-nucleotide segment absent from sca3-1b mRNA is underlined. An arrowhead indicates the single-nucleotide change found in sca3-1 genomic DNA. The cryptic splicing donor site within exon 16 used in sca3-1b is boxed. D, Alignment of the amino acid sequences corresponding to the C-terminal part of the protein products of the wild-type SCA3 and the mutant sca3-1 alleles of the SCA3 gene. An asterisk denotes a stop codon. E, Expression analysis of the SCA3 gene by RT-PCR in sca3/sca3 and wild-type individuals. The bands shown were obtained after PCR amplifications were performed using as a template genomic DNA (gDNA) or RNA extracted from 3-week-old plants and reverse transcribed, and the primers shown. The OTC gene was used as an internal control. F, Expression analysis of the SCA3 gene by RT-PCR in sca3-2/sca3-2 and Col-0 individuals. The bands shown were obtained after PCR amplifications performed using as a template gDNA or RNA extracted from 3-week-old plants and reverse transcribed, and the primers shown. G, Alignment of the amino acid sequences corresponding to the protein products of the wild-type SCA3 and the mutant sca3-2 alleles of the SCA3 gene. An asterisk denotes a stop codon.
Molecular Characterization of sca3 Alleles
We sequenced the At2g24120 transcription unit in the sca3-1 mutant and its wild-type Ler and found a G → T transversion at position 4,856, affecting the splicing donor site of the sixteenth exon (Fig. 2, B and C). To examine the effect of the sca3 mutations on SCA3 gene expression, total RNA was extracted from 3-week-old sca3/sca3, Ler, and Col-0 plants and subjected to reverse transcription (RT)-PCR, and the amplification products were sequenced. Using primers that hybridize upstream of both T-DNA insertions (At2g24120F7 and R7; Supplemental Table I; Fig. 2, B and E), a single band of the expected size (548 bp) was detected in all cases, corresponding to similar amounts of RT-PCR products from sca3-2/sca3-2, sca3-3/sca3-3, and Col-0 plants, whereas slightly fewer amplification products were obtained from sca3-1/sca3-1 than from Ler plants (Fig. 2E). Similar results were obtained using different primer set combinations.
Two different SCA3 cDNAs were found in the sca3-1 mutant (see “Materials and Methods;” Supplemental Fig. 1), one of which carries only a G → T change, and the other lacks 8 bp (Fig. 2C). As a consequence, translation of sca3-1 mRNAs would produce two proteins, one of which has no amino acid difference with the wild type and the other lacks 109 amino acids of its C-terminal region due to a frameshift and a premature stop codon (Fig. 2D). Primers flanking the T-DNA insertions (F2 and R3 for sca3-2; F4 and R9 for sca3-3) yielded amplification products only from Col-0, but not from sca3-2 and sca3-3 cDNA or genomic DNA (Fig. 2F). Sequencing of additional amplification products (obtained using the F2 and LBa1 primers; Fig. 2, B and F) indicated that a chimeric transcript is obtained in sca3-2, which includes 1,667 nucleotides of SCA3 mRNA and at least 272 nucleotides of the left border of the T-DNA insert. Translation of this chimeric transcript would produce a truncated protein of 389 amino acids, lacking 604 residues of the C-terminal part of the wild-type protein (Fig. 2G). We did not characterize sca3-3 mRNA, which was assumed to encode a protein lacking 50 amino acids in its C-terminal region and probably including some divergent amino acids, translated from the T-DNA insert.
Effects of sca3 Mutations on Expression of RpoT Nuclear Genes
The predicted product of the SCA3 gene is a protein of 993 amino acids with a molecular mass of 112.6 kD (http://www.arabidopsis.org/index.jsp) corresponding to the T7 phage-type RNAP, RpoTp, which is targeted exclusively to plastids (Hedtke et al., 1997, 1999). RpoTp is a single-subunit RNAP that contains 11 well-conserved domains (Fig. 3, I–XI; McAllister and Raskin, 1993). The frameshift caused by the sca3-1 mutation would remove the X and XI domains, including the so-called C-motif, which is part of the catalytic site of T7 RNAPs. The sca3-2 mutation would remove the III to XI domains (Fig. 3), which suggests that it abolishes RpoTp function.
Figure 3.
Predicted amino acid sequence of the Arabidopsis SCA3 protein (RpoTp; Y08463). Residues shaded in black are identical across the sequences of homologous gene products from H. vulgare (CAD45446), T. aestivum (AF091839), Z. mays (AF127022), O. sativa (AB120430), N. sylvestris (AJ302020), and tobacco (AJ416570). Residues with similar chemical properties conserved across all seven sequences are shaded gray. The alignment was obtained using ClustalX, version 1.5b. Continuous and dotted thin lines indicate the domains (I–XI) and the C-motif that are conserved among T7 phage-type NEP plastid RNAPs, respectively (McAllister and Raskin, 1993; Chang et al., 1999). The amino acid sequences deleted in the mutant protein products of sca3-1b (see Fig. 2), sca3-2, and sca3-3 are indicated by white boxes, thick discontinuous lines, and black boxes, respectively.
Using quantitative RT (qRT)-PCR, we studied the expression of the RpoTm, RpoTmp, and SCA3 (RpoTp) genes of Arabidopsis in 4-d-old seedlings (showing only expanded cotyledons), 12-d-old seedlings (showing expanded cotyledons and four expanding leaves), 3-week-old rosettes (see Fig. 1, G and H), and roots (Table I). The three genes were found to be expressed in all the organs and stages analyzed in wild-type and sca3-2/sca3-2 plants. In Col-0, RpoTm reached the highest level of expression in 12-d-old seedlings, where it was 1.9-, 1.5-, and 2.1-fold higher than in 4-d-old seedlings, rosettes, and roots, respectively. On the contrary, RpoTmp and SCA3 were predominantly expressed in rosettes, especially SCA3, whose transcripts accumulated 3.3-, 2.5-, and 2.8-fold higher than in 4- and 12-d-old seedlings and roots, respectively (data not shown). Compared with Col-0, we found small changes in the expression of the RpoT genes in sca3-2/sca3-2 seedlings, rosettes, and roots. In sca3-2/sca3-2 individuals, RpoTm and RpoTmp showed higher transcript levels than in Col-0 in 4-d-old seedlings and 21-d-old plants, whereas a reduction was found for both genes in 12-d-old seedlings (Table I). SCA3 was down-regulated in the mutant in all developmental stages studied, especially in 4-d-old seedlings. Root transcript levels of the RpoT genes were similar in sca3-2/sca3-2 and Col-0 plants, the only exception being SCA3, for which a 2-fold increase was detected in the mutant.
Table I.
qRT-PCR analyses of the expression of RpoT and plastid genes
Relative expression values were determined as 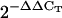 for each studied gene in the sca3-2 mutant after normalization with those of the OTC gene, and compared with those of wild-type Col-0, to which a value of 1 was given. Numbers in parentheses indicate the range of variation of
for each studied gene in the sca3-2 mutant after normalization with those of the OTC gene, and compared with those of wild-type Col-0, to which a value of 1 was given. Numbers in parentheses indicate the range of variation of 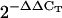 values of the studied gene obtained in two independent experiments using two different biological replicates and triplicate reactions. The range of variation of
values of the studied gene obtained in two independent experiments using two different biological replicates and triplicate reactions. The range of variation of 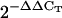 values in Col-0 for the RpoTm, RpoTmp, SCA3, rpoB, accD, clpP, rps18, and rpoC1 genes, respectively, were as follows: 0.69 to 1.44, 0.66 to 1.50, 0.78 to 1.27, 0.74 to 1.33, 0.67 to 1.42, 0.63 to 1.58, 0.68 to 1.45, and 0.66 to 1.50 in 4-d-old-seedlings; 0.72 to 1.38, 0.95 to 1.04, 0.55 to 1.81, 0.63 to 1.58, 0.97 to 1.02, 0.63 to 1.58, 0.87 to 1.14, and 0.98 to 1.01 in 12-d-old seedlings; 0.78 to 1.26, 0.46 to 2.15, 0.99 to 1.00, 0.59 to 1.67, 0.30 to 3.30, 0.82 to 1.20, 0.92 to 1.08, and 0.32 to 3.09 in rosettes. The range of variation for the RpoTm, RpoTmp, and SCA3 genes in Col-0 roots was 0.40 to 2.46, 0.33 to 3.00, and 0.40 to 2.48, respectively. N.D., Not determined.
values in Col-0 for the RpoTm, RpoTmp, SCA3, rpoB, accD, clpP, rps18, and rpoC1 genes, respectively, were as follows: 0.69 to 1.44, 0.66 to 1.50, 0.78 to 1.27, 0.74 to 1.33, 0.67 to 1.42, 0.63 to 1.58, 0.68 to 1.45, and 0.66 to 1.50 in 4-d-old-seedlings; 0.72 to 1.38, 0.95 to 1.04, 0.55 to 1.81, 0.63 to 1.58, 0.97 to 1.02, 0.63 to 1.58, 0.87 to 1.14, and 0.98 to 1.01 in 12-d-old seedlings; 0.78 to 1.26, 0.46 to 2.15, 0.99 to 1.00, 0.59 to 1.67, 0.30 to 3.30, 0.82 to 1.20, 0.92 to 1.08, and 0.32 to 3.09 in rosettes. The range of variation for the RpoTm, RpoTmp, and SCA3 genes in Col-0 roots was 0.40 to 2.46, 0.33 to 3.00, and 0.40 to 2.48, respectively. N.D., Not determined.
| Genes | Normalized Transcript Levels in the sca3-2 Mutant Relative to Col-0
|
|||
|---|---|---|---|---|
| 4-d-Old Seedlings | 12-d-Old Seedlings | Rosettes | Roots | |
| RpoTm | 1.97 (1.32–2.95) | 0.67 (0.49–0.92) | 1.50 (1.40–1.60) | 1.10 (0.34–3.60) |
| RpoTmp | 1.45 (0.99–2.12) | 0.74 (0.71–0.77) | 1.38 (0.68–2.80) | 1.03 (0.32–3.31) |
| SCA3 | 0.32 (0.31–0.33) | 0.95 (0.64–1.40) | 0.72 (0.66–0.80) | 2.07 (1.13–3.79) |
| rpoB | 0.19 (0.05–0.74) | 0.54 (0.31–0.95) | 0.72 (0.26–1.96) | N.D. |
| accD | 0.33 (0.23–0.48) | 1.42 (1.02–1.97) | 1.02 (0.36–2.87) | N.D. |
| clpP | 0.34 (0.15–0.77) | 2.12 (1.56–2.87) | 0.44 (0.36–0.54) | N.D. |
| rps18 | 0.57 (0.45–0.77) | 1.09 (0.97–1.23) | 0.87 (0.79–0.96) | N.D. |
| rpoC1 | 0.33 (0.22–0.50) | 0.78 (0.64–0.96) | 0.65 (0.27–1.55) | N.D. |
Effects of sca3 Mutations on Expression of Plastid Genes
Tobacco plastid genome genes have been classified into three classes, depending on which RNAP transcribes them: PEP only (class I), PEP and NEP (class II), or NEP only (class III; Hajdukiewicz et al., 1997). To our knowledge, such a study has yet to be made in Arabidopsis. We decided to ascertain whether the expression of plastid genes containing putative NEP-responsive sequences in their promoters was compromised in the sca3 mutants. qRT-PCR amplifications were performed on RNA extracted from 4- and 12-d-old seedlings and 21-d-old rosettes of Col-0 and the sca3-2 mutant (Table I) to quantify expression of the following plastid genes: clpP (assumed to be class II), rpoB, rpoC1, accD (assumed to be class III), and rps18 (not classified, given that its transcription initiation sites and promoters have not been mapped). These genes respectively encode the proteolytic subunit of the Clp ATP-dependent protease (Gray et al., 1990; Maurizi et al., 1990), the plastid β (Hu and Bogorad, 1990) and β′ core subunits of PEP (Shinozaki et al., 1986), a subunit of the acetyl-CoA carboxylase involved in lipid biosynthesis (Sasaki et al., 1993), and a ribosomal protein (Shinozaki et al., 1986).
RpoTp seemed to be required for the transcription of all these plastid genes, as indicated by their significant down-regulation in 4-d-old seedlings of the sca3-2 mutant. In this mutant, only rpoB and rpoC1 were down-regulated in all the developmental stages studied, whereas the transcript levels of accD and clpP were reduced in 4-d-old seedlings, but increased in 12-d-old seedlings. In contrast, clpP transcript levels were reduced in sca3-2/sca3-2 rosettes and those of rps18 only in 4-d-old seedlings. No significant changes were found for accD in rosettes.
To ascertain whether the sca3 mutations affect plastid rRNA levels and consequently plastid ribosome abundance, total RNA was extracted from 21-d-old wild-type and sca3-1/sca3-1 and sca3-2/sca3-2 mutant plants, and their rRNAs were quantified after being visualized in a denaturating agarose gel stained by ethidium bromide (Supplemental Fig. 2). We found reduced signal intensities for the chloroplastic rRNAs of the sca3-2 mutant compared to Col-0, but no significant differences between sca3-1 and Ler (Supplemental Fig. 2). This is consistent with the molecular nature of the sca3-2 allele and its stronger mutant phenotype.
Phenotype of sca3 Mutants
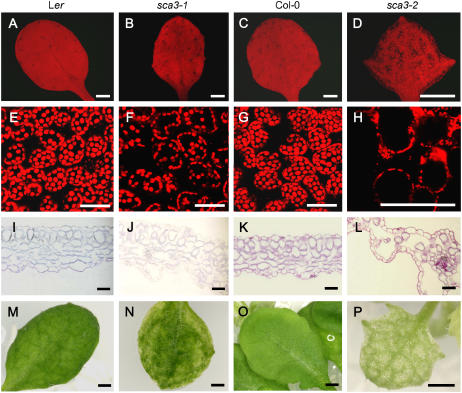
Col-0 and Ler genetic backgrounds had no visible effect on the phenotypes of the sca3 mutants, as deduced from their comparison to the phenotypically mutant F2 progeny of their intercrosses. The sca3 mutants displayed pale-green cotyledons and vegetative leaves, particularly the sca3-2 and sca3-3 homozygotes, which showed yellowish cotyledons and leaves (Fig. 1, A–D). Consistent with the paleness of the sca3 mutants, we found a significant reduction in their chlorophyll content compared to the wild types, the differences being more pronounced in the sca3-2 mutant, which also showed lower amounts of carotenoids (Supplemental Table II). The vegetative leaves of the sca3-1 mutant were rounded, slightly reduced in size and petiole length, and presented lateral teeth (Fig. 1, I–L). In the sca3-2 and sca3-3 mutants, leaf size was much more reduced and leaf margins displayed deep serrations corresponding to the positions of hydathodes.
sca3 mutants displayed pale-green organs and a general reduction of growth, as seen from their short stems and petioles and small cauline leaves and siliques (Fig. 1, M–Q). Apart from the paleness of the sepals, we found no other obvious alteration in their floral organs. All these phenotypic traits were more extreme in the sca3-2 and sca3-3 mutants. For instance, root length was 7.5 ± 1.7 cm for Ler, 6.1 ± 1.6 for sca3-1/sca3-1, 6.9 ± 2.0 for Col-0, and 3.6 ± 0.7 for sca3-2/sca3-2. In addition, sca3-2/sca3-2 plants exhibited almost no lateral roots (Fig. 1R).
We found no differences in the photomorphogenic response of sca3 mutants and their wild types grown for 2 weeks in the dark or after their return to normal light conditions (data not shown). This indicates that SCA3 is not required to complete the etiolated developmental program, nor, therefore, for the function that etioplasts perform in the dark, at least at the stage analyzed.
The phenotype of the three sca3 mutants studied here was cold sensitive. When grown at 16°C, they showed leaf chlorosis and were much smaller than the wild type (Supplemental Fig. 3). Growth at 26°C was similarly increased in wild-type and mutant plants compared to that observed at 20°C. In addition, the mutant phenotype of the sca3-1/sca3-1 plants was partially suppressed at 26°C, at which temperature the irregularities of the leaf margin almost completely disappeared. This was consistent with the ratio of SCA3 wild-type/mutant splice forms in sca3-1/sca3-1 plants, which was found higher at 26°C (3.5 ± 1.8) than at 20°C (1.7 ± 0.4). On the contrary, apart from the increased growth, the phenotypic traits of the sca3-2/sca3-2 plants grown at 26°C were not significantly different from that observed at 20°C (Supplemental Fig. 3).
Ultrastructure of sca3/sca3 Leaves
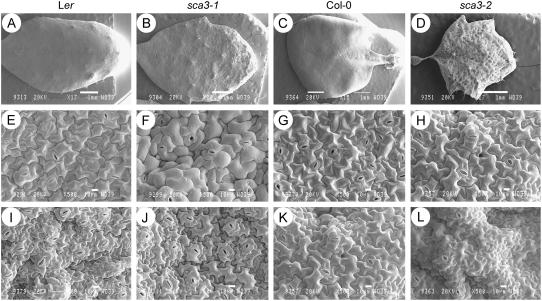
Scanning electron microscopy showed the surface of the mutant leaves to be wrinkled, extremely so in the case of the sca3-2 and sca3-3 homozygotes, whose lamina was completely crumpled (Fig. 4, A–D). Nevertheless, no obvious differences with the wild type were observed for the size and morphology of the sca3/sca3 adaxial and abaxial epidermal cells (Fig. 4, E–L).
Figure 4.
Scanning electron micrographs of sca3/sca3 leaves. Micrographs are shown of the adaxial surface (A–D) of whole leaves and details of their adaxial (E–H) and abaxial epidermises (I–L). All plants were homozygous for the mutations shown. Leaves were collected 28 d after sowing. Scale bars indicate 1 mm (A–D) and 10 μm (E–L).
We analyzed internal leaf anatomy by means of confocal microscopy of intact leaves (Fig. 5, A–H) and cross sections (Fig. 5, I–L) and found a reduced density of mesophyll cells in the sca3 mutants, particularly in sca3-2, whose interveinal areas and leaf margins were almost devoid of such cells. As a consequence, their leaf vascular network was distinguishable on a paler green background in intact leaves (Fig. 5, M–P). To ascertain whether this reduction was due to an increase in the frequency of cell death caused by sca3 mutations, we stained the mutants with trypan blue. No differences with the wild types were found (data not shown), which indicated that the number of dead cells was not increased by the sca3 mutations. In addition, mesophyll cells were irregularly shaped in the sca3-2 mutant (Fig. 5, K and L), making it impossible to distinguish between the palisade and spongy layers.
Figure 5.
Reduced cell density in the mesophyll of sca3/sca3 leaves. A to H, Confocal micrographs, showing fluorescing chlorophyll within mesophyll cells of whole third leaves (A–D), and details of the mesophyll (E–H). Transverse sections of leaves (I–L) and whole leaves (M–P). All plants were homozygous for the mutations shown. Leaves were collected 28 d after sowing. Scale bars indicate 1 mm (A–D), 50 μm (E–L), and 0.5 mm (M–P).
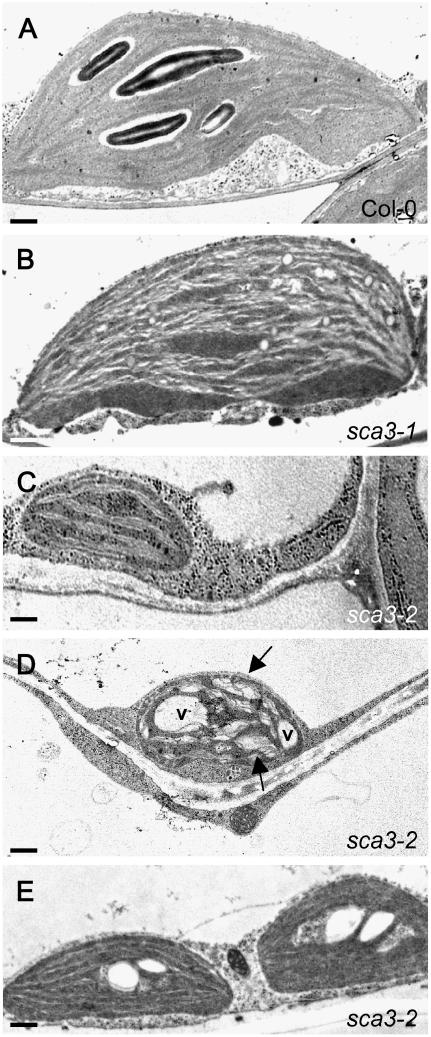
We examined leaf chloroplast ultrastructure in sca3-1/sca3-1, sca3-2/sca3-2, and wild-type individuals by transmission electron microscopy and found defects that paralleled the harshness of the morphological phenotype. Chloroplasts in the sca3-1 mutant displayed a reduced number of starch grains, but they were similar in size, morphology, and number to those of the wild type (Fig. 6, A and B). On the contrary, mesophyll cells in the sca3-2 mutant exhibited a large reduction in the number of chloroplasts, which, in turn, lacked starch grains, were smaller, and showed a less-developed thylakoid organization (Fig. 6, A and C). Some chloroplasts of the sca3-2 mutant displayed enlarged thylakoid lamellas and transparent vacuoles (Fig. 6, A and D), a trait never observed in the wild type. Consistent with the small morphological effect of sca3 mutations on the vascular network, chloroplasts in the cells surrounding the midvein were found to be more similar to those of the wild type than those of the mesophyll cells (Fig. 6, A and C–E).
Figure 6.
Transmission electron micrographs of sca3/sca3 chloroplasts. A to C, Representative chloroplasts of Col-0 and the sca mutants. Chloroplasts of Ler and Col-0 were indistinguishable and no differences were found between those of their mesophyll cells and those of cells neighboring the midvein. D, One of the chloroplasts occasionally seen in sca3-2/sca3-2 mesophyll cells, showing enlarged thylakoids (arrows) and vacuoles (v). E, Chloroplasts of a cell adjacent to the midvein of the sca3-2/sca3-2 mutant. All plants were homozygous for the mutations shown. All leaves were collected from the third node 21 d after sowing. Scale bars indicate 1.5 μm (A and B), 0.6 μm (C and D), and 1.2 μm (E).
Photoautotrophic Growth in sca3 Mutants
It has been proposed that the chloroplastic NEP indirectly promotes expression of plastid genome photosynthetic genes through the activation of genes such as rpoB, whose product is a PEP subunit (Hajdukiewicz et al., 1997). Together with the observed perturbation in plastid development, the pale-green pigmentation and reduction in growth of sca3 mutants suggest that SCA3 might be important for photoautotrophic growth. To test this hypothesis, we compared the growth of sca3 mutants in vitro in the presence or absence of 2% Suc in the culture medium. The absence of Suc did not perturb the growth of wild-type plants, but strongly affected that of mutants, especially sca3-2, which displayed pale-green expanded cotyledons and a first pair of leaves of reduced size, a stage of arrested development that remained unchanged even 4 weeks after sowing (Supplemental Fig. 4). Consistent with this, the growth of sca3-2/sca3-2 and sca3-3/sca3-3 plants was seriously impaired when the seeds were sown on soil. Whereas all wild-type and mutant seeds reached 100% germination on agar medium including Suc, all of the wild-type seeds germinated and developed properly on soil, but only 19.0% of the sca3-2/sca3-2 and 36.7% of the sca3-3/sca3-3 seeds did so. These results indicate that SCA3 is required early in development to establish autotrophic growth. Similar results have been reported previously for the reticulate mutant cue1 (Li et al., 1995) and the pale mutant pac (Reiter et al., 1994), whose leaf internal organization is perturbed.
Microarray Analysis of the sca3-2 Mutant
To examine the effect of sca3 mutations on the nuclear transcriptome of Arabidopsis, we performed a microarray analysis using RNA extracted from 3-week-old plants of the sca3-2 mutant and wild-type Col-0. Among the 26,173 genes represented on the DNA chip, 301 were found to be misregulated in the sca3-2 mutant. A total of 103 (34.2%) and 198 (65.8%) genes were at least 1.5-fold up-regulated or down-regulated, respectively (Supplemental Table III). Surprisingly, the most up-regulated gene found in the microarray was the MADS-box family member SEP3 (Mandel and Yanofsky, 1998; Pelaz et al., 2000). Consistent with the role of SCA3 in chloroplast biogenesis, 83 (27.6%) of the misregulated genes encoded proteins targeted to plastids. Given that the DNA chip contained the whole set of Arabidopsis nuclear genes, this proportion is significantly higher than that predicted for nuclear-encoded chloroplastic proteins, approximately 3,500 (13.7% of the genes in the array), according to the Arabidopsis Genome Initiative (Arabidopsis Genome Initiative, 2000). Among the genes encoding chloroplast-targeted proteins, the photosynthetic ones were the most widely represented (14; 16.8%), including genes encoding proteins of light-harvesting complexes (such as LHCB2; Standfuss and Kühlbrandt, 2004), as well as different subunits of the reaction center of PSI (such as PSAK; Jensen et al., 2000) and PSII. Interestingly, 84.3% of the nuclear genes encoding chloroplast-targeted proteins were down-regulated in sca3-2, which represents 35.0% of all the genes down-regulated, including all the photosynthetic genes and genes encoding FtsH proteases, the RNAP σ-subunit SigA (SIG1; Isono et al., 1997), ATP-binding cassette transporters, and proteins involved in isoprenoid biosynthesis. The latter group included CLA1 (encoding the 1-deoxy-d-xylulose-5-P synthase; Mandel et al., 1996), ABSCISIC ACID1 (ABA1; zeaxanthin epoxidase; Audran et al., 2001; Xiong et al., 2001), NCED4 and CHLOROPLAST BIOGENESIS 4 [CLB4; hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase], and CLB6 (Gutiérrez-Nava et al., 2004). On the contrary, among the up-regulated genes, only 12.6% corresponded to chloroplastic genes. We compared our results for the 20 genes that we found most down-regulated or up-regulated, as well as all the down-regulated photosynthetic genes, with those deposited at an Arabidopsis microarray database (http://www.genevestigator.ethz.ch). This comparison was made with the results obtained by previous authors in experiments performed using norflurazon, an inhibitor of carotenoid biosynthesis that blocks both chloroplast development (Susek et al., 1993) and expression of nuclear photosynthetic genes. All but one of the genes that we found down-regulated were also repressed in the presence of norflurazon. Other functional groups of genes identified in our microarray analysis included those involved in heat shock response (eight genes), protein modification (13), defense (14), electron transport (16), membrane transport (19), transcription regulation (31), and metabolism (34). The RpoT genes were not found to be misregulated in the microarray, which is consistent with the small variation found between Col-0 and sca3-2/sca3-2 plants using qRT-PCR (Table I). Of the 301 genes identified in our analysis, 60 (20.0%) are of unknown function.
To validate our microarray results, we used qRT-PCR to analyze the expression of five genes in 3-week-old sca3-2/sca3-2 and Col-0 plants. SEP3 was 7.0- and 2.7-fold up-regulated in sca3-2 compared with Col-0, as detected by qRT-PCR and microarray analysis, respectively. In the sca3-2 mutant, the LCHB2, PSAK, SIG1, and ABA1 genes were respectively found 6.4-, 2.8-, 2.1-, and 1.6-fold down-regulated by qRT-PCR, whereas microarray analyses showed 3.7-, 2.1-, 1.7-, and 1.7-fold levels.
Double-Mutant Analysis
To gain insight into the role of the RpoT genes in plant development, the sca3-2 mutant was crossed to the rpoT;2 mutant, which carries a T-DNA insertion in the RpoTmp gene (Baba et al., 2004). Both mutants were in a Col-0 genetic background. Because the rpoT;2 mutant was hard to distinguish from the wild type in our growth conditions, three phenotypic classes were found in the F2 progeny, conforming to an expected ratio of 12:3:1 (χ2 = 4.25; P = 0.05). This included 165 phenotypically wild-type plants, 27 that displayed the phenotype of the sca3-2 parent and 14 that were considered double mutants, which was confirmed by genotyping their T-DNA insertions. Development of the sca3-2 rpoT;2 double mutants was arrested early after germination, when they displayed pale-green or bleached expanded cotyledons, and only a few of them produced a few leaves, which were extremely abnormal and tiny (Fig. 7).
Figure 7.
Genetic interaction between the SCA3 and RpoTmp genes. sca3-2 (A), rpoT;2 single mutants (B), and sca3-2 rpoT;2 double mutants (C and D). Images were taken 21 d after sowing. Scale bars indicate 1 mm.
DISCUSSION
The SCA3 Gene Encodes the RpoTp Protein
We describe here the positional cloning of the SCABRA3 gene of Arabidopsis and the characterization of three recessive sca3 mutant alleles at the phenotypic, genetic, and molecular levels. We found that the protein product of SCA3 is RpoTp, a nuclear-encoded, plastid-targeted RNAP. SCA3 has two paralogs in the nuclear genome of Arabidopsis: RpoTm and RpoTmp, whose protein products are respectively targeted to mitochondria and to both plastids and mitochondria (Hedtke et al., 1997, 1999, 2000). A functional analysis has recently been reported for one of these genes, RpoTmp (Baba et al., 2004). Although homologs of the Arabidopsis SCA3 gene have been characterized in several plant species, to our knowledge, the role of RpoTp in plant development has not been studied at a functional level using a mutational approach.
RpoTp and RpoTmp Are Partially Redundant in Green and Nongreen Tissues
The sca3 mutants displayed rounded and pale-green vegetative leaves with marked irregularities on the surface and the margins of the lamina, although leaf epidermal cell size and morphology were not perturbed. All the phenotypic traits studied were much more conspicuous in sca3-2 and sca3-3 than in sca3-1, which is consistent with their molecular nature. The sca3-1 allele carries a point mutation and encodes both a truncated protein lacking 109 amino acids and a wild-type protein. Each of the sca3-2 and sca3-3 insertional alleles encode a single protein, lacking 604 and 50 amino acids, respectively, and including divergent amino acids translated from the T-DNA. Because the protein product of sca3-2 lacks many more amino acids than that of sca3-3, the latter might represent an example of a mutation perturbing a redundant functional activity, such as that of RpoTmp. This has been hypothesized for the recessive sleepy1-2 (sly1-2) and sly1-10 alleles of the Arabidopsis gibberellin-signaling SLEEPY1 gene, which interfere with the function of the homologous SNEEZY gene (Strader et al., 2004).
Internal structure is severely perturbed in sca3/sca3 leaves as a consequence of a strong reduction in the number of mesophyll cells and an increase in intercellular airspaces, which presumably cause the surface irregularities that characterize their external morphology. Nevertheless, their leaf veins remained almost unaltered, suggesting that sca3 mutations differentially affect vascular and mesophyll cell development. A transition from underdeveloped to normal mesophyll cells and from leaf margins to green sectors close to the midvein has been reported for the virescent cue6 mutant of Arabidopsis (López-Juez et al., 1998). Other Arabidopsis mutants, such as cue1 (Li et al., 1995; Streatfield et al., 1999) and plastid protein import2 (ppi2; Asano et al., 2004), show reduced cell density in the mesophyll and an associated alteration in leaf morphology. The reduction in the number of mesophyll cells in the sca3 mutants might be due to reduced cell proliferation or, alternatively, to increased cell death. The first of these two scenarios is more likely because no difference was observed in the frequency of cell death between sca3/sca3 and wild-type individuals.
Leaf epidermal cells seem unaffected in the sca3 mutants, which is remarkable given that plastids are the primary site of amino acid and lipid biosynthesis. Further research will be required to address the question of whether RpoTp plays a more active role in the mesophyll than in the epidermis, where RpoTmp might be more important.
Roots contain photosynthetically inactive but differentiated plastids such as leucoplasts and amyloplasts (Waters and Pyke, 2005). NEPs are assumed to transcribe genes in nonphotosynthetic plastids to maintain their metabolic activity, as well as being required in proplastids to keep them ready to differentiate into functional chloroplasts (Magee and Kavanagh, 2002). In addition, RpoTp has been found not only in leaf chloroplasts but also in nongreen root plastids (Hedtke et al., 1999). Consistent with a function for RpoTp in nongreen tissues, we detected SCA3 expression in roots and found a significant reduction in root length and secondary roots in the strong sca3-2 mutant. The rpoT;2 mutant also exhibits short roots (Baba et al., 2004), which, taken together with our results, suggests overlapping functions for RpoTmp and RpoTp in root plastids.
sca3 mutations perturb other aspects of plant development, causing a general reduction in size, stem and silique length, and a generalized loss of pigmentation. The small size of most organs of the sca3 mutants suggests a role for SCA3 in promoting cell proliferation not limited to leaf mesophyll cells. Reduced growth, delayed greening, and wrinkled leaf lamina are also characteristic traits of the rpoT;2 mutant (Baba et al., 2004). Given that RpoTmp and RpoTp are homologous and chloroplast targeted, our results suggest that they are involved in similar developmental events and that they are partially redundant. Consistent with this, we found that the double mutant sca3-2 rpoT;2 is seedling lethal.
The phenotype of sca3 mutants is enhanced by growth at low temperature, as previously described for mutations affecting chloroplast and mesophyll development, such as var2 (Chen et al., 2000; Takechi et al., 2000) and chilling-sensitive5 (chs5; Schneider et al., 1995), which is allelic to cla1 (Araki et al., 2000). Cold sensitivity of the sca3 mutants might be explained by a reduction in the activity of SCA3 mutant proteins at the restrictive temperature, which would require the existence of some degree of remnant activity in the protein product of the strong sca3-2 and sca3-3 alleles. However, this activity would not be increased at 26°C because the phenotype of the sca3-2/sca3-2 plants is not suppressed as in sca3-1/sca3-1 individuals. In the latter, the increased level of the wild-type splice form found at 26°C might explain the suppression of the mutant phenotype. Alternatively, the temperature sensitivity of these mutants could be due to decreased activity of a functionally redundant protein at the restrictive condition. We find the latter hypothesis more likely, given the extreme mutant phenotype, the molecular nature of the sca3-2 and sca3-3 alleles, and the fact that RpoTmp and RpoTp proteins share a substantial part of their structure and function. Given that other mutants affected in chloroplast development also show cold sensitivity, we cannot rule out the influence that enhanced photodynamic damage caused by the restrictive temperature might produce on the phenotype of the sca3 mutants.
Perturbation of Chloroplast Development in sca3 Mutants Impairs Mesophyll Cell Differentiation
Chloroplasts were dramatically reduced in number and not properly developed in leaves of sca3-2/sca3-2 plants, indicating that SCA3 activity is essential for chloroplast development and suggesting a role for RpoTp in transcribing the plastid genes required for the conversion of proplastids into functional chloroplasts. Arabidopsis variegated mutants such as im (Josse et al., 2000; Aluru et al., 2001), var1, and var2 (for review, see Sakamoto, 2003) also display abnormal, vacuolated plastids in white leaf sectors. As a likely consequence of the perturbation in plastid development, we found that photoautotrophic growth is severely impaired in the strong sca3-2 and sca3-3 mutants.
Several mutants with altered internal leaf anatomy and chloroplast biogenesis have been described, suggesting the existence of a putative plastid-to-nucleus developmental signal that controls mesophyll cell proliferation and differentiation. sca3 mutants provide further support for the hypothesis that perturbation of chloroplast biogenesis affects mesophyll cell differentiation and hence leaf morphogenesis. Although the existence of a plastid developmental signal controlling leaf morphogenesis is yet to be demonstrated, a study of the genomes uncoupled (gun) mutants of Arabidopsis identified magnesium-protoporphyrin IX as one of the plastid signals that regulates expression of nuclear photosynthetic genes (Mochizuki et al., 2001; Surpin et al., 2002; Strand et al., 2003; Strand, 2004). In our microarray analysis of the sca3-2 mutant, we found several photosynthetic genes significantly down-regulated, including LHCB2, whose expression is known to be reduced in other mutants with abnormal leaf anatomy (Rodermel, 2001). Other down-regulated genes were ABA1, CLA1, CLB4, and CLB6, which are related to isoprenoid biosynthesis, a group of molecules with essential functions in photosynthesis, plant growth, and development (Peñuelas and Munné-Bosch, 2005). Another misregulated gene was NCED4, whose function is unknown and belongs to the 9-cis-epoxycarotenoid dioxygenase family. Another member of this family, NCED3 (Tan et al., 2003), participates in ABA biosynthesis, which also requires ABA1 (Audran et al., 2001; Xiong et al., 2001). ABA is an abiotic stress response hormone (Zhu, 2002) that also acts as a positive regulator of plant growth (Cheng et al., 2002; González-Guzmán et al., 2002; Barrero et al., 2005) and seems to be involved in plastid differentiation (Rohde et al., 2000). On the other hand, the CLA1, CLB4, and CLB6 genes are required for chloroplast development and function (Estévez et al., 2000; Gutiérrez-Nava et al., 2004). Therefore, our results are consistent with those obtained in other Arabidopsis pigment mutants with abnormal leaf morphogenesis and suggest the existence of two different plastid-to-nucleus signaling pathways, one controlling cell differentiation and leaf development and the other controlling expression of nuclear photosynthetic genes (Rodermel, 2001). We considered as an alternative hypothesis that the reduction in cell viability and vigor found in the sca3 mutants might be due to a decrease in the expression of ycf1 and ycf2, the two largest open reading frames encoded by the chloroplast genome of dicotyledonous plants, which are cell essential (Drescher et al., 2000), but our expression analyses ruled this out (data not shown).
Expression of RpoT Genes Is Not Strongly Affected by sca3 Mutations
SCA3, RpoTm, and RpoTmp expression was detected in seedlings, roots, and rosettes of wild-type plants. We found small variations in RpoTm and RpoTmp expression between Col-0 and sca3-2/sca3-2 plants. Thus, the level of RpoTm and RpoTmp expression was slightly reduced in sca3-2 12-d-old mutant seedlings, but both genes were up-regulated earlier (4-d-old seedlings) and later (21-d-old rosettes) compared with the wild type. Consistent with this, an increase in the levels of RpoTm and SCA3 (RpoTp) transcripts in leaves of the Arabidopsis rpoT;2 mutant has been reported (Baba et al., 2004) and an accumulation of RpoTp and RpoTm transcripts has been found in ribosome-deficient leaves of the albostrians mutant of H. vulgare (Emanuel et al., 2004). These results would indicate the existence of a compensatory effect acting as a regulatory mechanism that would maintain chloroplast function in the presence of mutations impairing their development. Nevertheless, the differences found in the expression of the RpoT genes in the sca3-2 mutant do not seem to be sufficient to compensate for the lack of SCA3 function, given the pleiotropic phenotype of the sca3 mutants.
Expression of Plastid Genes Is Affected by sca3 Mutations
It has been proposed that orthologs of the Arabidopsis RpoTp gene control the expression of a subset of genes of the plastid genome, those containing NEP-type promoters (Allison et al., 1996; Hajdukiewicz et al., 1997; Magee and Kavanagh, 2002), such as some housekeeping genes and PEP core subunits. According to this model (Hajdukiewicz et al., 1997), nonphotosynthetic genes (such as accD, clpP, and others) and PEP components (such as the rpoB gene) are induced by NEP early in plastid differentiation. In later stages, light-dependent expression of sigma factors, such as SIG2, produces activation of PEP and triggers the differentiation of proplastids into functional chloroplasts (Hanaoka et al., 2005). At this stage, photosynthetic and housekeeping genes would be transcribed mainly by PEP.
Expression analysis of some plastid genes assumed to be representative of class II (transcribed by PEP and NEP) and III (transcribed by NEP) indicated that SCA3 is required early and later in development for the expression of some plastid genes, such as rpoB, rpoC1, and clpP in both seedlings and rosettes. We found the lowest level of plastid gene expression in sca3-2/sca3-2 4-d-old seedlings, indicating that RpoTp plays an important role very early in chloroplast and plant development. Nevertheless, we cannot rule out that other factors, such as a reduction in RNA stability and/or synthesis, might contribute to the observed decrease in the levels of the plastid genes studied.
We found increased accD and clpP transcript levels in sca3-2/sca3-2 12-d-old seedlings. Consistent with this, an accumulation of NEP-dependent plastid transcripts has been reported in seedlings of Arabidopsis mutants with altered PEP components, such as sig2 (Kanamaru et al., 2001; Nagashima et al., 2004) and sig6 (Ishizaki et al., 2005), or defective NEP transcriptional machinery (RpoT;2; Baba et al., 2004). Because the RpoTm and RpoTmp transcript levels are not increased in the sca3-2/sca3-2 12-d-old seedlings, the observed rise in accD and clpP transcript levels was probably not due to a compensatory up-regulation of the RpoT genes. Alternatively, an increase in the stability of the accD and clpP transcripts or PEP activity would compensate for the loss of RpoTp function in sca3-2/sca3-2 seedlings, at least in the case of clpP, which is PEP responsive. This compensatory mechanism would be absent or less efficient later in development because clpP mRNA levels decrease in sca3-2 mutant rosettes.
The strong sca3-2 mutation also causes a decrease in plastid rRNA levels and, probably, in the abundance of plastid ribosomes, which is consistent with the dramatic reduction in the number of chloroplasts found in this mutant. However, we cannot exclude the possibility that RpoTp might be required for the transcription of plastid rRNAs, given that mutants defective in chloroplast rRNA processing, maturation, or both, and displaying abnormalities in leaf development, have already been reported in Arabidopsis (Bellaoui et al., 2003; Bisanz et al., 2003; Kishine et al., 2004).
CONCLUSION
Mutations in SCA3 negatively affect the expression of both photosynthetic nuclear genes and plastid genes (the latter probably through the control of rpoB and rpoC1 by SCA3), resulting in abnormal chloroplast development and impaired photoautotrophic growth. In contrast to the role assigned to RpoTmp in early seedling development, characterization of sca3 mutants indicates that RpoTp is required both in early and late stages of vegetative development in Arabidopsis. Consistent with this, it has recently been found (Emanuel et al., 2005) that SCA3 (RpoTp) transcripts accumulated at similar levels early (cotyledons) and late (leaf tissue) during vegetative development, whereas RpoTm and RpoTmp were predominantly expressed early in Arabidopsis development. Our findings highlight the complexity of the mechanisms that control plastid gene transcription.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Cultures and crosses were performed as described in Ponce et al. (1998) and Berná et al. (1999), respectively. Seeds of the Arabidopsis (Arabidopsis thaliana L. Heynh.) wild-type accessions Ler and Col-0 were obtained from the Nottingham Arabidopsis Stock Centre (NASC). The sca3-1 mutant was isolated in a Ler background after EMS mutagenesis (Berná et al., 1999). Seeds of several T-DNA insertion lines, including N567191 (sca3-3) and N593884 (sca3-2), were provided by the NASC and are described at the SIGnAL Web site (Alonso et al., 2003; http://signal.salk.edu). Seeds of the rpoT;2 mutant were kindly provided by Julien Schmidt (Umea Plant Science Centre).
Growth Assays and Pigment Extraction
For temperature-sensitivity analyses, seeds were sown on agar plates as described above, incubated at 16°C, 20°C, or 26°C and observed 21 d after sowing. For root studies, seedlings were grown vertically on agar plates (15 seedlings/plate), photographed 15 d after sowing, and a Student's t test was applied to the data obtained. To study autotrophic growth, seeds were sown on petri dishes and plants grown in vitro in the presence or absence of 2% Suc in the culture medium, and the percentages of survival and arrested development were scored 21 d after sowing. To study the photomorphogenic response, seeds were sown on agar plates that were kept in the dark (wrapped in aluminum foil) for 2 weeks.
For pigment extraction, 3-week-old plants were harvested and frozen in liquid nitrogen. Eighty milligrams (fresh weight) from each sample (four to eight individuals) were ground and chlorophyll and carotenoids were extracted adding 3.5 mL of 80% acetone to each sample. Pigments were quantified as already described (Rabino and Mancinelli, 1986). A Student's t test was applied to the data obtained.
Morphological and Ultrastructural Analyses
Whole-rosette and single-leaf images were taken using a Leica MZ6 stereomicroscope equipped with a Nikon DXM1200 digital camera. Confocal imaging and trypan blue staining were performed as described in Pérez-Pérez et al. (2002) and Koch and Slusarenko (1990), respectively.
For light microscopy, plant material was fixed with formaldehyde-acetic acid/Triton (1.85% formaldehyde, 45% ethanol, 5% acetic acid, and 1% Triton X-100), as described in Serrano-Cartagena et al. (2000). Transverse sections of leaves (0.5-μm-thick) were cut on a microtome (HM350S; Microm International), stained with 0.1% toluidine blue, and observed using a Leica DMRB microscope equipped with a Nikon DXM1200 digital camera under bright-field illumination.
For scanning electron microscopy, plant material was prepared as described in Serrano-Cartagena et al. (2000). Micrographs were taken in a JSM-840 JEOL scanning electron microscope. For transmission electron microscopy, mutant and wild-type plant material was harvested at exactly the same time of the day and fixed with 3% (v/v) glutaraldehyde in 0.1 m cacodylate buffer (pH 7.2–7.4), washed with cacodylate-Suc buffer, and then postfixed in 1% (v/v) OsO4 (Wanson and Drochmans, 1968). The fixed specimens were dehydrated in an ethanol series (30%, 50%, 70%, and 100% ethanol; 30 min each) and embedded in LR White resin. Polymerized blocks were sectioned on a Reichert-Jung Ultracut E microtome, stained with uranyl acetate and lead citrate, and visualized at 60 kV using a Zeiss EM10C transmission electron microscope.
Positional Cloning and Molecular Characterization of sca3 Mutations
To positionally clone the SCA3 gene, simple sequence-length polymorphisms, single-nucleotide polymorphisms, and cleaved-amplified polymorphic sequence markers were designed according to the polymorphisms between the Ler and Col-0 described at the Monsanto Arabidopsis Polymorphism Collection database (http://www.arabidopsis.org). For allele sequencing, PCR products spanning the At2g24120 transcription unit were obtained using as a template wild-type and mutant genomic DNA and the oligonucleotide primers shown in Supplemental Table I. Sequencing reactions were carried out with ABI PRISM BigDye Terminator cycle sequencing kits in 5-μL reaction volumes. Sequencing electrophoreses were performed on an ABI PRISM 3100 genetic analyzer.
RNA Extraction and Analyses
Unless otherwise stated, all RNA extractions from plant material (50–100 mg), RTs using random primers, and PCR amplifications of first-strand cDNA were performed as described in Quesada et al. (1999). For chloroplast rRNA analysis, 8 μg of total RNA from each sample were fractionated in formaldehyde-agarose gels. For detection of the different SCA3 transcripts in the sca3-1 mutant, SCA3 expression was tested by using a fluorescence-based semiautomated method (Ponce et al., 2000). In brief, total RNA was extracted from 3-week-old Ler and sca3-1/sca3-1 plants grown at either 20°C or 26°C, and RT-PCR amplification products were obtained using oligonucleotides flanking the sca3-1 mutation (F9, which had been labeled with 6-FAM phosporamidite, and R10; Supplemental Table I) and electrophoresed in an ABI PRISM 3100 genetic analyzer (Supplemental Fig. 1).
qRT-PCR
Total RNA was extracted from 50 to 70 mg of 4- and 12-d-old seedlings and 3-week-old rosettes and roots (Col-0 and sca3-2) and DNase I treated using the Qiagen RNeasy plant mini kit, following the manufacturer's instructions. RNA was ethanol precipitated and resuspended in 40 μL of RNase-free water. Five micrograms from each sample were reverse transcribed using random primers as described by Quesada et al. (1999) and 1 μL of the resulting cDNA solution was used for qRT-PCR amplifications, which were carried out in an ABI PRISM 7000 sequence detection system (Applied Biosystems) as described in Pérez- Pérez et al. (2004). Primer pairs (Supplemental Table I) were designed to yield amplification products of approximately 100 bp. One of the primers of each pair contained the sequences of the ends of two contiguous exons so that genomic DNA could not be amplified. Each 25-μL reaction mix contained 12.5 μL of the SYBR-Green PCR master kit (Applied Biosystems), 0.4 μm of primers, and 1 μL of cDNA solution. Relative quantification of gene expression data was performed using the 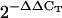 or comparative CT method (Livak and Schmittgen, 2001). Each reaction was performed in three replicates and levels of expression were normalized by using the CT values obtained for the housekeeping OTC gene (Quesada et al., 1999).
or comparative CT method (Livak and Schmittgen, 2001). Each reaction was performed in three replicates and levels of expression were normalized by using the CT values obtained for the housekeeping OTC gene (Quesada et al., 1999).
Microarray Analysis
Arabidopsis wild-type Col-0 and mutant sca3-2/sca3-2 3-week-old plants grown in vitro from six different sowings (80–100 mg/sample) were frozen in liquid N2 and ground by mortar and pestle. Total RNA was extracted as described in Quesada et al. (1999) and the six RNA samples obtained were pooled in pairs to generate three biological replicates. Ten micrograms of total RNA from each biological replicate were used for microarray hybridization and analysis. Superamine Telechem slides containing more than 26,000 spots corresponding to the Arabidopsis oligo set from Qiagen-Operon, obtained from David Galbraith (Arizona University; http://ag.arizona.edu/microarray), were hybridized by conventional methods with RNA probes labeled with either Cy3 or Cy5 Mono N-hydroxysuccinimide esters. Printed slides were rehydrated over a 65°C water bath for 10 s and dried on a 65°C heating block for 10 s. This hydration step was repeated three times. Oligonucleotides were fixed by 120 mJ of UV radiation. Slides were washed in 1% SDS for 5 min, in water for 5 min, and in absolute ethanol for 30 s. Finally, slides were dried by centrifugation at 141 g for 3 min.
Total RNA (1 μg of each sample) was amplified and aminoallyl labeled using a MessageAmp II amplified RNA (aRNA) kit (Ambion) and 5-(3-aminoallyl)-2′deoxyuridine-5′-triphosphate (Ambion), according to the manufacturer's instructions, which yielded 40 to 50 μg of aRNA. For each sample, 7.5 μg of aminoallyl-labeled aRNA were resuspended in 0.1 m Na2CO3 (pH 9.0), labeled with either Cy3 or Cy5 Mono N-hydroxysuccinimide ester, and purified with Megaclear (Ambion), following the manufacturer's instructions. For each hybridization, 200 pmol of Cy3- and Cy5-labeled probes were mixed, dried in a speed vac, and resuspended in 9 μL of RNase-free water. Labeled aRNA was fragmented by adding 1 μL of 10× fragmentation buffer (Ambion) and incubated at 70°C for 15 min. The reaction was stopped with 1 μL of Stop solution (Ambion). Integrity and average size of total RNA, aRNA, and fragmented aRNA was evaluated using Bioanalyzer 2100 (Agilent). Average size of aRNAs was about 1,000 nucleotides and of fragmented aRNAs 100 nucleotides. The probe was finally diluted to 100 μL in hybridization buffer. Prehybridization was performed at 42°C for 30 to 45 min in 6× SSC, 0.5% SDS, and 1% bovine serum albumin, and slides were rinsed five times with water. Cy5 and Cy3 aRNA fragmented probes were mixed (200 pmol of each label) with 20 μg of PolyA (Sigma) and 20 μg of yeast (Saccharomyces cerevisiae) tRNA (Sigma) in a final volume of 90 μL of hybridization buffer (50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt's). The probe was denatured at 95°C for 5 min and poured into the slide using a LifterSlip (Erie Scientific). Slides were then incubated at 37°C for 16 h in hybridization chambers (Array-It) and then sequentially washed in the following solutions: twice in 0.5× SSC, 0.1% SDS, twice in 0.5× SSC, and finally in 0.05× SSC for 5 min each. Slides were finally dried by centrifugation at 563g for 1 min before being scanned.
Images from the Cy3 and Cy5 channels were equilibrated and captured with a GenePix 4000B (Axon) and spots quantified using GenePix software (Axon). The data from each scanned slide were first escalated and normalized using the Lowess method and then log transformed to correct the artifacts inherent in labeling, hybridization, scanning, and quantification, and analyzed by using the SOLAR package (Bioalma; http://www.bioalma.com). Two statistical approaches were used to identify differentially regulated genes: a t test (Smyth et al., 2002) and a z score (Quackenbush, 2002). Only genes with a gene signal > 50, P value < 0.05, z-score > 1.96 or < −1.96, and fold change > 1.5 or < −1.5 were considered differentially expressed.
Supplementary Material
Acknowledgments
We wish to thank M.R. Ponce and P. Robles for useful comments on the manuscript, the NASC and Julien Schmidt for providing seeds, J.M. Barrero for ABA1 and SEP3 primers, R. Solano for microarray analyses, and J.M. Serrano and V. García-Sempere for excellent technical assistance.
This work was supported by the Ministerio de Educación y Ciencia of Spain (grant nos. BMC2002–02840 and BFU2005–01031 to J.L.M.) and the European Commission (contract no. HPRN–CT–2002–00267 [DAGOLIGN] to A.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José Luis Micol (jlmicol@umh.es).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.080069.
References
- Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82: 537–548 [DOI] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastid of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Araki N, Kusumi K, Masamoto K, Niwa Y, Iba K (2000) Temperature-sensitive Arabidopsis mutant defective in 1-deoxy-d-xylulose 5-phosphate synthase within the plastid non-mevalonate pathway of isoprenoid biosynthesis. Physiol Plant 108: 19–24 [Google Scholar]
- Asano T, Yoshioka Y, Machida Y (2004) A defect in Toc159 of Arabidopsis causes severe defects in leaf development. Genes Genet Syst 79: 207–212 [DOI] [PubMed] [Google Scholar]
- Audran C, Liotenberg S, Gonneau M, North H, Frey A, Tap-Waksman K, Vartanian N, Marion-Poll A (2001) Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Aust J Plant Physiol 28: 1161–1173 [Google Scholar]
- Baba K, Schmidt J, Espinosa-Ruiz A, Villarejo A, Shiina T, Gardestrom P, Sane AP, Bhalerao RP (2004) Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J 38: 38–48 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Fuangthong M, Van Montagu M, Inzé D, Kushnir S (1997) Efficient gene tagging in Arabidopsis using a gene trap approach. Proc Natl Acad Sci USA 94: 12722–12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL (2005) A mutational analysis of the ABA1 gene of Arabidopsis highlights the involvement of ABA in vegetative development. J Exp Bot 56: 2071–2083 [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Keddie JS, Gruissem W (2003) DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Mol Biol 53: 531–543 [DOI] [PubMed] [Google Scholar]
- Berná G, Robles P, Micol JL (1999) A mutational analysis of leaf morphogenesis in Arabidopsis. Genetics 152: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisanz C, Begot L, Carol P, Perez P, Bligny M, Pesey H, Gallois JL, Lerbs-Mache S, Mache R (2003) The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol Biol 51: 651–663 [DOI] [PubMed] [Google Scholar]
- Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M (1999) Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Sheen J, Bligny M, Niwa Y, Lerbs-Mache S, Stern DB (1999) Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11: 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C (1996) DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J 15: 4194–4207 [PMC free article] [PubMed] [Google Scholar]
- Chen M, Choi Y, Voytas DF, Rodermel S (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Emanuel C, von Groll U, Müller M, Börner T, Weihe A (2006) Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta 223: 998–1009 [DOI] [PubMed] [Google Scholar]
- Emanuel C, Weihe A, Graner A, Hess WR, Börner T (2004) Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J 38: 460–472 [DOI] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Romero C, Kawaide H, Jiménez LF, Kuzuyama T, Seto H, Kamiya Y, Leon P (2000) Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-d-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol 124: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JC, Hird SM, Dyer TA (1990) Nucleotide sequence of a wheat chloroplast gene encoding the proteolytic subunit of an ATP-dependent protease. Plant Mol Biol 15: 947–950 [DOI] [PubMed] [Google Scholar]
- Grevelding C, Suter-Crazzolara C, von Menges A, Kemper E, Masterson R, Schell J, Reiss B (1996) Characterisation of a new allele of pale cress and its role in greening in Arabidopsis. Mol Gen Genet 251: 532–541 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Nava M, Stewart Gillmor C, Jiménez LF, Guevara-García A, León P (2004) CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol 135: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagio M, Sakurai I, Sato S, Kato T, Tabata S, Wada H (2002) Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis. Plant Cell Physiol 43: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Fujiwara M, Takahashi H, Tanaka K (2005) Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep 6: 545–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277: 809–811 [DOI] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (2000) One RNA polymerase serving two genomes. EMBO Rep 1: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Legen J, Weihe A, Herrmann RG, Börner T (2002) Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J 30: 625–637 [DOI] [PubMed] [Google Scholar]
- Hedtke B, Meixner M, Gillandt S, Richter E, Börner T, Weihe A (1999) Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo. Plant J 17: 557–561 [DOI] [PubMed] [Google Scholar]
- Hess WR, Börner T (1999) Organellar RNA polymerases of higher plants. Int Rev Cytol 190: 1–59 [DOI] [PubMed] [Google Scholar]
- Hu J, Bogorad L (1990) Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci USA 87: 1531–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL, Kössel H (1992) The transcriptional apparatus of chloroplasts. CRC Crit Rev Plant Sci 10: 525–558 [Google Scholar]
- Ikeda TM, Gray MW (1999) Identification and characterization of T3/T7 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol Biol 40: 567–578 [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H (1997) Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of sigma70 factors of bacterial RNA polymerases in Arabidopsis. Proc Natl Acad Sci USA 94: 14948–14953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gilpin M, Knoetzel J, Scheller HV (2000) The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J Biol Chem 275: 24701–24708 [DOI] [PubMed] [Google Scholar]
- Josse EM, Simkin AJ, Gaffe J, Laboure AM, Kuntz M, Carol P (2000) A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Hashimoto K, Sato NS (2002) Identification and characterization of two phage-type RNA polymerase cDNAs in the moss Physcomitrella patens: implication of recent evolution of nuclear-encoded RNA polymerase of plastids in plants. Plant Cell Physiol 43: 245–255 [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H (2001) An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol 42: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Keddie J, Carrolll B, Jones J, Gruissem W (1996) The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J 15: 4208–4217 [PMC free article] [PubMed] [Google Scholar]
- Kishine M, Takabayashi A, Munekage Y, Shikanai T, Endo T, Sato F (2004) Ribosomal RNA processing and an RNase R family member in chloroplasts of Arabidopsis. Plant Mol Biol 55: 595–606 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Sugiera M, Niwa Y, Sugita M (2001) Genomic organization and organ-specific expression of a nuclear gene encoding phage-type RNA polymerase in Nicotiana sylvestris. Gene 279: 33–40 [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Gen Genet 263: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Kusumi K, Yara A, Mitsui N, Tozawa Y, Iba K (2004) Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol 45: 1194–1201 [DOI] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S (1993) The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc Natl Acad Sci USA 90: 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Culligan K, Dixon RA, Chory J (1995) CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López-Juez E, Jarvis RP, Takeuchi A, Page AM, Chory J (1998) New Arabidopsis cue mutants suggest a close connection between plastid-and phytochrome regulation of nuclear gene expression. Plant Physiol 118: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Magee AM, Kavanagh TA (2002) Plastid genes transcribed by the nucleus-encoded plastid RNA polymerase show increased transcript accumulation in transgenic plants expressing a chloroplast-localized phage T7 RNA polymerase. J Exp Bot 53: 2341–2349 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1998) The Arabidopsis AGL9 MADS-box gene is expressed in young flower primordial. Sex Plant Reprod 11: 22–28 [Google Scholar]
- Maurizi MR, Clark WP, Kim SH, Gottesman S (1990) Clp P represents a unique family of serine proteases. Biol Chem 265: 12546–12552 [PubMed] [Google Scholar]
- McAllister WT, Raskin CA (1993) The phage RNA polymerases are related to DNA polymerases and reverse transcriptases. Mol Microbiol 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Motohashi R, Seki M, Shinozaki K, Kanamaru K, Takahashi H, Tanaka K (2004) DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci Biotechnol Biochem 68: 694–704 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require sepallata MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Peñuelas J, Munné-Bosch S (2005) Isoprenoids: an evolutionary pool for photoprotection. Trends Plant Sci 10: 166–169 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL (2004) The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol 134: 101–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce MR, Pérez-Pérez JM, Piqueras P, Micol JL (2000) A multiplex reverse transcriptase-polymerase chain reaction method for fluorescence-based semiautomated detection of gene expression in Arabidopsis thaliana. Planta 211: 606–608 [DOI] [PubMed] [Google Scholar]
- Ponce MR, Quesada V, Micol JL (1998) Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J 14: 497–501 [DOI] [PubMed] [Google Scholar]
- Pyke KA, Zwobka M, Day A (2000) Marking cell layers in Brassica napus with spectinomycin provides a new tool for studying cell fate and demonstrates a requirement for chloroplasts in palisade cell differentiation. J Exp Bot 51: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32: 496–501 [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL (1999) OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis. FEBS Lett 461: 101–106 [DOI] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RS, Coomber SA, Bouret TM, Bartley GE, Scolnik PA (1994) Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell 6: 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Kiessling J, Hedtke B, Decker E, Reski R, Börner T, Weihe A (2002) Two RpoT genes of Physcomitrella patens encode phage-type RNA polymerases with dual targeting to mitochondria and plastids. Gene 290: 95–105 [DOI] [PubMed] [Google Scholar]
- Robles P, Micol JL (2001) Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol Genet Genomics 266: 12–19 [DOI] [PubMed] [Google Scholar]
- Rodermel S (2001) Pathways of plastid-to-nucleus signaling. Trends Plant Sci 6: 471–478 [DOI] [PubMed] [Google Scholar]
- Rohde A, De Rycke R, Beeckman T, Engler G, Van Montagu M, Boerjan W (2000) ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12: 35–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W (2003) Leaf-variegated mutations and their responsible genes in Arabidopsis. Genes Genet Syst 78: 1–9 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Tamura T, Hanba-Tomita Y, Murata M, Sodmergen (2002) The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7: 769–780 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R (1993) Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem 268: 25118–25123 [PubMed] [Google Scholar]
- Schneider JC, Nielsen E, Sommerville C (1995) Chilling-sensitive mutant of Arabidopsis is deficient in chloroplast protein accumulation at low temperature. Plant Cell Environ 18: 23–31 [Google Scholar]
- Serrano-Cartagena J, Candela H, Robles P, Ponce MR, Pérez-Pérez JM, Piqueras P, Micol JL (2000) Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156: 1363–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GH, Yang YH, Speed TP (2002) Statistical issues in cDNA microarray data analysis. In MJ Brownstein, AB Khodursky, eds, Functional Genomics: Methods and Protocols. Methods in Molecular Biology Series. Humana Press, Totowa, NJ [DOI] [PubMed]
- Sommer H, Carpenter R, Harrison BJ, Saedler H (1985) The transposable element Tam3 of Antirrhinum majus generates a novel type of sequence alteration upon excision. Mol Gen Genet 199: 225–231 [Google Scholar]
- Standfuss J, Kühlbrandt W (2004) The three isoforms of the light-harvesting complex II: spectroscopic features, trimer formation and functional roles. Biol Chem 279: 36884–36891 [DOI] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101: 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A (2004) Plastid-to-nucleus signalling. Curr Opin Plant Biol 7: 621–625 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Weber A, Kinsman EA, Hausler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flugge UI, Chory J (1999) The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11: 1609–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell (Suppl) 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W (2000) The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41: 1334.–1346 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Wanson JC, Drochmans P (1968) Rabbit skeletal muscle glycogen: a morphological and biochemical study of glycogen b-particles isolated by precipitation-centrifugation method. J Cell Biol 38: 130–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M, Pyke K (2005) Plastid development and differentiation. In S Moller, ed, Plastids. Blackwell Scientific Publications, Oxford, pp 30–59
- Weihe A, Hedtke B, Börner T (1997) Cloning and characterization of a cDNA encoding a bacteriophage-type RNA polymerase from the higher plant Chenopodium album. Nucleic Acids Res 25: 2319–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CM, Jiang CZ, Meehan LJ, Voytas DF, Rodermel SR (1994) Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J 6: 161–175 [DOI] [PubMed] [Google Scholar]
- Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S (1999) The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK (2001) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem 277: 8588–8596 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.