Abstract
Mutants with defects in the cytochrome (cyt) b6/f complex were analyzed for their effect on the expression of a subgroup of nuclear genes encoding plastid-localized enzymes participating in chlorophyll biosynthesis. Their defects ranged from complete loss of the cytb6/f complex to point mutations affecting specifically the quinone-binding QO site. In these seven mutants, light induction of the tetrapyrrole biosynthetic genes was either abolished or strongly reduced. In contrast, a normal induction of chlorophyll biosynthesis genes was observed in mutants with defects in photosystem II, photosystem I, or plastocyanin, or in wild-type cells treated with 3-(3′4′-dichlorophenyl)-1,1-dimethylurea or 2,5-dibromo-3-methyl-6-isopropyl benzoquinone. We conclude that the redox state of the plastoquinone pool does not control light induction of these chlorophyll biosynthetic genes. The signal that affects expression of the nuclear genes appears to solely depend on the integrity of the cytb6/f complex QO site. Since light induction of these genes in Chlamydomonas has recently been shown to involve the blue light receptor phototropin, the results suggest that cytb6/f activity regulates a plastid-derived factor required for their expression. This signaling pathway differs from that which regulates state transitions, since mutant stt7, lacking a protein kinase involved in phosphorylation of the light-harvesting complex II, was not altered in the expression of the chlorophyll biosynthetic genes.
Cells of plants and eukaryotic algae harbor, in addition to the nucleus, two DNA-containing organelles of endosymbiotic origin, the plastids, and the mitochondria. The vast majority of genetic information of these endosymbionts in the course of evolution was transferred to the nuclear genomes of their hosts (Rujan and Martin, 2001; Martin et al., 2002). However, the chloroplasts, as predicted for Arabidopsis (Arabidopsis thaliana), contain between 2,500 and 3,000 proteins, half of which are of cyanobacterial origin (Abdallah et al., 2000; Richly and Leister, 2004). The location of genes encoding organelle proteins in different compartments implies the existence of mechanisms that serve to integrate nuclear and organellar gene expression. The cross talk between organelles includes both anterograde (nucleus-to-organelle) and retrograde (organelle-to-nucleus) controls. Via anterograde mechanisms, nucleus-encoded proteins tightly control essential steps in gene expression within the organelles (Goldschmidt-Clermont, 1998; Leon et al., 1998; Choquet and Wollman, 2002). Retrograde signaling regulates the expression of a subset of nuclear genes (mostly those encoding organelle proteins) in response to the metabolic and developmental state of the organelle.
The first evidence for retrograde chloroplast signaling came from analysis of barley (Hordeum vulgare) mutants defective in plastidic ribosomes (Bradbeer and Börner, 1978). Subsequent analyses have so far provided genetic and/or molecular evidence for five chloroplast-to-nucleus signaling pathways: one that requires plastid protein synthesis, a second that is associated with chloroplast-generated singlet oxygen, a third that appears to employ hydrogen peroxide, a fourth that senses the redox poise of the plastoquinone pool, and a fifth that involves tetrapyrrole biosynthesis intermediates (for review, see Surpin et al., 2002; Gray, 2003; Gray et al., 2003; Pfannschmidt et al., 2003; Beck, 2005; Fey et al., 2005).
It is unclear, however, how these pathways interact to regulate gene expression. Photosynthetic activity appears to be an important factor in controlling gene expression, since in mutants that are deficient in the cytochrome (cyt) b6/f complex or lacking plastidic ATP synthase, about one quarter of more than 3,000 nuclear genes analyzed were expressed at significantly different levels from wild type (Maiwald et al., 2003; Richly et al., 2003). This has led to the hypothesis that physiological effects of mutations affecting the redox state of the plastoquinone pool and/or the transmembrane H+-gradient control nuclear gene expression. However, large differences were also observed between different mutants lacking PSI (Maiwald et al., 2003; Richly et al., 2003; Ihnatowicz et al., 2004), which suggests that the integrity of a photosynthetic complex, rather than its activity in electron transfer, could be the source of the signals that control nuclear gene expression. While a large collection of data is available on the genes whose expression is perturbed in response to alterations in the chloroplast (Biehl et al., 2005; Leister, 2005), the sites of signal generation within the chloroplast have not yet been identified.
In the analysis presented here, we focused on the role of the cytb6/f complex in signaling toward the nucleus and present an analysis of seven mutants defective in this complex. In all of these mutants, the light induction of a subset of chlorophyll biosynthetic genes was either abolished or strongly diminished, while the expression of these genes was not affected when photosynthetic electron flow and the redox state of the plastoquinone pool were altered by inhibitors or by mutations in other photosynthetic complexes.
RESULTS
Mutants Defective in the Cytb6/f Complex
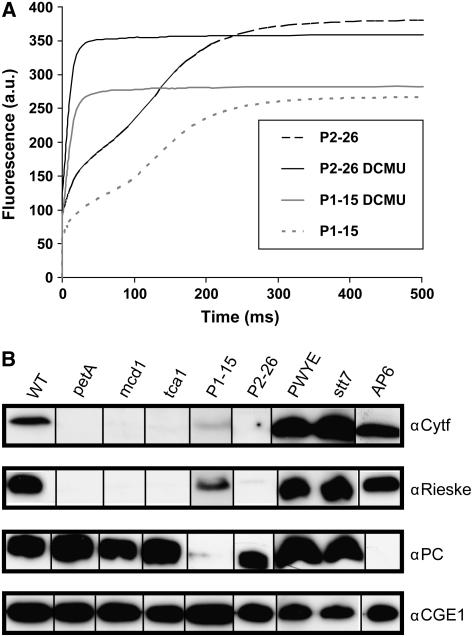
The mutants analyzed and their relevant characteristics are summarized in Table I. They include two mutants isolated in a screen aimed at the identification of genes that control light sensitivity (P1-15 and P2-26). These two strains as well as five mutants (ΔpetA, mcd1-F16, tca1-693, petD-PWYE, petC-Δ1, and clpP1-AUU) described before have defects in photosynthesis, i.e. they do not grow photoautotrophically and thus require acetate for growth. In addition, they are light sensitive, i.e. they grew on acetate-containing plates irradiated with a fluence rate of 20 μE·m−2·s−1 of white light but not when irradiated with a fluence rate of 80 μE·m−2·s−1 or 500 μE m−2·s−1 in the case of the P1-15 mutant. The P1-15 and P2-26 strains displayed fluorescence induction kinetics typical for cytb6/f mutants, i.e. a continued rise in fluorescence up to levels attained in the presence of 3-(3′4′-dichlorophenyl)-1,1-dimethylurea (DCMU; Fig. 1A). In P2-26, the maximum level of fluorescence obtained in the absence of DCMU was slightly higher than that seen in the presence of the herbicide, indicating a complete block in cytb6/f electron transfer. The higher level in the absence of DCMU may be explained by a full oxidation of plastoquinone, acting as a quencher of fluorescence (Joliot et al., 1998). Mutant P1-15, in contrast, resembles leaky cytb6/f mutants (see, for example, figure 4 in de Vitry et al., 1999), with a lower level of fluorescence in the absence of DCMU. Electron transfer through the cytb6/f complex can be revealed with high sensitivity by the slow phase of the flash-induced electrochromic shift measured at 520 nm (Joliot et al., 1998). In P1-15, a small absorption change was observed, suggesting that it is indeed capable of a slow rate of electron transfer via the cytb6/f complex (F. Rappaport, personal communication). This is consistent with a very weak signal observed in some immunoblots for cyt f and Rieske protein in extracts of the P1-15 strain (Fig. 1B).
Table I.
Mutants employed and their characteristics
LS, No growth at 80 μE m−2 s−1; HLS, no growth at 500 μE m−2 s−1; R, light resistant, i.e. growth at 500 μE m−2 s−1; cont., continued; WT, wild type; ND, not done.
| Mutant | Phenotypes
|
Mutational Defect | Reference | ||
|---|---|---|---|---|---|
| Growth without Acetate | Light Sensitivity | Fluorescence Induction | |||
| ΔpetA | − | LS | Cont. rise to Fmax | Deletion of chloroplast petA gene | Kuras and Wollman (1994) |
| mcd1-F16 | − | LS | Cont. rise to Fmax | Defect in a nuclear gene that controls petD mRNA stability | Drager et al. (1998) |
| tca1-693 | − | LS | Cont. rise to Fmax | Nuclear gene; encodes a translation activator for petA mRNA | Wostrikoff et al. (2001) |
| petD-PWYE | − | LS | Cont. rise to Fmax | A petD mutation that alters the QO pocket of the cytb6/f complex | Zito et al. (1999) |
| clpP1-AUU | + | R | WT | Mutation in clpP1 start codon resulting in low ClpP protease level | Majeran et al. (2000) |
| petC-Δ1 clpP1-AUU | − | LS | ND | 2-bp deletion in PETC; devoid of Rieske protein | Majeran et al. (2000) |
| P1-15 | − | HLS | Cont. rise to Fmax | Nuclear mutation | Dent et al. (2005) and this report |
| P2-26 | − | LS | Cont. rise to Fmax | Nuclear mutation | This report |
| stt7 | + | R | WT | Defect in a thylakoid-associated protein kinase involved in LHCII phosphorylation | Depège et al. (2003) |
| AP6 | − | HLS | ND | Mutation in nuclear gene PCY1 | Kindle and Lawrence (1998) |
| ΔpsbD | − | HLS | ND | Deletion in chloroplast psbD gene | Minai et al. (2006) |
| ΔpsaB | − | HLS | ND | Deletion in chloroplast psaB gene | Redding et al. (1999) |
Figure 1.
Analysis of mutants for fluorescence induction and two cytb6/f proteins as well as PC. A, Chlorophyll fluorescence induction curves of strains P1-15 and P2-26 measured in the absence or presence of 10 μm DCMU. Note that the Fmax level of P1-15 in the absence of the herbicide is lower than in its presence, indicating residual electron transfer. B, Total soluble proteins (15 μg per lane) were separated by SDS-PAGE (15% polyacrylamide). After transfer to membranes, protein blots were reacted sequentially with antisera directed against cyt f, the Rieske protein, and PC as described in “Materials and Methods.” Antibodies that recognized the plastidic GrpE homolog of Chlamydomonas (CGE1) served as a loading control.
Figure 4.
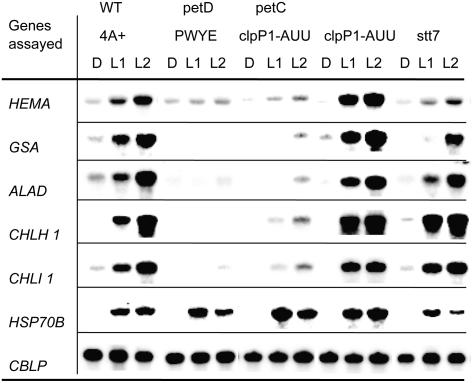
Expression of genes in mutants with defects in cytb6/f subunits or in state transitions. Mutant cultures, after incubation for 20 h in the dark (D), were shifted to light (40 μE m−2 s−1) for one (L1) and 2 h (L2). Samples were processed as described in the legend of Figure 2. Strain clpP1-AUU is a parent of the strain petC-Δ1 clpP1-AUU, and is used to show that the attenuation of ClpP is not responsible for an impairment in light induction.
Mutant ΔpetA has a deletion of the plastid genes for cyt f, while petD-PWYE, also a plastome mutation, carries substitutions in three residues of subunit IV, inactivating the QO site where plastoquinol oxidation occurs. In the other three previously characterized mutants, nuclear genes are affected: mcd1-F16 and tca1-693 lack nuclear-encoded factors necessary for stabilization of petD mRNA and translation of petA mRNA, respectively, while petC-Δ1 carries a short deletion in the PETC gene encoding the Rieske Fe-S protein, leading to a complete absence of this essential subunit. In the strain we have used, the latter mutation was combined with clpP1-AUU, a mutation reducing accumulation of the ClpP protease, resulting in the stabilization of a cytb6/f subcomplex lacking the Rieske protein (Majeran et al., 2000). The absence of cyt f was demonstrated by immunoblot analysis for mutant P2-26 and confirmed for mutants ΔpetA, mcd1-F16, and tca1-693 (Fig. 1B). Traces of Rieske protein could be observed in some of the mutants depending on the individual experiment, resulting from incomplete degradation when other subunits are missing (O. Vallon, unpublished data). In contrast, both cyt f and Rieske proteins accumulated normally in the petD-PWYE mutant, as described before (Zito et al., 1999). Also included in this analysis was the mutant stt7, which is defective in the protein kinase that is required for the phosphorylation of the major light-harvesting protein (LHCII), an essential requirement for state transitions to occur (Depège et al., 2003). This mutant has normal levels of cytb6/f (Fig. 1B), grows photoautotrophically, and is not light sensitive. Mutant AP6 has a defect in the PCY gene for plastocyanin (PC; Fig. 1B) and was used as a control. We observed a reduced level of PC in P1-15, which was shown to be caused by a second site mutation since it separated from the cytb6/f defect in crosses. This defect was not linked to the regulatory phenotype of mutant P1-15 (data not shown).
Mutants with Defects in the Cytb6/f Complex Exhibit Deregulation of Genes Involved in Chlorophyll Synthesis
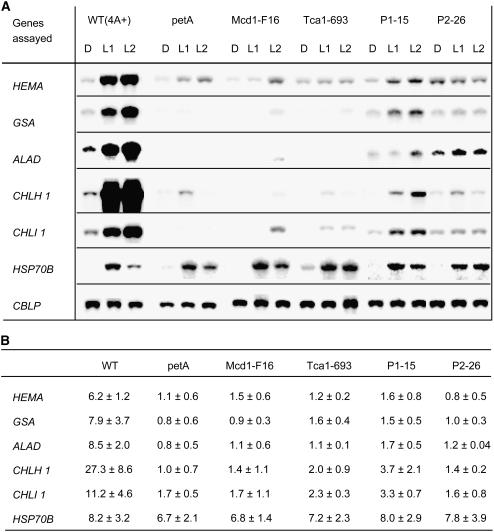
A number of Chlamydomonas reinhardtii genes involved in chlorophyll biosynthesis (HEMA, GSA, ALAD, CPXI, CHLD, CHLH1, CHLI1, and CTH1) have been shown to be induced in dark-adapted cultures by a shift from dark to light of a moderate fluence rate (40 μE·m−2·s−1; Vasileuskaya et al., 2004). The expression of five of these nuclear genes was analyzed at the RNA level in those mutants that either lacked the cytb6/f complex or possessed strongly reduced levels. In all cases, the patterns of mRNA accumulation following the shift to light were clearly different from that of the wild type, exhibiting a lack of light-induced accumulation (Fig. 2). In some mutants, mRNA levels for genes GSA, ALAD, and CHLI1 in the dark or after shift to light remained below the level of detection. In the P1-15 mutant, some residual light-induced mRNA accumulation was observed but this was distinctly lower than in the wild-type strain. In contrast to the five genes of tetrapyrrole biosynthesis, HSP70B, a nuclear gene encoding a chloroplast-localized chaperone (Schroda et al., 1999), exhibited normal light induction in the mutants. Also, expression of CBLP, encoding a Gβ-like protein (von Kampen et al., 1994) and used as a loading control, appeared not to be affected by defects in the cytb6/f complex. Thus, expression of a subset of nuclear genes encoding enzymes required for chlorophyll synthesis was specifically deregulated in mutants defective in the cytb6/f complex, i.e. showed no up-regulation in the light.
Figure 2.
Expression of selected genes of chlorophyll biosynthesis and HSP70B in mutants lacking cytb6/f complex components. A, RNA-blot analysis, details of which are given in “Materials and Methods.” Prior to light exposure, cultures were incubated in the dark for 20 h (D). Then, cultures were shifted to light (fluence rate 40 μE m−2·s−1) for 1 h (L1) and 2 h (L2). At these time points samples were taken for RNA extraction. For RNA-blot analyses, 10 μg of total RNA were hybridized with probes specific for the genes indicated. The constitutively expressed CBLP gene, encoding a Gβ-like protein (von Kampen et al., 1994), served as a loading control. B, Quantitative evaluation of RNA-blot data from at least three experiments. The relative induction of each gene with sem was normalized using the signal for the CBLP gene. The induction ratios given were determined by dividing the values of the L2 samples by those of the D samples.
Genetic Analysis of Cytb6/f Mutants
In C. reinhardtii, a haploid organism, second site mutations manifest themselves phenotypically and may falsely contribute to the phenotype of a first site mutation. It could be envisioned that mutations affecting gene regulation tend to accumulate over time in cytb6/f mutants, provided they enhance their survival under storage conditions. To address this question, mutants with defects in the cytb6/f complex were crossed with wild-type strains and random spores were picked among the progeny and scored for defective photosynthesis, light sensitivity, and light induction of chlorophyll biosynthesis genes (Table II). If the deregulation of the genes observed was due to a mutation different from that affecting the cytb6/f complex, independent segregation of the phenotypes acetate requirement/light sensitivity (ac−) and no light regulation of chlorophyll biosynthesis genes (lrc−) would be expected (unless the two lesions were closely linked).
Table II.
Analysis of progeny from crosses of mutants with wild type for cosegregation of the mutant phenotypes
ac−, Acetate requiring; LS, no growth at 80 μE m−2 s−1; HLS, no growth at 500 μE m−2 s−1; LR, light resistant; lrc, light regulation of chlorophyll biosynthesis gene expression.
| Crosses Performed | Phenotypes of Progeny Assayed on Plates | Regulatory Phenotypes of Eight Clonesa |
|---|---|---|
| ΔpetA (mt+) × 4A− (mt−) | 152ac−, LS/12ac+, LR | 4lrc−, ac−, mt−/4lrc+, ac+, mt+a |
| petD-PWYE (mt+) × 4A− (mt−) | 94ac−, LS/6ac+, LR | 4lrc−, ac−, mt−/4lrc+, ac+, mt+a |
| mcd1-F16 (mt−) × 4A+ (mt+) | 23ac−, LS/24ac+, LR | 4lrc−, ac−, LS/4lrc+, ac+, LR |
| tca1-693 (mt−) × 4A+ (mt+) | 21ac−, LS/27ac+, LR | 4lrc−, ac−, LS/4lrc+, ac+, LR |
| P1-15 (mt+) × 4A− (mt−) | 24ac−, HLS/23ac+, LR | 4lrc−, ac−, HLS/4lrc+, ac+, LR |
| P2-26 (mt+) × 4A− (mt−) | 20ac−, LS/27ac+, LR | 4lrc−, ac−, LS/4lrc+, ac+, LR |
To ensure the analysis of recombinants (by RNA-blot techniques) from these crosses, the lrc phenotype was assayed only in ac− progeny that was mt− and in ac+ progeny that was mt+.
For the chloroplast-born mutation ΔpetA, 93% of the progeny was acetate requiring and light sensitive, as expected from the largely uniparental transmission of chloroplast genes (Harris, 1989). Four each of the ac− mutant and ac+ progeny were tested for their ability to induce chlorophyll biosynthesis genes upon dark-to-light shift using RNA blots. To ensure that truly recombinant progeny was tested, as opposed to unmated parental gametes that would have survived the zygote selection process, mt+ clones were chosen for the ac+ progeny, and mt− clones for the ac− progeny. In the eight clones tested, strict correlation was observed between the photosynthesis defect/light sensitivity and absence of gene induction by light (Table II).
Crosses of the four nuclear mutants showed an approximately equal distribution of wild-type and ac− phenotypes (Table II), as expected for segregation of single nuclear mutations. As above, four randomly picked clones of each phenotypic class were analyzed for the lrc phenotype and here again, perfect correlation was observed between the ac and lrc phenotypes (Table II). In spite of the small number of progeny analyzed, the fact that this result was observed for five independent mutants carrying mutations in two distinct genomes clearly shows that the defect in light induction of the chlorophyll biosynthesis genes is a direct consequence of cytb6/f deficiency.
Light Induction of Genes Involved in Chlorophyll Synthesis Is Not Affected by Other Types of Photosynthesis Mutations or by Inhibitors of Electron Transport
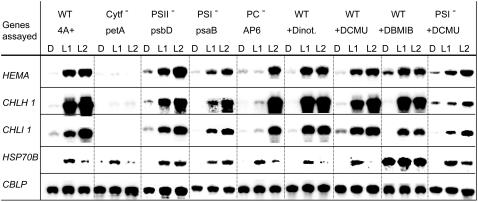
The deregulation phenotype of mutants defective in the cytb6/f complex raised the question whether interruption of photosynthetic electron transport by itself was sufficient to cause the alteration in gene expression observed. To address this question we made use of mutants defective in different steps of electron transport. A ΔpsbD mutant, defective in PSII, showed a wild-type pattern of gene expression (Fig. 3). Also mutant AP6, which lacks PC (Fig. 1B) and thus, in the absence of the alternative electron carrier cyt c6 that only is present when cells are deprived of copper (Merchant and Bogorad, 1986), is unable to transport electrons from the cytb6/f complex to PSI, showed light induction of the genes assayed (Fig. 3). The slight delay in light induction in the PC mutant was not observed with other PC-defective strains (data not shown). Lastly, mutant ΔpsaB, defective in PSI, was also not affected in the regulation of the genes assayed. We conclude that a block in photosynthetic electron transport by itself is not sufficient to prevent light induction of the nuclear genes analyzed. Importantly, while in PSII mutants the plastoquinone pool is rapidly oxidized during the dark-to-light shift, the reverse will occur in PSI and PC mutants where, just like in cytb6/f mutants, the plastoquinone pool will be overreduced. Thus, the deregulation phenotype of cytb6/f mutants cannot be attributed to an indirect effect on the redox state of the plastoquinone pool during illumination.
Figure 3.
Effect on gene expression patterns of mutations affecting PSII, PC, PSI, and of inhibitors that block electron transfer from PSII to the cytb6/f complex in wild-type cells. All cultures were incubated in the dark for 20 h (D). Cultures of the 4A+ strain and the four mutants were then exposed to white light for one (L1) and 2 h (L2). For inhibitor studies, dinoterb (final concentration 30 μm), DCMU (final concentration 3 μm), and DBMIB (final concentration 1.5 μm) were added to cultures 40 min prior to shift to light. Samples were processed as described in the legend of Figure 2. Note that DBMIB caused overaccumulation of HSP70B mRNA in the dark and in the light, an observation that was not pursued.
In accordance with these observations, the application of the PSII inhibitors DCMU and the phenolic herbicide dinoterb did not affect gene expression patterns (Fig. 3). These compounds interact with the QB binding site of the PSII complex and prevent reoxidation of the primary quinone acceptor by the plastoquinone pool.
More surprisingly, 2,5-dibromo-3-methyl-6-isopropyl benzoquinone (DBMIB), which inhibits the oxidation of plastoquinol at the QO site of the cytb6/f complex (Roberts et al., 2004), did not influence the light induction of the chlorophyll biosynthesis genes (Fig. 3). DBMIB binds at the QO site of cytb6/f, where plastoquinol is reoxidized, suggesting that plastoquinol binding and turnover at this site is not necessary for light induction to occur. To rule out any direct effect of light on the electron transfer chain, the PSI mutant was treated with DCMU. Here again, light induction was observed both at 3 μm DCMU (Fig. 3) and 6 μm DCMU (data not shown), in spite of the fact that inactivation of both photosystems would obviously prevent any light-induced redox changes in the plastoquinone pool and cytb6/f complex.
Analysis of Mutants with Discrete Defects in Cytb6/f Subunits or in Cytb6/f Signaling
These observations suggested that the abrogation of light induction of the chlorophyll biosynthesis genes was due to the lack of accumulation of the cytb6/f complex, rather than to the absence of electron transport through the complex. We therefore analyzed mutants that accumulated the cytb6/f complex, but carried more discrete defects preventing electron transfer. The petC-Δ1 mutant (de Vitry et al., 1999) is devoid of Rieske Fe-S protein, hence of a functional QO site, but accumulates almost normal levels of cyt f and other core subunits of the complex, at least when combined with a mutation that attenuates ClpP (Majeran et al., 2000). This strain clearly exhibited the same deregulated phenotype seen for the mutants that lack the cytb6/f complex (Fig. 4). We also analyzed the petD-PWYE mutant, which carries three substitutions in subunit IV and assembles the complex (including the Rieske protein), but is unable to bind plastoquinol in the QO pocket and therefore to transfer electrons (Zito et al., 1999). This mutant also exhibited no light induction of the five genes analyzed (Fig. 4). The petD-PWYE mutant was crossed to wild type, and the phenotype of the progeny was analyzed as described above. Again, a perfect correlation was observed between the ac− and lrc− phenotypes (Table II; Fig. 5).
Figure 5.
Analysis of lrc phenotypes among progeny from a cross between the petD-PWYE mutant and wild type. The data represent one example for the analyses performed with progeny from all crosses listed in Table II. The analyses were done as described in the legend of Figure 2. Above the columns, the phenotype of the strain analyzed with respect to acetate requirement and mating type is listed.
Together, these results indicate that normal light induction of chlorophyll biosynthesis genes not only requires the assembly of a complete cytb6/f complex, but also that its QO site is able to bind quinones. The QO site is also known to regulate state transitions, activating the LHCII kinase in a redox-dependent manner. In Chlamydomonas, the Stt7 kinase has been shown to be required for this process (Depège et al., 2003). In Arabidopsis, mutation of the orthologous gene Stn7 (Bellaflore et al., 2005) not only impairs state transitions, but also affects expression of nuclear genes, as does inactivation of the paralogous gene Stn8 (Bonardi et al., 2005). We therefore analyzed the Chlamydomonas stt7 mutant and found that it exhibited a pattern of gene expression similar to that of the wild-type strain (Fig. 4), indicating that the Stt7 kinase is not involved in light induction of chlorophyll biosynthesis genes.
DISCUSSION
The light induction of the five chlorophyll biosynthesis genes analyzed here has recently been shown to be mediated via the blue light receptor phototropin (Im et al., 2006). The fact that induction of these genes observed after a shift from dark to light is markedly diminished in strains with reduced levels of phototropin implies an essential role of this photoreceptor in the regulation of these genes. This signaling pathway is envisioned to act at the level of activation of transcription or, alternatively, via stabilization of the transcripts. For one gene (GSA), a partial characterization of the blue light signaling pathway has suggested the participation of a heterotrimeric G-protein and phospholipase C, an increase in the cytosolic Ca2+ concentration, and activation of calmodulin as well as of a calmodulin-dependent kinase (Im et al., 1996). Here we show that mutants lacking a functional cytb6/f complex are unable to carry out this light-induction process. This suggests that the light activation of these nuclear genes is in part controlled by signals originating from the chloroplast, where the gene products reside. However, none of the mechanisms hitherto described as mediating retrograde chloroplast-to-nucleus signaling (Beck, 2005; Nott et al., 2006) appear able to explain our results.
Intermediates of the tetrapyrrole biosynthetic pathway are known to participate in the regulation of chlorophyll biosynthesis genes in particular at the level of transcript accumulation. In Arabidopsis, Mg-Protoporphyrin IX accumulates in photodamaged chloroplasts, leading to repression of LHCB1 expression (Strand et al., 2003). The light-mediated activation of HEMA1 and, to a minor degree that of GSA, was impaired by chloroplast-damaging treatments such as the application of the phytoene desaturase-inhibitor norflurazon, by far-red light pretreatment, or by inhibitors of plastid protein synthesis (Kumar et al., 1996; McCormac and Terry, 2004). In contrast, the C. reinhardtii HEMA gene is induced by the feeding of Mg-Protoporphyrin IX (Vasileuskaya et al., 2005), which also activates expression of three nuclear HSP70 genes (Kropat et al., 1997; Vasileuskaya et al., 2004). It could be hypothesized that lack of light induction in cytb6/f mutants is related to a defect in the accumulation of Mg-protoporphyrin IX, or its release from the chloroplast. However, no link can be made at present between cytb6/f activity and the metabolism of tetrapyrroles. We note that HSP70B is normally induced in cytb6/f mutants, which suggests that the mechanism of tetrapyrrole signaling is not impaired.
We have also examined the possibility that reactive oxygen species, produced in the cytb6/f mutants upon exposure to light, would prevent the induction of the chlorophyll biosynthetic genes. Exogenous addition of hydrogen peroxide (2 mm) did not prevent the light induction of the genes assayed; neither did singlet oxygen generated by treatment with methylene blue in the light (Anthony et al., 2005), suggesting that reactive oxygen species are unlikely to play a role in this deregulation (data not shown).
Redox control has been invoked as a major player in the field of chloroplast-to-nucleus signaling in higher plants (for review, see Fey et al., 2005) as well as in green algae (Escoubas et al., 1995; Maxwell et al., 1995; Durnford and Falkowski, 1997). This is based mostly on experiments where the redox state of the plastoquinone pool was manipulated by illumination in the presence of DCMU or DBMIB, as well as by changes in the light regime. For a number of nuclear genes, the transcript levels have been shown to vary in a manner consistent with long-term regulation of photosystem stoichiometry and light-harvesting capacity. However, the chlorophyll biosynthesis genes that we have studied have not been identified as redox controlled in these studies. Here, we show that treatment with DCMU and DBMIB do not prevent Chlamydomonas wild-type cells from inducing the chlorophyll biosynthesis genes (Fig. 3), in spite of the fact that they will change drastically, and in opposite ways, the redox state of the plastoquinone pool (fully oxidized with DCMU, fully reduced with DBMIB). Moreover, PSII mutants on the one hand, and PSI as well as PC mutants on the other hand, do undergo changes in the redox state of the plastoquinone pool and of the high potential chain of the cytb6/f complex (cyt f and Rieske protein) when illuminated, but in opposite directions. Still, all showed normal light induction of the chlorophyll biosynthesis genes (Fig. 3). It could be argued that cyclic electron transport, which is still carried out in the presence of DCMU or in PSII mutants (Finazzi et al., 1999), can lead to reduction of the plastoquinone pool in response to light. But we have observed light induction in a DCMU-treated PSI mutant, where no redox change can be brought about by illumination (Fig. 3). Thus, we conclude that changes in the redox state of the plastoquinone pool play no role in the light induction of the chlorophyll biosynthesis genes. In this respect, the signaling we describe differs markedly both from the putative redox control of nuclear genes mentioned above, and from state transitions.
State transitions govern light-energy distribution between photosystems and the balance between linear and cyclic electron transport, and are redox controlled (for review, see Wollman, 2001). Cytb6/f plays a central role in this process: cytb6/f mutants are unable to perform this reorganization because they fail to activate the protein kinase responsible for LHCII phosphorylation. The kinase has been suggested to be physically associated with the cytb6/f complex. The complete loss of state transitions in the petD-PWYE mutant clearly demonstrated the involvement of the QO site in kinase activation (Zito et al., 1999). Based on inhibitor studies (Finazzi et al., 2001), a dynamic model has been proposed where activation of the kinase requires oscillation of the Rieske protein between its proximal and distal positions. The protein kinase Stt7 is required for LHCII phosphorylation since strains with defects in the STT7 gene are unable to undergo state transitions (Fleischmann et al., 1999). Here we show that the Stt7 kinase is not involved in the control of the chlorophyll biosynthesis genes, since an stt7 mutant exhibited a wild-type pattern of gene expression (Fig. 4). This does not necessarily mean that the signaling process under study has no component in common with state transitions, because the Stt7 kinase could simply be a downstream component of a cascade. However, the fact that the control of signaling is different, redox in one case, light in the other, argues for distinct mechanisms.
Still, the fact that mutants with defects in the QO site (petD-PWYE and petC-Δ1) are completely unable to induce the chlorophyll biosynthesis genes (Fig. 4), in spite of having an assembled (or partially assembled) cytb6/f complex, indicates a requirement for a functional QO site. How do we account for a light-signaling process that is at the same time dependent on the presence of a functional QO site, and independent on changes in its occupancy?
We examined the possibility that the cytb6/f complex itself senses light. A chlorophyll molecule is present in the cytb6/f complex (Stroebel et al., 2003), which in principle could act as a light sensor if its excited state could be transduced into a stable modification/conformational change of the complex. Its phytyl chain lies in the vicinity of the QO site, which led us to consider the possibility that its conformation or presence was affected in QO site mutants. However, a series of petD mutants lacking this chlorophyll molecule (kindly provided by F. Zito) still showed induction of the chlorophyll biosynthesis genes by light, ruling out this hypothesis. In contrast, a mutant carrying a petD deletion mutation was defective in induction (data not shown). No mutants are available that affect the other pigment of the complex, β-carotene, but the fact that its excited states are extremely short lived does not make it an attractive candidate for a signaling role.
We thus would like to propose another type of mechanism whereby an essential component of a light-dependent signal-generating system would interact with the cytb6/f complex and be stabilized/activated by this interaction. Light could act directly on the associated signaling component, if it was to harbor a chromophore. Alternatively, and possibly more likely, it could just be part of a light-signaling cascade in which phototropin serves as a light receptor. Not much is known about how phototropin regulates gene expression (Huang and Beck, 2003; Im et al., 2006). It can be hypothesized that the pathway that leads to activation of the chlorophyll biosynthesis genes has a branch located in the thylakoid membrane, which could act as a sensor of the state of the photosynthetic apparatus.
Our results are not in line with the assumption that physiological states conditioned by photosynthetic mutations represent the principal trigger for different modes of plastid signaling and nuclear response (Maiwald et al., 2003; Ihnatowicz et al., 2004; Biehl et al., 2005). Rather, the identification of a role for the cytb6/f complex in plastid signaling suggests that components of the photosynthetic machinery themselves may be involved. The results thus provide new routes to analyze signaling by the chloroplast. One of the principal questions to be addressed will have to focus on the molecular nature of the signal generated and its subsequent transduction to the cytosol/nucleus.
MATERIALS AND METHODS
Algal Strains
Chlamydomonas reinhardtii strain 4A+ (mt+) and mutant stt7 were obtained from J.-D. Rochaix (University of Geneva). Strain 4A− (mt−), a wild-type strain that is near isogenic to 4A+, was generated by backcrossing to 4A+ (Dent et al., 2005). Mutants P1-15 (CAL007.01.20) and P2-26 (CAL011.01.03) were generated in the 4A+ strain as described previously (Dent et al., 2005). In these two mutants, the phenotypes acetate requirement/light sensitivity were not linked to zeocin resistance, suggesting that these phenotypes were caused by separate mutational events (Dent et al., 2005; data not shown). Mutants defective in PSI (ΔpsaB), PC (AP6), PSII (ΔpsbD), cytb6/f (ΔpetA, mcd1-F16, tca1-693, petD- PWYE, and petC-Δ1clpP-AUU), or state transitions (stt7), have been described before (Table I).
Culture Conditions
Strains were grown heterotrophically or photoheterotrophically in Tris-acetate phosphate (TAP) media or photoautotrophically in minimal media (Harris, 1989). Mutant stocks were maintained on TAP agar medium at a very low light intensity of 5 μE m−2 s−1 at 23°C. Liquid cultures of wild-type cells were grown with continuous irradiation (40 μE m−2 s−1) at 23°C to a density of 3 to 5 × 106 cells per mL. For light induction, the cultures were then divided into subcultures of 50 mL each, and incubation continued in the dark. After 20 h of dark incubation, these cultures were exposed to white light (40 μE m−2 s−1) and samples were taken for RNA isolation (von Gromoff et al., 1989). Mutant cultures, due to their light sensitivity, were grown at very low light intensity and treated like wild-type cells after reaching a density of 3 to 5 × 106 cells per mL. For testing light-sensitivity phenotypes, mutants and wild type were incubated at a light intensity of 80 μE m−2 s−1 or 500 μE m−2 s−1 on TAP plates using a high light incubator with a cooling system.
For measurements of fluorescence induction at room temperature, dark-adapted cells were transferred to a 1 mL cuvette placed in a home-built fluorimeter (Joliot et al., 1998), in the absence or presence of DCMU (10 μm), and fluorescence induction was recorded over 1.5 s of illumination.
Genetic Analyses
The mutants were crossed with wild-type strains following standard protocols (Harris, 1989). For gamete generation, vegetative cells were resuspended in nitrogen-free (TAP-N) medium at a density of 1 × 107 cells/mL and incubated for 16 h in the light. Gametes of mt+ and mt− were mixed, incubated for 1 h, and then plated on TAP agar (4%) medium. These plates were incubated in the light for 24 h and then in the dark for at least 4 d. Zygotes adhering to the agar after nonmated cells had been scraped off were germinated by irradiation and random spores were spread on TAP agar plates (1.5%). For testing acetate requirement and light sensitivity, the progeny of random spores was irradiated with 80 μE m−2 s−1 or 500 μE m−2 s−1 on TAP or photoautotrophically in minimal plates. All plates were scored for cell growth and color after 10 to 14 d of incubation.
Immunoblot Analyses
Cells were sedimented by centrifugation (3,000g for 5 min) and resuspended in 0.1 m dithiothreitol/0.1 m Na2CO3. Then, 0.66 volumes of 5% SDS/30% Suc were added. In cases where the lysates were too viscous, samples were sonicated. Homogenization of the suspensions was achieved by rapid shaking at room temperature for 20 min. The protein concentration was determined by staining with amido black, using bovine serum albumin as a standard (Popov et al., 1975). Total soluble protein (15 μg) was separated on 15% polyacrylamide gels under denaturing (0.1% SDS) conditions. The proteins were transferred from the gels to polyvinylidene difluoride membranes (Hybond-P; Amersham Biosciences). Protein blots were reacted sequentially with antisera directed against cyt f, the Rieske Fe-S protein, PC (Li et al., 1996), and CGE1 (chloroplast GrpE homolog of Chlamydomonas; Schroda et al., 2001).
Peroxidase-conjugated anti-rabbit serum (Sigma-Aldrich) was used to detect the primary antibodies. For signal detection we used the enhanced chemiluminescence system (Amersham Biosciences).
RNA Gel-Blot Analyses
RNA extraction, electrophoretic separation of RNA, and hybridizations were performed as described previously (von Gromoff et al., 1989). Ten micrograms of total RNA per lane were separated on formaldehyde-containing agarose gels and transferred to nylon membranes (Hybond-N; Amersham). Prehybridization (2 h) and hybridization (18 h) were performed at 65°C in 0.1 m NaCl, 50 mm Tris HCl (pH 7.5), 0.1% sodium pyrophosphate, 10× Denhardt solution, 1% SDS, 10% dextran sulfate, 60% formamide, and 100 μg/mL of sheared, denatured herring sperm DNA. The probes were labeled with [α-32P]dCTP by the random priming protocol. After hybridization the membranes were washed twice in 2× standard saline citrate (SSC) at room temperature, once in 2× SSC, 1% SDS for 30 min at 65°C, and once in 0.2× SSC at room temperature.
Probes for Hybridization
The probes used for detection of HSP70B and HEMA transcripts have been described previously (Schroda et al., 2001; Vasileuskaya et al., 2004). Probes used to detect mRNAs of the other genes were a 1.6 kb EcoRI, SmaI cDNA fragment for GSA (Matters and Beale, 1994), a 1.7 kb cDNA fragment for ALAD (Matters and Beale, 1995), a 1.4 kb cDNA fragment for CHLH1 (H subunit of Mg-chelatase; Chekounova et al., 2001), and a 459 bp PCR fragment from genomic DNA for CHLI1 (I subunit of Mg-chelatase). For a loading control, CBLP encoding a Gβ-like protein (von Kampen et al., 1994) was used as a probe.
Acknowledgments
We thank members of the Paris laboratory, in particular Francis-André Wollman and Giovanni Finazzi, as well as Michael Schroda and Anja Krieger-Liszkay, for intellectual input, and Michael Schroda in addition for a critical reading of the manuscript. We also thank Yves Choquet, René Matagne, Janette Kropat, and Jean-David Rochaix for providing strains. Francesca Zito's willingness to share data and mutant strains prior to publication is greatly appreciated.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (to C.F.B.) and from the U.S. National Science Foundation (grant no. MCB–0235878 to K.K.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christoph F. Beck (beck@uni-freiburg.de).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081059.
References
- Abdallah F, Salamini F, Leister D (2000) A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci 5: 141–142 [DOI] [PubMed] [Google Scholar]
- Anthony JR, Warczak KL, Donohue TJ (2005) A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci USA 102: 6502–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CF (2005) Signaling pathways from the chloroplast to the nucleus. Planta 222: 743–756 [DOI] [PubMed] [Google Scholar]
- Bellaflore S, Barneche F, Peltier G, Rochaix J-D (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Biehl A, Richly E, Noutsos C, Salamini F, Leister D (2005) Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and coordination of nuclear and plastid gene expression. Gene 344: 33–41 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Bradbeer JW, Börner T (1978) Activities of glyceraldehyde-phosphate dehydrogenase (NADP+) and phosphoribulokinase in two barley mutants deficient in chloroplast ribosomes. In G Akoyunoglou, JH Argyrondi-Akoyunoglou, eds, Chloroplast Development. Elsevier/North-Holland Biomedical Press, Amsterdam, pp 727–732
- Chekounova E, Voronetskaya V, Papenbrock J, Grimm B, Beck CF (2001) Characterization of Chlamydomonas mutants defective in the H-subunit of Mg-chelatase. Mol Genet Genomics 266: 363–373 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Wollman F-A (2002) Translational regulations as specific traits of chloroplast gene expression. FEBS Lett 529: 39–42 [DOI] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N, Bellafiore S, Rochaix J-D (2003) Role of chloroplast protein kinase stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- de Vitry C, Finazzi G, Baymann F, Kallas T (1999) Analysis of the nucleus-encoded and chloroplast-targeted Rieske protein by classic and site-directed mutagenesis of Chlamydomonas. Plant Cell 10: 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager RG, Girard-Bascou J, Choquet Y, Kindle KL, Stern DB (1998) In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J 13: 85–96 [DOI] [PubMed] [Google Scholar]
- Durnford DG, Falkowski PG (1997) Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosynth Res 53: 229–241 [Google Scholar]
- Escoubas JM, Lomas M, La Roche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92: 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V, Wagner R, Bräutigam K, Pfannschmidt T (2005) Photosynthetic redox control of nuclear gene expression. J Exp Bot 56: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Finazzi G, Furia A, Barbagallo RP, Forti C (1999) State transitions, cyclic and linear electron transport and photophosphorylation in Chlamydomonas reinhardtii. Biochim Biophys Acta 1413: 117–129 [DOI] [PubMed] [Google Scholar]
- Finazzi G, Zito F, Barbagallo RP, Wollman F-A (2001) Contrasted effects of inhibitors of cytochrome b6f complex on state transitions in Chlamydomonas reinhardtii. J Biol Chem 276: 9770–9774 [DOI] [PubMed] [Google Scholar]
- Fleischmann MM, Ravanel S, Delosme R, Olive J, Zito F, Wollman F-A, Rochaix J-D (1999) Isolation and characterization of photoautotrophic mutants of Chlamydomonas reinhardtii deficient in state transition. J Biol Chem 274: 30987–30994 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177: 115–180 [DOI] [PubMed] [Google Scholar]
- Gray JC (2003) Chloroplast-to-nucleus signalling: a role for Mg-protoporphyrin. Trends Genet 19: 526–529 [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D (2003) Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci 358: 135–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory use. Academic Press, San Diego [DOI] [PubMed]
- Huang K, Beck CF (2003) Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 100: 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D (2004) Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Im C-S, Eberhard S, Huang K, Beck CF, Grossman A (2006) Phototropin involvement in expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J (in press) [DOI] [PubMed]
- Im C-S, Matters GL, Beale SI (1996) Calcium and calmodulin are involved in blue light induction of the gsa gene for an early chlorophyll biosynthetic step in Chlamydomonas. Plant Cell 8: 2245–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Beal D, Delosme R (1998) In vivo measurement of photosynthetic activity: methods. In J-D Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 433–449
- Kindle KL, Lawrence SD (1998) Transit peptide mutations that impair in vitro and in vivo chloroplast protein import do not affect accumulation of the gamma-subunit of chloroplast ATPase. Plant Physiol 116: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rüdiger W, Beck CF (1997) Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci USA 94: 14168–14172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Csankovszki G, Söll D (1996) A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol Biol 30: 419–426 [DOI] [PubMed] [Google Scholar]
- Kuras R, Wollman F-A (1994) The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D (2005) Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354: 110–116 [DOI] [PubMed] [Google Scholar]
- Leon P, Arroyo A, Mackenzie S (1998) Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Physiol Plant Mol Biol 49: 453–480 [DOI] [PubMed] [Google Scholar]
- Li HH, Quinn J, Culler D, Girard-Bascou J, Merchant S (1996) Molecular genetic analysis of plastocyanin biosynthesis in Chlamydomonas reinhardtii. J Biol Chem 271: 31283–31289 [DOI] [PubMed] [Google Scholar]
- Maiwald D, Dietzmann A, Jahns P, Pesaresi P, Joliot P, Joliot A, Levin JZ, Salamini F, Leister D (2003) Knock-out of the genes coding for the Rieske protein and the ATP-synthase δ-subunit of Arabidopsis: effects on photosynthesis, thylakoid protein composition, and nuclear chloroplast gene expression. Plant Physiol 133: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Wollman F-A, Vallon O (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matters GL, Beale SI (1994) Structure and light-regulated expression of gsa gene encoding the chlorophyll biosynthetic enzyme, glutamate 1-semialdehyde aminotransferase, in Chlamydomonas reinhardtii. Plant Mol Biol 24: 617–629 [DOI] [PubMed] [Google Scholar]
- Matters GL, Beale SI (1995) Structure and expression of the Chlamydomonas reinhardtii alad gene encoding the chlorophyll biosynthetic enzyme, delta-aminolevulinic acid dehydratase (porphobilinogen synthase). Plant Mol Biol 27: 607–617 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Laudenbach DE, Huner NPA (1995) Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol 109: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ (2004) The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signaling pathways during de-etiolation. Plant J 40: 672–685 [DOI] [PubMed] [Google Scholar]
- Merchant S, Bogorad L (1986) Regulation by copper of the expression of plasto-cyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol Cell Biol 6: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L, Wostrikoff K, Wollman F-A, Choquet Y (2006) Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Schütze K, Fey V, Sherameti I, Oelmüller R (2003) Chloroplast redox control of nuclear gene expression — a new class of plastid signals in interorganellar communication. Antiox Redox Signal 5: 95–101 [DOI] [PubMed] [Google Scholar]
- Popov N, Schmitt S, Matthices H (1975) Eine störungsfreie Mikromethode zur Bestimmung des Proteingehalts in Gewebshomogenaten. Acta Biol Germ 31: 1441–1446 [PubMed] [Google Scholar]
- Redding K, Cournac L, Vassiliev IR, Golbeck JH, Peltier G, Rochaix J-D (1999) Photosystem I is indispensable for photoautotrophic growth, CO2 fixation, and H2 photoproduction in Chlamydomonas reinhardtii. J Biol Chem 274: 10466–10473 [DOI] [PubMed] [Google Scholar]
- Richly E, Dietzmann A, Biehl A, Kurth J, Laloi C, Apel K, Salamini F, Leister D (2003) Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep 4: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Roberts AG, Bowman ML, Kramer DM (2004) The inhibitor DBMIB provides insight into the functional architecture of the QO site in the cytochrome b6f complex. Biochemistry 43: 7707–7716 [DOI] [PubMed] [Google Scholar]
- Rujan T, Martin W (2001) How many genes in Arabidopsis came from cyanobacteria? An estimate from 386 protein phylogenies. Trends Genet 17: 113–120 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman F-A (2001) The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13: 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman F-A, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D (2003) An atypical haem in the cytochrome b(6)f complex. Nature 426: 399–400 [DOI] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell (Suppl) 14: S327–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileuskaya Z, Oster U, Beck CF (2004) Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth Res 82: 289–299 [DOI] [PubMed] [Google Scholar]
- Vasileuskaya Z, Oster U, Beck CF (2005) Mg-protoporphyrinIX and heme control HEMA, the gene encoding the first specific step of tetrapyrrole biosynthesis, in Chlamydomonas reinhardtii. Eukaryot Cell 4: 1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff ED, Treier U, Beck CF (1989) Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol Cell Biol 9: 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kampen J, Nieländer U, Wettern M (1994) Stress-dependent transcription of a gene encoding a Gβ-like polypeptide from Chlamydomonas reinhardtii. J Plant Physiol 143: 756–758 [Google Scholar]
- Wollman F-A (2001) State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J 16: 3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Choquet Y, Wollman F-A, Girard-Bascou J (2001) TCA1, a single nuclear encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman F-A (1999) The QO site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J 18: 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]