Abstract
Plasmodium falciparum chloroquine resistance was first detected in Cambodia in the early sixties. Treatment with chloroquine was abandoned 20 years ago. In vitro chloroquine sensitivity monitoring indicates that all eastern Cambodian isolates were sensitive to chloroquine, whereas most isolates collected from western provinces displayed reduced susceptibility to chloroquine. This indicates that the rate of chloroquine resistance remains high and stable in this region in the absence of chloroquine pressure. Characterization of codons 72 to 78 and 218 to 220 of pfcrt revealed six distinct haplotypes, four of which had never been described. The frequency of each haplotype depended on the geographical origin of the samples. The CVIETIF//ISS haplotype was detected in 92% of western Cambodian isolates and in 11% of isolates collected from the eastern province, where CVMNKIF//ISA and CVIDTIF//ISS predominate. The detection of an intermediate haplotype from a susceptible area with 76T/220A, suggests that acquisition of chloroquine resistance might be a stepwise process, during which accumulation of point mutations modulates the response to chloroquine. The association of the K76T mutation with chloroquine resistance was not clear. The mutation was detected in resistant and susceptible samples, suggesting that additional factors are involved in chloroquine resistance. By contrast, the pfcrt D/N75E mutation was strongly associated with the in vitro chloroquine resistance in Cambodian isolates. The N86 allelic form of pfmdr1 was detected in all isolates, consistent with a poor association with resistance to chloroquine. This indicates that in vitro resistance to chloroquine was associated with accumulation of point mutations in pfcrt.
Cambodia has long been known to be an area of endemicity for malaria. According to the National Malaria Center, a total of 115,614 cases were treated by public health services during 2001, and 476 deaths were recorded. The reported morbidity and mortality are underestimated, as more than 80% of malaria cases are treated in the private sector, for which few to no data are available (23). The number of malaria cases has declined since 1995. This is due to a combination of several factors: improved malaria prevention and control activities, large-scale bed net distribution, increased political stability, reduced venturing into forests to collect timber, and reduction of population displacements and resettlement in forested areas (Ministry of Health, National Malaria Control Program, Background Papers 4th RBM Global Partners Meet. 2001, p. 19-22, 2001). Nevertheless, due to the scarcity of health care facilities in inland regions, morbidity and mortality in Cambodia remain unacceptably high and efforts must continue to bring the disease under control, especially in the mountainous areas that separate Cambodia from Vietnam and Thailand, where transmission is high.
Cambodia is one of the focal areas of emergence of multiresistant falciparum malaria in Southeast Asia. Chloroquine resistance was first detected in the early sixties (11). As in other areas of the Mekong subregion, the prevalence of multidrug-resistant malaria is now reaching alarming levels, increasing the need for regular monitoring of drug resistance (15, 17, 22, 24a, 29). Several surveys, carried out in Cambodia and Thailand during the last 20 years, have revealed marked resistance to pyrimethamine and chloroquine, as well as decreased sensitivity to mefloquine and quinine along the Thai border. However, in the Ratanakiri province located in the northeastern part of Cambodia, P. falciparum parasites are still sensitive to most drugs, including chloroquine, which has a reported cure rate of nearly 70% (7, 24a, 29). The first line of treatment in Cambodia currently consists of an association of artesunate plus mefloquine. This combination is used all over the country, even in the few northeastern areas where P. falciparum malaria is susceptible to conventional drugs (19, 24a, 29). The heterogeneous situation of Plasmodium falciparum drug resistance in Cambodia makes this region particularly attractive for studying the epidemiological and molecular aspects of drug resistance.
The pfcrt gene, which encodes a transmembrane protein located in the P. falciparum digestive vacuole, was recently characterized (13, 25). A perfect correlation was found between in vitro response to chloroquine and sequence polymorphism at the pfcrt locus (13, 18). In particular, in vitro chloroquine resistance was associated with the substitution of lysine for threonine at position 76 in field isolates and laboratory strains. In addition, haplotypes with specific combinations of mutations showed a specific geographical distribution, with distinct haplotypes in African and Asian isolates compared to South American or Papua New Guinean isolates, suggesting that pfcrt polymorphism is a useful tool for the public health surveillance of chloroquine resistance (4, 9, 10, 13, 21, 26, 27).
We have investigated here the pfcrt polymorphism in isolates from different areas of Cambodia with high and low rates of chloroquine resistance. Our aim was to determine whether a similar relationship exists between alleles of the pfcrt gene and the level of in vitro chloroquine resistance in Cambodia. Our aim was also to examine polymorphism of this gene after a long time period without chloroquine pressure, since treatment with this drug was abandoned 20 years ago in the country.
MATERIALS AND METHODS
Study areas.
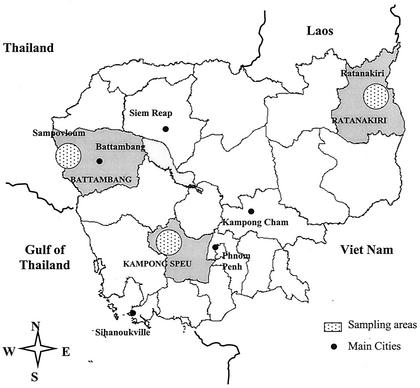
We studied clinical isolates of P. falciparum from three areas of endemic malaria differing by their epidemiological situation and their local rate of resistance to chloroquine (Fig. 1).
FIG. 1.
Map of Cambodia showing sampling areas in the Ratanakiri, Kampong Speu, and Battambang provinces.
The Ratanakiri province is a mountainous area, located in the northeastern part of Cambodia near the Vietnamese border. Several ethnic minorities with limited access to health facilities live in this area. Malaria transmission is intense, but the parasites in this region are still sensitive to most drugs, including chloroquine. An in vivo trial conducted in 1999 showed that only 29% of cases were RI/RII. There was only 4% RIII resistance to chloroquine (7, 24a, 29).
The other two study sites are located in regions with high levels of in vivo chloroquine resistance. The Kampong Speu province is located close to the Cardamoms chain in southwest Cambodia. Regular monitoring of the drug response of falciparum malaria in this province and neighboring areas indicated a high prevalence of chloroquine and sulfadoxine-pyrimethamine resistance (M. B. Denis, personal communication). Although the current level of drug resistance in this area is unknown since chloroquine is no longer used, earlier in vivo surveys have indicated that the rate of resistance to chloroquine was 60 to 80%. Sampovloum, in the Battambang province, is situated in the northwest along the Thai-Khmer border, in a mountainous area characterized by high malaria transmission mostly during the rainy season. The Pailin site, close to Sampovloum, in the Battambang province, is the first area of Cambodia in which resistance to chloroquine was reported in 1962. Since this time, parasites from this region have become resistant to nearly all the available antimalarial drugs. This area is characterized by continuous population movements due to gem mining and agricultural activities. Longitudinal surveys indicated that the rate of chloroquine-resistant P. falciparum increased continuously between 1988 and 1992, reaching nearly 100%. The results of recent in vivo monitoring confirmed this trend, showing that virtually no parasites from this region are susceptible to chloroquine or to sulfadoxine-pyrimethamine. (6, 7, 15, 24a, 29).
Sampling.
Samples were collected during active case detection surveys carried out between February and December 2001 by the National Center for Malaria Control. The survey protocols were approved by the Cambodian Ministry for Health. Briefly, 5-ml blood samples were collected in EDTA Vacutainers from patients diagnosed as having uncomplicated P. falciparum malaria on the basis of their clinical symptoms and by a rapid test (Paracheck F; Orchid Biomedical Systems). Immediately after blood collection, the patients were treated according to national policies. Blood samples were kept at 4°C and sent to Pasteur Institute (Phnom Penh) for in vitro drug sensitivity testing. An aliquot of each isolate was frozen at −80°C for molecular analysis.
In vitro chloroquine sensitivity testing.
In vitro chloroquine susceptibility of isolates in fresh samples was determined within 72 h of collection by use of a classical microtest method (8). As soon as the blood samples arrived at the laboratory, thin films were prepared and stained. Parasite density and species determinations were performed by microscopic examination. The isolates with P. falciparum as the single Plasmodium species and parasitemia higher than 0.1% were selected for in vitro testing. Parasites were incubated for 48 h at 37°C in complete RPMI medium containing [3H]hypoxanthine to assess parasite maturation. Each isolate was tested once in duplicate, in microplates precoated with serial dilutions of drugs. Drug response was quantified by monitoring [3H]hypoxanthine uptake in a MicroBeta Trilux counter (Perkin-Elmer). Only assays with significant differences in parasite counts between drug-free control wells containing uninfected or infected red blood cells, at least a 10-fold increase, were considered interpretable. The drug concentration at which 50% of the [3H]hypoxanthine was incorporated into the control wells in the absence of the drug (50% inhibitory concentrations [IC50]) was calculated by use of a log probit approximation. Samples exhibiting an IC50 of >100 nM were considered resistant to chloroquine.
DNA extraction.
Parasite DNA was extracted from frozen blood aliquots by the phenol-chloroform method (24). Briefly, samples were thawed at room temperature, and 2 volumes of cold 0.5× TEN buffer (50 mM Tris, 10 mM EDTA, 150 mM NaCl [pH 7.5]) was added. The hemolysates, containing red blood cell membranes and free parasites, were washed three times with cold TE buffer (10 mM Tris, 1 mM EDTA [pH 7.5]) and then resuspended in an extraction buffer consisting of TE plus 0.5% Triton X-100, 0.5% sodium dodecyl sulfate, and proteinase K (5 mg/ml). After 1 h at 37°C, samples were treated with phenol and chloroform. DNA was recovered by ethanol precipitation and centrifugation. The pellet was resuspended in 100 μl of TE buffer and stored at −20°C.
PCR analysis.
The genetic diversity of Cambodian isolates was determined by using three highly polymorphic markers (msp-1, msp-2, and glurp) according to a classical PCR-based strategy (5). The pfcrt gene was amplified by use of two sets of primers designed to amplify large regions of exons 1 to 2 (crt1, 5′AAATGACGAGCGTTATAGAG3′, and crt7, 5′ATAAAGTTGTGAGTTTCGGATG3′) and in exon 4 (crt4, 5′TTATACAATTATCTCGGAGCAG3′, and crt5, 5′CATGTTTGAAAAGCATACAGGC3′). The primers were selected on the basis of the sequence of the pfcrt gene of HB3 clone (accession number AF233068). PCR were performed in a PTC-100 thermal cycler (MJ Research, Merck Eurolab) under the following conditions: 50-μl final volume containing 2 μl of DNA (approximately 20 to 300 ng), a 200 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, 2 mM MgCl2, and 2 U of TaqI DNA polymerase (Promega) in the buffer supplied by the manufacturer. The amplification conditions were as follows for primers 4 and 5: 1 min at 95°C, 1 min 30 at 59°C, and 1 min at 72°C for 1 cycle; then, 30 s at 95°C, 1 min 30 at 59°C, and 2 min at 72°C for 4 cycles; followed by 10 s at 95°C, 1 min 30 at 59°C, and 2 min at 72°C for 35 cycles, and a final extension at 72°C for 10 min. Similar conditions were used for primers 1 to 7, except that the hybridization step was done at 60°C. The resulting 426- and 309-bp PCR products contained codons 76 and 220, respectively, as well as the introns located just downstream of exons 1 and 4.
The mdr1 gene was amplified as described previously (3) using primers 5′AGAGAAAAAAGATGGTAACCTCAG3′ and 5′ACCACAAACATAAATTAACGG3′. The 609-bp PCR fragment was then digested with AflIII (Roche) according to the manufacturer's recommendations. This enzyme should only digest fragments containing a Y at codon 86, which is associated with resistance to chloroquine.
All fragments were subjected to gel electrophoresis on agarose gels containing ethidium bromide. Prior to sequence analysis, the PCR products were purified by use of the Geneclean II kit (Biogene) either directly or after excision of the band from the gel. Then, they were sequenced using the fluorescent dye terminator method (Genomex).
Statistical analysis.
Chi-square or Fisher's exact two-tailed tests were used to evaluate the association between pfcrt polymorphisms at codons 76 and 75 and in vitro response to chloroquine. The Spearman rank test was used for correlation analysis. All statistical analyses were performed by using the STATA software (Stata Corporation).
RESULTS
In vitro chloroquine response.
The in vitro sensitivities to chloroquine of the isolates collected in the study areas were determined. Interpretable data were obtained for 88 of the 111 isolates tested. Initial parasitemia varied between 0.1 and 2.2%, with a mean of 0.39%. IC50 values ranged from 4.8 to 1,146 nM. As predicted, all 7 isolates from the Ratanakiri area producing interpretable data were chloroquine sensitive (mean IC50, 31.5 + 22.2 nM), whereas 15 of the 17 isolates (88%) from Kampong Speu (mean IC50, 269.5 ± 262.5 nM) and 56 of the 64 (87.5%) isolates from Sampovloum (mean IC50, 155.8 ± 125.6 nM) displayed reduced susceptibilities to chloroquine, as well as to pyrimethamine and mefloquine (data not shown) (6, 7, 24a).
pfcrt amplification and sequence analysis.
Thirty-five samples were selected on the basis of their in vitro sensitivities, including as many in vitro-chloroquine-sensitive parasites as possible from the two western regions (Table 1). The overall population polymorphism and the multiplicity of infection were first assessed using polymorphic, unlinked single-copy gene loci. Investigation of msp-1, msp-2, and glurp diversity (data not shown) highlighted a large P. falciparum population diversity with seven, five, and three distinct alleles, respectively. However, the three-locus typing showed that 77.1% (27 of 35) of the isolates were from subjects with single-genotype infections with only one allele detected for each locus (Table 1). The remaining samples contained two alleles at least at one of the three loci, indicating that these isolates contained mixed genotypes. For those patients (n = 35) there was an average number of 1.23 clones (standard deviation, 0.43) per isolate.
TABLE 1.
pfcrt haplotypes in relation to in vitro response of Cambodian P. falciparum isolates to chloroquinea
| Code isolate | Area of sampling
|
In vitro response
|
PfCRT amino acid (codon) encoded at position:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Position within regionb | IC50 (nM) | Out- comec | 72 (TGT) | 73 (GTA) | 74 (ATT) | 75 (GAA) | 76 (ACA) | 77 (ATT) | 78 (TTT) | 218 (ATT) | 219 (AGT) | 220 (TCC) | |
| 159d | Sampovloum | NW | 665 | R | C | V | I | E | T | I | F | I | S | S |
| 170d | Sampovloum | NW | 592 | R | C | V | I | E | T | I | F | I | S | S |
| 209d | Sampovloum | NW | 410 | R | C | V | I | E | T | I | F | I | S | S |
| 194d | Sampovloum | NW | 368 | R | C | V | I | E | T | I | F | I | S | S |
| 186d | Sampovloum | NW | 349 | R | C | V | I | E | T | I | F | I | S | S |
| 197d | Sampovloum | NW | 338 | R | C | V | I | E | T | I | F | I | S | S |
| 176d | Sampovloum | NW | 228 | R | C | V | I | E | T | T (ACT) | F | I | S | S |
| 164d | Sampovloum | NW | 206 | R | C | V | I | E | T | I | F | I | S | S |
| 183d | Sampovloum | NW | 73.8 | S | C | V | I | E | T | I | F | I | S | S |
| 182d | Sampovloum | NW | 71.3 | S | C | V | I | E | T | I | F | I | S | S |
| 201d | Sampovloum | NW | 65.0 | S | C | V | I | E | T | I | F | I | S | S |
| 172d | Sampovloum | NW | 42.3 | S | C | V | I | E | T | I | F | I | S | S |
| 173d | Sampovloum | NW | 27.8 | S | C | V | I | E | T | I | F | I | S | S |
| 174d | Sampovloum | NW | 27.0 | S | C | V | I | E | T | I | F | I | S | S |
| 200d | Sampovloum | NW | 18.0 | S | C | V | I | E | T | I | F | I | S | S |
| 171d | Sampovloum | NW | 18.0 | S | C | V | I | E | T (ACT) | I | F | I | S | S |
| 91 | Kampong Speu | W | 1,146 | R | C | V | I | E | T | I | F | I | S | S |
| 73d | Kampong Speu | W | 504 | R | C | V | I | E | T | I | F | I | S | S |
| 112d | Kampong Speu | W | 442 | R | C | V | I | E | T | I | F | I | S | S |
| 85 | Kampong Speu | W | 315 | R | C | V | I | E | T | I | F | I | S | S |
| 75 | Kampong Speu | W | 241 | R | C | V | I | E | T | I | F | I | S | S |
| 108d | Kampong Speu | W | 238 | R | C | V | I | D (GAT) | T | I | F | I | S | S |
| 110d | Kampong Speu | W | 210 | R | C | V | I | E | T | I | F | I | S | S |
| 128d | Kampong Speu | W | 118 | R | C | V | I | E | T | I | F | I | S | S |
| 103d | Kampong Speu | W | 95.3 | S | C | V | I | E | T | I | F | I | S | S |
| 125d | Kampong Speu | W | 4.8 | S | C | V | I | E | T | I | F | I | S | S |
| 46 | Rattanakiri | NE | 57.0 | S | C | V | M (ATG) | N (AAT) | T | I | F | I | S | A (GCC) |
| 56d | Rattanakiri | NE | 55.8 | S | C | V | I | D (GAT) | T | I | F | I | S | S |
| 32d | Rattanakiri | NE | 51.0 | S | C | V | I | D (GAT) | T | I | F | I | S | S |
| 60 | Rattanakiri | NE | 21.4 | S | C | V | I | D (GAT) | T | I | F | I | S | S |
| 36d | Rattanakiri | NE | 17.0 | S | C | V (GTC) | T (ACC) | N (AAT) | T (ACC) | I | F | I | S | S |
| 63d | Rattanakiri | NE | 9.3 | S | C | V | M (ATG) | N (AAT) | K (AAA) | I | F | I | S | A (GCC) |
| 42 | Rattanakiri | NE | 8.7 | S | C | V | M (ATG) | N (AAT) | K (AAA) | I | F | I | S | A (GCC) |
| 35 | Rattanakiri | NE | NDe | ND | C | V | I | E | T | I | F | I | S | S |
| 53 | Rattanakiri | NE | ND | ND | C | V | M (ATG) | N (AAT) | K (AAA) | I | F | I | S | A (GCC) |
| HB3 clone | Honduras | C (TGT) | V (GTA) | M (ATG) | N (AAT) | K (AAA) | I (ATT) | F (TTT) | I (ATT) | S (AGT) | A (GCC) | |||
The sequences at positions 72 to 78 and positions 218 to 220 of pfcrt isolates from the Sampovloum, Kampong Speu, and Ratanakiri areas of Cambodia were determined by sequencing fragments of amplified genomic DNA. The nucleotide sequence of the canonical resistant haplotype (CVIETIF//ISS) is indicated at the top of the table. The sequence of the HB3 sensitive haplotype is indicated at the bottom of the table.
Abbreviations: NW, northwestern; W, western; NE, northeastern.
In vitro chloroquine resistance (R) and sensitivity (S) are defined as an IC50 of >100 nM and <100 nM, respectively.
Single genotype.
ND, not determined.
pfcrt polymorphism was investigated by amplifying and sequencing two gene regions. The first one, yielding a 426-bp PCR product, was centered around codon 76 (from nucleotide 44 located in exon 1 to nucleotide 468 located in exon 2). The second one, yielding a 309-bp fragment, was centered around codon 220 (from nucleotide 1042 in exon 4 to nucleotide 1349 in exon 5) (13). All isolates tested yielded a single PCR fragment and produced legible sequences from intron 1 to codon 80 (in exon 2) and from codon 202 (in exon 4) to intron 5, respectively. Chromatograms were rechecked individually to verify signal homogeneity and absence of ambiguity at the polymorphic positions. This indicated that one single (or major) allele was amplified and sequenced from each isolate. Sequence analysis identified polymorphic and conserved bases. Codons 31 to 72 and 78 to 80 in exon 2 and 202 to 219, 221, and 222 in exon 4 were conserved in all isolates studied, whereas single-nucleotide polymorphism was detected at other positions.
Table 1 lists the observed polymorphisms at positions 74 to 77 and 220, and their flanking conserved residues. Compared to the sequence of the chloroquine-sensitive clone HB3, nucleotide changes were observed for codon 73 (GTA codon changed into GTC, silent mutation), codon 74 (ATG codon changed into ATT or ACC, resulting in substitution of M for I and T, respectively), codon 75 (AAT codon changed into GAT or GAA, resulting in substitution of N for D and E, respectively), codon 76 (AAA codon changed into ACA, ACT, or ACC, resulting in substitution of K for T), codon 77 (ATT codon changed into ACT, resulting in substitution of I for T), and codon 220 (GCC changed into TCC, resulting in substitution of A for S).
Six distinct allelic forms were observed for the 426-bp fragment containing codons 72 to 78 (among them, CVIETIF and CVMNKIF were previously described), whereas only two different forms, ISS and ISA (both described in the literature), were detected for the 309-bp fragment containing codons 218 to 220. For the 27 isolates with a three-locus typing pointing to a single-genotype infection, pfcrt haplotypes pairing results of the two PCRs can be established with a fair confidence. Twenty-one contained the CVIETIF//ISS haplotype (codes 159, 170, 209, 194, 186, 197, 164, 183, 182, 201, 172, 173, 174, 200, 171, 73, 112, 110, 128, 103, and 125), and three presented the CVIDTIF//ISS genotype (codes 108, 56, and 32), whereas CVMNKIF//ISA (code 63), CVTNTIF//ISS (code 36), and CVIETTF//ISS (code176) sequences were detected once each. Out of the eight isolates containing two genotypes, four displayed the CVIETIF//ISS haplotype (codes 91, 85, 75, and 35). The sequence CVMNKIF//ISA was found in two samples (codes 42 and 53), and haplotypes CVMNTIF//ISA (code 46) and CVIDTIF//ISS (code 60) were observed once each.
pfcrt polymorphism and geographic origin.
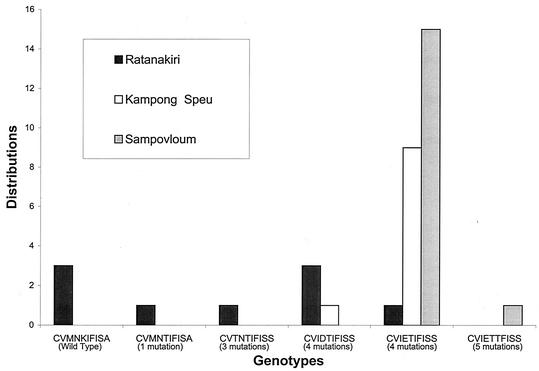
The pfcrt haplotypes were grouped by number of point mutations detected compared to the HB3 pfcrt sequence. The distribution of the various genotypes in relation to the sampling site is shown in Fig. 2. The CVMNKIF//ISA allele is similar to the HB3 haplotype. The CVMNTIF//ISA and CVTNTIF//ISS alleles are related to HB3 by one and three point mutations, respectively. All strains harboring these polymorphisms originated from the Ratanakiri area (northeast part of the country). In contrast, parasites presenting more divergent pfcrt genotypes—CVIDTIF//ISS, CVIETIF//ISS, and CVIETTF//ISS, which differed from HB3 by four or five changes—were detected in the majority of samples collected in the Kampong Speu and Sampovloum areas (northwestern and western parts of the country, respectively). The CVIETIF//ISS haplotype was detected in 92% of isolates from western Cambodia but in only 11% of isolates collected from the Ratanakiri province. Therefore, the genotypes showed a strong association with the geographical origin of the samples. The number of point mutations increased from east to west and was correlated with the transition from chloroquine sensitivity to chloroquine resistance phenotypes (Fig. 1 and 2).
FIG. 2.
Geographic distribution of pfcrt alleles. Shown is the distribution of the various pfcrt alleles identified in Cambodian isolates according to their geographical origin. The number of point mutations detected in each haplotype compared to pfcrt sequence of the HB3 chloroquine-sensitive clone is indicated.
pfcrt polymorphism and response to chloroquine.
We examined the predictive value of pfcrt gene analysis as a molecular tool for monitoring chloroquine susceptibility. The relationship between the pfcrt polymorphism and in vitro chloroquine susceptibility was first evaluated by the Spearman rank test. A linear model was fitted to determine the extent to which pfcrt polymorphisms at positions 74 to 77 and 220 (i.e., number of mutations) affect in vitro susceptibility to chloroquine. This showed that the number of point mutations (mean, 3.2; range, 0 to 5) was positively correlated with the mean chloroquine IC50 (Spearman's rank test, P = 0.013). This indicates that in vitro resistance was associated with accumulation of point mutations in pfcrt.
Previous studies have pointed out that polymorphic positions in pfcrt are not all of equal importance in predicting chloroquine resistance. As the pfcrt-76T allele is to date the best predictor of chloroquine resistance (4, 9, 13, 18, 21, 26), the association between the pfcrt-76T allele and in vitro chloroquine response was further tested. Scores were compared to those obtained for the pfcrt-75E allele. Both mutations were highly sensitive (100 and 93.7%, respectively) in detecting in vitro-resistant strains, although specificity was higher when the 75E mutation was used as a marker for in vitro resistance. The specificity of the 75E mutation was 41.2%, compared to 11.7% for the K76T mutation (Table 2). More importantly, the 75E mutation positive predictive value for in vitro chloroquine resistance was 60.0%, compared to 51.6% for the K76T mutation. The pfcrt-75E mutation was therefore significantly associated with the in vitro chloroquine response phenotype (Fisher's exact test, P = 0.024), whereas the association between the 76T mutation and in vitro response to chloroquine was not significant (Fisher's exact test, P > 0.05).
TABLE 2.
Association between polymorphisms in the pfcrt marker and in vitro response to chloroquined
| pfcrt mutation | na | % Sensitivity (95% CIb) | % Specificity (95% CI) | Pc |
|---|---|---|---|---|
| N/D75E | 33 | 93.7 (69.8-99.8) | 41.2 (18.4-67.1) | 0.024 |
| K76T | 33 | 100.0 (79.4-100.0) | 11.7 (1.5-36.4) | 0.258 |
Number of paired results.
CI, confidence interval.
probability (P) refers to the significance level of the two-tailed Fisher's exact test (P < 0.05).
Association between the occurrence of pfcrt-76T and pfcrt-75E alleles and in vitro response to chloroquine in Cambodian isolates as determined by Fisher's exact test (P). Sensitivity and specificity of 76T and 75E allele mapping in detecting in vitro chloroquine resistance phenotype (IC50 >100 nm) were calculated from data presented in table 1.
Genetic polymorphism at the pfmdr1 locus.
Some studies have reported an association between decreased chloroquine sensitivity and an N-to-Y mutation at codon 86 in the mdr1 gene of P. falciparum (14; I. S. Adagu, F. Dias, L. Pinheiro, L. Rombo, V. do Rosario, and D. C. Warhust, Letter, Trans. R. Soc. Trop. Med. Hyg. 90:90-91, 1996). To determine whether this correlation exists in Cambodian isolates, a 609-bp fragment was amplified and digested with AflIII. The mutant 86Y codon was visualized by the presence of a 232-bp fragment and a 377-bp fragment on an agarose gel, whereas the wild-type sequence was not digested. All 35 samples tested had the N86 sensitive allele, and one sample (code 60) from Ratanakiri carried mixed alleles as shown by the presence of both digested and undigested fragments. A control DNA sample (FUP/CB strain) (12) was completely digested under similar conditions, indicating that the restriction patterns observed after electrophoresis did not result from the incomplete digestion of the templates due to poor efficiency of the reaction. These data are consistent with previous studies, indicating that the N86 codon is highly prevalent in Cambodian isolates while displaying reduced sensitivity to antifols and chloroquine (2).
DISCUSSION
The high level of drug-resistant malaria in Southeast Asia justifies the need to develop new combined therapeutic strategies for the treatment of multiresistant malaria. At the same time, it is equally important to look for trends for a declining chloroquine resistance rate in Cambodia, which eventually might lead to consideration of the reintroduction of treatment with this molecule. A better knowledge of how resistance patterns change in the absence of chloroquine pressure would provide useful information for this region, as well as for other countries where chloroquine is still used as the first-line treatment.
Resistance to chloroquine was first reported in Cambodia on the Thai border. It subsequently spread elsewhere so that, by 1980, most Cambodian provinces were affected. Based on this finding, the use of chloroquine for the treatment of falciparum malaria was officially abandoned in Cambodia about 20 years ago. Some in the private sector may have continued distributing chloroquine, but certainly use of this drug has dramatically diminished over the years as a result of its poor efficacy and as artemisinin-based alternatives flooded the market. Several clinical trials since performed by the National Malaria Center (29) have confirmed the high degree of chloroquine resistance in Cambodia, apart from that in the northeastern region. Our in vitro data are consistent with these observations, since all isolates collected in the northeastern region were sensitive to chloroquine, whereas 71 out of 81 (88%) isolates collected in western Cambodia, in the Kampong Speu and Sampovloum areas, were chloroquine resistant. This suggests that the rate of chloroquine resistance remains high in these areas. Thus, it appears that reversion to chloroquine susceptibility is slow, even in areas where drug pressure has been withdrawn for many years. The actual level of drug pressure is difficult to assess. Indeed, as chloroquine is still being used to treat P. vivax infections in Southeast Asia, it may contribute to maintaining P. falciparum chloroquine resistance. However, the small number of reported P. vivax treatments in Cambodia suggests that their impact on P. falciparum populations is rather limited. All together, these data suggest that the long-term maintenance of P. falciparum chloroquine resistance in Cambodia likely results from the lack of loss of fitness associated with the chloroquine resistance phenotype rather than from continued drug pressure.
pfcrt polymorphism was studied by amplification followed by sequencing to examine sequence diversity at positions 72, 74, 75, 76, and 220, reported as mutated in chloroquine-resistant isolates (13). All attempts at amplifying single PCR fragments carrying both codons 76 and 220 were unsuccessful, as they resulted in producing complex banding patterns on agarose gels. This probably reflects impaired progression of the enzyme across the long polyadenylated stretch located within the third intron. As a consequence, the gene regions containing codons 72 to 76 and 218 to 220 were amplified separately. Four distinct allelic forms were detected for codon 76, three were detected for codons 74 and 75, and two were detected for codons 73, 77, and 220. The other codons of this region sequenced were strictly conserved. Identification of haplotypes by pairing sequence data from distinct PCR fragments could be done with complete confidence only for those isolates typed as containing single-genotype parasites. Multiple infections were detected in only 8 out of 35 isolates examined. The fact that all sequences produced were homogeneous, with no superimposition of signals at polymorphic positions, indicates that only major parasite populations were amplified and sequenced. This strengthens our conclusions, but of course, the possibility of pairing together two distinct alleles present in one isolate cannot be completely ruled out. This strategy identified six distinct pfcrt genotypes at codons 72 to 78 and 218 to 220. Four of these—CVTNTIF//ISS, CVIDTIF//ISS, CVMNTIF//ISA, and CVIETTF//ISS—had never been observed before, indicating that the extent of pfcrt polymorphism is larger than reported so far.
K76T has been reported as the key pfcrt mutation implicated in the chloroquine resistance phenotype (13, 27). In agreement with previous observations, we found a correlation between the in vitro response to chloroquine and the mutated sequence at codon 76, which was present in all resistant isolates examined. However, this correlation was far from perfect, as the K76T mutation was also detected in 15 of 17 samples classified as susceptible to chloroquine in vitro. Detection of the K76T mutation in 13 of 15 susceptible samples cannot be attributed to the presence of multiple genotypes within the same isolate, since the three-locus typing pointed to single-genotype infections. Therefore, it unlikely reflects misinterpretation of the in vitro data due to the presence of mixed parasite populations with opposed phenotypes. This also is unlikely due to threshold IC50 value used to interpret in vitro tests. The IC50 for chloroquine is still a matter a debate (10). We have here used a threshold of 100 nM to classify chloroquine susceptibility, as proposed recently (27), which may lead to an overestimation of the prevalence of in vitro chloroquine susceptible isolates. But even for a cutoff of approximately 60 nM, reported to best fit with the presence of the pfcrt K76T mutation (10), the prevalence of K76T mutation among the sensitive isolates would remain high, with then 11 of 17 sensitive isolates that harbor the K76T mutation. Interestingly, for the panel of isolates studied here, the 75E mutation had a better predictive value for in vitro chloroquine resistance. By contrast, consistent with previous observations (2, 3, 16, 28) we failed to detect a positive correlation between the pfmdr1 codon 86 polymorphism and in vitro chloroquine phenotype.
The observed correlation (and the discordances) between the in vitro IC50 and the pfcrt sequence has to be qualified, due to our deliberate bias in sampling for sequencing. Our in vitro drug susceptibility assays on a large panel of samples showed that multiresistant falciparum malaria was highly prevalent in western Cambodia and that, conversely, only a small proportion of isolates from the Ratanakiri province presented a decreased chloroquine susceptibility in vitro. Given the high prevalence of chloroquine-resistant isolates in western Cambodia, we decided not to select the samples to be genotyped based on their in vitro IC50, so that as many in vitro-sensitive isolates as possible were included. The discordance between in vitro IC50 and the pfcrt sequence for isolates 125, 171,172, 173, 174, and 200 suggests that additional factors or mutations in other regions of the gene influence chloroquine sensitivity of these parasites.
A positive association was found between pfcrt genotypes and the sampling area. All isolates from Sampovloum and most samples from Kampong Speu had the CVIETIF//ISS haplotype, whereas five distinct pfcrt haplotypes were observed in Ratanakiri, a region of chloroquine susceptibility. These differences are striking, as these areas are very close on a geographical scale. Isolates presenting a monomorphic CVIETIF//ISS allele were further characterized for msp-1, msp-2, and glurp polymorphism (data not shown). A considerable degree of heterogeneity between isolates was observed, consistent with each isolate consisting of a unique combination of alleles. Thus, despite the high selection pressure resulting from drug use, parasite populations in Cambodia consist of a large number of clonal types. This suggests that chloroquine drug pressure, rather than other epidemiological factors, accounts for the pfcrt genetic clustering of the parasites circulating on the western border. The genetic structure of these populations is consistent with a critical role for pfcrt polymorphism in chloroquine resistance (1). Recent data indicate that resistance to chloroquine has been acquired independently in different parts of the world following natural pfcrt sequence evolution, and spread out by recombination from a limited number of original foci (30). Consistent with this, we found that the microsatellite markers located just downstream of exons 1 (AT repeats) and 4 (TA repeats) are less polymorphic in the 26 isolates originating from areas of multiresistance (only 16 or 17 AT repeats and 15 or 16 TA repeats) than the 9 susceptible isolates (11, 16, 17, or 18 AT repeats and 12, 14, 15, or 16 TA repeats) collected in area of chloroquine susceptibility (data not shown). This suggests that a single CVIETIF//ISS resistant haplotype has spread over western Cambodia and is still present to date.
The isolates from the Ratanakiri area displayed a larger sequence polymorphism than the parasites collected in western areas. All but two parasites from western areas, characterized by a high level of resistance to chloroquine, carried the CVIETIF//ISS haplotype. These two isolates displayed one mutation each, at position 75 and 77, respectively. In contrast the CVIETIF//ISS haplotype was observed in only one isolate from the Ratanakiri area. The detection of an intermediate CVMNTIF//ISA haplotype, in which 76T is associated with 220A and which presents an increased IC50 compared to the CVMNKIF//ISA wild haplotype, suggests that acquisition of chloroquine resistance is a stepwise process, during which the accumulation of point mutations might modulate the response to chloroquine. This is consistent with the observed broad spectrum of chloroquine resistance, ranging from slightly decreased susceptibility to high resistance, and with the observed trend for an increased in vitro IC50 with an increasing number of mutations.
Taken together, these findings are consistent with the current view of a critical implication of pfcrt polymorphism in resistance to chloroquine. Mutations at position 76, and possibly at position 75, are necessary but not sufficient to predict chloroquine resistance. Whether additional mutations or the level of pfcrt expression are involved in the regulation of chloroquine susceptibility remains to be determined. This possibility should be further examined in field isolates, as it may also explain why molecular events underlying the resistance of P. vivax and P. falciparum to chloroquine appear to be so different (20).
Acknowledgments
This work was funded by the World Health Organization (Regional Office for the Western Pacific) and the French Academy of Sciences.
We thank Nimol Khim for her skillful technical assistance in typing parasites with mdr1, msa1, msa2, and glurp markers. We are indebted to Philippe Glaziou for his assistance with the statistical analysis and to Jean Louis Sarthou for constant interest. We also thank the National Malaria Center staff for participation in collecting parasitized blood samples in the field and especially Yi Poravuth and Soun Seila as well as Khieng Heng and Va Soch.
REFERENCES
- 1.Anderson, T. J. C., B. Haubold, J. T. Williams, J. G. Estrada-Franco, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, N. French, J. Whitworth, I. D. Velez, A. H. Brockman, F. Nosten, M. U. Ferreira, and K. Day. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. [DOI] [PubMed] [Google Scholar]
- 2.Basko, L. K., P. E. de Pécoulas, J. Lebras, and M. W. Wilson. 1996. Plasmodium falciparum: characterization of multidrug-resistant Cambodian isolates. Exp. Parasitol 82:97-103. [DOI] [PubMed] [Google Scholar]
- 3.Basco, L. K., and P. Ringwald. 1997. Pfmdr1 gene mutation and clinical response to chloroquine in Yaoundé, Cameroon. Trans. R. Soc. Trop. Med. Hyg. 91:210-211. [DOI] [PubMed] [Google Scholar]
- 4.Basco, L. K., and P. Ringwald. 2001. Analysis of the key pfcrt point mutation and in vitro and in vivo response to chloroquine in Yaoundé, Cameroon. J. Infect. Dis. 183:1828-1831. [DOI] [PubMed] [Google Scholar]
- 5.Contamin, H., T. Fandeur, S. Bonnefoy, F. Skoury, F. Ntoumi, and O. Mercereau-Puijalon. 1995. PCR typing of field isolates of Plasmodium falciparum. J. Clin. Microbiol. 33:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis, M. B. 1986. Etude préliminaire in vitro de la sensibilité de P. falciparum à la chloroquine, la méfloquine et à la quinine en 1985 au Kampuchéa. Bull. Soc. Path. Ex. 79:360-367. [PubMed] [Google Scholar]
- 7.Denis, M. B. 1999. Evolution de la Chimiorésistance de Plasmodium falciparum aux traitements antipaludiques au Cambodge (1991-1997). Doctoral thesis 449 in medicine. Université des Sciences de la Santé, Phnom Penh, Cambodia.
- 8.Desjardins, R. E., C. J. Canfield, J. D. Haynes, and J. D. Chulay. 1979. Quantitative assessment of antimalarial activity in vitro by an automated microdilution technique. Antimicrob. Agents Chemother. 16:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djimdé, A., O. K. Doumbo, J. F. Cortese, K. Kayento, S. Doumbo, Y. Diourté, A. Dicko, X.-Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, and C. V. Plowe. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 3344:257-263. [DOI] [PubMed] [Google Scholar]
- 10.Durand, R., S. Jafari, J. Vauzelle, J. F. Delabre, Z. Jesic, and J. Le Bras. 2001. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 114:95-102. [DOI] [PubMed] [Google Scholar]
- 11.Eyles, D. E., C. C. Hoo, M. C. Warren, and A. A. Sandosham. 1963. Plasmodium falciparum resistant to chloroquine in Cambodia. Am. J. Trop. Med. Hyg. 12:840-843. [DOI] [PubMed] [Google Scholar]
- 12.Fandeur, T., S. Bonnefoy, and O. Mercereau-Puijalon. 1991. In vivo and in vitro derived Palo alto lines of Plasmodium falciparum are genetically unrelated. Mol. Biochem. Parasitol. 47:167-178. [DOI] [PubMed] [Google Scholar]
- 13.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naudé, K. W. Deitsch, X.-Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein pfcrt and evidence of their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foote, S. J., D. E. Kyle, R. K. Martin, A. M. J. Oduola, K. Forsyth, D. J. Kemp, and A. F. Cowman. 1990. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255-258. [DOI] [PubMed] [Google Scholar]
- 15.Giboda, M., and M. B. Denis. 1988. Response of Kampuchean strains of Plasmodium falciparum to antimalarials: in vivo assessment of quinine and quinine plus tetracycline; multiple drug resistance in vitro. J. Trop. Med. Hyg. 91:205-211. [PubMed] [Google Scholar]
- 16.McCutchean, K. R. G., J. A. Freese, J. A. Frean, B. L. Sharp, and M. B. Markus. 1999. Two mutations in the multidrug-resistance gene homologue of Plasmodium falciparum, pfmdr, are not useful predictors of in vivo or in vitro chloroquine resistance in Southern Africa. Trans. R. Soc. Trop. Med. Hyg. 93:300-302. [DOI] [PubMed] [Google Scholar]
- 17.Meek, S. R., E. B. Doberstyn, B. A. Gauzère, C. Thanapanich, E. Nordlander, and S. Phuphaisan. 1986. Treatment of falciparum malaria with quinine and tetracycline or combined mefloquine/sulfadoxine/pyrimethamine on the Thai-Kampuchean Border. Am. J. Trop. Med. Hyg. 35:246-250. [DOI] [PubMed] [Google Scholar]
- 18.Mehlotra, R. K., H. Fujioka, P. D. Roepe, O. Janneh, L. M. Ursos, V. Jacobs-Lorena, D. T. McNamara, M. J. Bockarie, J. W. Kazura, D. E. Kyle, D. A. Fidock, and P. A. Zimmerman. 2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc. Natl. Acad. Sci. USA 98:12689-12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ministry of Health, National Malaria Control Program. 2000. Treatment guideline for malaria in the Kingdom of Cambodia. National Malaria Center, Phnom Penh, Cambodia.
- 20.Nomura, T., J. M. R. Carlton, J. K. Baird, H. A. Del Portillo, D. J. Fryauff, D. Rathore, D. A. Fidock, X.-Z. Su, W. Collins, T. F. McCutchan, J. C. Wootton, and T. E. Wellems. 2000. Evidence for different mechanisms of chloroquine resistance in two Plasmodium species that cause malaria. J. Infect. Dis. 183:1653-1661. [DOI] [PubMed] [Google Scholar]
- 21.Pillai, D. R., A.-C. Labbé, V. Yanaviseth, B. Hongvangthong, S. Phomphida, S. Inkathone, K. Zhong, and K. C. Kain. 2001. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J. Infect. Dis. 183:789-795. [DOI] [PubMed] [Google Scholar]
- 22.Plasai, V., and A. Spelman. 1996. Mefloquine insusceptibility of malaria in Thailand not promoted by nonregulated drug-use. Acta Trop. 60:281-289. [DOI] [PubMed] [Google Scholar]
- 23.Rozendaal, J. 2001. Fake antimalaria drug in Cambodia. Lancet 357:890.. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24a.Singhasivanon, P. 1999. Mekong malaria. Malaria, multi-drug resistance and economic development in the greater Mekong subregion of Southeast Asia. Southeast Asian J. Trop. Med. Public Health 30(Suppl. 4):1-101. [PubMed] [Google Scholar]
- 25.Su, X.-Z., L. S. Kirkman, and T. E. Wellems. 1997. Complex polymorphisms in a ∼330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 91:593-603. [DOI] [PubMed] [Google Scholar]
- 26.Viera, P. P., M. Das Craças Alecrim, L. H. P. Da Silva, I. Gonzalez-Jilmenez, and M. G. Zalis. 2001. Analysis of the Pfcrt K76T mutation in Plasmodium falciparum isolates from the Amazon region of Brazil. J. Infect. Dis. 183:1832-1833. [DOI] [PubMed] [Google Scholar]
- 27.Wellems, T., and C. Plowe. 2001. Chloroquine-resistant malaria. J. Infect. Dis. 184:770-776. [DOI] [PubMed] [Google Scholar]
- 28.Wellems, T. E., L. J. Panton, I. Y. Gluzman, and D. E. do Rosario. 1990. Chloroquine resistance is not linked to mdr-like genes in Plasmodium falciparum cross. Nature 345:253-255. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organization. 2000. Report on the informal consultation on monitoring resistance to antimalarial drugs in the Mekong region. World Health Organization, Geneva, Switzerland.
- 30.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X.-Z. Su. 2002. Widespread use of antimalarial agents can profoundly influence the evolution of the human malaria parasite Plasmodium falciparum. Nature 418:320-323. [DOI] [PubMed] [Google Scholar]