Abstract
Gemifloxacin, a novel quinolone with potent activity against Staphylococcus aureus, was 8- to 16-fold more active against wild-type S. aureus than ciprofloxacin. The two- to fourfold increase in the MIC of gemifloxacin in genetically defined grlBA mutants and the twofold increase in a single gyrA mutant, supported by the low frequency of selection of resistant mutants at twice the MIC (7.4 × 10−11 to 1.1 × 10−10), suggested similar targeting of the two enzymes by gemifloxacin. Dual mutations in both gyrase and topoisomerase IV caused a 64- to 128-fold increase in the MIC of gemifloxacin, similar to that seen with ciprofloxacin. Gemifloxacin also had similar activity in vitro against topoisomerase IV and gyrase purified from S. aureus (50% inhibitory concentrations of 0.25 and 0.31 μg/ml, respectively). This activity was 10- to 20-fold higher than that of ciprofloxacin for topoisomerase IV and 33-fold higher than that for gyrase. In contrast to the in vitro findings, only topoisomerase IV mutants were selected in first-step mutants. Overexpression of the NorA efflux pump had a minimal effect on resistance to gemifloxacin, and a mutation in the promoter region of the gene for NorA was selected only in the sixth step of serial selection of mutants. Our data show that although gemifloxacin targets purified topoisomerase IV and gyrase similarly in vitro, topoisomerase IV is the preferred target in the bacteria. Selection of novel resistance mutations in grlA requires further expansion of quinolone-resistance-determining regions, and their study may provide increased insight into enzyme-quinolone interactions.
Quinolones act by forming ternary complexes with DNA gyrase and/or topoisomerase IV, thereby blocking DNA replication and triggering events leading to cell death (7, 21, 22). Stepwise accumulation of chromosomal mutations leads to quinolone resistance, with the first mutation usually occurring in the primary or more sensitive enzyme target (23). Quinolone structure affects the target preference of quinolones (1, 34). In Staphylococcus aureus, topoisomerase IV has been shown to be the primary target for most quinolones, although recently nadifloxacin and sparfloxacin have been suggested to target gyrase primarily (46), and garenoxacin (BMS-284756) (26) and other nonfluorinated quinolones (41) appear to target both enzymes similarly. In cases of similar targeting of gyrase and topoisomerase IV, the frequency of selection of resistant mutants and the increment in resistance following selection of a mutation in one of the drug targets are low (16, 38). In the ideal setting of equal dual targeting, mutations in both enzymes would be needed for phenotypic expression of resistance.
Gemifloxacin is a novel fluoronaphthyridone with a C-7 pyrrolidinyl substituent (6). It has been reported to have enhanced activity compared to that of older-generation fluoroquinolones against gram-positive aerobic bacteria (15, 19, 32) while retaining an activity similar to that of ciprofloxacin against gram-negative species (6, 31). Similar to other quinolones, it is less potent against methicillin-resistant staphylococci (19), but recent work has shown that it might retain activity against some ciprofloxacin-resistant S. aureus strains (30, 44). Gemifloxacin has exceptional activity against another gram-positive coccus, Streptococcus pneumoniae. It has been shown to be effective against not only penicillin-resistant strains but also against multidrug-resistant strains, including many that are ciprofloxacin resistant (16, 17). This potent antipneumococcal activity of gemifloxacin has been shown to be due to dual targeting of topoisomerase IV and gyrase in vitro, with enhanced stabilization of the mutant gyrase and topoisomerase IV complexes on DNA (16, 50). While gemifloxacin targets both enzymes similarly in vitro in S. pneumoniae, it has a preference for gyrase in vivo, selecting for gyrase mutations in first-step mutants (16). The reported potent activity of gemifloxacin against S. aureus, even against ciprofloxacin-resistant strains, might also suggest dual targeting of topoisomerase IV and gyrase in S. aureus. Therefore, in this study we sought to determine the mechanism of activity of gemifloxacin in S. aureus. We determined the effect of mutations in topoisomerase IV and gyrase in genetically defined mutants of S. aureus, determined the frequency of selection of resistant mutants, and characterized these mutants to determine the primary target of gemifloxacin in S. aureus. In vitro inhibition of S. aureus topoisomerase IV and DNA gyrase by gemifloxacin in comparison to ciprofloxacin was also studied by using enzymes purified from wild-type S. aureus and a GrlA mutant (Ser80Phe).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are shown in Table 1. Escherichia coli strains were grown in Luria-Bertani medium, and S. aureus strains were grown in brain heart infusion (BHI) medium. All strains were grown at 37°C except S. aureus strains with the thermosensitive plasmids derived from pCL52.2, which were grown at 30°C. Spectinomycin and ampicillin were used at a concentration of 50 μg/ml, and tetracycline was used at a concentration of 5 μg/ml, except during integration of the plasmid pCL52.2 into the chromosome, when it was used at a concentration of 3 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotypea or description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| ISP794 | 8325 pig-131 | 45 |
| MT5 | 8325 nov (gyrB142) hisG15 pig-131 | 10, 47 |
| SS1 | 8325 pig-131 gyrB142 (Ile102Ser, Arg144Lys) gyrA (Ser84Leu) | 5 |
| MT5224c2 | 8325 nov (gyrB142) hisG15 pig-131 grlA552 | 36 |
| MT5224c3 | 8325 nov (gyrB142) hisG15 pig-131 grlA547 | 47 |
| MT5224c4 | 8325 nov (gyrB142) hisG15 pig-131 grlA542 (Ser80Phe) | 47 |
| MT5224c9 | 8325 nov (gyrB142) hisG15 pig-131 grlB543 (Asn470Asp) | 47 |
| MT23142 | 8325 pig-131 flqB Ω(chr::Tn916)1108 | 36 |
| EN1252a | 8325 nov (gyrB142) hisG15 pig-131 grlA542 gyrA Ω1051 (Erm) Nov+ | 5 |
| P4 | ISP794 grlA563 (Arg43Cys) | 24 |
| P10 | ISP794 grlA555 (Ala176Thr) | 24 |
| P21 | ISP794 grlA556 (Asp69Tyr) | 24 |
| GB | ISP794 grlB560 (Pro25His) | 25 |
| CS33 | ISP794 grlB566 (Asn470Ile); spontaneous gemifloxacin resistance | This study |
| GM3 | ISP794 grlA567 (insertion of Asn between codons 271 and 272); spontaneous gemifloxacin resistance | This study |
| GM9 | ISP794 grlA568 (Ile490Thr); spontaneous gemifloxacin resistance | This study |
| GMS1 | ISP794 (unidentified mutation); serial-passage mutant | This study |
| GMS2-3 | ISP794 grlA569 (Asn327Lys); serial-passage mutant | This study |
| GMS4-5 | ISP794 grlA569 (Asn327Lys) gyrB (Va1464A1a); serial-passage mutant | This study |
| GMS6-7 | ISP794 grlA569 (Asn327Lys) gyrA(Ser84Leu) gyrB (Val464Ala); serial-passage mutant | This study |
| GMS8-9 | ISP794 grlA569 (Asn327Lys) grlA (Glu84Lys) gyrA(Ser84Leu) gyrB (Val464Ala); serial-passage mutant | This study |
| GMS10 | ISP794 grlA569 (Asn327Lys) grlA (Glu84Lys) gyrA(Ser84Leu) gyrB (Val464Ala) flqB (ATAGA insertion); serial-passage mutant | This study |
| GMS11 | ISP794 grlA569 (Asn327Lys) grlA (Glu84Lys) gyrA(Ser84Leu) gyrB (Val464Ala) flqB (ATAGA insertion); serial-passage mutant | This study |
| DIGM3 | ISP794 grlA567 (insertion of Asn between codons 271 and 272); allelic exchange mutant | This study |
| DIGMS2 | ISP794 grlA569 (Asn327Lys); allelic exchange mutant | This study |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA hsdR17 (rK− mK−) supE44λ−thi-1 gyrA96 relA1 | GIBCO-BRL |
| Plasmids | ||
| PGEM3-zf(+) | 3,199-bp cloning vector, Apr | Promega |
| pCL52.2 | 8,119-bp plasmid containing the replicon of pGB2, Spr (E. coli), temperature-sensitive replicon of pE194, Tcr | 43 |
| pTrcHisA, B, and C | 4.4-kb expression vector, Apr | Invitrogen |
Abbreviations: Ap, ampicillin; Sp, spectinomycin; Tc, tetracycline.
Drug susceptibility determinations.
Gemifloxacin was supplied by GlaxoSmithKline, and ciprofloxacin was supplied by Bayer Corp. Stock solutions of gemifloxacin and ciprofloxacin were prepared in 0.1 N sodium hydroxide and distilled water, respectively, and was stored at −70°C. Nalidixic acid, novobiocin, ethidium bromide, and reserpine were purchased from Sigma Chemical Co. (St. Louis, Mo.). MICs of all antimicrobials and chemicals except reserpine were determined in duplicate at least twice, and in the condition of a twofold increase three times, on Trypticase soy agar containing serial twofold dilutions of antibiotics, and growth was scored after 24 and 48 h of incubation at 37°C. The effect of reserpine on quinolone susceptibility was determined by broth dilution in Penassay broth supplemented with 0.4% glucose (35). MICs of nalidixic acid were used to screen for gyrA mutations, MICs of novobiocin for grlB mutations (11), and MICs of ethidium bromide to screen for NorA overexpression (29).
Frequency of selection of mutants.
Overnight cultures of S. aureus ISP794 were diluted or concentrated appropriately in normal saline before being plated on BHI agar without any antibiotic or containing gemifloxacin or ciprofloxacin at one-, two-, and fourfold above the MIC of each drug. For selection with gemifloxacin, approximately 1011 to 1012 CFU of ISP794 were plated on 150 by 15 mm petri dishes containing BHI agar with appropriate gemifloxacin concentrations. Selection plates were incubated at 37°C. Frequency of selection of resistant mutants was calculated as the ratio of the number of resistant colonies at 48 h to the number of cells plated. To purify, selected colonies were restreaked once on BHI agar plates containing the selecting concentration of gemifloxacin and, if necessary, once more on BHI agar without any antibiotic and then were stored at −70°C in 10% glycerol in BHI broth. Selection of mutants was repeated three times.
Stepwise selection of resistant mutants.
S. aureus ISP794 was serially passaged on BHI agar containing twofold-increasing concentrations of gemifloxacin to select for increasingly resistant mutants. Selection began at the MIC of gemifloxacin for ISP794. At each step, several mutant colonies were restreaked on BHI agar plates containing the selecting concentration of gemifloxacin before being stored at −70°C and being passaged on a twofold-higher antibiotic concentration.
Sequence analysis.
Following lysis of various single-step and multiple-step mutants of S. aureus ISP794 with lysostaphin (Ambi, Lawrence, N.Y.) at a concentration of 0.1 mg/ml, chromosomal DNA extraction was performed by using the Easy-DNA kit (Invitrogen, Carlsbad, Calif.), and it was used as template for PCR. PCR for the quinolone-resistance-determining regions (QRDRs) of grlA, grlB, gyrA, gyrB, and the promoter region of norA was performed by using platinum Pfx DNA polymerase (Life Technologies, Rockville, Md.) as described previously (24, 25). Automated DNA sequencing of the PCR products was performed by using automated ABI 3100 DNA sequencers (Tufts University Core Facility, Boston, Mass.). For strains in which no mutation was found in these regions, the entireties of the grlA, grlB, gyrA, and gyrB genes were amplified by PCR and were sequenced by the same method.
Cloning and allelic exchange.
A 1,096-bp internal fragment of grlA was amplified by PCR with the upstream primer containing an engineered EcoRI site (5′-GAAGGAATTCAAGCAACAC-3′; 5′ nucleotide at position 2931 in the sequence published by Yamagishi et al. [51]) and the downstream primer containing an engineered BamHI site (5′-CTGGATCCGTATCTTGCGT-3′; 5′ nucleotide at position 4026). The annealing temperature was 50°C, and the extension time was 70 s for this PCR. Following gel extraction with the Compass kit (American Bioanalytical, Natick, Mass.), PCR products were ligated into the EcoRI and BamHI sites of pGEM3-zf(+) and the recombinant plasmids were electroporated into E. coli DH5α. The insert of each construct was sequenced to confirm that new mutations were not introduced by the DNA polymerase. The inserts were then ligated into pCL52.2, a thermosensitive shuttle vector, and the plasmids were again electroporated into E. coli DH5α, followed by electroporation into S. aureus RN4220 and subsequently to S. aureus ISP794, as previously described (37). Allelic exchange was performed as previously described (24, 25). The resulting colonies were screened for susceptibility to tetracycline at a concentration of 3 μg/ml and reduced susceptibility to ciprofloxacin at a concentration of 0.25 μg/ml. MICs of ciprofloxacin and gemifloxacin were determined for tetracycline-susceptible colonies that grew on the ciprofloxacin plate. Direct DNA sequencing of the PCR product of the appropriate region amplified from chromosomal DNA was used to confirm the presence of the expected mutations.
Purification of recombinant proteins and inhibition of topoisomerase IV and DNA gyrase.
Cloning of S. aureus grlA, grlB, gyrA, and gyrB from wild-type S. aureus ISP794 as well as the grlA subunit from the grlA mutant with the Ser80Phe mutation, strain MT5224c4, into pTrcHis A, B, or C vectors (Invitrogen, Carlsbad, Calif.), and overexpression and purification of recombinant proteins containing N-terminal His6 have been reported earlier (26).
Inhibition of topoisomerase IV and DNA gyrase.
Conditions for reconstitution of S. aureus topoisomerase IV and DNA gyrase and assay conditions for inhibition of kinetoplast DNA (kDNA) (from Crithidia fasciculata; TopoGEN Inc., Columbus, Ohio) decatenation by topoisomerase IV and DNA supercoiling by gyrase have been published previously, except that 200 ng of kDNA was used for decatenation assays (26).
RESULTS
Activities for genetically defined mutants.
To understand which quinolone resistance mutations affected susceptibility to gemifloxacin, we determined the MIC of gemifloxacin for our genetically defined mutants of S. aureus and compared the results with those of ciprofloxacin (Table 2). The MICs determined in this study for ciprofloxacin were identical to or within one dilution of those previously published (24, 25). Gemifloxacin was 8- to 16-fold more active than ciprofloxacin against wild-type S. aureus ISP794, in accordance with previously published results (6, 19, 30). A single mutation in either subunit of topoisomerase IV or gyrA caused a two- to fourfold increase in the MIC of gemifloxacin, indicating that single mutations in either enzyme might contribute to resistance. This pattern was different from that observed for ciprofloxacin, which has been shown primarily to target topoisomerase IV (9, 36, 51), for which mutations in either subunit of topoisomerase IV (the more sensitive enzyme) caused a four- to eightfold increase in the MIC of ciprofloxacin but for which a single mutation in gyrA (the less sensitive enzyme) caused a less than twofold increase. Mutations in both enzymes together caused a 64- to 128-fold increase in the MIC of gemifloxacin while causing a 128- to 256-fold increase in the MIC of ciprofloxacin. Gemifloxacin was determined to be a poor substrate for the NorA efflux pump, with its MIC increasing at most twofold for the genetically defined mutant overexpressing NorA (MT23142), while the MIC of ciprofloxacin for that mutant increased four- to eightfold.
TABLE 2.
Activity of gemifloxacin and ciprofloxacin against genetically defined mutants of S. aureus
| Strain | Mutation | MICs (μg/ml)
|
|
|---|---|---|---|
| Gemifloxacin | Ciprofloxacin | ||
| ISP794 | Wild-type (parent) | 0.016 | 0.125-0.25 |
| MT5 | gyrB142 (Ile102Ser, Arg144Ile) | 0.016 | 0.125-0.25 |
| SS1 | gyrA (Ser84Leu) | 0.032 | 0.25 |
| MT5224c2 | grlA (Ala116Pro) | 0.032 | 1.0-2.0 |
| MT5224c3 | grlA (Ala116Glu) | 0.032 | 2.0 |
| MT5224c4 | grlA (Ser80Phe) | 0.032-0.064 | 1.0-2.0 |
| MT5224c9 | grlB (Asn470Asp) | 0.032-0.064 | 2.0 |
| P4 | grlA (Arg43Cys) | 0.032 | 1.0 |
| P21 | grlA (Asp69Tyr) | 0.032 | 1.0 |
| P10 | grlA (Ala176Thr) | 0.032-0.064 | 1.0 |
| GB | grlB (Pro25His) | 0.032-0.064 | 1.0 |
| MT23142 | flqB (NorA overexpression) | 0.016-0.032 | 0.5-1.0 |
| EN1252a | grlA (Ser80Phe) gyrA (Ser84Leu) | 1.0-2.0 | 32 |
Frequency of selection of mutants.
Since mutations in either gyrase or topoisomerase IV caused a slight increase in the MIC of gemifloxacin, suggesting dual targeting, we expected the frequency of selection of resistant mutants with greater than twofold increments in MIC to be low. Therefore, we performed selection at the MIC (0.016 μg/ml) as well as at twice and four times the MIC of gemifloxacin and plated approximately 1011 to 1012 CFU for selection of resistant mutants. Although the frequency of selection was 1.5 × 10−5 to 2.4 × 10−5 at the MIC, it decreased dramatically at two times the MIC, to 7.4 × 10−11 to 1.1 × 10−10 (Table 3). At four times the MIC, no mutants could be selected with gemifloxacin. As previously published, mutants could be selected at two and four times the MIC of ciprofloxacin, and the frequency of selection at four times the MIC of ciprofloxacin was 3 logs higher than the frequency at twice the MIC of gemifloxacin.
TABLE 3.
Frequency of selection of resistant mutants for S. aureus ISP794
| Drug | Selecting drug concna | Frequency of selection of mutants |
|---|---|---|
| Gemifloxacin | 1 (0.016) | 1.5 × 10−5-2.4 × 10−5 |
| 2 (0.032) | 7.4 × 10−11-1.1 × 10−10 | |
| 4 (0.064) | <7.4 × 10−12 | |
| Ciprofloxacin | 2 (0.5) | 2.8 × 10−6-1.5 × 10−5 |
| 4 (1.0) | 3.0 × 10−8-6.1 × 10−8 |
Drug concentrations are given as factors of the MICs, and the numbers in parentheses are the MICs in micrograms per milliliter.
Target preference for topoisomerase IV in single-step mutants.
Since the MICs of genetically defined mutants suggested dual targeting of both gyrase and topoisomerase IV but did not identify a preferred target, we characterized first-step mutants selected with gemifloxacin (Table 4). For the three single-step mutants studied, each of which was selected in an independent experiment, the MIC of gemifloxacin increased two- to fourfold (up to 0.032 to 0.064 μg/ml), and that of ciprofloxacin increased four- to eightfold (up to 1.0 μg/ml). The MIC of novobiocin decreased fourfold for mutant CS33 but did not change for the other two mutants. There was at most a twofold change in the MICs of nalidixic acid and ethidium bromide for all three mutants. Sequencing of the QRDRs of grlA, grlB, gyrA, and gyrB revealed a mutation (Asn470Ile) in a hot spot of grlB in only one of these mutants (CS33), consistent with other grlB mutations that alter novobiocin susceptibility (11). For the other two single-step mutants, which also had increased resistance to ciprofloxacin and which grew more slowly than the wild-type strain (data not shown), we first sequenced the entire grlBA genes in search of a resistance mutation and identified two novel mutations in grlA. Interestingly, one of these mutations encoded the insertion of an amino acid (asparagine) between amino acids 271 and 272 in mutant GM3, and the other was a conservative mutation (Ile490Thr) in the other mutant, GM9. For these two mutants, the QRDRs of gyrBA were sequenced to exclude the possibility of other mutations, and none was found.
TABLE 4.
Characteristics of single-step mutants of S. aureus ISP794 selected with gemifloxacin
| Strain | MIC (μg/ml)a
|
Mutationb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GEM | CIP | NOV | NAL | EB | grlA | grlB | gyrA | gyrB | |
| ISP794 | 0.016 | 0.125-0.25 | 0.064 | 64 | 2.0 | ||||
| CS33 | 0.064 | 1.0 | 0.016 | 128 | 2.0 | Nonec | Asn470Ilec | Nonec | Nonec |
| GM3 | 0.064 | 1.0 | 0.064 | 128 | 2.0 | Insertion of Asn between codons 271 and 272 | None | Nonec | Nonec |
| GM9 | 0.032 | 1.0 | 0.032-0.064 | 128 | 4.0 | Ile490Thr | None | Nonec | Nonec |
GEM, gemifloxacin; CIP, ciprofloxacin; NOV, novobiocin; NAL, nalidixic acid; EB, ethidium bromide.
The indicated mutations resulted in the following nucleotide changes: Asn470Ile in grlB, AAT → ATT; insertion of Asn between codons 271 and 272 in grlA, insertion of AAC; Ile490Thr in grlA, ATT → ACT.
No mutation was found in the QRDR, which was the only region sequenced.
Sequential acquisition of mutations in serial-passage mutants.
A fourth single-step mutant selected at the MIC of gemifloxacin (0.016 μg/ml) was serially passaged on twofold-increasing concentrations of gemifloxacin to determine the order of acquisition of resistance mutations. Results of MIC determinations and DNA sequencing are shown in Table 5. All MIC results that showed only a twofold difference were determined at least three times for reproducibility. The MICs of gemifloxacin and ciprofloxacin were increased only twofold for this first-step mutant. The MICs of nalidixic acid increased twofold, and those of novobiocin and ethidium bromide did not change. For the second-step mutant (GMS2-3), the MIC of gemifloxacin increased another twofold for both quinolones, whereas the MICs of nalidixic acid, novobiocin, and ethidium bromide did not change. Sequencing of the entire grlBA gene in addition to the QRDRs of gyrBA revealed a novel mutation in grlA, Asn327Lys, which was not found in the first-step mutant. At the next step of selection (GMS4-5) the MIC of gemifloxacin increased fourfold, with no increase in that of ciprofloxacin. Sequencing of gyrBA revealed a novel mutation in gyrB (Val464Ala). At the fourth step of selection, we identified a common mutation in gyrA (Ser84Leu) which was associated with an additional fourfold increase in the MICs of both gemifloxacin and ciprofloxacin followed by a common mutation in grlA (Glu84Lys) at the fifth step when the QRDR was sequenced. The MICs of nalidixic acid, novobiocin, and ethidium bromide changed only twofold for these mutants. There was a fourfold increase in the MIC of ethidium bromide in the sixth-step mutant (GMS10), suggesting overexpression of either the NorA or another efflux pump. Sequencing of the NorA promoter region in this and the subsequent mutant (GMS11) revealed duplication of the sequence ATAGA (nucleotides −99 and −94 before the ATG start codon) right after the −10 consensus sequence, a change that was absent in the preceding mutants. In addition, the MICs of gemifloxacin, ciprofloxacin, and ethidium bromide for GMS10 and GMS11 decreased twofold in the presence of the NorA inhibitor reserpine, whereas the presence of reserpine did not cause a decrement in the MICs of these two fluoroquinolones for the wild-type strain ISP794 or the serial-passage mutants without the duplication (data not shown). However, we did not identify any other mutation in the promoter region of the gene coding for NorA in mutant GMS11 and we do not know the mechanism responsible for the additional fourfold increment in resistance (from 8 to 32 μg/ml) to ethidium bromide in this last-step mutant. Thus, stepwise accumulation of mutations caused an approximately 2,000-fold increase in the MICs of both quinolones in the seventh-step mutant, the highest level of resistance achievable.
TABLE 5.
Characteristics of serial-passage mutants of S. aureus ISP794 selected with gemifloxacin
| Strain (selecting concn in μg/ml) | MIC (μg/ml)a
|
Mutation
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GEM | CIP | NOV | NAL | EB | grlA | grlB | gyrA | gyrB | norAc | |
| ISP794 | 0.008-0.016 | 0.125-0.25 | 0.064-0.128 | 64-128 | 2.0-4.0 | |||||
| GMS1 (0.016) | 0.016-0.032 | 0.25-0.5 | 0.064 | 256 | 4.0 | |||||
| GMS2-3 (0.032-0.064) | 0.064 | 1.0 | 0.064 | 256 | 4.0 | Asn327Lys | None | None | None | |
| GMS4-5 (0.125-0.25) | 0.25 | 1.0 | 0.064 | 256 | 4.0 | None | Val464Ala | |||
| GMS6-7 (0.5-1.0) | 1.0 | 4.0 | 0.064 | 256 | 2.0 | Asn327lysb | Noneb | Ser84Leub | Noneb | |
| GMS8-9 (2.0-4.0) | 4.0 | 32 | 0.064 | 256 | 2.0 | Glu84Lys, Asn327Lysb | Noneb | Ser84Leub | None | |
| GMS10 (8.0) | 8.0 | 64 | 0.032-0.064 | 256 | 8.0 | Duplication of ATAGAd | ||||
| GMS11 (16.0) | 32 | 256 | 0.064 | 256 | 32.0 | Duplication of ATAGAd | ||||
GEM, gemifloxacin; CIP, ciprofloxacin; NOV, novobiocin; NAL, nalidixic acid; EB, ethidium bromide.
None in the QRDR.
The promoter region of norA.
Duplication of the 5-bp (94th and 99th nucleotides) upstream of the ATG start codon of norA. The indicated mutations in grlA resulted in the following nucleotide changes: Asn327Lys, AAT → AAA; Glu84Lys, GAA → AAA. The indicated mutations in gyrA and gyrB resulted in the following mutations, respectively: Ser84Leu, TCA → TTA; Val464Ala, GTT → GCT.
Confirmation of the role of novel mutations by allelic exchange.
The Asn470Asp mutation in grlB has previously been shown to cause resistance (11). The mutation in mutant CS33, occurring at the same codon (Asn470Ile), has previously also been selected with ciprofloxacin (D. Ince and D. C. Hooper, unpublished results), strongly suggesting that this mutation is responsible for the resistance phenotype. For mutant GM3 with the previously unencountered insertion of an asparagine between codons 271 and 272, 1 of 300 colonies screened in the allelic exchange experiment was found to be tetracycline sensitive and to have reduced susceptibility to ciprofloxacin. For this allelic exchange mutant, DIGM3, the MICs of gemifloxacin (0.032 μg/ml) and ciprofloxacin (0.5 μg/ml) were within one dilution of the MICs for the original mutant, GM3 (0.064 and 1.0 μg/ml, respectively). MIC determination for the allelic exchange mutant (DIGMS2) and the original mutant (GMS2) containing the grlA Asn370Lys mutation revealed identical MICs of ciprofloxacin (1.0 μg/ml) and a twofold-lower MIC of gemifloxacin for DIGMS2 (0.032 versus 0.064 μg/ml). MIC determinations were performed three times for reproducibility of the twofold differences, with the same results being obtained each time. The presence of the expected mutations in DIGM3 and DIGMS2 was confirmed by DNA sequencing. We were unable to identify an allelic exchange mutant with the grlA (Ile490Thr) mutation despite three attempts and screening of approximately 3,000 colonies. Allelic exchange was not performed for the gyrB mutation (Val464Ala).
Activities of gemifloxacin and ciprofloxacin against purified topoisomerase IV and gyrase.
The individual subunits of topoisomerase IV and gyrase purified as histidine-tagged proteins were >90% homogenous by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). Specific activities of each subunit of gyrase and topoisomerase IV were determined in the presence of an excess of the complementing wild-type subunit. One unit was defined as the amount of enzyme that produces half-maximal decatenation of 0.2 μg of kDNA for topoisomerase IV and as the amount of enzyme that produces half-maximal supercoiling of 0.5 μg of relaxed pBR322 DNA for gyrase. The specific activities of wild-type GrlA and GrlB were found to be 1.8 × 105 and 5.3 × 105 U/mg, respectively, and of wild-type GyrA and GyrB were found to be 1.1 × 104 and 5.2 × 103 U/mg, respectively. The specific activity of GrlA from mutant MT5224c4 carrying the Ser80Phe alteration in GrlA was 3.6 × 105 U/mg.
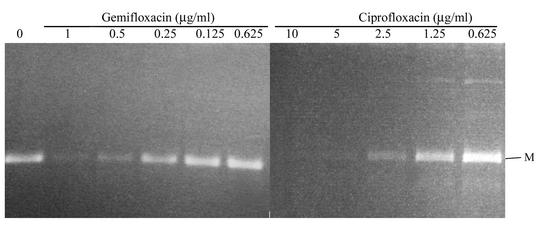
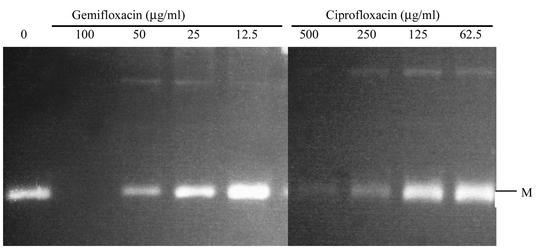
We examined and compared the effects of gemifloxacin and ciprofloxacin on the activities of these S. aureus topoisomerases. Decatenation activity was used to assess the inhibition of topoisomerase IV and DNA supercoiling for the activity of DNA gyrase. For decatenation assays, the 50% inhibitory concentration (IC50) is defined as the concentration that reduces the intensity of the DNA band representing decatenated kDNA minicircles by half. IC50 was determined in the absence and presence of increasing concentrations of gemifloxacin and ciprofloxacin by using just-sufficient concentrations of GrlA and GrlB to fully decatenate the kDNA minicircles (usually 2 U). Both quinolones inhibited decatenation in a dose-dependent manner. The IC50 of gemifloxacin was 0.25 μg/ml, 10- to 20-fold less than that of ciprofloxacin, which was 2.5 to 5.0 μg/ml, in part accounting for the greater potency of gemifloxacin (Fig. 1; Table 6). The IC50 of both gemifloxacin and ciprofloxacin increased substantially for the grlA mutant (to 50 μg/ml for gemifloxacin and 250 μg/ml for ciprofloxacin) (Fig. 2).
FIG. 1.
Decatenation of kDNA by wild-type GrlA and GrlB in the presence of gemifloxacin and ciprofloxacin. Assays were performed as described in Materials and Methods. The letter M indicates minicircles.
TABLE 6.
Inhibitory effects of gemifloxacin and ciprofloxacin on wild-type and mutant (grlA-deficient) topoisomerase IV and wild-type gyrase
| Enzyme | Subunits
|
IC50 (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| GrlA | GrlB | GyrA | GyrB | Gemifloxacin | Ciprofloxacin | |
| Topoisomerase IV | Wild type | Wild type | 0.25 | 2.5-5.0 | ||
| Topoisomerase IV | Phe80 | Wild type | 50 | 250 | ||
| Gyrase | Wild type | Wild type | 0.31 | 10 | ||
FIG. 2.
Decatenation of kDNA by GrlA (Ser80Phe) and wild-type GrlB in the presence of gemifloxacin and ciprofloxacin. The letter M indicates minicircles.
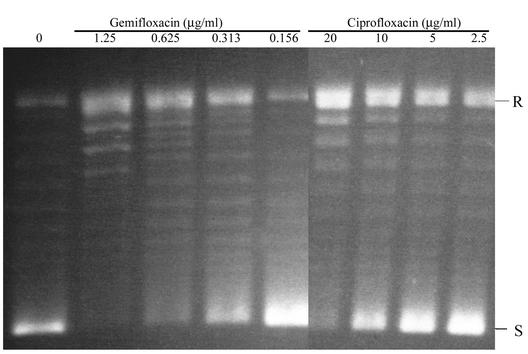
For DNA supercoiling activity of DNA gyrase, the IC50 was defined as the concentration of drug that reduces the intensity of the most supercoiled DNA band by half and was determined in the presence of just-sufficient amounts of GyrA and GyrB to fully supercoil the relaxed substrate DNA (2 U). DNA supercoiling inhibition was also dose dependent, and the IC50 of gemifloxacin for the wild-type gyrase (0.31 μg/ml) was similar to its IC50 for topoisomerase IV (0.25 μg/ml) and was approximately 30-fold less than that of ciprofloxacin for the wild-type gyrase (10 μg/ml) (Fig. 3).
FIG. 3.
DNA supercoiling activity of gyrase in the presence of gemifloxacin and ciprofloxacin. R and S indicate relaxed and supercoiled DNA, respectively.
DISCUSSION
In this study, gemifloxacin was found to be 8- to 16-fold more active against wild-type S. aureus than ciprofloxacin. This result is comparable to those of previous studies, which have shown 8- to 32-fold greater activity for clinical isolates of methicillin-sensitive S. aureus (6, 14, 15, 19, 20, 28, 30-32). The fact that in genetically defined mutants of S. aureus single mutations in either subunit of topoisomerase IV caused a two- to fourfold increase and a mutation in DNA gyrase caused a twofold increase in the MIC of gemifloxacin suggested similar targeting of the two enzymes. In support of this hypothesis, selection of single-step mutants proved to be difficult at twice the MIC (Table 3), similar to results obtained with garenoxacin (26) as well as with other nonfluorinated quinolones (41) with dual targeting in S. aureus. Thus, similar to its mechanism of action in S. pneumoniae (16, 50), gemifloxacin appears to target both topoisomerase IV and DNA gyrase in S. aureus. However, analysis of four single-step mutants, one of which was used for selection of serial-passage mutants obtained from three different selection experiments, revealed mutations in either the grlA or the grlB subunit of topoisomerase IV, suggesting an in vivo preference for topoisomerase IV, in contrast to the in vivo preference of gemifloxacin for gyrase in S. pneumoniae (16). Similar to results with other quinolones, stepwise selection of mutants studied in one series of mutants showed stepwise accumulation of resistance mutations, with the first change appearing in a subunit of topoisomerase IV, GrlA, as in the other single-step mutants selected with gemifloxacin, followed by a novel mutation in gyrB, a common mutation in gyrA, and, consecutively, another mutation in grlA.
Interestingly, only one of the mutations in topoisomerase IV was in the QRDR of grlB, at codon 470. An Asn470Asp alteration at this site has previously been associated with resistance to quinolones and increased susceptibility to novobiocin (11) as well as reduced enzyme catalytic activity (X. Zhang and D. C. Hooper, unpublished result). We predict the Asn470Ile alteration may also affect catalytic action of the enzyme similarly.
One of the novel mutations selected with gemifloxacin was the insertion of an amino acid, asparagine, between Lys271 and Arg272 in GrlA. The only other report of an insertional mutation in the structural topoisomerase genes, to our knowledge, is the insertion of an alanine and an arginine between residues 378 and 379 in GyrB in an E. coli strain that is deficient in topoisomerase I (18). The strain with this mutation, however, was more sensitive to the bacteriostatic and bactericidal action of quinolones, and the mutant enzyme was more sensitive to quinolones than the wild-type enzyme and had reduced DNA supercoiling, DNA relaxing, ATPase, and quinolone-mediated DNA cleavage activities, hypothesized to be due to loss of activity due to misfolding of the protein (18). Mutant GM3 with the insertion of asparagine was fourfold more resistant to gemifloxacin and four- to eightfold more resistant to ciprofloxacin, and the role of this novel type of mutation in resistance to quinolones was proved by allelic exchange. The mechanism by which this mutation causes resistance is yet to be defined, although it might be hypothesized to cause a misfolding of GrlA that perturbs either DNA complex formation or a conformation necessary for quinolone binding.
Another novel mutation in the gene that codes for GrlA that we could demonstrate to cause resistance is the Asn327Lys alteration in mutant GMS2. Alignment to E. coli gyrase (33) and Saccharomyces cerevisiae topoisomerase II (3) maps this mutation to the β14 sheet, far from the α4 helix on which the commonly occurring mutations in the QRDR predicted to be involved in quinolone binding (2, 49) are located. The amino acid at this position is not a conserved amino acid in either E. coli gyrase (33) or S. cerevisiae topoisomerase II (3).
The final novel change in GrlA (Ile490Thr) lies near the midportion of grlA, also far from the QRDR, which is near the 5′ end of the gene. This portion of the gene is not included in the crystal structures of E. coli gyrase (33) or yeast topoisomerase II (3, 8); therefore, we cannot predict, by alignment, the means by which it might be associated with resistance. Allelic exchange studies were not successful in providing genetic evidence for the role of this mutation in resistance. Another method to confirm its role would be comparison of quinolone inhibition of wild-type topoisomerase IV and that reconstituted with mutant GrlA (Ile490Thr), experiments that have not yet been performed.
An interesting observation was the absence of an increase in the MIC of ciprofloxacin in contrast to a fourfold increase in the MIC of gemifloxacin for mutant GMS4-5. Since an allelic exchange experiment was not performed for the GyrB mutant (Val464Ala), we do not know its role in resistance. Previously, however, Weigel et al. have reported that the effects of specific mutations varied significantly depending on the quinolone tested (48). Further studies are needed to clarify the role of this mutation in resistance to gemifloxacin and ciprofloxacin.
Biochemical studies also showed greater activity of gemifloxacin against S. aureus topoisomerase IV and DNA gyrase. Gemifloxacin was 10- to 20-fold more active against wild-type S. aureus topoisomerase IV and 30-fold more active against wild-type gyrase than ciprofloxacin (Table 6). The interesting feature was the similar inhibitory activity of gemifloxacin for two target enzymes, despite the selection of grlBA mutations in vivo in first-step mutants, suggesting that experimental conditions in vitro do not always represent the conditions inside the bacterium, a phenomenon also reported with S. pneumoniae topoisomerases (16, 39). Another striking feature was the 200-fold increase in the IC50 of gemifloxacin and the 50- to 100-fold increase in that of ciprofloxacin for topoisomerase IV reconstituted with mutant GrlA (Phe80). The MIC of gemifloxacin and ciprofloxacin for the strain with the same mutation (MT5224c4) increased only two- to fourfold and four- to eightfold, respectively. This finding supports the concept that the increment in resistance in the bacterium in the presence of an alteration in one target enzyme (in this case, topoisomerase IV) is additionally affected by the sensitivity of the second target enzyme (gyrase) (23).
This study also shows that gemifloxacin is a poor substrate for the NorA efflux pump, with resistance increasing at most twofold for a strain overexpressing NorA and with apparent efflux pump mutants not being selected until the sixth step in stepwise selection of mutants. The hydrophobicity of quinolones and the bulkiness of the C-7 substituent have been implicated in the efficiency of efflux by NorA (4, 40). The increased bulkiness of gemifloxacin with substitution of a 3-aminomethyl and 4-methoxyimino at the 7-pyrrolidine ring might be one of the reasons for decreased efflux. We do not yet know, however, if gemifloxacin, like sparfloxacin, binds to NorA at a site different from those of ciprofloxacin and norfloxacin (52). The mutation predicted to cause overexpression of the efflux pump in the serial passage mutant was also interesting. It was found to be in the 5′-untranslated region of norA, close to the previously identified point mutation which has been shown to be associated with NorA overexpression (35) due to increased stability of the norA mRNA (12). However, the mutation in this strain was a duplication of five nucleotides, similar to the mutations seen in the same region in mutants selected with gatifloxacin and premafloxacin, which had different insertions of four nucleotides (24, 25). We do not yet know the means by which these mutations might effect resistance or alter norA expression.
This study, besides exhibiting the greater activity of gemifloxacin against S. aureus compared to that of ciprofloxacin, gives biochemical evidence of dual targeting of topoisomerase IV and gyrase by gemifloxacin in a genetically defined S. aureus strain, ISP794, and gives genetic evidence for in vivo preference for topoisomerase IV. The identification of novel resistance mutations requires further expansion of the QRDR for grlA to include codons 271 and 327. It also suggests that in the laboratory setting, when the cost of a resistance mutation might not be as important as it is in the clinical setting, novel mutations can be identified more easily. Recently, we and others have identified many novel mutations in both subunits of topoisomerase IV outside the QRDRs in S. aureus and S. pneumoniae and have proved the role of some of these mutations in resistance to quinolones (24, 25, 27, 42, 48). In contrast, the range of mutations reported for DNA gyrase appears to be less, with only a few novel mutations in or outside the QRDR isolated, and proved to be responsible for quinolone resistance in S. aureus (26, 42), S. pneumoniae (38, 48), and E. coli (13). These findings suggest that a smaller range of tolerated mutations occur on DNA gyrase and that the study of various topoisomerase IV mutants may provide additional insights into enzyme-DNA-quinolone interactions.
Acknowledgments
This work was supported by a grant from the U.S. Public Health Service, National Institutes of Health (R01 AI23988 to D.C.H.), and a grant from GlaxoSmithKline.
REFERENCES
- 1.Alovero, F. L., X. S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, J. M., S. J. Gamblin, S. C. Harrison, and J. C. Wang. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379:225-232. [DOI] [PubMed] [Google Scholar]
- 4.Beyer, R., E. Pestova, J. J. Millichap, V. Stosor, G. A. Noskin, and L. R. Peterson. 2000. A convenient assay for estimating the possible involvement of efflux of fluoroquinolones by Streptococcus pneumoniae and Staphylococcus aureus: evidence for diminished moxifloxacin, sparfloxacin, and trovafloxacin efflux. Antimicrob. Agents Chemother. 44:798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisognano, C., P. E. Vaudaux, D. P. Lew, E. Y. W. Ng, and D. C. Hooper. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormican, M. G., and R. N. Jones. 1997. Antimicrobial activity and spectrum of LB20304, a novel fluoronaphthyridone. Antimicrob. Agents Chemother. 41:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica, K., and X. L. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 6:322-326. [DOI] [PubMed] [Google Scholar]
- 9.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 10.Fournier, B., and D. C. Hooper. 1998. Effects of mutations in GrlA of topoisomerase IV from Staphylococcus aureus on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42:2109-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, B., and D. C. Hooper. 1998. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier, B., Q. C. Truong-Bolduc, X. M. Zhang, and D. C. Hooper. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J. Bacteriol. 183:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, S. M., T. Lu, and K. Drlica. 2001. Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother. 45:2378-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonullu, N., Z. Aktas, M. Salcioglu, C. Bal, and O. Ang. 2001. Comparative in vitro activities of five quinolone antibiotics, including gemifloxacin, against clinical isolates. Clin. Microbiol. Infect. 7:499-503. [DOI] [PubMed] [Google Scholar]
- 15.Hardy, D., D. Amsterdam, L. A. Mandell, and C. Rotstein. 2000. Comparative in vitro activities of ciprofloxacin, gemifloxacin, grepafloxacin, moxifloxacin, ofloxacin, sparfloxacin, trovafloxacin, and other antimicrobial agents against bloodstream isolates of gram-positive cocci. Antimicrob. Agents Chemother. 44:802-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heaton, V. J., J. E. Ambler, and L. M. Fisher. 2000. Potent antipneumococcal activity of gemifloxacin is associated with dual targeting of gyrase and topoisomerase IV, an in vivo target preference for gyrase, and enhanced stabilization of cleavable complexes in vitro. Antimicrob. Agents Chemother. 44:3112-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaton, V. J., C. E. Goldsmith, J. E. Ambler, and L. M. Fisher. 1999. Activity of gemifloxacin against penicillin- and ciprofloxacin-resistant Streptococcus pneumoniae displaying topoisomerase- and efflux-mediated resistance mechanisms. Antimicrob. Agents Chemother. 43:2998-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heddle, J. G., T. Lu, X. L. Zhao, K. Drlica, and A. Maxwell. 2001. gyrB-225, a mutation of DNA gyrase that compensates for topoisomerase I deficiency: investigation of its low activity and quinolone hypersensitivity. J. Mol. Biol. 309:1219-1231. [DOI] [PubMed] [Google Scholar]
- 19.Hoban, D. J., S. K. Bouchillon, J. L. Johnson, G. G. Zhanel, D. L. Butler, L. A. Miller, and J. A. Poupard. 2001. Comparative in vitro activity of gemifloxacin, ciprofloxacin, levofloxacin and ofloxacin in a North American surveillance study. Diagn. Microbiol. Infect. Dis. 40:51-57. [DOI] [PubMed] [Google Scholar]
- 20.Hoban, D. J., S. K. Bouchillon, J. L. Johnson, G. G. Zhanel, D. L. Butler, L. A. Miller, and J. A. Poupard. 2001. Comparative in vitro potency of gemifloxacin and fluoroquinolones against recent European clinical isolates from a global surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 20:814-819. [DOI] [PubMed] [Google Scholar]
- 21.Hooper, D. C. 1998. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin. Infect. Dis. 27:S54-S63. [DOI] [PubMed] [Google Scholar]
- 22.Hooper, D. C. 1999. Mechanisms of quinolone resistance. Drug Resist. Updates 2:38-55. [DOI] [PubMed] [Google Scholar]
- 23.Hooper, D. C. 2001. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 32:S9-S15. [DOI] [PubMed] [Google Scholar]
- 24.Ince, D., and D. C. Hooper. 2000. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob. Agents Chemother. 44:3344-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ince, D., and D. C. Hooper. 2001. Mechanisms and frequency of resistance to gatifloxacin in comparison to AM-1121 and ciprofloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2002. Dual targeting of DNA gyrase and topoisomerase IV. Target interactions of garenoxacin (BMS-284756, T-3811ME), a new desfluoroquinolone. Antimicrob. Agents Chemother. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janoir, C., E. Varon, M. D. Kitzis, and L. Gutmann. 2001. New mutation in ParE in a pneumococcal in vitro mutant resistant to fluoroquinolones. Antimicrob. Agents Chemother. 45:952-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, R. N., M. A. Pfaller, and M. E. Erwin. 2000. Evaluation of gemifloxacin (SB-265805, LB20304a): in vitro activity against over 6000 gram-positive pathogens from diverse geographic areas. International J. Antimicrob. Agents 15:227-230. [DOI] [PubMed] [Google Scholar]
- 29.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King, A., J. May, G. French, and I. Phillips. 2000. Comparative in vitro activity of gemifloxacin. J. Antimicrob. Chemother. 45:1-12. [DOI] [PubMed] [Google Scholar]
- 31.Lopez, H., D. Stepanik, V. Vilches, S. Scarano, B. Sarachian, G. Mikaelian, J. Finlay, and A. Sucari. 2001. Comparative in vitro activity of gemifloxacin against gram-positive and gram-negative clinical isolates in Argentina. Diag. Microbiol. Infect. Dis. 40:187-192. [DOI] [PubMed] [Google Scholar]
- 32.McCloskey, L., T. Moore, N. Niconovich, B. Donald, J. Broskey, C. Jakielaszek, S. Rittenhouse, and K. Coleman. 2000. In vitro activity of gemifloxacin against a broad range of recent clinical isolates from the USA. J. Antimicrob. Chemother. 45:13-21. [DOI] [PubMed] [Google Scholar]
- 33.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 34.Morris, J. E., X. S. Pan, and L. M. Fisher. 2002. Grepafloxacin, a dimethyl derivative of ciprofloxacin, acts preferentially through gyrase in Streptococcus pneumoniae: role of the C-5 group in target specificity. Antimicrob. Agents Chemother. 46:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1994. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob. Agents Chemother. 38:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship of the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 38.Pan, X. S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan, X. S., G. Yague, and L. M. Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson, L. R. 2001. Quinolone molecular structure-activity relationships: what we have learned about improving antimicrobial activity. Clin. Infect. Dis. 33:S180-S186. [DOI] [PubMed] [Google Scholar]
- 41.Roychoudhury, S., C. E. Catrenich, E. J. McIntosh, H. D. McKeever, K. M. Makin, P. M. Koenigs, and B. Ledoussal. 2001. Quinolone resistance in staphylococci: activities of new nonfluorinated quinolones against molecular targets in whole cells and clinical isolates. Antimicrob. Agents Chemother. 45:1115-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roychoudhury, S., T. L. Twinem, K. M. Makin, M. A. Nienaber, C. Y. Li, T. W. Morris, B. Ledoussal, and C. E. Catrenich. 2001. Staphylococcus aureus mutants isolated via exposure to nonfluorinated quinolones: detection of known and unique mutations. Antimicrob. Agents Chemother. 45:3422-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sau, S., J. W. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulte, A., and P. Heisig. 2000. In vitro activity of gemifloxacin and five other fluoroquinolones against defined isogenic mutants of Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 46:1037-1038. [DOI] [PubMed] [Google Scholar]
- 45.Stahl, M. L., and P. A. Pattee. 1983. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J. Bacteriol. 154:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takei, M., H. Fukuda, R. Kishii, and M. Hosaka. 2001. Target preference of 15 quinolones against Staphylococcus aureus, based on antibacterial activities and target inhibition. Antimicrob. Agents Chemother. 45:3544-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trucksis, M., J. S. Wolfson, and D. C. Hooper. 1991. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J. Bacteriol. 173:5854-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weigel, L. M., G. J. Anderson, R. R. Facklam, and F. Tenover. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:3517-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willmott, C. J., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yague, G., J. E. Morris, X. S. Pan, K. A. Gould, and L. M. Fisher. 2002. Cleavable-complex formation by wild-type and quinolone-resistant Streptococcus pneumoniae type II topoisomerases mediated by gemifloxacin and other fluoroquinolones. Antimicrob. Agents Chemother. 46:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamagishi, J. I., T. Kojima, Y. Oyamada, K. Fujimoto, H. Hattori, S. Nakamura, and M. Inoue. 1996. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, J.-L., L. L. Grinius, and D. C. Hooper. 2002. NorA functions as a multidrug transporter in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. J. Bacteriol. 184:1370-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]