INTRODUCTION
The flowering plants comprise some 250,000 species and are tremendously diverse in growth habit, environmental adaptation, and nuclear genome structure. Plant genomes tend to be large and complex, varying in size from ∼38 Mb (1C) for the crucifer Cardamine amara to >87,000 Mb for Fritillaria assyriaca, a member of the Lilliaceae (Flavell et al., 1974; Bennett and Leitch, 1995). Despite this diversity, plant geneticists have recently found that plants exhibit extensive conservation of both gene content and gene order (Bennetzen and Freeling, 1993). Moreover, comparative genetic analyses have begun to show that different plant species often use homologous genes for very similar functions (Fatokun et al., 1992; Ahn et al., 1993; Paterson et al., 1995; Lagercrantz et al., 1996).
The advent of DNA marker technology not only facilitated the rapid generation of detailed plant genetic maps but also allowed map comparisons when common DNA markers were employed. Initial comparisons of the chromosome sets within polyploid species (Helentjaris et al., 1988; Chao et al., 1989) and of diploid genomes between closely related species (Bonierbale et al., 1988; Hulbert et al., 1990) indicated extensive colinearity of genetic maps. These comparisons have been greatly extended, particularly in the grasses (reviewed in Gale and Devos, 1998), revealing significant conservation of gene content and order across plant species that diverged from a common ancestor <50 million years ago (Crepet and Feldman, 1991). These comparative studies show that, within the limits of sequence divergence (i.e., evolutionary time) that permit cross-hybridization, the large majority (>90%) of plant genes have close homologs within most other plant genomes. Gene order is somewhat more variable, however. Comparative genetic maps based on shared restriction fragment length polymorphism probes indicate many large chromosomal rearrangements, some arising specifically during the origin of a particular family of plants (Moore et al., 1995; Gale and Devos, 1998). These large rearrangements are most commonly observed, as one might expect, between distantly related species (Paterson et al., 1996) but can also occur in a relatively short period of time within some lineages (Devos et al., 1993; Zhang et al., 1998).
However, there are problems in the interpretation and use of comparative genetic maps based on recombinational mapping of DNA markers. The two chief difficulties arise from marker density and sequence orthology. Because only a subset of DNA probes hybridize efficiently and reveal polymorphism within mapping populations of two different plant species, most genetic map comparisons employ 200 or fewer shared markers. Hence, marker density averages <1 per 10 centimorgans, guaranteeing that small exceptions to colinearity (e.g., inversions of less than a few centimorgans) will not be observed. Even more problematic is the fact that most genes are represented as multiple homologs within a given plant genome, which makes it difficult to determine whether truly orthologous loci are being compared. Orthologous loci are those that show common vertical descent from an ancestral gene, whereas paralogous genes are derived from amplification of the original ancestral gene, an event that may predate or postdate the independent derivation of any two compared species.
In many cases, a restriction fragment length polymorphism probe will hybridize to several gel blot bands from each of two species, but only one may be polymorphic and mapped in each species. If they appear to map in colinear positions, then the researcher often assumes that they are orthologous. However, if they map on nonsyntenic linkage groups, the researcher commonly assumes that paralogs have been mapped. Although this circular reasoning often is correct, it can lead to a bias toward promoting colinear cases and ignoring exceptions to colinearity. Further confusing this issue is the fact that many paralogs (derived from tandem duplications) are commonly found within a few centimorgans of each other, thus making mere synteny a less-than-perfect argument for orthology. All of these problems are intensified by comparative maps that rely on small numbers of analyzed progeny. If the accuracy of placement of a particular marker is only within 5 to 10 centimorgans, then it is difficult to determine whether an apparent local exception to colinearity is really due to a small chromosomal rearrangement or to imprecise map positions for the markers involved.
Partly because of the limitations of genetic maps based on DNA markers, several laboratories have begun to investigate and compare the structure of small regions of plant genomes by DNA sequence analysis. These studies will provide our first views into the detailed composition and organization of numerous plant genomes. Moreover, such comparative sequence analyses will help determine how and at what rates plant genomes change. Although many studies have focused on the timing and nature of gene evolution in plants (reviewed in Doyle and Gaut, 2000), a lack of genomic sequencing capability has meant that these investigations primarily target gene sequences themselves, without any reference to the different contexts in which these genes are located. The remainder of this article discusses recent findings that have begun to uncover the relationship between plant gene and genome evolution in the nucleus.
PLANT GENOME SEQUENCES
Over the last several years, a large-scale effort has been under way to sequence the genome of a model plant, Arabidopsis. Arabidopsis was chosen for this ambitious effort because of its exceptionally small genome (∼130 Mb) (Arumuganathan and Earle, 1991), its powerful array of genetic tools, and its outstanding research community (Meinke et al., 1998). Currently, ∼80% of the Arabidopsis genome is completely sequenced, with the rest due to be finished this year, 4 years ahead of schedule. Analysis of the Arabidopsis genome sequence has shown that gene density throughout most of the genome is very high, approximately one gene per 4 to 5 kb, suggesting a total gene content of 25,000 to 30,000 (Kaneko et al., 1999; Lin et al., 1999). Repetitive DNAs are relatively rare, comprising ∼10% of the Arabidopsis genome (Leutweiler et al., 1984). Many of these repeats are transposable elements, found as single copies interspersed between genes (Lin et al., 1999). Most of the repeats, however, are sequestered within centromeric regions, pericentromeric heterochromatin, and knobs. These repeat-rich, gene-poor chromosome segments contain tandem satellite repeats, some apparently involved in centromere function, as well as large quantities of transposable elements (Copenhaver et al., 1999; Fransz et al., 2000; McCombie et al., 2000).
Plants other than Arabidopsis have received much less attention at the level of DNA sequence analysis. Several large segments of the rice (Oryza sativa) genome have now been subjected to sequence analysis, revealing at least one gene per 10 kb, with scattered transposable elements making up 25% or less of the total DNA (Chen and Bennetzen, 1996; Han et al., 1999; Tarchini et al., 2000). Two relatively large segments of the maize (Zea mays) genome have been sequenced, a 225-kb segment around the adh1 gene (SanMiguel et al., 1996; Tikhonov et al., 1999) and a 78-kb segment that contains a 22-kD zein gene cluster (Llaca and Messing, 1998). In both maize segments, genic regions were interspersed with large blocks of repetitive DNA. The repetitive DNA was found to be composed mainly of retrotransposons with long terminal repeats (LTRs), of which most were found to be inserted into each other (SanMiguel et al., 1996). The LTR retrotransposon blocks are highly methylated and presumably heterochromatic in most or all adult tissues, and range in size from a few kilobases up to ∼200 kb (Springer, 1992; Bennetzen et al., 1994). Overall, these LTR retrotransposons are estimated to comprise 50 to 80% of the DNA in the maize nucleus (SanMiguel and Bennetzen, 1998).
Small, multigene segments of a few other plant species have also been sequenced, including a 60-kb region around the barley mlo locus (Panstruga et al., 1998), a 23-kb region on wheat chromosome 1 (Feuillet and Keller, 1999), a 78-kb segment flanking a sorghum adh gene (Tikhonov et al., 1999), and a 26-kb region of Capsella rubella, a close relative of Arabidopsis (A. Acarkan, M. Rossberg, M. Koch, and R. Schmidt, personal communication). These regions all exhibit high gene densities, with only the occasional single transposable element. Thus, most of the plant nuclear genomes that have been subjected to sequence analysis have a gene and repetitive DNA composition like that of the genic regions of Arabidopsis, whereas maize appears to be exceptional in having genic regions that look more like the pericentromeric heterochromatin of Arabidopsis or the β heterochromatin of Drosophila melanogaster (Holmquist et al., 1998; McCombie et al., 2000). Given that so few plant genomes have been investigated, it is not clear whether the structure and composition of maize genic regions are due to exceptional mechanisms of genome alteration or whether they might be due to the relatively short period of time since most of these LTR retroelements have amplified, a mere 2 to 6 million years (SanMiguel et al., 1998). It is possible that many increases in plant genome size are due to transposable element amplification, perhaps in very narrow temporal windows, while a slow but steady process of genome size reduction counteracts this trend (Petrov et al., 1996; Bennetzen and Kellogg, 1997; SanMiguel and Bennetzen, 1998).
GENOMIC AND SEGMENTAL DUPLICATIONS
One universal observation of all fine-scaled analyses of eukaryotic genomes has been the great number of duplicated genes. In some cases, these duplications may be due to ancient polyploidizations that were subsequently obscured by variation in the pairing, segregation, or structure of the different chromosome sets within a single nucleus. For instance, even the simple yeast genome appears to be derived from a primordial tetraploid (Wolfe and Shields, 1997), and ancient duplications, including polyploidy, have been well documented in the evolution of vertebrates (Pebusque et al., 1998). It should not be surprising that plants, given their wide sexual promiscuity and potential for vegetative reproduction, are particularly prone to polyploidization. Many current “diploid” species, like maize and those in the Brassicaceae (Helentjaris et al., 1988; Lagercrantz, 1998), have an ancient polyploid origin, whereas many other plant species currently behave as true polyploids (reviewed in Wendel, 2000).
Detailed mapping and sequence analyses have now shown that even simple plant genomes contain tremendous amounts of segmental duplication. Nearly saturated genetic maps of rice, Arabidopsis, and other species have revealed that whole chromosome arms and smaller segments are duplicated (Kishimoto et al., 1994; Nagamura et al., 1995). By sequence criteria, the genome of Arabidopsis appears to be at least 60% duplicated, suggesting a possible tetraploid history (Blanc et al., 2000). Previous studies had also shown that within the already ancient tetraploid genome of maize, frequent “distantly tandem” duplications serve to complicate genetic mapping studies (Sanz-Alferez et al., 1995). Hence, these regional or genomic duplications can create tremendous opportunities for novel analyses and, conversely, for confusion as to what is actually being analyzed. For instance, sequence comparisons can be conducted between homologous regions within a species' chromosome complement, indicating the different types and rates of change that can occur to comparable segments within a single nucleus. Yet, in comparisons between species, it may be consistently unclear whether an orthologous or paralogous comparison is being made.
GENOMIC SEQUENCE COMPARISONS
Analysis of sequences derived from two different plant species becomes more and more tenuous as the degree of relatedness decreases. For instance, sequence comparisons between Arabidopsis and any of the grasses have shown that few colinear segments can be found, even at the level of adjacent genes (Tikhonov et al., 1999; van Dodeweerd et al., 1999; K. Devos, personal communication; W. Ramakrishna and J.L. Bennetzen, unpublished observations). However, these sequences might have diverged so greatly that the question of orthology is almost impossible to resolve. On the other hand, comparisons between very closely related species, or populations within a species, can only reveal changes that have been fixed within the last few thousands to millions of years.
One 29-kb segment of the rice genome, containing genes orthologous to the a1 and sh2 loci of maize, has been sequenced and analyzed on clones from the japonica and indica races (Chen, 1998; M. Chen, J. Lucas, and J.L. Bennetzen, unpublished observations). The two orthologous regions appear to have evolved independently for ∼1 million years with conservation in order and orientation of all genes. Intragenic and intergenic regions exhibited very different rates and types of sequence divergence, however. Chen found that exons and introns evolved most slowly (except those introns that contained a simple sequence repeat), whereas most intergenic DNA diverged approximately threefold more rapidly. However, miniature inverted repeat transposable elements (MITEs) (Wessler et al., 1995) within the region diverged approximately threefold faster than did the rest of the intergenic DNA (Chen, 1998; M. Chen, J. Lucas, and J.L. Bennetzen, unpublished observations). In fact, the MITEs seemed to be changing in sequence so rapidly, both through single base-pair alterations and deletions, that they should become unrecognizable within a few million years. This may explain why no two MITEs have been seen at a conserved location in sequence comparisons of orthologous regions of the genomes of plant species, like maize and sorghum (Tikhonov et al., 1999), that have diverged for >15 million years (Gaut and Doebley, 1997).
Studies of gene sequence polymorphism suggest that C. rubella and Arabidopsis have evolved independently for ∼6 to 10 million years and that they diverged from the Brassicaceae ∼12 to 20 million years ago (A. Acarkan, M. Rossberg, M. Koch, and R. Schmidt, personal communication). In a 26-kb region, the same five genes were present and in the same order in the two species. However, one of the genes had undergone a tandem duplication in C. rubella after its divergence from Arabidopsis. In this time frame, no conservation was seen for intergenic spaces, but intron number and location were highly conserved. The coding sequences within exons were most conserved, but orthologous introns exhibited short stretches of conserved sequence (A. Acarkan, M. Rossberg, M. Koch, and R. Schmidt, personal communication). Further studies may indicate whether these short conserved segments are related to any functional component of an intron.
Sequence analyses of sorghum, maize, and rice regions represent the most distant comparisons of clearly orthologous regions yet performed in plants. Maize and sorghum appear to have diverged ∼15 to 20 million years ago, whereas their common ancestor diverged from a shared ancestor with rice ∼50 million years ago (Gaut and Doebley, 1997). Not surprisingly, intergenic regions reveal no identified sequence conservation among these species. In particular, none of the mobile DNAs present at a location in one species can be found in the orthologous location in another species that is this distantly related (Chen et al., 1998; Tikhonov et al., 1999). However, in a comparison of 29 kb surrounding the sh2/a1-orthologous regions of rice and sorghum, Chen and coworkers found that all of the genes were conserved in both order and orientation. In this region, one putative transcription factor was found to differ by a particular exon, suggesting that these otherwise conserved genes might have diverged in function (Chen et al., 1998).
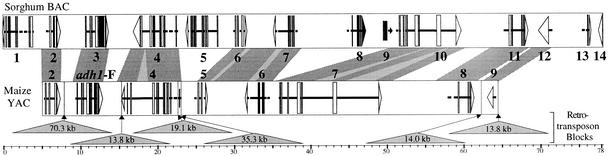
In stark contrast to the similarities observed in the sh2/a1-orthologous regions of maize, sorghum, and rice, adh-orthologous regions have undergone significant gene rearrangements (Tikhonov et al., 1999; Tarchini et al., 2000). In a comparison of maize and sorghum, Tikhonov and colleagues found that nine genes were shared in colinear order, but the maize region contained two apparent deletions that had removed three genes (Tikhonov et al., 1999) (Figure 1). The rice adh1 region exhibited no colinearity with the maize or sorghum adh1 region beyond the adh1 gene itself (Tikhonov et al., 1999; Tarchini et al., 2000), suggesting that adh1 had moved to its current nonsyntenic location in rice as a single-gene translocation.
Figure 1.
Comparison of adh-Orthologous Regions of Sorghum and Maize.
Numbers above the bars indicate genes that are homologous to cDNAs or genomic genes from plants and other species. The arrows within the bars show genes, including exons (thick) and introns (thin), and their apparent transcriptional orientation. The shading between the two bars indicates regions with extensive sequence identity. The lower bar has been greatly shortened (∼80%) by removal of the numerous blocks of nested LTR retrotransposons (SanMiguel et al., 1996, 1998) represented by triangles below the bar. This figure is based on data and analyses generated by Tikhonov et al. (1999). BAC, bacterial artificial chromosome; YAC, yeast artificial chromosome.
More distant comparisons between plant genomes, crossing the divide between monocotyledonous and dicotyledonous plants, have not yielded many solid conclusions. Genetic mapping studies suggest that some colinear regions might exist between Arabidopsis and the grasses (Paterson et al., 1996), but a more detailed study comparing the rice and Arabidopsis genomes does not offer evidence for significant degrees of conserved colinearity (Devos et al., 1999). Limited genome sequence comparisons also have not revealed any consistent genetic colinearity across the monocot/dicot divergence (Tikhonov et al., 1999; K. Devos, personal communication; W. Ramakrishna and J.L. Bennetzen, unpublished observations). However, all of these studies may be complicated by issues of orthology and paralogy. Perhaps only sophisticated statistical analyses of the completed sequences of a monocot and a dicot genome will allow resolution of this issue.
PATTERNS AND VARIATIONS IN PLANT GENOME ORGANIZATION
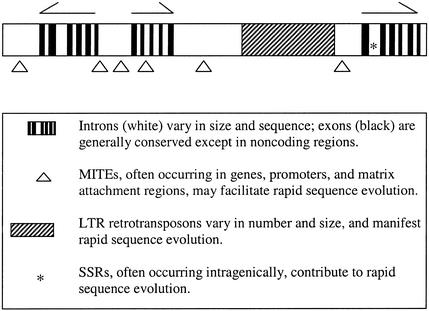
Despite their great differences in both size and nongenic composition, plant genomes exhibit some consistent organizational patterns. Ubiquitous but exceptional structures such as nucleolar organizers, centromeres (Jiang et al., 1996; Copenhaver et al., 1999), knobs (Ananiev et al., 1998; Fransz et al., 2000; McCombie et al., 2000), and telomeres may have very similar organizations in very different plant species. Genic regions also exhibit some routine patterns, as depicted in Figure 2.
Figure 2.
A Model for the Genic Regions of Plant Genomes.
Arrows above the bar indicate genes (with exons represented by black vertical lines below each gene) and their directions of transcription. The cross-hatched region represents an LTR retrotransposon block. The frequency and sizes of these blocks appear to be quite variable between genomes and perhaps between regions of the same genome. Other features of the model are described in the key. A simple sequence repeat (SSR) is represented by an asterisk.
In all sequence analyses to date, plant genes have been found to be rather compact, with small introns. It is not unusual for two genes, transcribed in opposite orientations, to share an upstream region of <1 kb. In most cases, genes do not appear to be clustered by function or tissue/timing of expression, except in the case of tandemly duplicated loci. Aside from the genes, numerous classes of transposable elements are found in genic regions. Some of these classes, like MITEs and some retrotransposons, are found preferentially near or within genes (Cresse et al., 1995; Hirochika et al., 1996; Tikhonov et al., 1999). Others, like most highly repetitive LTR retrotransposons, appear to associate preferentially with other repetitive DNAs (Pelissier et al., 1996; SanMiguel et al., 1996; Ananiev et al., 1998; McCombie et al., 2000).
We have no solid understanding of the reasons for the differences in genome composition or organization among plants. It is clear that the enormous size variation is largely an outcome of polyploidy and/or transposable element amplification, but it is not clear why some species seem to have undergone these processes more often than others. This may be purely a function of chance, or it might be that some species benefit from or are less resistant to genome size increases (Bennetzen and Kellogg, 1997). It is interesting, however, that all comparative investigations to date have revealed this great variation in genome size to be unassociated with any initial effect on genic colinearity in orthologous regions.
THE EPISODIC NATURE OF MICROCOLINEARITY: MANY SMALL REARRANGEMENTS
Comparative studies of orthologous regions have revealed that microcolinearity is often observed even between distantly related grass species. As might be expected, colinearity is most likely between closely related species. For instance, colinearity is greater between the adh-orthologous regions of maize and sorghum than it is between those regions from either of these species and rice (Tikhonov et al., 1999). However, there are some significant exceptions to this rule. In genetic map comparisons within the Triticeae, Devos and coworkers have shown that wheat and barley exhibit greater colinearity than wheat does with rye, despite the fact that rye is a closer relative (Devos et al., 1993). Thus, some lineages of plants appear to have more active processes of rearrangement than do others.
Some analyses of the frequency of large chromosomal rearrangements have argued for a consistent rate for the generation and fixation of such events during the evolution of the grasses (Paterson et al., 1996), albeit with some dramatic exceptions (Devos et al., 1993, 2000; Zhang et al., 1998). However, it is not at all clear whether small rearrangements might occur at comparable or proportional rates. That is, comparisons of the structures of large clones of orthologous regions (Dunford et al., 1995; Kilian et al., 1997) and of genomic sequences (Chen et al., 1998; Tikhonov et al., 1999) nearly always reveal small rearrangements, and tandem duplications or their reciprocal deletions appear to be particularly common. Thus, it appears that small rearrangements are orders of magnitude more frequent than are large chromosomal rearrangements. It is not clear whether small local events and large chromosomal rearrangements would be generated by the same mechanisms. Whether or not there are different mechanisms, it is entirely plausible that an organism undergoing a high rate of small rearrangements might not undergo a proportionally high rate of large rearrangements or vice versa. It is certain that we do not know nearly enough about the modes or periodicity of genome rearrangement, at any level, in any family of species.
PLANT GENOME REARRANGEMENT: RATES, CONSTRAINTS, AND OUTCOMES
Very few plant genomes have been studied at the DNA sequence level, and these only in a few regions. Hence, it is hazardous to make general conclusions about how and why genomes have assumed these forms. However, a few patterns have emerged early in the study of plant genome structure and evolution.
Plant Genomes Can Change Rapidly
Studies of chromosome number and genome size in closely related plant species have long suggested drastic changes in genome structure within a short evolutionary time span. Comparative sequence analyses of genes and of repetitive DNAs have shown that these evolve at different rates, with the genes being more highly conserved (Chen, 1998; SanMiguel et al., 1998). A few genes, like some disease resistance loci, are selected for rapid change (reviewed in Michelmore and Meyers, 1998) and thus may evolve more rapidly than do some repeats. However, the epigenetic inactivation of many repeats and the mutagenic effects of their transposition apparently cause most of these sequences to change even more rapidly than do genes under selective pressures.
Are There Restraints on Plant Genome Content or Arrangement?
The tremendous variation in genome size and repetitive DNA content (Flavell et al., 1974) implies that neither of these factors has much bearing on plant fitness. Alternatively, the subtle changes associated with differences in genome size and repetitive DNA content might offer a selective advantage under some circumstances (Sparrow and Miksche, 1961; Jasienski and Bazzaz, 1995). McClintock proposed that the presence of a great array of normally quiescent transposable elements would be useful under dire circumstances of “genomic shock” in which their activation, along with the subsequent massive genome rearrangement, would be of value in producing a new plant that could survive a severe new environment (McClintock, 1984). However, it has been argued that it is difficult to see how such a genomic shock mechanism could be maintained without being used for thousands of generations and that it is unlikely that a huge number of rearrangements could ever yield a surviving individual even in the absence of a severe environment (Bennetzen, 2000). Moreover, closely related plant species do show huge variations in genome size, but not in gene content, structure, or order, despite the fact that McClintock's model suggested that changes in gene content, structure, and order would be the necessary positive outcome of the genomic shock phenomenon. In short, it seems simpler to propose that the mobile DNAs and tandem repeats that make up most of the repetitive DNA in genomes are selfish or parasitic entities that survive primarily because they have no drastic detrimental effects upon their hosts (Doolittle and Sapienza, 1980; Orgel and Crick, 1980).
Some types of genome rearrangement, including large deletions and heterozygous inversions and translocations, have been amply demonstrated to be detrimental. Each of these types of rearrangement can greatly reduce gametic and/or organismal viability. Hence, natural selection will work toward the removal of these events at a rate that is proportional to their deleterious nature.
One expects the gene content of the plant genome to be under strong selective pressure. By definition, the homozygous deletion of any essential gene will be lethal in a diploid, and the loss of important but nonessential loci could also greatly decrease fitness. Increases in gene number might also impair plant fitness, particularly if these increases are segmental, thereby altering genic balance. Polyploidy allows an increase in gene number without a drastic alteration in genic balance (with the exception of those differences, usually in regulatory genes, that might differentiate the two chromosome sets in an allopolyploid).
In an incipient polyploid, all genes are present in multiple copies, so that one copy of an essential gene could drift into a new form/function without total loss of the original gene function. Alternatively, extraneous gene copies may be either selectively lost if the genetic function is incompatible with the polyploid state or selectively retained if the genetic function is advantageous to the new polyploid. Significantly, the loss of an extra copy of a gene may be of negligible effect if it occurs before the functional divergence of homeologous copies.
In hexaploid wheat, a polyploid generated within the last 10,000 years, each of the A, B, and D genomes appears to carry one copy of an orthologous locus (Chao et al., 1989). Hence, with a few exceptions (Devos et al., 1995), the A, B, and D genomes within hexaploid wheat have not undergone extensive gene loss or rearrangement since polyploid formation. In more ancient polyploids, like maize, many differences between the homeologous chromosome sets have begun to emerge. Some of these involve major rearrangements (Moore et al., 1995), but most appear to be associated with small local events. Tikhonov et al. (1999) determined that three genes located in the sorghum adh region were absent from the orthologous maize region. However, by hybridization analysis, these three genes were found to be present elsewhere in the maize genome. It will be particularly interesting to discover if the three genes are linked and/or colinear within the maize genome.
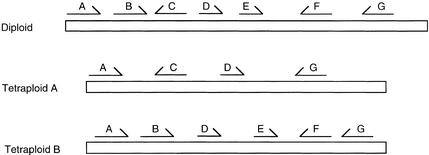
Figure 3 depicts a possible model for the structure of the orthologous regions of a diploid genome and the tetraploid genome of a related species. Because there are extra copies of each gene in the tetraploid, there is greater tolerance for loss or divergence of each of these genes to another function. In the example shown, all genes are present and all are colinear in the tetraploid, but in reality one would have to investigate both orthologous segments from the tetraploid to see such conservation.
Figure 3.
A Model for Orthologous Gene Relationships in a Diploid and Its Tetraploid Relative.
Genes are indicated by arrows, and ortholog representations share common letters. In this model, all of the ancestral genes are present in the diploid region depicted, but three genes (two apparent events) have been deleted in generation of the tetraploid A genome and one gene from the tetraploid B genome. Note that copies of all of the genes are retained in the degenerating tetraploids, but some genes (e.g., B, C, E, and F) are now present only as diploid loci.
Is There Selection for or against Gene Order?
The apparent lack of microcolinearity between the genomes of Arabidopsis and the grasses (Bennetzen et al., 1998; Devos et al., 1999; Tikhonov et al., 1999) is a rather amazing observation that suggests actual selection against colinearity. If many grass genomes have retained extensive colinearity over 50 million years or more, then why should colinearity be completely lost over a time frame that is only a few-fold greater (Crepet and Feldman, 1991)? It is difficult to imagine how selection against colinearity could be manifested on a global, full-genome scale. Perhaps it is simpler to postulate that a very high level of genome rearrangement took place in dicots very early after their divergence from monocots, resulting in a low level of colinearity (van Dodeweerd et al., 1999). Alternatively, high rates of rearrangement may have occurred specifically in the ancestors of Arabidopsis and its close relatives. Further DNA sequence comparisons between Arabidopsis and other dicots should distinguish between these two possibilities (Grant et al., 2000).
It is possible to imagine selection either for or against tight linkages among certain genes. In the case of some plant disease resistance genes, for instance, tight linkage can allow rapid evolution of new specificities by a process of unequal recombination (Sudupak et al., 1993; reviewed in Richter and Ronald, 2000). However, this same process of frequent unequal recombination guarantees the rapid, random removal of even the best alleles, thus selecting for movement of very successful resistance alleles to new locations that lack adjacent resistance genes of the same type. This may account for the fact that many disease resistance genes do not appear in orthologous locations among grass species (Leister et al., 1998). However, the linkage of genes that have coevolved a particular specificity would be favored, thereby providing a haplotype that is inherited as a single segment.
What Are the Possible Uses of Plant Genome Colinearity?
Although more data are desperately needed, we can now conclude that genomic microcolinearity in plants will be a useful tool for plant gene identification and study. The most obvious use for genomic microcolinearity is in the map-based isolation of genes by chromosome walking within small colinear genomes of representative species (Bennetzen and Freeling, 1993). However, the effectiveness of this approach relies not on overall genome colinearity but rather on local microcolinearity at sites where reference genes are located. Largely because of the high frequency of small rearrangements between distantly related grass species, this approach has yet to prove successful. However, several projects are very close to succeeding, and even instances of interrupted microcolinearity can provide a tremendous number of DNA probes tightly linked to targeted loci (Kilian et al., 1997).
Comparative sequence analysis can provide a useful tool for sequence annotation. In comparisons of genomes that have diverged over 5 million years or more, it appears that only the genes are extensively conserved. Hence, any sequences that are shared between two orthologous regions are likely to be genes (Avramova et al., 1996; Tikhonov et al., 1999), even when they are not identified by a gene-finding program or by homology to any other known gene or cDNA.
For the long term, the most interesting and valuable use of comparative sequencing and mapping will be to determine the nature of the evolved functions that make one species different from another (Bennetzen and Freeling, 1997). Because colinear map positions allow a solid determination of orthology, the investigator can associate changes in the sequence and functional properties of a locus with the different outcomes of mutation and selection of a particular orthologous gene in two or more different species. When compared across the entire plant kingdom, the potential properties of a single orthologous gene can be fully determined. This will reveal not only how a gene can change but also what functions it may attain—central questions in understanding the nature of evolution and the orchestrated function of all of the genes in a genome.
CONCLUSIONS
Comparative sequence analyses have begun to uncover the generalities and peculiarities of plant genome structure. Several patterns are beginning to emerge regarding the properties of repetitive DNAs, including their associations with genes and with heterochromatin.
Microcolinearity is apparent in comparisons of many plant genomes, but there are also many small exceptions. Small rearrangements, including frequent insertions of transposable elements and duplications or deletions of genes, occur without significantly rearranging most adjacent sequences. The timing of these events may be punctuated (SanMiguel et al., 1998), perhaps related to the environmental or ploidy status of the plant. Small exceptions to colinearity are much more frequent than are the large rearrangements detected by traditional cytogenetics and low-resolution genetic maps. Plant genome microcolinearity can be exploited by plant geneticists and genetic engineers. Indeed, the most valuable information to be gleaned from microcolinearity will likely be the definition of orthology that will allow for clear correlation between the structural nature and functional outcome of plant genome evolution.
References
- Ahn, S., Anderson, J.A., Sorrells, M.E., and Tanksley, S.D. (1993). Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241, 483–490. [DOI] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Complex structure of knob DNA on maize chromosome 9: Retrotransposon invasion into heterochromatin. Genetics 149, 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. [Google Scholar]
- Avramova, Z., Tikhonov, A., SanMiguel, P., Jin, Y.-K., Liu, C., Woo, S.-S., Wing, R.A., and Bennetzen, J.L. (1996). Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 10, 1163–1168. [DOI] [PubMed] [Google Scholar]
- Bennett, M.D., and Leitch, I.J. (1995). Nuclear DNA amounts in angiosperms. Ann. Bot. 76, 113–176. [Google Scholar]
- Bennetzen, J.L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42, 251–269. [PubMed] [Google Scholar]
- Bennetzen, J.L., and Freeling, M. (1993). Grasses as a single genetic system: Genome composition, collinearity and compatibility. Trends Genet. 9, 259–261. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Freeling, M. (1997). The unified grass genome: Synergy in synteny. Genome Res. 7, 301–307. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Kellogg, E.A. (1997). Do plants have a one-way ticket to genomic obesity? Plant Cell 9, 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L., SanMiguel, P., Chen, M., Tikhonov, A., Francki, M., and Arromova, Z. (1998) Grass genomes. Proc. Natl. Acad. Sci. USA 95, 1975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L., Schrick, K., Springer, P.S., Brown, W.E., and SanMiguel, P. (1994). Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37, 565–576. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis thaliana genome. Plant Cell 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale, M.D., Plaisted, R.L., and Tanksley, S.D. (1988). RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, S., Sharp, P.J., Worland, A.J., Warham, E.J., Koebner, R.M.D., and Gale, M.D. (1989). RFLP-based genetic maps of wheat homoeologous group 7 chromosomes. Theor. Appl. Genet. 78, 495–504. [DOI] [PubMed] [Google Scholar]
- Chen, M. (1998). The Structure of sh2/a1-Homologous Regions in Maize, Rice and Sorghum. Ph.D. Dissertation (West Lafayette, IN: Purdue University).
- Chen, M., and Bennetzen, J.L. (1996). Sequence composition and organization in the Sh2/A1-homologous region of rice. Plant Mol. Biol. 32, 999–1001. [DOI] [PubMed] [Google Scholar]
- Chen, M., SanMiguel, P., and Bennetzen, J.L. (1998). Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics 148, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G.P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- Crepet, W.L., and Feldman, G.D. (1991). The earliest remains of grasses in the fossil record. J. Bot. 78, 1010–1014. [Google Scholar]
- Cresse, A.D., Hulbert, S.H., Brown, W.E., Lucas, J.R., and Bennetzen, J.L. (1995). Mu1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics 140, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M., Atkinson, M.D., Chinoy, C.N., Harcourt, R.L., Koebner, R.M.D., Liu, C.J., Masojc, P., Xie, D.X., and Gale, M.D. (1993). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85, 673–680. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Dubcovsky, J., Dvorak, J., Chinoy, C.N., and Gale, M.D. (1995). Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor. Appl. Genet. 91, 282–288. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Beales, J., Nagamura, Y., and Sasaki, T. (1999). Arabidopsis–Rice: Will colinearity allow gene prediction across the eudicot–monocot divide? Genome Res. 9, 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M., Pittaway, T.S., Reynolds, A., and Gale, M.D. (2000). Comparative mapping reveals a complex relationship between the pearl millet genome and those of foxtail millet and rice. Theor. Appl. Genet. 100, 190–198. [Google Scholar]
- Doolittle, W.F., and Sapienza, C. (1980). Selfish genes, the phenotype paradigm and genome evolution. Nature 284, 601–603. [DOI] [PubMed] [Google Scholar]
- Doyle, J.F., and Gaut, B.F. (2000). Plant Molecular Evolution. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Dunford, R.P., Kurata, N., Laurie, D.A., Money, T.A., Minobe, Y., and Moore, G. (1995). Conservation of fine-scale DNA marker order in the genomes of rice and the Triticeae. Nucleic Acids Res. 23, 2724–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun, C.A., Menacio, D.I., Danesh, D., and Young, N.D. (1992). Evidence for orthologous seed weight genes in cowpea and mungbean, based upon RFLP mapping. Genetics 132, 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., and Keller, B. (1999). High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96, 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell, R.B., Bennett, M.D., Smith, J.B., and Smith, D.B. (1974). Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 12, 257–269. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H.. (2000). Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100, 367–376. [DOI] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Plant comparative genetics after 10 years. Science 282, 656–659. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 88, 2060–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, D., Cregan, P., and Shoemaker, R.C. (2000). Genome organization in dicots. I. Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA, 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., Kilian, A., Chen, J.P., Kudrna, D., Steffenson, B., Yamamoto, K., Matsumoto, T., Sasaki, T., and Kleinhofs, A. (1999). Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome 42, 1071–1076. [DOI] [PubMed] [Google Scholar]
- Helentjaris, T., Weber, D.L., and Wright, S. (1988). Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist, G.P., Kapitonov, V.V., and Jurka, J. (1998). Mobile genetic elements, chiasmata, and the unique organization of beta-heterochromatin. Cytogenet. Cell Genet. 80, 113–116 [DOI] [PubMed] [Google Scholar]
- Hulbert, S.H., Richter, T.E., Axtell, J.D., and Bennetzen, J.L. (1990). Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc. Natl. Acad. Sci. USA 87, 4251–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasienski, M., and Bazzaz, F.A. (1995). Genome size and high CO2. Nature 376, 559–560. [Google Scholar]
- Jiang, J., Nasuda, S., Dong, F., Scherrer, C.W., Woo, S.-S., Wing, R.A., Gill, B.S., and Ward, D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, T., Katoh, T., Sato, S., Nakamura, Y., Asamizu, E., Kotani, H., Miyajima, N., and Tabata, S. (1999). Structural analysis of Arabidopsis thaliana chromosome 5. IX. Sequence features of the regions of 1,011,550 bp covered by seventeen P1 and TAC clones. DNA Res. 6, 183–195. [DOI] [PubMed] [Google Scholar]
- Kilian, A., Chen, J., Han, F., Steffenson, B., and Kleinhofs, A. (1997). Towards map-based cloning of the barley stem rust resistance genes Rpg1 and rpg4 using rice as an intergenomic cloning vehicle. Plant Mol. Biol. 35, 187–195. [PubMed] [Google Scholar]
- Kishimoto, N., Higo, H., Abe, K., Arai, S., Saito, A., and Higo, K. (1994). Identification of the duplicated segments in rice chromosomes 1 and 5 by linkage analysis of cDNA markers of known functions. Theor. Appl. Genet. 88, 722–726. [DOI] [PubMed] [Google Scholar]
- Lagercrantz, U. (1998). Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U., Putterill, J., Coupland, G., and Lydiate, D. (1996). Comparative mapping in Arabidopsis and Brassica, fine scale genome collinearity and congruence of genes controlling flowering time. Plant J. 9, 13–20. [DOI] [PubMed] [Google Scholar]
- Leister, D.M., Kurth, J., Laurie, D.A., Yano, M., Sasaki, T., Devos, K.M., Graner, A., and Schulze-Lefert, P. (1998). Rapid reorganisation of resistance gene homologues in cereal genomes. Proc. Natl. Acad. Sci. USA 95, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutweiler, L.S., Hough-Evans, B.R., and Meyerowitz, E.M. (1984). The DNA of Arabidopsis thaliana. Mol. Gen. Genet. 194, 15–23. [Google Scholar]
- Lin, X.Y., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768. [DOI] [PubMed] [Google Scholar]
- Llaca, V., and Messing, J. (1998). Amplicons of maize zein genes are conserved within genic but expanded and constricted in intergenic regions. Plant J. 15, 211–220. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. [DOI] [PubMed] [Google Scholar]
- McCombie, W.R., et al. (2000). The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100, 377–386. [PubMed] [Google Scholar]
- Meinke, D.W., Cherry, J.M., Dean, C., Rounsley, S.D., and Koorneef, M. (1998). Arabidopsis thaliana: A model plant for genome analysis. Science 282, 679–682. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Moore, G., Devos, K.M., Wang, Z., and Gale, M.D. (1995). Grasses, line up and form a circle. Curr. Biol. 5, 737–739. [DOI] [PubMed] [Google Scholar]
- Nagamura, Y., Inoue, T., Antonio, B.A., Shimano, T., Kajiya, H., Shomura, A., Lin, S.Y., Kuboki, Y., Harushima, Y., Kurata, N., Minobe, Y., Yano, M., and Sasaki, T. (1995). Conservation of duplicated segments between rice chromosomes 11 and 12. Breed. Sci. 45, 373–376. [Google Scholar]
- Orgel, L.E., and Crick, F.H.C. (1980). Selfish DNA: The ultimate parasite. Nature 284, 604–607. [DOI] [PubMed] [Google Scholar]
- Panstruga, R., Buschges, R., and Schulze-Lefert, P. (1998). A contiguous 60 kb genomic stretch from barley provides molecular evidence for gene islands in monocot genomes. Nucleic Acids Res. 26, 1056–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Lin, Y.-R., Li, Z., Schertz, K.F., Doebley, J.F., Pinson, S.R.M., Liu, S.-C., Stansel, J.W., and Irvine, J.E. (1995). Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269, 1714–1718. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., et al. (1996). Toward a unified genetic map of higher plants, transcending the monocot–dicot divergence. Nature Genet. 14, 380–382. [DOI] [PubMed] [Google Scholar]
- Pebusque, M.-J., Coulier, F., Birnbaum, D., and Pontarotti, P. (1998). Ancient large-scale genome duplications: Phylogenetic and linkage analyses shed light on chordate genome evolution. Mol. Biol. Evol. 15, 1145–1159. [DOI] [PubMed] [Google Scholar]
- Pelissier, T., Tutois, S., Tourmente, S., Deragon, J.M., and Pickard, G. (1996). DNA regions flanking the major Arabidopsis thaliana satellite are principally enriched in Athila retroelement sequences. Genetica 97, 141–151. [DOI] [PubMed] [Google Scholar]
- Petrov, D.A., Lozovskaya, E.R., and Hartl, D.L.. (1996). High intrinsic rate of DNA loss in Drosophila. Nature 384, 346–349. [DOI] [PubMed] [Google Scholar]
- Richter, T.E., and Ronald, P.C.. (2000). The evolution of disease resistance genes. Plant Mol. Biol. 42, 195–204. [PubMed] [Google Scholar]
- SanMiguel, P., and Bennetzen, J.L. (1998). Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82, 37–44. [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.-K., Motchoulskaia, N., Zakharov, D., Melake Berhan, A., Springer, P.S., Edwards, K.J., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nature Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- Sanz-Alferez, S., Richter, T.E., Hulbert, S.H., and Bennetzen, J.L. (1995). The Rp3 disease resistance gene of maize: Mapping and characterization of introgressed alleles. Theor. Appl. Genet. 91, 25–32. [DOI] [PubMed] [Google Scholar]
- Sparrow, A.H., and Miksche, J.P. (1961). Correlations of nuclear volume and DNA content with higher plant tolerance to chronic radiation. Science 134, 182–183. [DOI] [PubMed] [Google Scholar]
- Springer, P.S. (1992). Genomic Organization of Zea mays and Its Close Relatives. Ph.D. Dissertation (West Lafayette, IN: Purdue University).
- Sudupak, M.A., Bennetzen, J.L., and Hulbert, S.H. (1993). Unequal exchange and meiotic instability of the Rp1 region disease resistance genes in maize. Genetics 133, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dodeweerd, A.M., Hall, C.R., Bent, E.G., Johnson, S.J., Bevan, M.W., and Bancroft, I. (1999). Identification and analysis of homoeologous segments of the genomes of rice and Arabidopsis thaliana. Genome 42, 887–892. [PubMed] [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249. [PubMed] [Google Scholar]
- Wessler, S.R., Bureau, T.E., and White, S.E. (1995). LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5, 814–821. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., and Shields, D.C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Jia, J., Gale, M.D., and Devos, K.M. (1998). Relationship between the chromosomes of Aegilops umbellulata and wheat. Theor. Appl. Genet. 96, 69–75. [Google Scholar]