Abstract
Rhizobium nodulation (Nod) factors are lipo-chitooligosaccharides that act as symbiotic signals, eliciting several key developmental responses in the roots of legume hosts. Using nodulation-defective mutants of Medicago truncatula, we have started to dissect the genetic control of Nod factor transduction. Mutants in four genes (DMI1, DMI2, DMI3, and NSP) were pleiotropically affected in Nod factor responses, indicating that these genes are required for a Nod factor–activated signal transduction pathway that leads to symbiotic responses such as root hair deformations, expressions of nodulin genes, and cortical cell divisions. Mutant analysis also provides evidence that Nod factors have a dual effect on the growth of root hair: inhibition of endogenous (plant) tip growth, and elicitation of a novel tip growth dependent on (bacterial) Nod factors. dmi1, dmi2, and dmi3 mutants are also unable to establish a symbiotic association with endomycorrhizal fungi, indicating that there are at least three common steps to nodulation and endomycorrhization in M. truncatula and providing further evidence for a common signaling pathway between nodulation and mycorrhization.
INTRODUCTION
Symbiotic bacteria of the genera Rhizobium, Bradyrhizobium, Azorhizobium, and Sinorhizobium, collectively referred to as rhizobia, are able to elicit on their leguminous hosts the formation of specialized organs, called nodules, capable of fixation of atmospheric nitrogen. Among the earliest visible manifestations of symbiotic development are the infection of root hair cells by bacteria, contained in plant-derived infection structures called infection threads, and the induction of cortical divisions to form the nascent nodule primordium (Mylona et al., 1995; Schultze and Kondorosi, 1998).
Genetic analysis of rhizobia has led to identification of nod genes, which are involved in the control of host specificity, infection, and nodulation. nod gene expression is under the control of plant signals, essentially flavonoids, excreted in the rhizosphere. Once activated, nod genes specify the synthesis of nodulation (Nod) factors, which are lipo-chitooligosaccharides (Dénarié et al., 1996). Purified Nod factors are capable of eliciting in the roots of the legume hosts many of the plant responses characteristic of the bacteria themselves (Dénarié and Cullimore, 1993): root hair deformations, activation of the plant genes that are specifically induced during early stages of nodulation (early nodulin genes), initiation of cortical cell division, and triggering of a plant organogenic program that leads to nodule formation. Nod factors thus act as symbiotic signaling molecules for initiating nodule development. They also play a key role in the control of specificity of infection and nodulation, the result of particular substitutions present on the basic backbone of the Nod factor molecule (reviewed in Long, 1996; Cohn et al., 1998; Schultze and Kondorosi, 1998).
One of the most challenging areas of research in the study of the Rhizobium–legume symbiosis is dissecting the mechanisms by which the Nod factor signal is perceived and transduced by host plants. Because biological responses are induced by very low Nod factor concentrations (10–9 to 10–12 M) and have precise Nod factor structural requirements, Nod factors are probably perceived by specific receptors. A high-affinity binding site for Sinorhizobium meliloti Nod factors, NFBS2, has recently been identified in plasma membrane–enriched fractions of Medicago cell suspension cultures (Gressent et al., 1999), and a lectin nucleotide phosphohydrolase isolated from roots of the legume Dolichos biflorus also has been reported to have Nod factor binding activity (Etzler et al., 1999). However, whether either of these Nod factor binding components plays a role in Nod factor signaling has not been established.
After the perception of Nod factors, numerous rapid plant responses in root hairs have been detected (reviewed in Downie and Walker, 1999). Within seconds of Nod factor addition, fluxes of Ca2+, H+, Cl–, and K+ ions occur (Felle et al., 1996, 1998, 1999a), together with membrane depolarization (Ehrhardt et al., 1992; Felle et al., 1995, 1998; Kurkdjian, 1995). Minutes after addition of Nod factors, changes in cytoplasmic Ca2+ concentrations are observed, including transient and plateau-like increases (Gehring et al., 1997; Cárdenas et al., 1999; Felle et al., 1999a) and calcium spiking (Ehrhardt et al., 1996), and evidence suggests a role for Ca2+ as a secondary messenger in signal transduction of Nod factors (Felle et al., 1999b). Nod factor–induced changes to the actin cytoskeleton also occur in root hairs and may, in association with certain Ca2+ concentration changes, be involved in the inhibition and subsequent reinitiation of root hair tip growth (Cárdenas et al., 1998; de Ruijter et al., 1999; Miller et al., 1999). Gene expression is induced by Nod factors in different cell layers of host roots and with different temporal expression patterns. For example, the early nodulin genes ENOD11, ENOD12, and rip1 are induced in the epidermis within 2 hr of addition of Nod factors, whereas ENOD20 and ENOD40 are induced later in the cortex and in both the pericycle and cortex, respectively (Asad et al., 1994; Crespi et al., 1994; Journet et al., 1994; Cook et al., 1995; Vernoud et al., 1999; D. Barker, J.L. Pingret, M. Chaboud, and E.P. Journet, unpublished results). Nod factor–induced changes in gene expression are spatially and temporally correlated with cellular and developmental changes in the plant. For example, Nod factors trigger reorganization of the microtubular cytoskeleton and induce cell divisions, first in the root pericycle and then in the cortex (Timmers et al., 1999). These events are correlated with induction of ENOD40 in the root pericycle and the subsequent induction of several nodulin genes (e.g., ENOD20, rip1, and ENOD40) in the nascent nodule primordium (Peng et al., 1996; Charon et al., 1997, 1999).
The high level of complexity involved in Nod factor signal transduction, leading as it does to cellular, molecular, and developmental responses in several cell layers of the root, means that a genetic approach is needed to determine the sequence of events and fully dissect the mechanisms involved. Genetic analysis of pea nodulation mutants has identified one element of a Nod factor–induced signal transduction pathway, the gene SYM8, which is essential for the induction of two epidermal nodulin genes, PsENOD5 and PsENOD12A (Albrecht et al., 1998). To identify further genes involved in Nod factor perception and signal transduction, we have exploited nodulation mutants of the model legume Medicago truncatula (Barker et al., 1990; Cook et al., 1997; Cook, 1999). Our strategy was to characterize mutants that were pleiotropically affected in Nod factor responses. Because the majority of such responses are associated with events occurring before infection by Rhizobium, we first set out to identify mutants that were blocked at early stages of the symbiotic interaction. After large-scale mutagenesis of M. truncatula with ethyl methanesulfonate (EMS) (Penmetsa and Cook, 2000) and γ-ray (Sagan et al., 1995, 1998), we identified mutants that were devoid of both rhizobial infection and induction of nodule primordia. Here, we report the results of further mutant screens and the genetic assignment of nine independent mutants into four complementation groups, each of which is required for early symbiotic responses. After detailed analysis of Nod factor–induced responses (root hair deformation, cortical cell divisions, and early nodulin gene expression in the root epidermis, pericycle, and cortex), we have determined that each of these mutants is simultaneously affected in multiple Nod factor responses, which is consistent with a role for the corresponding genes in perception/transduction of the Nod factor signal.
RESULTS
Identification of Infection-Defective Mutants of M. truncatula
To identify mutants of M. truncatula Jemalong blocked at early stages of the symbiotic interaction, we first selected mutants that were unable to form nodules (Nod−) after rhizobial inoculation—an easy, visual screen that can be performed on large numbers of plants. After an initial round of screening ∼50,000 EMS-mutagenized plants, ∼50 putative Nod− mutants were selected, of which 25 were confirmed by subsequent testing (see Methods). Nod− mutants were then characterized cytologically to define the block. Thus, infection studies of these 25 mutants and of 26 other EMS-induced Nod− mutants (R.V. Penmetsa and D. Cook, unpublished results), using a strain of S. meliloti carrying a constitutively expressed lacZ gene (GMI6526), revealed two main groups of mutants: 26 mutants completely blocked for infection initiation and 25 mutants showing aborted infection, in which infection threads aborted either in root hair cells or in cortical cells. Close examination of mutants in the first group revealed that eight mutants showed limited cortical cell divisions and that the remaining 18 mutants were defective for cortical cell divisions (Ccd−). In the second group of mutants, however, Ccd was in certain cases extensive, giving rise to visible bumps.
The mutants chosen for further study were those blocked at the earliest stage of the symbiotic interaction, that is, the mutants of group 1 that were defective for induction of cortical cell divisions. The final choice of mutants was based both on results of genetic analysis, which enabled us to eliminate possible siblings (allelic mutants from the same bulk), and on practical considerations, such as the stage of backcrossing. The analysis of six mutants from group 1 (B85, B129, C54, C71, P1, and Y6) is described in this article, along with three previously described γ-ray Nod− mutants of M. truncatula Jemalong (TR25, TR26, and TRV25) (Table 1; Sagan et al., 1995, 1998). The shoot and root systems of plants of all nine Nod− mutants developed normally when grown in the presence of added nitrate (data not shown).
Table 1.
Bacterial Strains, Plasmids, and Plants Used in This Studya
| Designation | Relevant Characteristics | Reference/Source |
|---|---|---|
| S. meliloti | ||
| ABS7 | Nod+Fix+ on M. truncatula | Bekki et al. (1987) |
| GMI6526 | 2011(pXLGD4), Nod+Fix+ on M. truncatula | Ardourel et al. (1994) |
| GMI6390 | 2011(pMH682) | Roche et al. (1991b) |
| GMI3198 | 2011(pGMI3194)(pXLGD4) | This study |
| GMI6628 | 2011Δ(nodF)13nodL::Tn5, PmRa | Ardourel et al. (1994) |
| GMI6630 | 2011Δ(nodF)13nodL::Tn5(pXLGD4) | Ardourel et al. (1994) |
| GMI3125 | 2011Δ(nodF)13nodL::Tn5nodC::Sp | This study |
| GMI6702 | 2011nodA::Tn5#2208(pXLGD4) | Debellé et al. (1986) |
| Plasmids | ||
| pXLGD4 | pGD499 prime (IncP) carrying a hemA::lacZ fusion, TcR | Leong et al. (1985) |
| pMH682 | pWB85a prime (IncP) carrying nodD3 and syrM of S. meliloti, TcR | Honma et al. (1990) |
| pGMI1394 | pML132 prime (IncQ) carrying nodD1 of S. meliloti, GmR | Demont et al. (1994) |
| pRmM57 | pRMSL26 (IncP) nodC::Sp, TcR | Mulligan and Long (1985) |
| pR751-pMG2 | IncP plasmid, Tra+, GmR | Jacoby et al. (1976) |
| pRK2013 | Helper plasmid for mobilization of IncP and IncQ plasmids, KmR | Ditta et al. (1980) |
| M. truncatula | ||
| Jemalong A17 | Wild type, Nod+Fix+ with S. meliloti | Penmetsa and Cook (1997) |
| Jemalong (MtENOD11–GUS) | Transgenic wild type carrying a fusion between the promoter of MtENOD11 and the β-glucuronidase (GUS) gene | D.G. Barker, J.-L. Pingret, M. Chabaud, and E.-P. Journet, unpublished results |
| TR25 | γ-ray Nod− mutant of Jemalong | Sagan et al. (1995) |
| TR26 | γ-ray Nod− mutant of Jemalong | Sagan et al. (1995) |
| TRV25 | γ-ray Nod− mutant of Jemalong | Sagan et al. (1998) |
| B85 | EMS Nod− mutant of Jemalong | R.V. Penmetsa and D. Cook, unpublished results |
| B129 | EMS Nod− mutant of Jemalong | R.V. Penmetsa and D. Cook, unpublished results |
| C54 | EMS Nod− mutant of Jemalong | R.V. Penmetsa and D. Cook, unpublished results |
| C71 | “domi,” EMS Nod− mutant of Jemalong | Penmetsa and Cook (1997) |
| P1 | EMS Nod− mutant of Jemalong | This study |
| Y6 | EMS Nod− mutant of Jemalong | This study |
R, resistant.
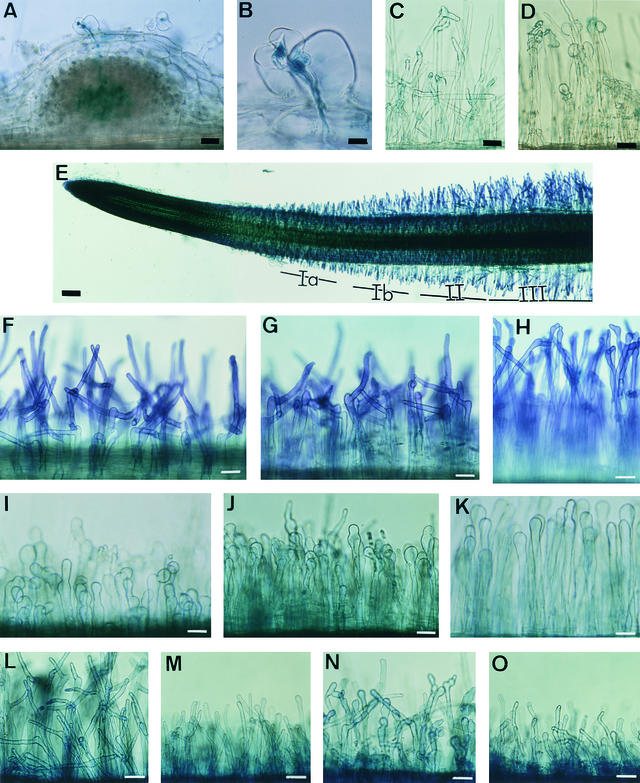
Wild-type M. truncatula plants grown on agar medium showed marked root hair curling (Hac+) and infection thread formation (Inf+) within 4 days of inoculation by S. meliloti GMI6526 (Figures 1A and 1B) and were nodulated (Nod+) within 7 days. When mutants were studied in the same conditions at different times up to 3 weeks after inoculation by this strain, no marked root hair curling (Hac−), no infection threads (Inf−), and no nodules could be seen on any of the mutants. The absence of induced cortical cell divisions in these mutants was confirmed by examining semi-thin sections of roots spot-inoculated by S. meliloti GMI6526 (data not shown). However, root hair deformations were induced in all mutants within 4 days of inoculation. For B85 and C54, only a few root hairs were deformed, and those showed branched or moderately curled root hair tips (Figure 1C); all the other mutants showed a large proportion of swollen root hair tips (Figure 1D).
Figure 1.
Root Hair Deformation Phenotypes of Wild-Type and Mutant Plants.
To exclude the possibility that the M. truncatula mutants were Nod− because of an inability to secrete rhizobial nod gene–inducing flavonoids, we inoculated plant mutants with the constitutive Nod factor–producing strain GMI6390 (S. meliloti carrying the regulatory genes nodD3 and syrM; Table 1). No nodules were observed on roots of any of the mutants 3 weeks after inoculation (data not shown). To determine whether the infection and nodulation deficiency of Nod− mutants resulted from decreased sensitivity to Nod factors, we inoculated mutants with the Nod factor–overproducing strain GMI3198 (S. meliloti carrying the regulatory gene nodD1 for Nod factor overproduction and a constitutively expressed lacZ reporter gene for bacterial visualization; see Table 1). No infection-related events could be detected in any of the Nod− mutants 7 days after spot inoculation, although mutants B85 and C54 showed more root hair deformations than they did after inoculation with a wild-type S. meliloti strain (data not shown).
Identification of Four Complementation Groups of M. truncatula Infection-Defective Mutants
The mutations responsible for the Nod− phenotypes of TR25, TR26, TRV25, and C71 are monogenic and recessive (Sagan et al., 1995, 1998; R.V. Penmetsa and D. Cook, unpublished results). The remaining five mutants (B85, B129, C54, P1, and Y6) were crossed to wild-type plants (see Methods). As shown in Table 2, F1 plants of these crosses all had a wild-type Nod+ phenotype, indicating that all the mutations were recessive, and the F2 segregation data are consistent with the mutations all being nuclear and monogenic. The results of allelism tests, performed to define complementation groups, are presented in Table 3. TR25, TR26, and TRV25 had already been shown to fall into two complementation groups, one consisting of TR25 and TR26, the other TRV25 (Sagan et al., 1998); here, we determined that P1 is in the same complementation group as TR25, but no mutants could be placed in the same complementation group as TRV25. All the other five mutants fell into two new complementation groups: one containing B129, C71, and Y6; the other, B85 and C54. Pending further characterization of mutants, the four complementation groups were designated locus 1 (B129, C71, and Y6), locus 2 (TR25, TR26, and P1), locus 3 (TRV25), and locus 4 (B85 and C54).
Table 2.
Genetic Analysis of Nod− Mutants
| F1b
|
F2b,c
|
||||
|---|---|---|---|---|---|
| Mutantsa | Nod+ | Nod− | Nod+ | Nod− | χ2d |
| B85 | 16 | 0 | 76 | 24 | 0.02 |
| B129 | 6 | 0 | 134 | 43 | 0.05 |
| C54 | 5 | 0 | 74 | 21 | 0.01 |
| P1 | 6 | 0 | 86 | 28 | 0.01 |
| Y6 | 2 | 0 | 73 | 27 | 0.21 |
Mutants were crossed to Jemalong (MtENOD11–GUS) Nod+ plants, which were used in place of wild-type plants to provide a marker for crosses (see Methods).
Nodulation was scored in plants of the F1 and F2 generations, 3 weeks after inoculation with S. meliloti GMI6526; numbers indicate the number of plants found to be Nod+ or Nod−. In most cases, F1 results presented are from two or more independent crosses.
F2 results are given for a single F2 population but were confirmed in each case by analyzing at least one additional, independent F2 population derived from a different F1 plant.
χ2 calculated for a 3:1 ratio of Nod+/Nod− phenotype; P > 0.05 when χ2 < 3.84.
Table 3.
Allelism Tests on Nod− Mutantsa
| Group 1 | Group 2 | Group 3 | Group 4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y6 | B129 | C71 | P1 | TR25 | TRV25 | B85 | C54 | |||||
| Y6
|
− (22)
|
− (7)
|
|
+ (4)
|
+ (14)
|
|
+ (8)
|
|
+ (11)
|
NDb
|
||
| B129 | − (9) | + (10) | + (11) | + (19) | + (12) | + (7) | ||||||
| C71 | + (9) | + (7) | + (11) | ND | ND | |||||||
| P1 | − (12) | + (15 ) | + (5) | ND | ||||||||
| TR25 | +c | + (14) | + (8) | |||||||||
| TRV25 | + (13) | + (12) | ||||||||||
| B85 | − (17) | |||||||||||
Represented are the nodulation phenotype of F1 plants and the number of F1 individuals tested (figures in parentheses). (+), Nod+; (−), Nod−.
ND, not determined.
The Nod+ phenotype of F1 individuals from a cross between TR25 and TRV25 was demonstrated by Sagan et al. (1998).
Nod Factors Induce a Wild-Type Root Hair Deformation Phenotype in Mutants in Locus 4
The absence of any infection-related events (no Hac or Inf) in all the mutants studied suggested impaired abilities to respond to Nod factors. An early, characteristic response to Nod factors is root hair deformation, that is, swelling of the root hair tip and subsequent reinitiation of polar growth from the swelling (Heidstra et al., 1994; de Ruijter et al., 1998). Results presented in Table 4 show that homologous S. meliloti sulfated Nod factors (at 10–7 to 10–11 M), but not nonsulfated Nod factors, were able to induce root hair deformations, in the form of branched root hairs (the Hab phenotype, for hair branching), in wild-type M. truncatula plants. Deformation started by a swelling of the root hair tip, visible within 1 hr after treatment; a new outgrowth produced after 2 to 3 hr continued to grow, giving rise to branches that were observed after 16 hr (Figures 1F to 1H).
Table 4.
Effect of Sulfated and Nonsulfated S. meliloti Nod Factors at Different Concentrations on Root Hair Deformation of Wild Type and Mutants
| Nod Factor Concentration
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sulfated
|
Nonsulfated
|
||||||||
| Plant | Phenotypea | 10−7 M | 10−8 M | 10−9 M | 10−10 M | 10−11 M | 10−12 M | 10−7 M | 10−9 M |
| Wild type | Hab | +b | + | + | + | + | − | − | − |
| B129 | Has | + | + | + | + | + | − | − | − |
| B85 | Hab | NDc | + | + | + | − | − | − | − |
| C54 | Hab | ND | + | + | + | + | − | − | − |
Hab indicates root hair branching; Has indicates root hair swelling.
Samples were scored for the percentage of plants showing deformed root hairs and then compared with control samples. (+) indicates a significantly (P = 0.01) higher percentage of responding plants; and (−) indicates no significant difference (P = 0.01) compared with control plants. At least two independent experiments were performed on 10 to 15 plants/sample, all of which were scored double blindly 16 hr after Nod factor treatment. Analysis of variance was conducted with Fisher's Exact test (Kendall and Stuart, 1976).
ND, not determined.
To facilitate comparison with previous work performed on vetch (Heidstra et al., 1994), we adopted the same three root zones they used, corresponding to different stages of root hair growth: (I) growing, (II) terminating growth, and (III) fully grown root hairs (Figure 1E). As in vetch, root hairs of zone II exhibited the Hab phenotype in response to Nod factors, and those of zone III did not. However, in contrast to vetch, in M. truncatula the whole root hair–growing region responded by branching. To refine the analysis of the Hab phenotype, therefore, we divided zone I into Ia and Ib, given the clear differences in the forms of branched root hairs between zones Ia and Ib (Figure 1): In zone Ia, long branches are produced on very short root hairs (Figure 1F), whereas in zone Ib, shorter branches are produced on longer root hairs (Figure 1G). In zone II, root hairs show very short branches or, less frequently, have swollen tips without branches (Figure 1H). Thus, branch length decreased progressively from root hairs that were the shortest at the time of treatment to those that had almost reached their full-grown length when Nod factors were added.
To determine whether the Nod− mutants still respond to Nod factors by root hair branching, we observed root hairs of mutants 16 hr after Nod factor treatment. Root hair growth and morphogenesis were normal in untreated roots of all mutants (data not shown). For mutants in locus 4, sulfated but not nonsulfated Nod factors induced the Hab phenotype in zones Ia, Ib, and II, the same as for the wild type (data not shown). We then tested the sensitivity of this response; as the results in Table 4 show, for C54, the Hab phenotype could be detected for concentrations of Nod factors down to 10–11 M, whereas for B85, it could be detected for concentrations as low as 10–10 M. Except for this slight difference in sensitivity, the root hair branching response of mutants in locus 4 was therefore no different from the wild-type response.
Lasting Inhibition of Tip Growth Is Observed in Response to Nod Factors in Mutants in Loci 1 to 3
In contrast to wild-type plants and mutants in locus 4, root hair tips of mutants in loci 1, 2, and 3 all became swollen in the presence of 10–8 M sulfated Nod factors, without producing the long branches typical of the wild-type Hab phenotype (Figures 1I to 1K). To distinguish this new phenotype from the Hab phenotype, we have adopted the term Has (for hair swelling). This Has phenotype seen after Nod factor treatment resembled, but was more pronounced than, the swollen root hair tip phenotype seen in these same mutants after inoculation with S. meliloti (Figure 1D). As with the Hab phenotype in the wild-type plants, zones Ia, Ib, and II exhibited the Has phenotype in mutants in loci 1, 2, and 3. The youngest root hairs in zone Ia had a tendency for swollen tips and short, stumpy branches (Figure 1I); zone Ib root hairs were a combination of root hairs with swollen tips and root hairs with short, irregular-shaped deformations (Figure 1J); and the majority of root hairs in zone II showed swollen tips without such branches (Figure 1K). We then tested the Nod factor specificity and sensitivity of this response by looking at the ability of nonsulfated Nod factors to induce the Has phenotype in all mutants of loci 1 to 3 and by testing Nod factor dilutions. For one representative of the Has phenotype, B129 (locus 1), a range of sulfated Nod factor concentrations were tested (10–7 to 10–12 M); for all other mutants, sulfated Nod factors were tested at 10–9 or 10–10 M. Results in Table 4, and data not shown, revealed that the Has phenotype was dependent on sulfated Nod factors and was just as sensitive to low Nod factor concentrations as the Hab phenotype in wild-type plants.
To further characterize the Nod factor structural requirements of the Has phenotype, we investigated the ability of non-O-acetylated Nod factors with a modified acyl substituent to induce root hair deformations in mutants in loci 1 to 3. We therefore compared the response of mutants to the double nodFL mutant of S. meliloti, GMI6630 (Table 1), which produces such Nod factors and induces marked root hair deformations on wild-type plants (Ardourel et al., 1994), with the response induced by a nodFL mutant rendered incapable of producing Nod factors by introduction of a mutation in the nodC gene (GMI3125; see Table 1 and Methods). One mutant representative of each locus was tested: B129 (locus 1), TR25 (locus 2), and TRV25 (locus 3). The Has plant mutants responded to GMI6630 but not to GMI3125 by swollen root hair tips (data not shown), indicating that the Has response is not stringent with respect to modifications at the nonreducing end of the Nod factor molecule.
These data show that the Hab response in the wild type and the Has response in mutants of loci 1, 2, and 3 occurred in the same root hair zones, was observed in the same Nod factor concentration range (10–7 to 10–11 M), and had the same Nod factor structural requirements. These findings strongly suggest that the same mechanism for perception of Nod factors is involved in the induction of the Hab and Has phenotypes. In Has mutants, polar tip growth appears to be inhibited, the process of effective reinitiation of polar root hair tip growth is defective, and root hairs become markedly swollen, producing only short, deformed branches—indicating that root hairs of Has mutants are no longer capable of sustained polar growth after treatment with Nod factor. Furthermore, root hairs remained swollen for at least 1 week after treatment (data not shown), showing that this response has a lasting effect.
Finally, the symbiotic significance of the Has phenotype was investigated by testing the effect of combined nitrogen, which is inhibitory for nodulation and certain Nod factor–induced responses, including root hair deformation (Carroll and Mathews, 1990; Heidstra et al., 1994, 1997). In the case of wild-type M. truncatula grown in the presence of 10 mM ammonium nitrate, nodule formation was completely inhibited (data not shown) and the Hab response was greatly reduced, although root hairs were shorter after treatment with Nod factor, indicating that some inhibition of polar growth still occurs (Figures 1L and 1M). Similarly, for B85 (Hab+) and B129 (Has) mutants, Hab and Has phenotypes were also greatly decreased by growing the plants in the presence of 10 mM ammonium nitrate (illustrated for B129 in Figures 1N and 1O). We note that this inhibition was observed after 7 days of growth in the presence of ammonium nitrate but not after 24 hr. A preincubation period >24 hr is also needed in vetch (Heidstra et al., 1994).
All Four Loci Are Involved in Controlling Epidermal Nod Factor–Induced Nodulin Gene Expression
Nodulin genes are molecular markers induced during the symbiotic interaction (Mylona et al., 1995). In contrast to root hair deformation, nodulin gene expression is detected in different cell layers and with different temporal expression patterns. Thus, the study of their expression in mutants should help elucidate whether we have identified genes involved in Nod factor signal transduction. Two early epidermal nodulin genes expressed at the preinfection stage and for which expression can be induced by purified Nod factors were studied: MtENOD11 (D. Barker, J.L. Pingret, M. Chaboud, and E.P. Journet, unpublished results) and rip1 (Cook et al., 1995).
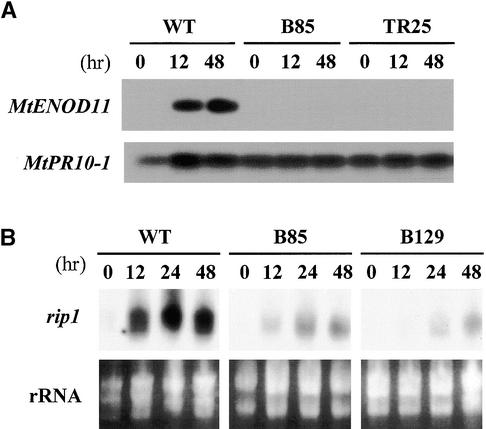
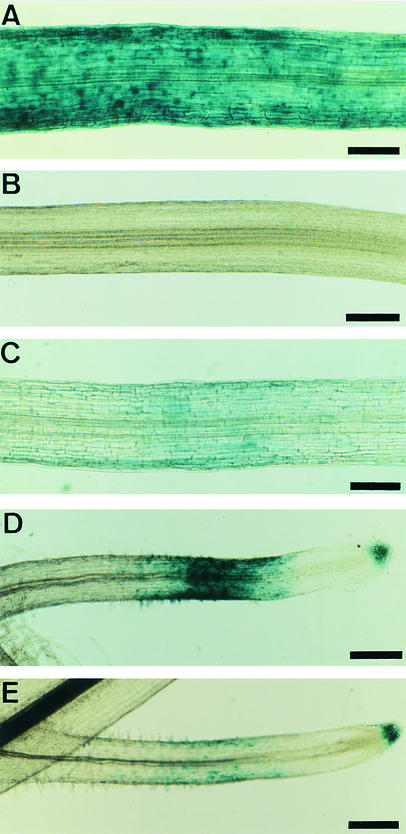
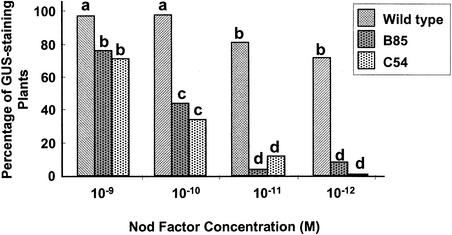
To determine whether Nod factors could induce MtENOD11 in mutants, we treated plants with S. meliloti Nod factors (10–8 M) and analyzed MtENOD11 expression by reverse transcription–polymerase chain reaction (RT-PCR). No MtENOD11 expression was detected in mutants in locus 1 (B129 and C71), locus 2 (TR25 and TR26), locus 3 (TRV25), or locus 4 (B85 and C54) (illustrated for TR25 and B85 in Figure 2A). Because transgenic plants carrying promoter–reporter gene fusions are a very sensitive means of analyzing gene expression, we analyzed transgenic plants carrying a fusion between the promoter of MtENOD11 and the β-glucuronidase (GUS) reporter gene. These mutant transgenic plants were generated by crossing mutants to wild-type plants carrying the construction MtENOD11–GUS (D.G. Barker, J.-L. Pingret, M. Chabaud, and E.-P. Journet, unpublished results; see Methods and Table 1). Strong induction of MtENOD11 was observed in wild-type transgenic plants in response to 10–8 M S. meliloti Nod factors (Figure 3A), but no MtENOD11 expression could be detected in the epidermis in response to 10–8 M S. meliloti Nod factors in mutants in locus 1 (B129, C71, and Y6) or locus 2 (P1) (illustrated for B129 in Figure 3B). As a control, GUS activity could be detected in root caps, lateral root primordia, and aerial parts of treated and nontreated plants, exactly as for wild-type plants (Figure 3D; data not shown), indicating that the nonsymbiotic pattern of MtENOD11 expression (D. Barker and V. Gianinazzi, unpublished results) was unaffected in these mutants. These results are similar to those found for mutants in other locus 2 alleles (TR25 and TR26) and for the locus 3 mutant TRV25 (D.G. Barker, J.-L. Pingret, M. Chabaud, and E.-P. Journet, unpublished results). For mutants in locus 4 (B85 and C54), analysis of transgenic plants revealed that MtENOD11 was induced in the epidermis by S. meliloti Nod factors but to a lesser extent than in wild-type plants and with a smaller responding zone (Figures 3A and 3C to 3E). This difference was quantified by testing transgenic plants with a series of Nod factor dilutions (from 10–9 to 10–12 M). Results presented in Figure 4 show that the MtENOD11 expression response is at least 100-fold less sensitive to Nod factors in B85 and C54 (locus 4) than in wild-type plants.
Figure 2.
Analysis of Early Nodulin Gene Expression in Wild-Type and Mutant Plants in Response to Nod Factors.
(A) RT-PCR analysis of MtENOD11 expression in wild type (WT) and mutant (B85, locus 4, and TR25, locus 2) roots treated with Nod factors at 10–8 M and harvested at the indicated times (see Methods). MtPR10-1 RNA was amplified as a control for the quality and quantity of the RNA samples.
(B) RNA gel blot of rip1 expression in wild type (WT) and mutant (B85, locus 4, and B129, locus 1) roots treated with S. meliloti Nod factors at 10–8 M and harvested at the indicated times (see Methods). rRNA concentrations are shown to control for equal loading.
Figure 3.
Histochemical Localization of GUS Activity in Roots of Transgenic Plants Carrying a Fusion between the Promoter of MtENOD11 and the GUS Reporter Gene after Treatment with S. meliloti Nod Factors for 6 hr.
Primary roots ([A] to [C]) and secondary roots ([D] and [E]), which generally show more intense GUS staining than do the primary roots.
(A) Wild type treated with 10–8 M.
(B) B129 (locus 1) treated with 10–8 M.
(C) C54 (locus 4) treated with 10–8 M.
(D) Wild type treated with 10–9 M.
(E) B85 (locus 4) treated with 10–9 M.
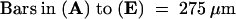 .
.
Figure 4.
Effect of Different Nod Factor Concentrations on MtENOD11 Expression in Transgenic Lines of Wild Type, B85, and C54 (Both at Locus 4).
At least two independent experiments were performed with 15 to 20 plants per sample, all of which were scored 6 hr after treatment with S. meliloti Nod factors. Values with different letters (a, b, and c) differ significantly at 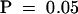 . d, values that were not significantly different from control, untreated plants at
. d, values that were not significantly different from control, untreated plants at 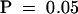 . Analysis of variance was conducted with Fisher's Exact test (Kendall and Stuart, 1976).
. Analysis of variance was conducted with Fisher's Exact test (Kendall and Stuart, 1976).
The ability of Nod factors to induce expression of another early nodulin gene, rip1, was studied by RNA gel blot analysis in the following mutants: B129 and C71 (locus 1), TR25 (locus 2), TRV25 (locus 3), and B85 and C54 (locus 4). We found that although rip1 was strongly induced in wild-type plants within 12 hr after 10–8 M S. meliloti Nod factors was added, induction of rip1 was substantially reduced in all of the mutants tested (illustrated for B129 and B85 in Figure 2B).
Taken together, these results indicate that all four loci are required for Nod factor–induced expression of MtENOD11 and rip1 in the root epidermis and are consistent with the function of the respective proteins in perception or transduction of the Nod factor ligand.
All Four Loci Are Involved in Controlling Nod Factor–Induced MtENOD40 Expression in the Pericycle and Cortex
ENOD40 is an early nodulin gene induced by Nod factors, first in the pericycle and subsequently in dividing cortical cells and in all differentiating cells of the growing nodule primordium (Asad et al., 1994; Crespi et al., 1994; Gamas et al., 1996; Fang and Hirsch, 1998). Accumulating evidence supports an important role for ENOD40 in primordium formation during nodulation. In particular, overexpression and bombardment of ENOD40 induce dedifferentiation and division of root cortical cells in legumes (Charon et al., 1997); transgenic lines of M. truncatula having reduced amounts of MtENOD40 transcripts form only a few modified, nodule-like structures (Charon et al., 1999). The absence of cortical cell divisions in mutants in the four loci provided an opportunity to examine the correlation between MtENOD40 induction and cortical cell division. The ability of Nod factors to induce MtENOD40 expression was studied by comparing plants inoculated with either a wild-type S. meliloti strain (GMI6526) or an S. meliloti nodA::Tn5 mutant (GMI6702) unable to produce Nod factors (Table 1).
MtENOD40 expression was analyzed by in situ hybridization after spot inoculation of wild-type and mutant plants. Although a very low background amount of MtENOD40 expression was detected throughout the pericycle of many roots, MtENOD40 was clearly induced in wild-type plants by GMI6526, in dividing cells of the pericycle, the inner cortex at 24 hr (data not shown). At 48 hr, when the infection process is just starting, MtENOD40 expression was observed in dividing cells of the pericycle and both the inner and middle cortex and in overlying cells of the external cortex (Figures 5A and 5B). No induction of MtENOD40 expression by the S. meliloti nodA::Tn5 mutant could be detected (data not shown), indicating that MtENOD40 expression in M. truncatula depends on the presence of Nod factors.
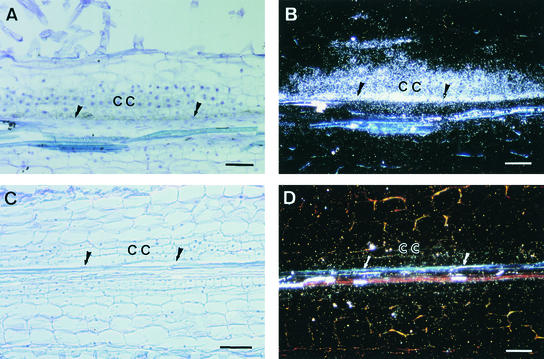
Figure 5.
In Situ Hybridization Analysis of MtENOD40 mRNA in Roots of Wild-Type and B85 (Locus 4) Plants in Response to S. meliloti.
Roots were hybridized with α-35S-UTP–labeled MtENOD40 RNA probes 48 hr after spot inoculation with S. meliloti GMI5626. Data are presented only for the antisense probes; no hybridization signals were detected with sense probes.
(A) and (C) Bright-field microscopy; hybridization signals are visible as dark spots.
(B) and (D) Dark-field microscopy; hybridization signals are visible as white dots.
(A) and (B) Longitudinal section of a wild-type root showing MtENOD40 RNA localized in the cortical and pericycle dividing cells of a nodule primordium.
(C) and (D) Longitudinal section of a B85 root, showing neither cell divisions nor MtENOD40 expression.
CC, cortical cells; arrows, pericycle.  .
.
MtENOD40 gene expression was then analyzed in mutants in locus 1 (B129), locus 2 (TR25), locus 3 (TRV25), and locus 4 (B85 and C54). No induction was detected in the pericycle or cortex of any of these mutants 48 hr after inoculation by the wild-type S. meliloti strain (illustrated for B85 in Figures 5C and 5D). RNA gel blot analysis has also shown that Nod factors can no longer induce MtENOD40 expression in the mutant C71 (locus 1) (S.K. Ramu, R.V. Penmetsa, and D. Cook, unpublished results). The requirement for Nod factor–producing rhizobia for MtENOD40 induction, together with the absence of MtENOD40 induction in mutants in loci 1 to 4, indicates that these four loci are required for the Nod factor signal transduction that leads to MtENOD40 expression.
Single Mutations Are Responsible for the Pleiotropic Defects in Nod Factor Responses
The preceding results show that all the nine mutants we have studied are affected in more than one Nod factor response. Four mutants (B129, C71, TR25, and TRV25) are affected for root hair branching; MtENOD11, rip1, and MtENOD40 expression; and cortical cell divisions. Three mutants (Y6, TR26, and P1) are affected for root hair branching, MtENOD11 expression, and cortical cell division. In the remaining two (B85 and C54), Nod factors are no longer able to induce MtENOD40 expression, cortical cell divisions are absent, and induction of MtENOD11 and rip1 is greatly reduced. Because most, if not all, mutagenic treatments commonly used on plants, including the γ-ray and EMS mutageneses used in this study, can induce more than one mutation in each plant (Feldmann et al., 1994), it was important to provide evidence that single mutations were responsible for these pleiotropic phenotypes. Such genetic evidence can be provided either by studying independent allelic mutants or by analyzing the cosegregation of two or more phenotypes in an F2 population of plants.
For three of the loci described, we studied more than one allelic mutant. Thus, for locus 1 we studied B129, C71, and Y6; for locus 2, TR25, TR26, and P1; and for locus 4, B85 and C54. The allelic mutants were found to be altered in Nod factor responses and in infection all in the same manner (see previous results), strongly indicating that they were the result of a mutation in the same gene in each case. Interestingly, only one case of allelic variation was detected, with B85 and C54 (locus 4) differing slightly for the Hab phenotype at low Nod factor concentrations. For locus 1, we supplemented the evidence that a single mutated gene is responsible for the pleiotropic defects in Nod factor responses by using cosegregation analysis. We thus checked an F2 population of B129 homozygous for the transgene MtENOD11–GUS for the cosegregation of three phenotypes: Nod factor induction of the Has phenotype, absence of Nod factor–induced GUS activity (MtENOD11−), and absence of nodulation (Nod−). In a population of 284 F2 plants, the three phenotypes cosegregated completely; that is, all 67 Nod− plants displayed the Has phenotype and were MtENOD11−, whereas all 217 Nod+ plants were Hab+ MtENOD11+.
Only a single allele, TRV25, has been isolated for locus 3. Therefore, we analyzed the cosegregation of the same three phenotypes (absence of nodulation, absence of MtENOD11 induction, and the Has response) to test whether the phenotypes were linked and were therefore likely to result from the same mutation. Because this was done in an F2 population of 201 plants showing segregation for the transgene MtENOD11–GUS, only those individuals carrying the transgene (GUS+) were scored for MtENOD11 expression in response to Nod factors. All Nod+ GUS+ plants were MtENOD11+, and all Nod− GUS+ plants were MtENOD11−. Among this population, 106 plants were scored for root hair branching; of these, 88 were Nod+, all of which were Hab+, and 18 were Nod− and all displaying the Has phenotype. These cosegregation data suggest that a single mutation in locus 3 is responsible for the pleiotropic phenotype of TRV25 but does not exclude the possibility that the mutation affects more than one gene. However, we hypothesize that a single mutated gene is responsible for the pleiotropic phenotype of TRV25, given that we were unable to distinguish the pleiotropic phenotype of TRV25 from those of mutants in loci 1 and 2, for which we did have strong evidence for the involvement of single genes.
The gene corresponding to mutant C71 (domi; Penmetsa and Cook, 1997) has been given the name dmi1 (for doesn't make infections) (R.V. Penmetsa and D. Cook, unpublished results). This name is based on the fact that C71 not only is defective for infection by rhizobia but also is unable to establish a symbiotic interaction with endomycorrhizal fungi (Myc− phenotype). The two other mutants (B129 and Y6) in this gene, which corresponds to locus 1, have also been found to be Myc− (M. Harrison, personal communication, for B129; V. Gianinazzi-Pearson and D. Morandi, personal communication, for Y6). Mutants in loci 2 and 3 are similarly Myc− (Sagan et al. [1995, 1998] for TR25, TR26, and TRV25; V. Gianinazzi-Pearson and D. Morandi, personal communication, for P1). Because of this, and because the mutants in loci 1, 2, and 3 are indistinguishable phenotypically, the corresponding genes have been named dmi2 (for locus 2 mutants) and dmi3 (for locus 3 mutants). Mutants in locus 4 (B85 and C54), however, can establish a normal, effective endomycorrhizal symbiotic interaction (M. Harrison, personal communication). The gene corresponding to this mutant locus has been named nsp (for nodulation signaling pathway) to reflect that this gene appears to control a step of Nod factor transduction specifically involved in the signaling leading to nodulation. The different alleles of these four genes have been distinguished as follows: dmi1-1 (C71); dmi1-2 (B129); dmi1-3 (Y6); dmi2-1 (TR25); dmi2-2 (TR26); dmi2-3 (P1); dmi3-1 (TRV25); nsp-1 (B85); and nsp-2 (C54).
DISCUSSION
Although many individual Nod factor–induced responses have been characterized, the mechanisms by which Nod factors induce these responses and the symbiotic significance of many of the responses are far from understood. The dissection of Nod factor perception/transduction has so far consisted of pharmacological approaches, identifying potential elements of a Nod factor signal transduction pathway that leads to expression of the early nodulin gene MtENOD12 (Pingret et al., 1998) or the characterization of certain plant mutants. Thus, two genetic loci of pea and one of Lotus japonicus have been studied for their role in Nod factor perception: SYM8 of pea, which is involved in controlling PsENOD5 and PsENOD12A gene expression in response to Nod factors (Albrecht et al., 1998); SYM2A of pea, which is involved in the specific recognition of Nod factors leading to infection but does not control Nod factor–induced root hair deformation or PsENOD12 expression (Geurts et al., 1997); and NIN of L. japonicus, which does not control the perception of Nod factors that leads to root hair curling and deformation (Schauser et al., 1999). In addition, an alfalfa Nod− mutant has been characterized that is deficient for calcium spiking in response to Nod factors, suggesting that this mutant is blocked in an early stage of Nod factor signal transduction (Ehrhardt et al., 1996).
A strategy aimed specifically at identifying genes involved in signal transduction is to isolate mutants that are altered pleiotropically in their response to the signal of interest. This strategy has been used very successfully, for example, to dissect genetically the ethylene signal transduction pathway by isolating “multiple response mutants” and has enabled both identification of the genes controlling steps in the pathway and elucidation of the sequence of events (Chang and Shockey, 1999). Here, we have used this strategy to initiate the genetic dissection of Nod factor transduction in M. truncatula. We isolated multiple response mutants by first screening for Nod− mutants and then searching among these Nod− mutants for those pleiotropically altered in their responses to Nod factors. The aim of this study was thus to deliberately identify legume genes involved in Nod factor transduction.
Nine Nod− mutants of M. truncatula were characterized and assigned to four loci. All nine mutants were defective in infection by the normally compatible rhizobial strain S. meliloti, and this deficiency could not be overcome by using Nod factor–overproducing strains. According to their response to Nod factors, mutants could be divided into two phenotypic classes. One, which included mutants in loci 1, 2, and 3, showed no induction of the early nodulin genes MtENOD11 and MtENOD40, had greatly reduced rip1 induction, responded with no cell divisions, and were affected in Nod factor–induced root hair branching. The other included mutants in locus 4, which were similarly defective for Nod factor–induced MtENOD40 induction and cell divisions and showed a greatly reduced presence of MtENOD11 and rip1 induction. By being defective in different responses—transcription of early nodulin genes in different tissues (the epidermis, the cortex, and the pericycle) and cortical cell divisions—these mutants clearly can be called pleiotropic and thus they correspond to multiple response mutants. Furthermore, the analysis of different alleles and cosegregation studies provided genetic evidence that a single mutated gene in each locus was responsible for the multiple defects. We have thus hypothesized that these mutations alter genes involved in a signal transduction pathway activated by Nod factors and leading to nodulation. The strategy adopted for identifying such genes has therefore been successful and should enable fine dissection of the nodulation signaling pathway, made possible because nodulation is a facultative trait for legume growth, in the presence of combined nitrogen. Moreover, the identification of such genes in the model legume M. truncatula will facilitate their cloning and detailed functional analysis.
A Model for Nod Factor Signal Transduction That Leads to Induction of Symbiotic Responses
Given that several of the symbiotic responses (induction of MtENOD11, rip1, and MtENOD40; cortical cell divisions; and nodulation) are altered by all the mutations we characterized in DMI1, DMI2, DMI3, and NSP, we propose a model in which these four genes intervene in the same Nod factor transduction pathway that leads to nodulation (Figure 6A). The four genes are good candidates to code for components of this signal transduction pathway, although we cannot exclude the possibility that products of these genes could allow the functioning of a signal transduction mechanism without their activity being modulated by signal perception itself. We also cannot exclude that any of the genes code for a Nod factor receptor, although all of the mutants described in this study are still able to specifically perceive Nod factors. Within the pathway, DMI1, DMI2, and DMI3 cannot be ordered because mutations in these three genes result in the same phenotype. Further work will aim at distinguishing dmi mutants, for example, by exploring the calcium-spiking response (Ehrhardt et al., 1996) or the induction of MtENOD11 or MtENOD12 by Nod factor agonists such as mastoparan (Pingret et al., 1998). nsp mutants could be clearly distinguished from the three dmi mutants: nsp mutants exhibit a wild-type root hair branching phenotype, whereas dmi mutants are defective in their ability to reinitiate root hair tip growth after treatment with Nod factor. These data indicate that NSP acts downstream of the three DMI genes in the pathway and are consistent with the finding that normal expressions of MtENOD11 and rip1 are not needed for root hair branching in M. truncatula. Expression of VsENOD5, VsENOD12, and VsLb1 in vetch similarly is not required for root hair deformation (Vijn et al., 1995; Heidstra et al., 1997). Although MtENOD11 and rip1 expressions are clearly diminished in nsp mutants, they are not completely eliminated, which suggests either the involvement of additional regulatory mechanisms or our possible characterization of leaky alleles of nsp.
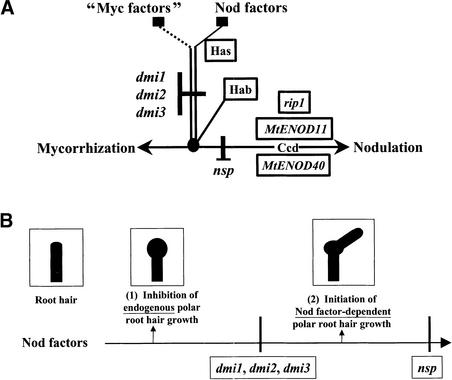
Figure 6.
Models for the Intervention of DMI1, DMI2, DMI3, and NSP1 in Nod Factor Signal Transduction.
(A) Model for roles in a Nod factor signal transduction pathway leading to MtENOD11, rip1, and MtENOD40 expression; cortical cell division; and nodulation. As deduced by the phenotypes of dmi1, dmi2, dmi3, and nsp1 mutants, DMI1, DMI2, and DMI3 presumably intervene at one or more steps of the pathway downstream of Has and upstream of both Hab and NSP. The part of the signaling pathway controlled by DMI1, DMI2, and DMI3 would be induced both by Nod factors and by potential mycorrhization signals (“Myc factors”) and would play a role in the preparation of the plant for infection by both rhizobia and endomycorrhizal fungi. Ccd, cortical cell division; Hab, root hair branching; Has, root hair swelling.
(B) Model for roles in Nod factor–induced root hair tip growth changes. Nod factors would (1) inhibit endogenous polar root hair growth and (2) initiate Nod factor–dependent growth. dmi1, dmi2 and dmi3 mutants uncouple these two events, which indicates either that DMI1, DMI2, and DMI3 are required for an element or elements of Nod factor signal transduction downstream of (1) and upstream of (2) or that Nod factors trigger (1) and (2) by different pathways. nsp mutants are able to initiate polar growth in response to Nod factors, indicating that NSP is involved in a component of Nod factor signal transduction downstream of (2). The root hair swelling response is shown as a phenotype associated with the inhibition of endogenous polar root hair growth; the root hair branching response is shown as a phenotype associated with the initiation of Nod factor–dependent root hair growth. Root hair branching is the phenotype observed when purified Nod factors are applied, but in the normal Rhizobium–legume interaction, when Nod factor–producing rhizobia are present, our model predicts that this initiation of Nod factor–dependent root hair growth contributes to marked root hair curling, a key step in the initiation of rhizobial infection.
The signaling pathway controlled by DMI1, DMI2, DMI3, and NSP may coincide, at least in part, with that identified by the gene SYM8 of pea, which controls the Nod factor signal transduction leading to epidermal expression of nodulin genes (Albrecht et al., 1998). However, we have extended the pathway to include root hair deformation, cortical cell divisions, and expression of MtENOD11 and rip1 in the epidermis and ENOD40 in the pericycle and cortex. Furthermore, the fact that in each mutant a single mutation apparently results in defective infection, the absence of nodulation, and defects in expression of MtENOD11, rip1, and MtENOD40 provides genetic evidence that these early nodulin genes, identified by a molecular approach, are indeed involved in the symbiotic process. Finally, the signaling pathway in which DMI1, DMI2, DMI3, and NSP intervene is probably part of a host-specific response to Nod factor recognition, given our demonstration that, just as for nodulation of host plants by S. meliloti (Lerouge et al., 1990), induction of two of the responses characterized (the Has and the Hab responses) depends on the sulfate group of S. meliloti Nod factors.
Dual Role of Nod Factors on Root Hair Tip Growth
Just as in vetch (Heidstra et al., 1994), the Nod factor–induced root hair branching response, or Hab phenotype, in M. truncatula starts by transient swelling of root hair tips and is followed by the formation of a branch. In contrast to vetch, however, in which only root hairs of zone II have been reported to deform, root hairs throughout the root hair elongation zones I and II in M. truncatula respond to Nod factors. Furthermore, the root hair branching phenotype of M. truncatula clearly differs from root hair deformation in vetch, being characterized in zones Ia and Ib by a very marked angle of branching and relatively long branches.
dmi mutants apparently perceive Nod factors by way of the same Nod factor perception mechanism that leads to root hair branching in wild-type plants, but they respond by root hair swelling without the production of branches. These mutants thus uncouple two stages in branch formation, the suppression of polar tip growth still being effectively induced by Nod factors but the second stage of reinitiation of tip growth being inhibited. Nod factors, in fact, have a spectacular effect on root hairs of dmi mutants, which become extensively swollen, as if the inhibition of polar tip growth is lasting instead of transient, while active, diffuse growth continues. Swollen root hair deformation phenotypes have also been reported for a Nod− mutant of pea, R72 (Markwei and Larue, 1992), and one of Melilotus alba, BT62 (Utrup et al., 1993), in response to inoculation by compatible rhizobial strains. Interestingly, both mutants, like dmi1, dmi2, and dmi3 mutants, are blocked for root hair infection.
Although our data clearly show that root hair swelling is a response to Nod factors, two alternative explanations can be proposed to explain the subsequent reinitiation of polar growth in M. truncatula: (1) the spontaneous restoration of normal, endogenous root hair tip growth after a transient arrest or (2) a novel Nod factor–controlled tip growth. Our data support the second hypothesis for the following reasons. In dmi mutants, adding Nod factors produces a lasting inhibition of polar tip growth. This suggests that Nod factors inhibit endogenous polar root hair growth through a component of the Nod factor transduction pathway that is upstream of DMI1, DMI2, and DMI3, whereas the reinitiation of polar growth depends on a component of Nod factor signal transduction that is downstream of the DMI genes (Figure 6B). The fact that nsp mutants are able to reinitiate polar growth also suggests that a component of the Nod factor transduction pathway required for polar growth reinitiation is upstream of NSP. Furthermore, if possibility (1) were correct, we might expect the reinitiation of growth leading to branch formation to be in the same direction as the original direction of root hair growth. Treatments that change the endogenous polarity of root hair growth, for example, do so only temporarily, after which growth returns to the original direction (Bibikova et al., 1997). In the case of Nod factor–induced root hair branching, that is not what was observed, indicating that Nod factors, by controlling the new tip growth, actually modify the polarity of root hair growth. Thus, as previously reported (de Ruijter et al., 1998), Nod factors seem to play two contradictory functions on root hair tip growth: inhibition and reinitiation. Our data allow us to propose the following interpretation of this apparent contradiction. Nod factors inhibit the endogenous tip growth of root hairs and initiate Nod factor–controlled polar growth. That Nod factors have the intrinsic ability to elicit tip growth is directly supported by the observation that their addition can elicit the formation, on cognate legume hosts, of elongated root hairs in cortical cells, a cell type not ordinarily programmed for tip growth. The production of such root hairs of cortical origin has been observed in vetch (van Brussel et al., 1992; van Spronsen et al., 1994) and alfalfa (G. Truchet, personal communication).
What could be the biological significance of this change in control of root hair tip growth? One key step in the initiation of Rhizobium infection is the formation of a marked curl of root hairs, called shepherd's crooks, which entrap bacterial cells in a pocket, from which they enter into the plant by forming an infection thread. Marked curling involves a modification of root hair growth by rhizobial cells that requires the production of Nod factors, because Rhizobium mutants unable to produce cognate Nod factors are unable to elicit this response. Van Batenburg et al. (1986) proposed that rhizobia induce marked root hair curling by redirection of tip growth. Our data are consistent with the following mechanism for this “hijacking” of root hair growth: by way of Nod factor secretion, rhizobia both inhibit endogenous tip growth and initiate a Nod factor–dependent polar growth (Figure 6B). Interestingly, nsp mutants respond to Nod factors by initiating root hair growth but are defective for shepherd's crook formation in the presence of rhizobia, suggesting that one or more additional mechanisms controlled by the Nod factor signaling pathway characterized here are involved in shepherd's crook formation.
Numerous studies of tip growth in root hairs of Arabidopsis, in fungal mycelia, and in pollen tubes, essentially in Lilium longiflorum and Agapanthus umbellatus, have shown that a tip-focused calcium gradient, the result of a localized calcium influx, controls tip growth and orientation (reviewed in Malhò, 1998; Yang, 1998; Sanders et al., 1999). For example, tip swelling in pollen tubes after treatments that disrupt polar growth is preceded by loss of the tip-focused calcium gradient and coincides with the reestablishment of this gradient (Malhò et al., 1995). Reinitiation of root hair tip growth, after Nod factor treatment, also involves the establishment of a tip-focused calcium gradient (de Ruijter et al., 1998), whereas changes in actin filaments have been associated both with the stage at which Nod factors abolish tip growth and with the reinitiation of growth (Cárdenas et al., 1998; de Ruijter et al., 1999; Miller et al., 1999). Future studies of Nod factor influence on legume root hair polar growth will aim at further dissecting the Nod factor transduction pathway to identify what components are required for both inhibiting endogenous root hair tip growth and initiating a Nod factor–dependent tip growth. For example, cell biology studies of Nod factor responses of wild-type and dmi mutants should allow us to identify the role of calcium and of various cellular components (e.g., the cytoskeleton, Golgi vesicles, and the nucleus). The finding that a lasting inhibition on tip growth is observed in dmi mutants in response to Nod factors may also be a useful tool for studying the mechanisms underlying polar growth. For example, in contrast to the transient suppression of tip growth by various chemicals, such as cytochalasin D (Miller et al., 1999), or by mechanical constraints (Bibikova et al., 1997), a lasting inhibition of tip growth is observed in dmi mutants by a defined ligand at low, nontoxic concentrations (10–8 to 10–11 M).
DMI1, DMI2, and DMI3 Control Steps of a Pathway Common to Nodulation and Endomycorrhization
dmi1, dmi2, and dmi3 mutants are not only defective for symbiosis with Rhizobium but are also unable to establish a symbiotic association with arbuscular endomycorrhizal fungi. Furthermore, these mutants are blocked at an early stage of both interactions, with mycorrhizal infection being limited in each case to formation of appressoria. Such Nod−Myc− mutants were first identified in pea (Pisum sativum) and faba bean (Vicia faba), where their ability to interact with soil pathogens was shown to be unaffected (Duc et al., 1989; Gollotte et al., 1993). Four genes essential for both the rhizobial and the endomycorrhizal symbioses have now been identified in pea (SYM8, SYM9, SYM19, and SYM30; Albrecht et al., 1999), and Nod−Myc− mutants have been characterized in other legume species also, including bean (Phaseolus vulgaris) (Shirtliffe and Vessey, 1996), L. japonicus (Szczyglowski et al., 1998; Wegel et al., 1998), M. sativa (Bradbury et al., 1991), and M. truncatula (Sagan et al., 1995, 1998; this study). Also, in pea, mycorrhizal mutants have been identified that both are affected at the stage of arbuscule development and show a delayed nodulation phenotype (Gianinazzi-Pearson, 1996).
Besides controlling both the Rhizobium–legume and the endomycorrhizal symbioses, the gene SYM8 of pea controls a step of a signal transduction pathway, induced both by Nod factors and by endomycorrhizal fungi and leading to PsENOD5 and PsENOD12A expression (Albrecht et al., 1998). Like SYM8 of pea, DMI1, DMI2, and DMI3 control both Nod factor signal transduction (leading to nodulin gene expression) and endomycorrhizae formation, and possibly one of the M. truncatula genes described in this work is the ortholog of SYM8 of pea. Nevertheless, we can say that M. truncatula has at least three common steps to the establishment of nodulation and endomycorrhization, all of which are involved in a Nod factor–activated signal transduction pathway leading to the induction of symbiotic responses. Previous data have suggested a possible conservation of signal transduction pathways between these two symbioses (van Rhijn et al., 1997; Albrecht et al., 1998), and the activation by both symbiotic partners of a genetic program that paves the way for accepted infection has been proposed as the welcoming “red carpet” (Gianinazzi-Pearson and Dénarié, 1997).
Unlike mutations in DMI1, DMI2, and DMI3, mutations in NSP do not affect the ability of plants to establish a symbiotic association with arbuscular endomycorrhizal fungi. Our data therefore indicate that we have identified a component required for Nod factor–activated signal transduction that is specific to the Rhizobium–legume symbiosis. In contrast to the Rhizobium–legume association, arbuscular endomycorrhizal symbioses show very little host specificity (for reviews on endomycorrhizae, see Gianinazzi-Pearson, 1996; Smith and Read, 1997; Harrison, 1999). Given this difference, it is perhaps surprising that genes intervening at early steps of the Nod factor–activated pathway should be common to both endosymbiotic associations. However, by analogy to the role of Nod factors in activating this pathway, it has been proposed that mycorrhizal signals also exist (Figure 6A, “Myc factors”) and activate the pathway during the endomycorrhizal symbiosis (Albrecht et al., 1998, 1999). As a next step toward understanding the mechanisms involved in the establishment of rhizobial and endomycorrhizal symbioses, we have initiated map-based cloning projects for DMI1, DMI2, and DMI3 in collaboration with the group of T. Huguet (INRA-CNRS, Toulouse, France).
METHODS
Bacterial Stains
Bacterial strains and plasmids are described in Table 1. Conditions used for bacterial growth and conjugation experiments have been described previously (Truchet et al., 1985). The strain GMI3198 was constructed by introducing the plasmid pGMI1394, carrying the regulatory gene nodD1, into the strain Sinorhizobium meliloti 2011(pXLDG4) by triparental mating using the helper plasmid pRK2013. The plasmid pGMI1394 confers the ability to overproduce nodulation (Nod) factors in various genetic backgrounds (Demont et al., 1994). The strain GMI3125 was constructed by marker exchange between the plasmid pRmM57, which carries a spectinomycin resistance (Sp) cassette inserted in the S. meliloti nodC gene, and the megaplasmid pSymA of S. meliloti GMI6628, which carries mutations in the nodF and nodL genes. pRmM57 was introduced into GMI6628 by triparental mating using the helper plasmid pRK2013, and phleomycin- (Pm; 20 μg/mL) and tetracycline- (Tc; 10 μg/mL) resistant colonies were selected. pRmM57 was then excluded from this strain by introducing the incompatible plasmid pR751-pMG2. Gentamicin- (Gm; 50 μg/mL) and Sp- (100 μg/mL) resistant colonies from this conjugation were found to be Tc-sensitive, indicating loss of pRm57. Homologous recombination in nodC was confirmed by DNA gel blot analysis (data not shown).
Plant Growth Conditions
Freshly collected seeds of Medicago truncatula enter dormancy within ∼1 week after pod falling; the germination rate is then very low for 3 to 4 months. To avoid the entry into dormancy, the majority of freshly collected seeds were stored at 4°C. To germinate them, the seeds were removed from their pods, dried overnight at 30°C, scarified with sandpaper, and then surface-sterilized with 12% sodium hypochlorite for 2 min. Seeds that had been stored at room temperature for >3 months were scarified directly by treatment with concentrated sulfuric acid for 7 min and then sterilized as described above. Both types of seeds were germinated on 1% deionized water agar plates at 14°C for 16 to 24 hr; plates with freshly collected seeds were first stored for at least 5 days at 4°C. When seeds were germinated to grow plants for seed production, sterilized seeds were vernalized for 10 days at 4°C before germinating them at 14°C.
Plants were all axenically grown on Fahraeus medium (0.132 g/L CaCl2, 0.12 g/L MgSO4.7H2O, 0.1 g/L KH2PO4, 0.075 g/L Na2HPO4.2H2O, 5 mg/L Fe-citrate, and 0.07 mg/L each of MnCl2.4H2O, CuSO4.5H2O, ZnCl2, H3BO3, and Na2MoO4.2H2O, adjusted to pH 7.5 before autoclaving), except in aeroponic culture. The composition of the low-nitrogen aeroponic medium was 1 mM CaCl2, 0.25 mM MgSO4.7H2O, 1.7 mM KH2PO4, 3.8 mM K2HPO4, 0.5 mM K2SO4, 0.05 mM FeSO4.7H2O, 0.05 mM Na2EDTA, 30 μM H3BO3, 10 μM MnSO4, 0.7 μM ZnSO4, 1 μM Na2MoO4.2H2O, 0.04 μM CoCl, and 0.2 μM CuSO4.5H2O (Lullien et al., 1987). Certain plants were grown on Fahraeus agar (1.5%), either as slopes (Truchet et al., 1985) for nodulation tests or in Petri dishes for histological and cytological studies, root hair deformation tests, and MtENOD40 expression studies. Others were grown on liquid medium, either in aeroponic culture in large plastic trash bins—modified by the addition of a motor to vaporize the growth medium and a lid adapted with 1000 small holes to enable the roots of seedlings to be pushed through for nodulation tests and MtENOD11–GUS transgenic studies—or in growth pouches (Journet et al., 1994) for nodulin gene expression studies. Plants were grown in the absence of added nitrate, except for studies on the effect of nitrate on root hair deformation and for cosegregation studies, in which the plants were first grown for 7 days in high-nitrogen aeroponic medium containing 5 mM NH4NO3 and then starved of nitrogen for 2 days before inoculation. Plants grown in aeroponic culture were placed in a growth chamber at 20°C with a 14-hr photoperiod at 70 μE sec–1 m–2. All other axenic plants were placed in a growth chamber at 20°C with a 16-hr photoperiod at 65 μE sec–1 m–2. For allelism tests and seed production, plants were grown in pots in a mixture of sterile sand/compost (1:2) in a growth room at 25/20°C, a 16-hr photoperiod, and 230 μE sec–1 m–2. Plants for allelism tests were grown in 9 × 9 × 8-cm pots and allowed to spread horizontally. Plants for seed production were grown in 8 × 8 × 7-cm pots and trained to grow vertically inside inverted 1.5-liter plastic, mineral water bottles. This system was essentially a large Aracon system (Beta-Tech, Gent, Belgium), as used for Arabidopsis thaliana. Initially, a single bottle was inserted into an Aracon, and young plants were trained to grow into the bottle; subsequently, as plants grew, a second and a third neckless bottle were added. In this way, as many as 100 plants could be grown per square meter, thus maximizing the use of growth room space. These vertically grown plants started flowering within 10 days, producing at least 300 seeds per plant over two to three months.
Bacterial Inoculation and Nod Factor Treatment of Plants
Plants were inoculated with 0.5 mL of a suspension of 108 bacteria/mL in water covering the root system, except for in situ hybridizations and cytology studies, when Petri dish–grown plants were spot-inoculated with 0.2 μL of a bacterial suspension in the region of root hair emergence. Inoculation was performed at the following times after germination: 3 days for nodulation tests in agar slopes, 5 days for nodulation tests in aeroponic culture, 7 days for histological and cytological characterization and in situ hybridizations, and 9 days for cosegregation studies in aeroponic culture. For Nod factor tests on root hair deformation, plants were treated with Nod factors (prepared as previously described by Roche et al. [1991a]) diluted in water at 24 hr after germination. When studying the effect of nitrate on root hair deformation, however, we treated plants 7 days after germination, and for nodulin gene expression studies we treated them 5 days after germination (transgenic plants) or 7 to 11 days after germination (for reverse transcription–polymerase chain reaction [RT-PCR] and RNA gel blot analysis). Nod factors were added directly to plants growing on agar for root hair deformation studies, directly to plants growing in growth pouches for RT-PCR, and as described by Pingret et al. (1998) and Cook et al. (1995) for transgenic plants and RNA gel blot analysis, respectively. Root hair deformation tests were compared between the seedlings grown first on agar and then transferred to Fahraeus slides, as described by Heidstra et al. (1994), and the seedlings growing on agar. Because this comparison gave similar results (data not shown), all reported results are those for studies performed directly on seedlings grown on agar.
Isolation of Nod− Mutants and Genetic Analysis
Seedlings from ∼50,000 ethyl methanesulfonate (EMS)–mutagenized M2 seeds of M. truncatula Jemalong (Penmetsa and Cook, 2000) were grown in aeroponic culture conditions (∼700 seedlings per container) and scored for absence of nodules 10 days after inoculation with S. meliloti ABS7 (Table 1). Putative Nod− mutants were planted and left to set seed. Nodulation tests were performed on 20 M3 plants from each mutant inoculated with S. meliloti GMI6526 (Table 1). Mutants for which all 20 plants were Nod− 3 weeks after inoculation were considered as confirmed Nod− mutants. Three γ-ray Nod− mutants of M. truncatula were included in this study (kindly provided by G. Duc, INRA, Dijon, France). All mutants were crossed manually (by using method 3 of Pathipanawat et al. [1994], except that female flowers were emasculated by removing anther sacs with forceps and crosses were performed at any time of the day) to transgenic Nod+ M. truncatula plants expressing a fusion between the MtENOD11 promoter and the reporter gene encoding β-glucuronidase (GUS) (D.G. Barker, J.-L. Pingret, M. Chabaud, and E.-P. Journet, unpublished results). In these crosses, transgenic plants were used as the source of pollen, which was transferred to flowers of emasculated mutant plants. The presence of the transgene in the F1 progeny (detected by nonsymbiotic expression of MtENOD11–GUS in root caps, lateral root primordia, and aerial parts of plants) confirmed that these plants were true hybrids. Nodulation tests (with S. meliloti GMI6526) were performed on F1 plants to determine the recessive/dominant nature of the mutations. F1 plants were then left to set seed, and nodulation tests (with S. meliloti GMI6526) were performed on F2 plants to determine whether the mutations were inherited monogenically. Phenotypic characterization was performed on F3 mutant plants (i.e., backcrossed once). In general, several different F3 families (each derived from a single Nod− F2 plant) were checked for the Nod− phenotype, and then a single F3 family was selected for detailed characterization. To be able to study Nod factor induction of the MtENOD11–GUS fusion, we checked nonsymbiotic expression of MtENOD11–GUS (in root caps, lateral root primordia, and aerial parts of plants) in 16 to 20 F3 plants to confirm that they were derived from F2 plants homozygous for the transgene. For allelism tests, mutants were crossed to each other. Whenever possible, each mutant was used as both a male and a female partner in each pairwise cross, with the male partner carrying the transgene MtENOD11–GUS as a marker (see above). A wild-type Nod+ phenotype of F1 plants was scored as indicating complementation. Mutants were considered allelic when three or more independent crosses, all confirmed by the presence of the marker, yielded only F1 plants with the mutant Nod− phenotype.
Microscopic Methods
Plants were observed for infection events by histochemical staining for β-galactosidase activity expressed by the plasmid pXLGD4, as previously described (Ardourel et al., 1994). Plants were observed for root hair deformations after staining with methylene blue as previously described (Vasse and Truchet, 1984). Transgenic MtENOD11–GUS plants were stained and observed for GUS activity as described by Pingret et al. (1998), except that plants were stained at 5 days after germination. To look for cortical cell divisions, we embedded spot-inoculated root sections in polyethylene glycol as described by Timmers et al. (1999) and prepared semithin sections (15 to 20 μm thick) by using a microtome (model 2040; Reichert-Jung, Vienna, Austria). One mutant was studied for each locus (B129, TR25, TRV25, and B85), and between 20 and 30 individuals were scored for each mutant. For bright- and dark-field microscopy, plants were viewed with Zeiss (Jena, Germany) Axiophot I and II microscopes.
RT-PCR Assay
At 0, 12, and 48 hr after Nod factor addition, reactive root zones (3 cm from the primary root tip) of ∼30 seedlings that were 7 to 11 days old were frozen in liquid nitrogen, and the total RNA was extracted as described by Jackson and Larkins (1976). cDNA synthesis and RT-PCR were performed as described by Pingret et al. (1998), except that cDNA samples were purified by passage through Sephadex G-50 columns (Amersham Pharmacia Biotech, Saclay, France), precipitated, and resuspended to ∼5 ng/μL, of which 10-μL aliquots were used for PCR amplification (20 or 25 cycles at 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min).
MtENOD11 cDNA was amplified (25 cycles) between positions 82 and 519 by using the forward primer 5′-CTCCATCCCACAATATGCCTCCA-3′ and the reverse primer 5′-ATCGATGCTAGGTGGAGGCT-3′. To control for equivalent cDNA contents, we also performed PCR amplifications for MtPR10-1, a constitutively and strongly expressed gene of roots (Gamas et al., 1998). The MtPR10-1 cDNA was amplified (20 cycles) between positions 149 and 433 by using the forward primer 5′-CTCCATCCCACAATATGCCTCCA-3′ and the reverse primer 5′-ATCGATGCTAGGTGGAGGCT-3′. To control that the amount of amplified product was proportional to the input of cDNA, we also performed PCR with cDNA dilutions, and the results were analyzed with a PhosphoImager (model Storm 840; Molecular Dynamics, Sunnyvale, CA). PCR products were analyzed by DNA gel blotting with the 450-bp MaeI/BstEII fragment of MtENOD11 (E.P. Journet, C. Calantzis, V. Vernoud, F. de Billy, G. Truchet, M. Pichon, A. Dedieu, D. Morandi, V. Gianinazzi-Pearson, and D. Barker, unpublished results) and with the complete cDNA of MtPR10-1 (Gamas et al., 1998).
RNA Gel Blot Analysis
At 0, 12, 24, and 48 hr after Nod factor addition, reactive root zones (3 cm from the primary root tip) of ∼30 seedlings 7 to 11 days old were frozen in liquid nitrogen. Total RNA and RNA gel blot analysis for rip1 expression was performed as described in Cook et al. (1995).
In Situ Hybridization Assay
In situ hybridizations were performed on 7-μm-thick sections essentially as described by Gamas et al. (1998). Approximately 10 plants were analyzed per mutant. An MtENOD40 cDNA clone (Gamas et al., 1996) was used to make MtENOD40 RNA probes with T3 (sense) or T7 (antisense) RNA polymerase (Promega) and α-35S-UTP (DuPont–New England Nuclear). In vitro transcription was performed at 37°C for 1 hr, followed by treatment with 1 unit of RQ1 RNase-free DNase (Promega) for 15 min and extraction with phenol-chloroform. Unincorporated ribonucleotides were removed by passage of the probes through MicroSpin S-200 columns (Pharmacia Biotech); the probes were then partially hydrolyzed by incubation in 60 mM Na2CO3, 40 mM NaHCO3, and 7.5 mM DTT in the presence of 80 units of a ribonuclease inhibitor (RNasin; Promega) at 60°C for 40 min. This period was calculated to reduce the probes from full length (700 bp) to ∼150 bp. Hydrolysis was stopped by adding acetic acid to a final concentration of 0.1 M, after which the probes were ethanol-precipitated in the presence of 2 μg of yeast tRNA. Probe sizes were determined by analyzing aliquots taken before and after hydrolysis on formaldehyde gels with an RNA ladder (Gibco BRL). Incorporation of radiolabel was measured with a Packard Tri-Carb 2100TR liquid scintillation analyzer (Meriden, CT), and each slide was hybridized with 6 × 106 count/min.
Acknowledgments
We are very grateful to David Barker for kindly providing us with transgenic lines of M. truncatula carrying fusions between the MtENOD11 promoter and the GUS reporter gene, which were provided before publication. We are also extremely grateful to Gérard Duc for providing us with seeds of TR25, TR26, and TRV25. We thank Maria Harrison, Vivienne Gianinazzi-Pearson, and Dominique Morandi for providing information on the mycorrhization phenotypes of mutants. Thanks also to Frédéric Debellé for his precious help in constructing the strain GMI3125 and for helpful discussions. The MtENOD40 cDNA clone used for in situ hybridizations was provided by Pascal Gamas. R.C. and C.G. were financed by the French Ministère de l'Enseignement Supérieur et de la Recherche. Financial support for the project was provided from the Human Frontier Science Program Organization (Grant No. RG0327/1998-M) and from the Centre National de la Recherche Scientifiqe (Genome program, 1998–2000) and the Institut National de la Recherche Agronomique (Genomes and Functions program, 1998–1999).
References
- Albrecht, C., Geurts, R., Lapeyrie, F., and Bisseling, T. (1998). Endomycorrhizae and rhizobial Nod factors both require Sym8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J. 15, 605–614. [DOI] [PubMed] [Google Scholar]
- Albrecht, C., Geurts, R., and Bisseling, T. (1999). Legume nodulation and mycorrhizae formation; two extremes in host specificity meet. EMBO J. 18, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel, M., Demont, N., Debellé, F., Maillet, F., de Billy, F., Promé, J.C., Dénarié, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6, 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asad, S., Fang, Y.W., Wycoff, K.L., and Hirsch, A.M. (1994). Isolation and characterization of cDNA and genomic clones of MsENOD40: Transcripts are detected in meristematic cells of alfalfa. Protoplasma 183, 10–23. [Google Scholar]
- Barker, D.G., et al. (1990). Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium–legume symbiosis. Plant Mol. Biol. Rep. 8, 40–49. [Google Scholar]
- Bekki, A., Trinchant, J.-C., and Rigaud, J. (1987). Nitrogen fixation (C2H2 reduction) by Medicago nodules and bacteroids under sodium chloride stress. Physiol. Plant. 71, 61–67. [Google Scholar]
- Bibikova, T.N., Zhigilei, A., and Gilroy, S. (1997). Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta 203, 495–505. [DOI] [PubMed] [Google Scholar]
- Bradbury, S., Peterson, R.L., and Bowley, S.R. (1991). Interactions between three alfalfa nodulation genotypes and two Glomus species. New Phytol. 119, 115–120. [DOI] [PubMed] [Google Scholar]
- Cárdenas, L., Vidali, L., Dominguez, J., Pérez, H., Sánchez, F., Hepler, P.K., and Quinto, C. (1998). Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol. 116, 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, L., Feijó, J.A., Kunkel, J.G., Sánchez, F., Holdaway-Clarke, T., Hepler, P.K., and Quinto, C. (1999). Rhizobium nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J. 19, 347–352. [DOI] [PubMed] [Google Scholar]
- Carroll, B.J., and Mathews, A. (1990). Nitrate inhibition of nodulation in legumes. In The Molecular Biology of Symbiotic Nitrogen Fixation, P.M. Gresshoff, ed (Boca Raton, FL: CRC Press), pp. 159–180.
- Chang, C., and Shockey, J.A. (1999). The ethylene-response pathway: Signal perception to gene regulation. Curr. Opin. Plant Biol. 2, 352–358. [DOI] [PubMed] [Google Scholar]
- Charon, C., Johansson, C., Kondorosi, E., Kondorosi, A., and Crespi, M. (1997). enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc. Natl. Acad. Sci. USA 94, 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon, C., Sousa, C., Crespi, M., and Kondorosi, A. (1999). Alteration of enod40 expression modifies Medicago truncatula root nodule development induced by Sinorhizobium meliloti. Plant Cell 11, 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, J., Day, R.B., and Stacey, G. (1998). Legume nodule organogenesis. Trends Plant Sci. 3, 105–110. [Google Scholar]
- Cook, D., Dreyer, D., Bonnet, D., Howell, M., Nony, E., and Vandenbosch, K. (1995). Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D.R. (1999). Medicago truncatula—A model in the making! Curr. Opin. Plant Biol. 2, 301–304. [DOI] [PubMed] [Google Scholar]
- Cook, D.R., Vandenbosch, K., de Bruijn, F.J., and Huguet, T. (1997). Model legumes get the nod. Plant Cell 9, 275–281. [Google Scholar]
- Crespi, M.D., Jurkevitch, E., Poiret, M., d'Aubenton-Carafa, Y., Petrovics, G., Kondorosi, E., and Kondorosi, A. (1994). enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13, 5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé, F., Rosenberg, C., Vasse, J., Maillet, F., Martinez, E., Dénarié, J., and Truchet, G. (1986). Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J. Bacteriol. 168, 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demont, N., Ardourel, M., Maillet, F., Promé, D., Ferro, M., Promé, J.-C., and Dénarié, J. (1994). The Rhizobium meliloti regulatory nodD3 and syrM genes control the synthesis of a particular class of nodulation factors N-acylated by (ω-1)-hydroxylated fatty acids. EMBO J. 13, 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié, J., and Cullimore, J. (1993). Lipo-oligosaccharide nodulation factors: A new class of signalling molecules mediating recognition and morphogenesis. Cell 74, 951–954. [DOI] [PubMed] [Google Scholar]
- Dénarié, J., Debellé, F., and Promé, J.C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: Signalling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65, 503–535. [DOI] [PubMed] [Google Scholar]
- de Ruijter, N.C.A., Rook, M.B., Bisseling, T., and Emons, A.M.C. (1998). Lipochito-oligosaccharides re-initiate root hair tip growth in Vicia sativa with high calcium and spectrin-like antigen at the tip. Plant J. 13, 341–350. [Google Scholar]
- de Ruijter, N.C.A., Bisseling, T., and Emons, A.M.C. (1999). Rhizobium Nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol. Plant-Microbe Interact. 12, 829–832. [Google Scholar]
- Ditta, G., Stanfield, S., Corbin, D., and Helinski, D.R. (1980). Broad host range DNA cloning system for Gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie, J.A., and Walker, S.A. (1999). Plant responses to nodulation factors. Curr. Opin. Plant Biol. 2, 483–489. [DOI] [PubMed] [Google Scholar]
- Duc, G., Trouvelot, A., Gianinazzi-Pearson, V., and Gianinazzi, S. (1989). First report of non-mycorrhizal plant mutants (Myc-) obtained in pea (Pisum sativum L.) and faba bean (Vicia faba L.). Plant Sci. 60, 215–222. [Google Scholar]
- Ehrhardt, D.W., Atkinson, E.M., and Long, S.R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256, 998–1000. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85, 673–681. [DOI] [PubMed] [Google Scholar]
- Etzler, M.E., Kalsi, G., Ewing, N.N., Roberts, N.J., Day, R.B., and Murphy, J.B. (1999). A nod factor binding lectin with apyrase activity from legume roots. Proc. Natl. Acad. Sci. USA 96, 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., and Hirsch, A.M. (1998). Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 116, 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K.A., Malmberg, R.L., and Dean, C. (1994). Mutagenesis in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 137–172.
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1995). Nod signal–induced plasma membrane potential changes in alfalfa root hairs are differentially sensitive to structural modifications of the lipochitooligosaccharide. Plant J. 7, 939–947. [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1996). Rapid alkalinization in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 10, 295–301. [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1998). The role of ion fluxes in Nod factor signalling in Medicago sativa. Plant J. 13, 455–463. [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1999. a). Nod factors modulate the concentration of cytosolic free calcium differently in growing and non-growing root hairs of Medicago sativa L. Planta 209, 207–212. [DOI] [PubMed] [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1999. b). Elevation of the cytosolic free [Ca2+] is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiol. 121, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas, P., Niebel, F.D.C., Lescure, N., and Cullimore, J.V. (1996). Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant-Microbe Interact. 9, 233–242. [DOI] [PubMed] [Google Scholar]
- Gamas, P., de Billy, F., and Truchet, G. (1998). Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol. Plant-Microbe Interact. 11, 393–403. [DOI] [PubMed] [Google Scholar]
- Gehring, C.A., Irving, H.R., Kabbara, A.A., Parish, R.W., Boukli, N.M., and Broughton, W.J. (1997). Rapid, plateau-like increases in intracellular free calcium are associated with Nod-factor–induced root-hair deformation. Mol. Plant-Microbe Interact. 10, 791–802. [Google Scholar]
- Geurts, R., Heidstra, R., Hadri, A.E., Downie, J.A., Franssen, H., Van Kammen, A., and Bisseling, T. (1997). Sym2 of pea is involved in a nodulation factor–perception mechanism that controls the infection process in the epidermis. Plant Physiol. 115, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson, V. (1996). Plant cell response to arbuscular mycorrhizal fungi: Getting to the roots of the symbiosis. Plant Cell 8, 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson, V., and Dénarié, J. (1997). Red carpet genetic programmes for root endosymbiosis. Trends Plant Sci. 2, 371–372. [Google Scholar]
- Gollotte, A., Gianinazzi-Pearson, V., Giovannetti, M., Sbrana, C., Avio, L., and Gianinazzi, S. (1993). Cellular localization and cytochemical probing of resistance reactions to arbuscular mycorrhizal fungi in a “locus a” mutant of Pisum sativum L. Planta 191, 112–122. [Google Scholar]
- Gressent, F., Drouillard, S., Mantegazza, N., Samain, E., Geremia, R.A., Canut, H., Niebel, A., Driguez, H., Ranjeva, R., Cullimore, J., and Bono, J.J. (1999). Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic Nod factors in Medicago cell suspension cultures. Proc. Natl. Acad. Sci. USA 96, 4704–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, M.J. (1999). Molecular and cellular aspects of the arbuscular mychorrhizal symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 361–389. [DOI] [PubMed] [Google Scholar]
- Heidstra, R., Geurts, R., Franssen, H., Spaink, H.P., Van Kammen, A., and Bisseling, T. (1994). Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 105, 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra, R., Nilsen, G., Martinezabarca, F., Vankammen, A., and Bisseling, T. (1997). Nod factor–induced expression of leghemoglobin to study the mechanism of NH4NO3 inhibition on root hair deformation. Mol. Plant-Microbe Interact. 10, 215–220. [DOI] [PubMed] [Google Scholar]
- Honma, M.A., Asomaning, M., and Ausubel, F.M. (1990). Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J. Bacteriol. 172, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.D., and Larkins, B.A. (1976). Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol. 57, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, G.A., Jacob, A.E., and Hedges, R.W. (1976). Recombination between plasmids of incompatibility groups P1 and P2. J. Bacteriol. 127, 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet, E.P., Pichon, M., Dedieu, A., de Billy, F., Truchet, G., and Barker, D.G. (1994). Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J. 6, 241–249. [DOI] [PubMed] [Google Scholar]
- Kendall, M.G., and Stuart, A. (1976). The Advanced Theory of Statistics. (London: Charles Griffin Ltd).
- Kurkdjian, A. (1995). Role of the differentiation of root epidermal cells in Nod factor (from Rhizobium meliloti)–induced root hair depolarization of Medicago sativa. Plant Physiol. 107, 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, S.A., Williams, P.H., and Ditta, G.S. (1985). Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 13, 5965–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Promé, J.C., and Dénarié, J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344, 781–784. [DOI] [PubMed] [Google Scholar]
- Long, S.R. (1996). Rhizobium symbiosis: Nod factors in perspective. Plant Cell 8, 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lullien, V., Barker, D.G., De Lajudie, P., and Huguet, T. (1987). Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol. Biol. 9, 469–478. [DOI] [PubMed] [Google Scholar]
- Malhò, R. (1998). Expanding tip-growth theory. Trends Plant Sci. 3, 40–42. [Google Scholar]
- Malhò, R., Read, N.D., Trewavas, A.J., and Pais, M.S. (1995). Calcium channel activity during pollen tube growth and reorientation. Plant Cell 7, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwei, C.M., and Larue, T.A. (1992). Phenotypic characterization of sym8 and sym9, two genes conditioning non-nodulation in Pisum sativum “Sparkle.” Can. J. Microbiol. 38, 548–554. [Google Scholar]
- Miller, D.D., deRuijter, N.C.A., Bisseling, T., and Emons, A.M.C. (1999). The role of actin in root hair morphogenesis: Studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 17, 141–154. [Google Scholar]
- Mulligan, J.T., and Long, S.R. (1985). Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc. Natl. Acad. Sci. USA 82, 6609–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona, P., Pawlowski, K., and Bisseling, T. (1995). Symbiotic nitrogen fixation. Plant Cell 7, 869–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathipanawat, W., Jones, R.A.C., and Sivasithamparam, K. (1994). An improved method for artificial hybridization in annual Medicago species. Aust. J. Agric. Res. 45, 1329–1335. [Google Scholar]
- Peng, H.M., Dreyer, D.A., Vandenbosch, K.A., and Cook, D. (1996). Gene structure and differential regulation of the Rhizobium-induced peroxidase gene rip1. Plant Physiol. 112, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530. [DOI] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (2000). Production and characterization of diverse developmental mutants in Medicago truncatula. Plant Physiol 123, 1387.–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingret, J.L., Journet, E.P., and Barker, D.G. (1998). Rhizobium Nod factor signaling. Evidence for a G protein–mediated transduction mechanism. Plant Cell 10, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, P., Lerouge, P., Ponthus, C., and Promé, J.C. (1991. a). Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti–alfalfa symbiosis. J. Biol. Chem. 266, 10933–10940. [PubMed] [Google Scholar]
- Roche, P., Debellé, F., Maillet, F., Lerouge, P., Faucher, C., Truchet, G., Dénarié, J., and Promé, J.C. (1991. b). Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67, 1131–1143. [DOI] [PubMed] [Google Scholar]
- Sagan, M., Morandi, D., Tarenghi, E., and Duc, G. (1995). Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn) after γ-ray mutagenesis. Plant Sci. 111, 63–71. [Google Scholar]
- Sagan, M., de Larambergue, H., and Morandi, D. (1998). Genetic analysis of symbiosis mutants in Medicago truncatula. In Biological Nitrogen Fixation for the 21st Century, C. Elmerich, A. Kondorosi, and W.E. Newton, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 317–318.
- Sanders, D., Brownlee, C., and Harper, J.F. (1999). Communicating with calcium. Plant Cell 11, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195. [DOI] [PubMed] [Google Scholar]
- Schultze, M., and Kondorosi, A. (1998). Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32, 33–57. [DOI] [PubMed] [Google Scholar]
- Shirtliffe, S.J., and Vessey, J.K. (1996). A nodulation (Nod+/Fix−) mutant of Phaseolus vulgaris L. has nodule-like structures lacking peripheral vascular bundles (Pvb−) and is resistant to mycorrhizal infection (Myc−). Plant Sci. 118, 209–220. [Google Scholar]
- Smith, S.E., and Read, D.J. (1997). Mycorrhizal Symbiosis. (London: Academic Press).
- Szczyglowski, K., Shaw, R.S., Wopereis, J., Copeland, S., Hamburger, D., Kasiborski, B., Dazzo, F.B., and de Bruijn, F.J. (1998). Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant-Microbe Interact. 11, 684–697. [Google Scholar]
- Timmers, A.C., Auriac, M.C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium–Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 12, 3617–3628. [DOI] [PubMed] [Google Scholar]
- Truchet, G., Debellé, F., Vasse, J., Terzaghi, B., Garnerone, A.M., Rosenberg, C., Batut, J., Maillet, F., and Dénarié, J. (1985). Identification of a Rhizobium meliloti pSym2011 region controlling the host specificity of root hair curling and nodulation. J. Bacteriol. 164, 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrup, L.J., Cary, A.J., and Norris, J.H. (1993). Five nodulation mutants of white sweetclover (Melilotus alba Desr) exhibit distinct phenotypes blocked at root hair curling, infection thread development, and nodule organogenesis. Plant Physiol. 103, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Batenburg, F.H.D., Jonker, R., and Kijne, J.W. (1986). Rhizobium induces marked root hair curling by redirection of tip growth, a computer simulation. Physiol. Plant. 66, 476–480. [Google Scholar]
- van Brussel, A.A.N., Bakhuizen, R., van Spronsen, P.C., Spaink, H.P., Tak, T., Lugtenberg, B.J.J., and Kijne, J.W. (1992). Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257, 70–72. [DOI] [PubMed] [Google Scholar]
- van Rhijn, P., et al. (1997). Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc. Natl. Acad. Sci. USA 94, 5467–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen, P.C., Bakhuizen, R., van Brussel, A.A.N., and Kijne, J.W. (1994). Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process. Eur. J. Cell Biol. 64, 88–94. [PubMed] [Google Scholar]
- Vasse, J., and Truchet, G. (1984). The Rhizobium–legume symbiosis: Observation of root infection by bright-field microscopy after staining with methylene blue. Planta 161, 487–489. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Journet, E.P., and Barker, D. (1999). MtENOD20, a Nod-factor–inducible molecular marker for root cortical cell activation. Mol. Plant-Microbe Interact. 12, 604–614. [Google Scholar]
- Vijn, I., Martinez-Abarca, F., Yang, W.C., das Neves, L., van Brussel, A., Van Kammen, A., and Bisseling, T. (1995). Early nodulin gene expression during Nod factor–induced processes in Vicia sativa. Plant J. 8, 111–119. [DOI] [PubMed] [Google Scholar]
- Wegel, E., Schauser, L., Sandal, N., Stougaard, J., and Parniske, M. (1998). Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol. Plant-Microbe Interact. 11, 933–936. [Google Scholar]
- Yang, Z. (1998). Signaling tip growth in plants. Curr. Opin. Plant. Biol. 1, 525–530. [DOI] [PubMed] [Google Scholar]