Abstract
Arabidopsis mismatch repair genes predict MutS-like proteins remarkably similar to eukaryotic MutS homologs—MSH2, MSH3, and MSH6. A novel feature in Arabidopsis is the presence of two MSH6-like proteins, designated AtMSH6 and AtMSH7. Combinations of Arabidopsis AtMSH2 with AtMSH3, AtMSH6, or AtMSH7 proteins—products of in vitro transcription and translation—were analyzed for interactions by analytical gel filtration chromatography. The AtMSH2 protein formed heterodimers with AtMSH3, AtMSH6, and AtMSH7, but no single proteins formed homodimers. The abilities of the various heterodimers to bind to mismatched 51-mer duplexes were measured by electrophoretic mobility-shift assays. Similar to the behavior of the corresponding human proteins, AtMSH2•AtMSH3 heterodimers bound “insertion–deletion” DNA with three nucleotides (+AAG) or one nucleotide (+T) looped out much better than they bound DNA with a base/base mispair (T/G), whereas AtMSH2•AtMSH6 bound the (+T) substrate strongly, (T/G) well, and (+AAG) no better than it did a (T/A) homoduplex. However, AtMSH2•AtMSH7 showed a different specificity: moderate affinity for a (T/G) substrate and weak binding of (+T). Thus, AtMSH2•AtMSH7 may be specialized for lesions/base mispairs not tested or for (T/G) mispairs in special contexts.
INTRODUCTION
Most organisms maintain genomic stability by using highly conserved protein machines which correct errors that result from DNA replication. Mitotic plant cells in particular might need efficient post-replication correction of errors. Because plants lack a reserved germ line, the meristematic cells that finally give rise to the gametes have divided more or less continually during the life of the plant, potentially accumulating both spontaneous and environmentally induced mutations. In all eukaryotes examined, as well as in most eubacteria, highly conserved mismatch repair systems correct DNA replication errors that escape mechanisms for polymerase base selection and proofreading fidelity, typically further reducing spontaneous mutation rates by factors of 102 to 103 (Kornberg and Baker, 1992; Modrich and Lahue, 1996). Recent identifications of Arabidopsis homologs of mismatch repair genes (Culligan and Hays, 1997; Bevan et al., 1998; Ade et al., 1999) suggest that similar systems function in plants.
Mismatch repair functions extend well beyond simple post-replication error correction. Although mismatch repair systems may promote gene conversion during homologous recombination by correcting occasional mismatches in heteroduplex intermediates, they can also recognize excessive heterozygosity and abort the exchange process. This might help prevent chromosomal “scrambling” resulting from recombination between diverged members of gene families (Petit et al., 1991), maintain barriers against massive genetic exchange (Rayssiguier et al., 1989), or prevent productive meiosis in interspecies hybrids (Hunter et al., 1996). Mismatch repair proteins recognize a surprising diversity of damaged DNA bases (Kat et al., 1993; Drummond et al., 1996; Duckett et al., 1996; Li et al., 1996; Wang et al., 1999). In the case of UV light photoproducts, binding appears directed to photoproduct/base mispairs (Mu et al., 1997; Wang et al., 1999), providing a potential mechanism for antagonism of UV mutagenesis (Liu et al., 2000). Independently of processes for error correction, certain mismatch repair proteins play positive roles in the promotion of meiosis, including meiotic recombination, such that deficiencies in these proteins reduce fertility (Ross-Macdonald and Roeder, 1994; Hollingsworth et al., 1995; de Vries et al., 1999).
The Escherichia coli paradigm illustrates the essential features of mismatch repair: error recognition, identification of strand-specificity signals, specific excision of nascent—presumably error-encoding—DNA strands, gap-filling DNA synthesis, and ligation (Modrich and Lahue, 1996). After MutS protein homodimers bind to DNA base-mismatches (e.g., T/G) or to DNA containing insertion–deletion “loopouts” (IDLs; e.g., TTTT/AAA), MutL and MutS protein homodimers together activate MutH protein to specifically nick the unmethylated strand at hemimethylated GATC sequences in a process involving ATP-dependent protein–DNA translocation between mismatches and GATC sequences (Allen et al., 1997). Alternatively, as suggested for human MutS homolog hMSH2•hMSH6 heterodimers (Gradia et al., 1997, 1999), binding of ATP to MutS may “switch” the ATP-MutS to a “sliding clamp” that initiates downstream repair events. Because methylation of adenines in newly synthesized GATC sequences is delayed a few minutes after DNA replication, the nicks in unmethylated strands thus direct excision to error-containing nascent strands. Depending on the orientation of the nicks at GATC sequences relative to mismatches, excision requires 3′ to 5′ or 5′ to 3′ exonucleases. Notably, gap-filling synthesis involves replicative polymerase holoenzymes rather than DNA “repair” polymerases.
During the evolution of eukaryotes, mutS genes of eubacterial endosymbionts in primitive eukaryotes appear to have given rise, through a series of duplication/specialization events, to a set of MutS homolog (MSH) proteins with specialized functions (Culligan et al., 2000). MSH1 proteins (most probably the immediate descendents of bacterial MutS proteins) can recognize mismatches and are required for mitochondrial stability (Reenan and Kolodner, 1992; Chi and Kolodner, 1994). MSH2, MSH3, and MSH6 proteins mediate error correction (see below) (Acharya et al., 1996; Marsischky et al., 1996; Umar et al., 1998). MSH4 and MSH5 proteins, however, which play essential roles in meiosis, lack mismatch-recognition domains (Ross-Macdonald and Roeder, 1994; Hollingsworth et al., 1995; Culligan et al., 2000). Similarly, the bacterial mutL gene has given rise to at least four function-specific homologs in yeast and four or more in higher eukaryotes, designated MLH (for mutL homolog) or PMS (for postmeiotic segregation) (Kolodner and Marsischky, 1999).
In eukaryotes, the bacterial MutHLS paradigm has been conserved in many respects, but with important changes. Thus, mismatch recognition is now accomplished by heterodimers with different but overlapping specificities: MSH2•MSH6 heterodimers, sometimes designated as MutSα, recognize base/base mismatches and short IDLs, whereas MSH2•MSH3 (MutSβ) recognizes loopouts of various sizes but not base/base mismatches. The primary MutL function is accomplished by MutLα, an MLH1•PMS2 heterodimer (equivalent to MLH1•PMS1 in yeast) (Prolla et al., 1994; Li and Modrich, 1995; Kolodner and Marsischky, 1999). As in eubacteria, preexisting nicks 5′ or 3′ to a mismatch can be utilized for efficient excision and correction of errors in extracts; this error correction is dependent on MutSα/β and MutLα (Fang and Modrich, 1993; Habraken et al., 1998). However, the mechanism for strand-specific incision/excision in vivo remains unknown, given that eukaryotes (and some bacteria) lack both MutH homologs and adenine methylation. The ends of the replicating strands themselves might be utilized to initiate excision, or proteins in the replication apparatus, such as the proliferating-cell nuclear antigen (PCNA), may provide strand specificity (Johnson et al., 1996; Umar et al., 1996; Chen et al., 1999).
Having previously identified the Arabidopsis AtMSH2 gene, we sought to determine whether the eukaryotic pattern of interaction of MSH2 with other MSH proteins, to form heterodimers having different substrate specificities, is conserved in plants. A search for mutS homologs revealed not only AtMSH3 and AtMSH6 but also a homolog thus far unique to plants, designated AtMSH7. Interaction and binding studies demonstrate the potential of the respective AtMSH proteins to form three different heterodimers, each with a different substrate specificity.
RESULTS
Identification of Arabidopsis mutS-like Genes
We previously reported (Culligan and Hays, 1997) the cDNA and genomic sequences of an Arabidopsis gene that predicts a protein highly similar to other eukaryotic MSH2 proteins and the sequence of a polymerase chain reaction (PCR) product encoding a polypeptide fragment similar to eukaryotic MSH6 proteins. We now designate the gene containing this latter sequence AtMSH7 (GenBank accession number AF193018), for reasons described below. Two additional mutS-like genes were identified in the Arabidopsis genome database (GenBank accession numbers AL022197 and AF001308), and corresponding complete cDNAs were obtained by reverse transcription–PCR (RT-PCR; see Methods). The first proved highly similar to eukaryotic MSH3 genes, and the second was highly similar to MSH6 genes. The sequences of these AtMSH3 and AtMSH6 cDNAs agree with cDNA sequences recently deposited in GenBank (GenBank accession numbers AJ007791 and AJ245967, respectively) (Ade et al., 1999).
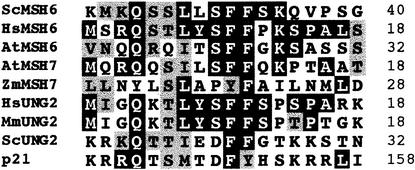
We isolated a full-length AtMSH7 cDNA by screening an Arabidopsis cDNA library. Figure 1 shows the extensive similarity of human and yeast MSH6 proteins to Arabidopsis AtMSH6, AtMSH7, and a recently identified MSH6-like protein sequence from maize, previously designated MUS2 (GenBank accession number AJ238786), which we now designate ZmMSH7 (see below). AtMSH7 and ZmMSH7 are slightly shorter than the other MSH6 proteins, by 57 to 250 amino acids, and share ∼30% identity with AtMSH6. In addition to three highly conserved domains previously identified in MSH proteins (Culligan et al., 2000; Figure 1), we identify an additional N-terminal domain (distinct from the N-terminal mismatch binding domain) present in both AtMSH6 and AtMSH7 proteins. Figure 2 shows the similarity of this domain to the N-terminal domains of uracil-DNA glycosylase2 (UNG2) and p21 proteins. A recent study identified this domain in UNG2 and p21 as a site of interaction with PCNA and replication protein A (RPA) (Otterlei et al., 1999).
Figure 1.
Alignment of MSH6-like Protein Sequences.
Black boxes highlight identical amino acids and gray boxes highlight similar amino acids in at least three of the sequences, based on Dayhoff's PAM250 matrix. Dashes denote gaps. The sequence prefixes At-, Hs-, Sc-, and Zm- denote Arabidopsis thaliana, Homo sapiens, Saccharomyces cerevisiae, and Zea mays, respectively. Amino acid positions are shown at right. The lines above the alignment denote conserved regions found in MSH proteins: line 1, the putative N-terminal PCNA/RPA interaction domain; line 2, the N-terminal mismatch recognition domain; line 3, the middle-conserved domain; and line 4, the highly conserved C-terminal domain.
Figure 2.
Alignment of the N-Terminal PCNA/RPA Interaction Domain of MSH6, MSH7, UNG2, and Human p21 Proteins as in Figure 1.
Mm, Mus musculus.
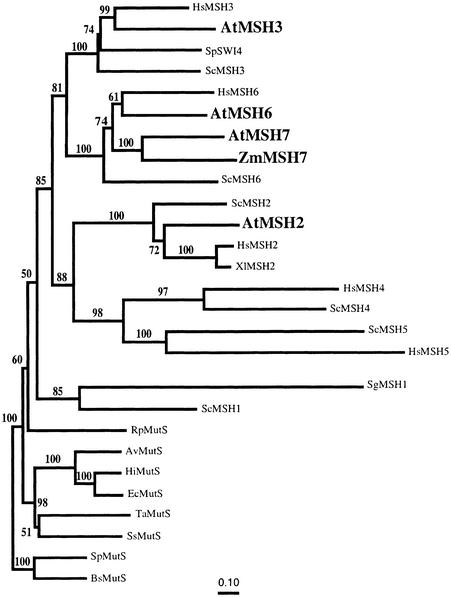
Figure 3 presents a phylogenetic tree for a representative set of MutS/MSH protein sequences, obtained by comparing their entire amino acid sequences, as described elsewhere (Culligan et al., 2000) but now including AtMSH3, AtMSH7, and ZmMSH7. The AtMSH2, AtMSH3, and AtMSH6 protein sequences branch with their respective eukaryotic homolog subfamilies, but AtMSH7 and ZmMSH7 form a separate subgroup within the MSH6 radiation. This suggests a common ancestor for MSH7 genes within the MSH6 subfamily, rather than an origin for AtMSH7 and ZmMSH7 from some other MSH gene. The marked divergence of the MSH6 and MSH7 sequences, indicated by their relatively long branch lengths, suggests a duplication/specialization event early, rather than recently, in the evolution of eukaryotes. The branching pattern of the MSH6 and MSH7 protein sequences in this tree, as well as in a phylogenetic tree produced by analysis of only MSH6 and MSH7 protein sequences (data not shown), suggests that MSH7 diverged from MSH6 before the divergence of plants and animals. However, a more definitive study of a more diverse set of sequences will be needed to determine when the MSH7 gene subfamily may have diverged from the MSH6 subfamily.
Figure 3.
Neighbor-Joining Tree for Dayhoff PAM Distances among a Representative Set of Complete MutS/MSH Protein Sequences.
Gaps and regions of ambiguous alignment were excluded from the analysis. Numbers above each branch represent the number of times the branch was found in 100 bootstrap replicas. The Gram-positive Bacillus subtilis and Streptococcus pneumoniae MutS protein sequences were used as an outgroup. All eukaryotic MutS homologs are encoded by the nuclear genome except for the mitochondrially encoded SgMSH1. At, A. thaliana; Av, Azotobacter vinelandii; Bs, B. subtilis; Ec, E. coli; Hi, Haemophilus influenzae; Hs, H. sapiens; Rp, Rickettsia prowazekii; Sc, S. cerevisiae; Sg, Sarcophytom glaucum; Sp (SW14), Saccharomyces pombe; Sp (MutS), S. pneumoniae; Ss, Synechocystis sp; Ta, Thermus aquaticus; Xl, Xenopus leavis; Zm, Z. mays.
Interactions between MSH Proteins in Translation Mixtures
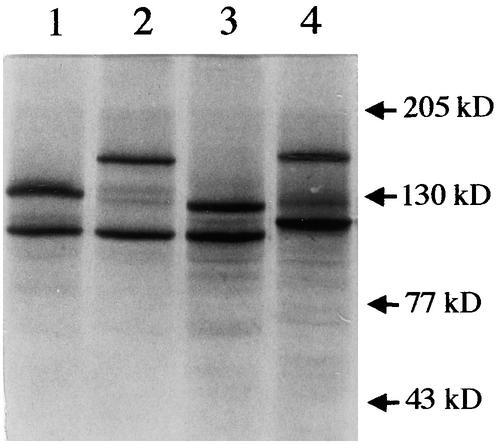
Because eukaryotic MSH proteins typically act as heterodimers (Acharya et al., 1996; Modrich and Lahue, 1996), rigorous biochemical analysis of their properties would require purification from cells in which they may not be abundant (nonmeristematic plant tissues, for example) or from heterologous cells in which simultaneous high expression of the respective recombinant cDNAs was carefully balanced. To make an initial comparison of AtMSH7 with other Arabidopsis MSH proteins, we synthesized the respective polypeptides by in vitro transcription and translation. Figure 4 shows electrophoretic analysis of 35S-methionine–labeled products of transcription–translation reactions optimized for balanced synthesis of pairs of polypeptides expected to form 1:1 heterodimers.
Figure 4.
SDS-PAGE Analysis of Human and Arabidopsis Cosynthesis Reaction Mixtures Used.
Lane 1 contains AtMSH2 and AtMSH3; lane 2, AtMSH2 and AtMSH6; lane 3, AtMSH2 and AtMSH7; and lane 4, hMSH2 and hMSH6. Numbers at the right denote molecular mass.
Physical interactions between different MSH polypeptides and aggregation of individual polypeptides to form homodimers, heterodimers, or higher order complexes, as well as possible interactions between MSH polypeptides and other proteins present in the translation reaction mixtures, were assayed by gel filtration chromatography, as shown in Figures 5 to 7. Because the respective contributions to various chromatographic fractions of two different polypeptides that are synthesized and radiolabeled together cannot be distinguished by bulk measurements of incorporated 35S-methionine, fractions of interest were further analyzed by SDS-PAGE.
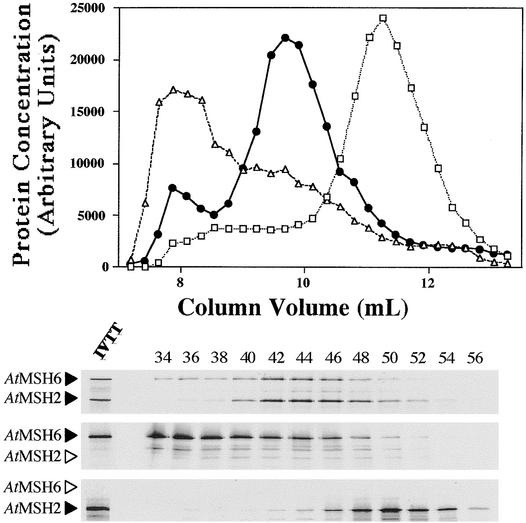
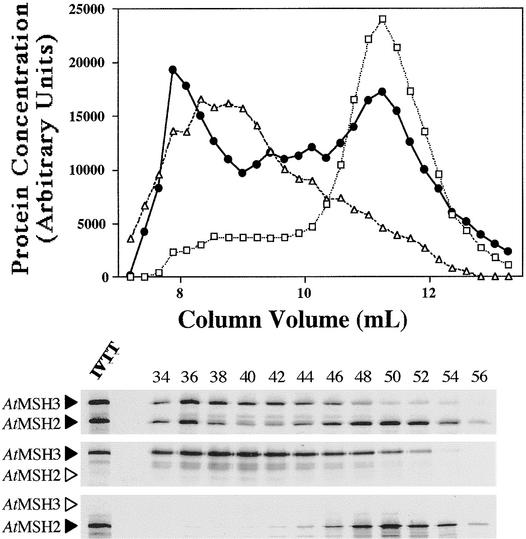
Figure 5.
Gel Filtration Chromatography Analysis of Arabidopsis 35S-Labeled Proteins.
Fifty-microliter synthesis mixture samples were layered onto the gel filtration column, fractionated, and analyzed by liquid scintillation and SDS-PAGE (see Methods).
(Top) Elution profiles for AtMSH2 (open squares), AtMSH6 (open triangles), and AtMSH2•AtMSH6 (closed circles) synthesis mixtures. Fractions 32 to 59 for each of the three mixtures are shown (elution profile).
(Bottom) Corresponding SDS-PAGE autoradiographs of the eluted fractions (even numbers 34 to 56). A small amount of the original transcription–translation synthesis reaction mixture is shown in the leftmost lane at bottom (IVTT lane). Arrowheads to the left of the gel panels denote the position expected for each polypeptide. Arrowheads in outline designate theoretical positions of polypeptides not actually present.
Figure 7.
Gel Filtration Chromatography Analysis of Arabidopsis 35S-Labeled Proteins.
Shown are the elution profiles and the corresponding SDS-PAGE autoradiographs of the eluted fractions for AtMSH2 (open squares), AtMSH3 (open triangles), and AtMSH2•AtMSH3 (closed circles) synthesis mixtures. See Figure 5 for details.
When synthesized alone, AtMSH2 eluted at a position near that expected for the predicted monomer, whereas most AtMSH6 eluted in the exclusion volume, the position expected for an aggregate of ∼1000 kD (Figure 5). However, when approximately equal amounts of the two polypeptides were synthesized together (Figure 4, lane 2), almost all radioactivity eluted in intermediate fractions, corresponding roughly to a peak at 400 kD and containing both AtMSH2 and AtMSH6 polypeptides (Figure 5). Human polypeptides hMSH2 and hMSH6 showed similar heterodimerization when synthesized together (Figure 4, lane 4; data not shown). However, hMSH6 synthesized alone eluted at a position corresponding to its expected monomer molecular mass. When the specificity of these interactions was tested by synthesis of hMSH2 with AtMSH6 or of AtMSH2 with hMSH6, no significant heterodimerization was apparent, that is, the respective polypeptides eluted as they did when synthesized alone (data not shown). Interestingly, gel filtration analysis of equimolar mixtures of separately synthesized AtMSH2 and AtMSH6 polypeptides showed only ∼20% as much interaction as when the polypeptides were synthesized together, as if heterodimerization were more efficient if the polypeptides were able to interact before completing post-translational intra-chain folding (or before interacting with other proteins in the mixtures).
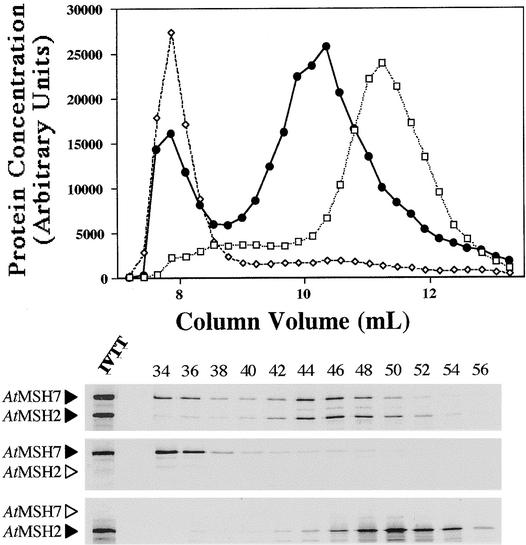
Figure 6 shows that interaction between AtMSH2 and AtMSH7 proteins is similar to that between AtMSH2 and AtMSH6. All of the AtMSH7 peptide synthesized alone eluted in the exclusion volume (apparent molecular mass >1000 kD), but when AtMSH2 and AtMSH7 were synthesized together (Figure 4, lane 3), most protein labeled with 35S-methionine and most AtMSH2 and AtMSH7 polypeptides eluted near the position expected for a 270-kD heterodimer. In contrast, AtMSH3 synthesized alone eluted as a very broad peak (Figure 7), corresponding to apparent molecular masses ranging from 102 to 103 kD. Furthermore, when AtMSH2 and AtMSH3 were synthesized and analyzed together (Figure 4, lane 1), no distinct heterodimer peak was apparent (Figure 7), although there was an increase in the amount of radioactivity in fractions between the peak shown by AtMSH2 alone and the position for the preponderance of AtMSH3 synthesized alone. However, the shift in AtMSH2 polypeptide in the presence of AtMSH3 to fractions corresponding to greater molecular masses suggests that AtMSH2• AtMSH3 heterodimers, and perhaps complexes of greater molecular mass containing the two polypeptides, were formed. Although these results indicate that AtMSH3 polypeptides react with AtMSH2 to form specific heterodimers more weakly—relative to interactions with itself or with other proteins present in the translation mixtures—than do AtMSH6 and AtMSH7, the AtMSH2•AtMSH3 mixture showed good substrate binding activity (see below). A similar phenomenon might account in part for the low amounts of hMSH2• hMSH3, relative to hMSH2•hMSH6, typically found in human cells (Modrich and Lahue, 1996).
Figure 6.
Gel Filtration Chromatography Analysis of Arabidopsis 35S-Labeled Proteins.
Shown are the elution profiles and the corresponding SDS-PAGE autoradiographs of the eluted fractions for AtMSH2 (open squares), AtMSH7 (open diamonds), and AtMSH2•AtMSH7 (closed circles) synthesis mixtures. See Figure 5 for details.
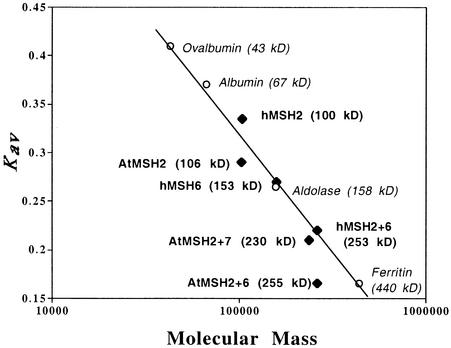
In the plot of relative elution position (denoted as the partition coefficient, Kav) versus protein molecular mass shown in Figure 8, the positions of some expected monomer and heterodimer positions do not fall on the line defined by a wide range of putative globular protein standards. Thus, hMSH2 alone elutes at a slightly lower apparent molecular mass (∼90 kD), and the AtMSH2 monomer elutes at a greater mass (∼125 kD) than those predicted by their amino acid sequences (100 and 106 kD, respectively). Although AtMSH2•AtMSH6 elutes at ∼400 kD, well beyond the position predicted by its 255-kD size, hMSH2•hMSH6 elutes approximately as expected, as does AtMSH2•AtMSH7. The symmetric peaks shown by the anomalously eluting Arabidopsis proteins as well as the contrasting behavior of their human homologs suggest that their positions reflect specific asymmetries that affect their hydrodynamic behavior, but we cannot rigorously rule out less specific interactions with other proteins in the translation mixture.
Figure 8.
Kav versus Log Molecular Mass Plot of Standards and MSH Polypeptides Analyzed by Gel Filtration Chromatography.
Open circles denote globular standards used to calibrate the gel filtration column. Black diamonds denote MSH polypeptides used in this study. The predicted (theoretical) molecular mass is shown in parentheses to the right of each monomeric or heterodimeric polypeptide.
Binding of AtMSH Proteins to Mismatched DNA
We used electrophoretic mobility shift assays to compare the abilities of MSH proteins synthesized in vitro to bind 51-mer homoduplexes (T/A) or heteroduplexes (T/G, C/C base/base mispairs, or “loopout” insertions of T or AAG in the top strand). The apparent relative intensities of 35S-methionine labeling and the number of known methionine residues present in each protein were used to select synthesis mixtures in which the pairs of peptides were equimolar. Approximately equal theoretical amounts of the respective heterodimers were added to the various binding reactions, and the apparent yields of DNA–protein complexes were normalized for the amounts of heterodimer predicted to form (see Methods). Figure 9 displays representative electropherograms for three Arabidopsis heterodimers and one human heterodimer, and Table 1 summarizes the results of corresponding phosphorimager measurements. The upper bands in each case appear to reflect specific binding by MSH proteins, because their intensities depend on the natures of both the DNA substrate and the particular MSH protein heterodimer. The lower bands, which might reflect binding by proteins specific for double-stranded DNA ends in general (Acharya et al., 1996), were seen in every gel lane, even when unprogrammed translation mixtures or mixtures containing only vector DNA (pGEM-3Z) were analyzed (data not shown). Polypeptides synthesized alone, namely, AtMSH2, AtMSH3, AtMSH6, AtMSH7, hMSH2, and hMSH6, showed no specific binding to any of the substrates tested (data not shown).
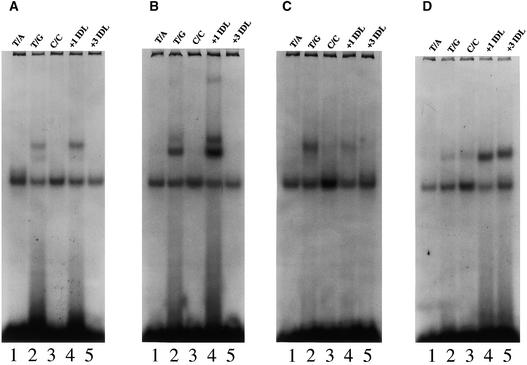
Figure 9.
Representative Mobility Shift Assays of Cosynthesis Reaction Mixtures of Human and Arabidopsis Polypeptides.
Cosynthesis mixtures were incubated with 32P-labeled homoduplex DNA (T/A, lane 1, all panels), or heteroduplex DNA (T/G, C/C, +1 IDL [insertion–deletion DNA with one T nucleotide looped out], and +3 IDL [insertion–deletion DNA with AAG nucleotides looped out], lanes 2 to 5, respectively, all panels) and analyzed on nondenaturing PAGE (see Methods).
(A) Representative mobility shift assay of hMSH2•hMSH6 cosynthesis mixtures.
(B) Representative mobility shift assay of AtMSH2•AtMSH6 cosynthesis mixtures.
(C) Representative mobility shift assay of AtMSH2•AtMSH7 cosynthesis mixtures.
(D) Representative mobility shift assay of AtMSH2•AtMSH3 cosynthesis mixtures.
Table 1.
Relative Binding of MSH Heterodimers to Oligomer Substrates
| Heterodimer | Oligomer Substrate 32P-Labeled | Relative Binding (Arbitrary Units)a | Binding Ratio (T/G):(+T IDL)b |
|---|---|---|---|
| hMSH2•hMSH6 | T/A | <0.10 | 0.59 ± 0.05 |
| T/G | 1.00 | ||
| C/C | <0.10 | ||
| +T IDL | 1.51 | ||
| +AAG IDL | <0.10 | ||
| AtMSH2•AtMSH6 | T/A | <0.10 | 0.57 ± 0.05 |
| T/G | 1.08 | ||
| C/C | <0.10 | ||
| +T IDL | 2.39 | ||
| +AAG IDL | <0.10 | ||
| AtMSH2•AtMSH7 | T/A | <0.10 | 5.0 ± 0.5 |
| T/G | 0.93 | ||
| C/C | <0.10 | ||
| +T IDL | 0.23 | ||
| +AAG IDL | <0.10 | ||
| AtMSH2•AtMSH3 | T/A | <0.10 | 0.19 ± 0.03 |
| T/G | 0.27 | ||
| C/C | 0.39 | ||
| +T IDL | 1.11 | ||
| +AAG IDL | 1.20 |
Substrate binding values correspond to areas of shifted bands (Figure 9) of the respective 32P-labeled oligomer substrates, as determined using a phosphorimager, for approximately equal amounts of MSH polypeptides. To facilitate comparisons, all phosphorimager values were arbitrarily divided by the value for (T/G) substrate binding by hMSH2•hMSH6.
Mean ratios (±se) are based on densitometry measurements from three independent mobility shift assay experiments (including the experiments corresponding to values in column 3), calculated by dividing the total densitometry band area of the shifted 32P-labeled (T/G) oligomer substrate by the total (+T IDL) substrate band area, for each respective heterodimer.
Both the Arabidopsis and human MSH2•MSH6 heterodimers showed the expected preference for a (T/G) base/base mispair and a one-nucleotide (+T) loopout (Figure 9). As has been shown for the hMSH2•hMSH6 heterodimer, the AtMSH2•AtMSH6 heterodimer bound the (+T) loopout with greater efficiency (almost twofold here) than the (T/G) base/base mispair (Figure 9 and Table 1) and showed negligible binding to homoduplex (T/A) DNA, to the (C/C) heteroduplex, or to the three-nucleotide (+AAG) loopout. Similarly, the AtMSH2•AtMSH3 heterodimer showed the same preference for loopout substrates over base/base mispair substrates as that reported for its human counterpart, hMSH2•hMSH3: the AtMSH2•AtMSH3 heterodimer bound best to the (+AAG) loopout and the (+T) loopout and weakly to the (C/C) and (T/G) base mispairs. Again, there was little affinity for (T/A) homoduplex DNA. However, the AtMSH2• AtMSH7 heterodimer showed a novel substrate specificity—preference for (T/G) base/base mispairs (perhaps slightly weaker in absolute terms than (T/G) binding by MSH2• MSH6 heterodimers) over (+T) loopouts and essentially no affinity for other substrates (Figure 9 and Table 1).
DISCUSSION
Searches for Arabidopsis MSH genes have revealed close homologs of genes in eukaryotic organisms—AtMSH2, AtMSH3, and AtMSH6—and an additional novel MSH6-like gene, designated here as AtMSH7. Analysis of interactions between polypeptides translated in vitro demonstrates conservation of the eukaryotic pattern of heterodimerization of MSH2 with other MSH polypeptides, here including AtMSH7. By analogy with the previous designations of MutSα and MutSβ for MSH2•MSH6 and MSH2•MSH3, respectively, we designate the MSH2•MSH7 heterodimer as MutSγ. Assays of binding to a representative set of mismatched double-stranded DNA oligomers show that the substrate specificities of AtMutSα and AtMutSβ are very similar to those of their eukaryotic counterparts, but the specificity of AtMutSγ appears to differ considerably from either of these.
The existence of Arabidopsis MutS-like proteins that resemble their eukaryotic homologs in both primary structure and activity in vitro suggests that plants use classic long-patch mismatch repair systems to enhance genomic stability. This inference is strengthened by the identification, by way of the Arabidopsis genome project, of MSH1, MLH1, and PMS2 genes, the cDNAs of which predict polypeptides highly similar to their eukaryotic counterparts (G. Meyer-Gauen, A. Torres, J. Leonard, K.M. Culligan, and J.B. Hays, unpublished observations). Furthermore, a putative MSH4-like gene is present in the Arabidopsis genome (K.M. Culligan and J.B. Hays, unpublished observations). Whether plants use MSH proteins to promote meiosis remains to be determined.
Our phylogenetic analysis suggests that MSH7 diverged from MSH6 early in eukaryotic evolution, possibly before the divergence of plants and animals. However, the yeast genome encodes no second MSH7-like protein, and none has been identified in other organisms, Caenorhabditis elegans, for example (K.M. Culligan and J.B. Hays, unpublished observations). MSH7 may thus have a role in maintaining genomic stability that is unique to plants, but we cannot rule out the possibility that some other eukaryotes also encode MSH7 proteins.
MSH7 shows a pattern of charged and aromatic residues in the N-terminal mismatch binding domain similar to that of MSH6 proteins (Malkov et al., 1998; Culligan et al., 2000) and forms heterodimers with AtMSH2, as do AtMSH3 and AtMSH6 proteins. Additionally, AtMSH6 and AtMSH7 contain N-terminal interaction domains, previously identified in several proteins that are involved in DNA metabolism, including hMSH6 (Nicolaides et al., 1996). These domains have been shown to be putative sites of interaction with PCNA or RPA (Otterlei et al., 1999). Furthermore, several other studies in yeast and human cells have implicated PCNA as a factor involved in both pre- and postexcision steps in mismatch repair (Johnson et al., 1996; Umar et al., 1996; Chen et al., 1999). Thus, both AtMSH6 and AtMSH7 proteins may interact in some way with the DNA replication machinery. The AtMSH2•AtMSH7 heterodimer (AtMutSγ) failed to bind strongly to DNA containing (+T) loopout mismatches, even though these DNAs are good substrates for both MutSα and MutSβ. Moreover, the affinity of AtMutSγ for (T/G) DNA appeared slightly weaker than that of AtMutSα. This suggests that a major function of AtMutSγ might be a specialized recognition of DNA lesions or of (T/G) mispairs in specialized contexts.
Purified human MutSα (Mu et al., 1997; Wang et al., 1999) and bacterial MutS (H. Wang and J.B. Hays, unpublished observations) bind with considerable affinity to photoproduct base mismatches, such as A-G opposite T-T cyclobutanepyrimidine (CPD) dimers (T<>T) or [6-4] photoproducts (T[6-4]T), but show no preference for T<>T/AA or T[6-4]T/AA moieties. Thus, mismatch repair might correct potential mutations resulting from insertion of incorrect bases opposite photoproducts during DNA replication, as genetic evidence suggests is the case in E. coli (Liu et al., 2000). Such a mechanism might be important for plants, whose DNA can be efficiently replicated even when photoproduct quantities are as much as one per kilobase (Draper and Hays, 2000). Furthermore, optimal transcription-coupled repair of CPDs and, most likely, bases damaged by oxyradicals (particularly thymine glycol and possibly 8-oxoguanine) requires MSH and MLH proteins in yeast and mammalian cells (Leadon and Avrutskaya, 1997, 1998; Le Page et al., 2000). Because plants are typically subject to UV irradiation and to byproducts of oxidative metabolism, they might utilize MSH7 proteins to facilitate transcription-coupled repair. With respect to (T/G) mispairs, one case of great potential interest for plants might involve the spontaneous deamination products of 5-methylcytosine, specifically at (G/C) pairs in CpG and CpNpG sequences, in which cytosines are frequently methylated in plants (Gruenbaum et al., 1981; Richards, 1997).
The differences between the gel filtration behavior of various Arabidopsis MSH proteins when synthesized alone or together in transcription–translation mixtures may reflect their properties in vivo. For example, the appearance of discrete stable monomer peaks of hMSH2 or AtMSH2, when synthesized alone, appears consistent with the lack of detectable hMSH2 homodimers in human cells (Drummond et al., 1997). These monomers show no affinity for mismatched DNA when synthesized in vitro by rabbit reticulocyte lysates (this study; Acharya et al., 1996). Although synthesis of AtMSH2 in the presence of an equal amount of AtMSH6 or AtMSH7 shifts all of the AtMSH2 into a heterodimer peak, synthesis of AtMSH3 alone, AtMSH6 alone, and AtMSH7 alone results in products that elute in the exclusion volume, suggesting a high degree of aggregation. Furthermore, both AtMSH3 and AtMSH6 show a long heterogeneous tail extending down to the monomer molecular mass. These data suggest that when AtMSH3, AtMSH6, or AtMSH7 polypeptides are synthesized alone, nonspecific interactions within the translation mixture may predominate. Thus, proper folding of AtMSH3, AtMSH6, and AtMSH7 polypeptides during synthesis, and perhaps resistance to aggregation and degradation, may require the presence of AtMSH2 to engender immediate heterodimerization. This notion is consistent with the inefficient interaction seen when polypeptides synthesized separately were mixed together. Human cells appears to have no free hMSH3 or hMSH6 protein not heterodimerized with hMSH2, and heterodimerization of hMSH2 with hMSH6 appears more efficient than with hMSH3, which is consistent with the preponderance of hMutSα over hMutSβ in human cells (Drummond et al., 1997; Marra et al., 1998).
Elucidation of the roles of plant MSH2, MSH3, MSH6, and MSH7 in maintaining genomic stability will require construction of plants that lack the respective proteins and application of assays for various mismatch repair functions, including binding to DNA-containing lesions and mismatches in various sequence contexts.
METHODS
Isolation of cDNAs
To obtain Arabidopsis thaliana cDNAs corresponding to the coding regions of AtMSH3 and AtMSH6 genes, available through the Arabidopsis genome project, we used reverse transcription–polymerase chain reaction (RT-PCR). Purified mRNA (Culligan and Hays, 1997) was amplified by using an mRNA PCR Amplification Kit (Clontech) as specified by the manufacturer. First-strand cDNA synthesis products were used as templates in standard PCR reactions to amplify 5′ and 3′ regions of AtMSH3. Primer 5P-MSH3 (5′-GGGGTACCATGG-GCAAGCAAAAGCAGC-3′), which overlaps the initiating ATG and encodes a KpnI site and Kozak consensus sequence, and primer MSH3-6 (5′-AAATCTCAGAAACAGCATCAAG-3′), corresponding to position +1456 of the cDNA, were used to amplify the 5′ region of AtMSH3 cDNA. Similarly, primer 3P-MSH3 (5′-ACGCGTCGACTCAAAATGAACAAGTTGG-3′), which overlaps the 3′ termination codon and encodes a SphI site, and primer MSH3-2 (5′-AATCAGCGCAGGTAACTTAGAAG-3′), corresponding to position +1048 of the cDNA, were used to amplify the 3′ end of AtMSH3 cDNA. DNA samples from individual clones were sequenced by the Oregon State University Central Services Laboratory, using a Taq dye-primer/dye-terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and compared with genomic sequences to determine the intron/exon boundaries and to verify the accuracy of the PCR amplifications. Correct 5′ and 3′ cDNA sequences were joined through use of a unique BamHI site present in the overlapping regions, and the product was inserted into the KpnI and SphI sites of plasmid pGEM-3Z (Promega, Madison, WI).
AtMSH6 cDNA sequences were obtained as described for AtMSH3, using primer 5P-MSH6 (5′-GTCGGATCCGCCATGGCTCCGTCTCGCCGA-3′), which overlaps the initiating ATG and encodes a BamHI site and Kozak consensus sequence, and primer MSH6-9 (5′-ACTTTGCAAATCTAAGCAGACTCTA-3′), corresponding to position +2074 of the cDNA, to amplify the 5′ region. Primer 3P-MSH6 (5′-CTGGTCGACTTAGTTGGTTAACCGGAG-3′), which overlaps the termination codon and contains a SalI site, and primer MSH6-8 (5′-GCTAAGGTGTTGAGTTATGCAACAG-3′), corresponding to position +1741 of the cDNA, were used to amplify the 3′ region of AtMSH6. Correct 5′ and 3′ clones were joined by way of a unique HindIII site in the overlapping region, and the product was inserted into the BamHI and SalI sites of pGEM-3Z.
A full-length AtMSH7 cDNA was isolated by probing an Arabidopsis cDNA library (size range 3 to 6 kb) in phage λZAPII (Kieber et al., 1993; Arabidopsis Biological Resource Center), with a 352-bp 32P-labeled PCR fragment of AtMSH7 (Culligan and Hays, 1997; GenBank accession number AF009657). The AtMSH7 cDNA was inserted into pGEM-3Z by using unique XbaI and HindIII sites flanking the cDNA. The cDNA was further altered to encode a Kozak consensus sequence at the 5′ end by PCR amplification with primer 5P-MSH7 (5′-CGGGATCCTATACCATGGTGCAGCGCCAGAGATCG-3′), which overlaps the initiating ATG and encodes a BamHI site, and primer MSH7-10 (5′-CGAGCTAATAGCTTTTGCACTGC-3′), corresponding to position +1019 of the cDNA. The 5′ portion of the cDNA was replaced with this PCR product, utilizing unique restriction sites.
The AtMSH2 cDNA (Culligan and Hays, 1997) was inserted into the EcoRI and SphI sites of pGEM-3Z. Primer 5P-MSH2 (5′-AGCAATTGTATACCATGGAGGGTAATTTCGAG-3′), which overlaps the initiating ATG and encodes a MunI site and Kozak consensus sequence, and primer MSH2-1300 (5′-ACCTCAGAGAAGCTGGTAACGTC-3′), corresponding to position +1763 of the cDNA, were used to modify the 5′ end as described above for AtMSH7.
Human MSH2 and MSH6 cDNAs, inserted into plasmids pET-3d and -29a, respectively, were generously provided by Dr. Richard Fishel (Acharya et al., 1996).
Sequence Alignment and Phylogenetic Analysis
Alignments of complete protein sequences and construction of the phylogenetic tree were performed as described previously (Culligan et al., 2000).
In Vitro Transcription and Translation
Plasmids containing cDNAs (see above) were purified from Escherichia coli lysates by using a Plasmid Maxi Kit (Qiagen). In vitro protein synthesis using TnT Quick (rabbit reticulocyte lysate) kits (Promega) was performed, as specified by the manufacturer, in 50-μL reaction volumes containing 35S-methionine (New England Nuclear, Beverly, MA). Each reaction was optimized by varying the amount of template DNA from 0.5 to 2 μg and varying the Mg2+ and K+ ion concentrations. Reactions were incubated at 30°C for 90 min. For cosynthesis (in vitro transcription–translation of two different DNA templates in the same mixture), a series of mixtures containing slightly different ratios of the two templates was incubated; the products were analyzed by SDS-PAGE and visualized by autoradiography. Mixtures containing equimolar ratios of each polypeptide, as determined by densitometry with normalization for the respective numbers of methionines, were used for further analyses. Mixtures were diluted 20-fold in buffer A (50 mM potassium phosphate, pH 7.5, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 5% glycerol) and centrifuged with a Centricon-100 microconcentrator (Millipore, Bedford, MA) until volumes were reduced 20-fold. The reconcentrated mixtures were again analyzed by SDS-PAGE and densitometry to determine the respective relative amounts of pairs of polypeptides (potential heterodimers; see Figure 4) and used for subsequent gel filtration or mobility-shift assay experiments.
Gel Filtration Chromatography
Mixtures for in vitro protein synthesis were applied to a 0.7 × 50-cm column of Sephacryl S300 (Amersham-Pharmacia, Piscataway, NJ) previously equilibrated with buffer B (50 mM potassium phosphate, pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 5% glycerol) and calibrated with ovalbumin (43 kD), albumin (67 kD), aldolase (158 kD), and ferritin (440 kD). Fractions (225 μL) were collected at an elution rate of 0.08 mL/min. Aliquots were used to determine the amount of incorporated 35S-methionine present by liquid scintillation analysis or were analyzed by SDS-PAGE and autoradiography.
Preparation of Oligomer Duplexes
DNA substrates were prepared and purified as described previously (Wang et al., 1999). Briefly, purified top-strand 51-mers (5′-AATGGTTAGCAATCATAGTGGCAAGTTGGAGTCAATCGTCTCTCGTTA-TTC-3′) were 5′-end labeled with γ-32P-ATP in 50-μL reactions containing 20 pmol of oligomer, 40 pmol of γ-32P-ATP, 10 units of T4 polynucleotide kinase, and 1 × kinase buffer (Gibco BRL, Gaithersburg, MD) and incubated for 30 min at 37°C. The reaction was terminated by heating to 70°C for 10 min. We added to the 32P-labeled top strand 20 pmol of a particular bottom strand (51-mer T/A, 5′-GAATAACGAGAGACGATTGACTCCAACTTGCCACTATGATTGCTAACC-ATT-3′; 51-mer T/G, 5′-GAATAACGAGAGACGATTGACTCCGACTTGCCACTATGATTGCTAACCATT-3′; 51-mer C/C, 5′-GAATAA-CGAGAGACGATTGACTCCAACTTCCCACTATGATTGCTAACCATT-3′; 50-mer +T, 5′-GAATAACGAGAGACGATTGACTCCACTTGCCAC-TATGATTGCTAACCATT-3′; or 48-mer +AAG, 5′-GAATAACGAGAGACGATTGACTCCAAGCCACTATGATTGCTAACCATT-3′), heated the mixtures to 85°C for 5 min, and allowed them to cool to room temperature over a period of at least 6 hr. After addition of 0.2 volumes of benzoylated naphthoylated DEAE cellulose, 5 M NaCl was added to a final concentration of 1 M. Mixtures were incubated for 5 min and then layered onto Sephadex G-50 Nick Spin Columns (Amersham Pharmacia Biotech, Cleveland, OH) previously equilibrated with TE buffer (10 mM Tris-HCl, pH 7.6, and 1 mM EDTA); samples were recovered as specified by the manufacturer. To confirm the absence of single-stranded unannealed oligomers, a small aliquot of the recovered oligomer sample was re-treated with polynucleotide kinase and 32P-ATP and electrophoresed on 15% polyacrylamide gels, under conditions that resolve double-stranded from single-stranded 51-mers.
Mobility Shift Assays
Mobility shift assays were performed essentially as described previously (Acharya et al., 1996) in 30-μL reactions containing 50 mM potassium phosphate, pH 7.5, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 5 mM AMP, 10% glycerol, 2 μg of poly (dI•dC), 0.5 pmol of 32P-labeled oligonucleotides, and 8 to 12 μL of in vitro transcription–translation mixtures. These mixtures were incubated at 25°C for 30 min, loaded onto 4% polyacrylamide gels containing 2% glycerol and TAE buffer (0.04 M Tris-acetate and 2 mM EDTA), and electrophoresed at 50 mA for 4 hr with buffer recirculation. The gels were dried onto 3MM Whatman (Whatman Inc., Clifton, NJ) paper and visualized by autoradiography at room temperature or analyzed with a Molecular Dynamics PhosphorImager.
Acknowledgments
We thank Dr. Jeffery Leonard for isolating the AtMSH7 cDNA and for assistance. We also thank Adela Torres and Dr. Manuel Perez Alonso for providing the AtMSH3 sequence before publication. This work was supported by National Science Foundation Grant No. 9631048-MCB to J.B.H. This is contribution No. 11605 from the Oregon Agricultural Experiment Station.
References
- Acharya, S., Wilson, T., Gradia, S., Kane, M.F., Guerrette, S., Marsischky, G.T., Kolodner, R., and Fishel, R. (1996). hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. USA 93, 13629–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade, J., Belzile, F., Philippe, H., and Doutriaux, M.P. (1999). Four mismatch repair paralogues coexist in Arabidopsis thaliana: AtMSH2, AtMSH3, AtMSH6–1 and AtMSH6–2. Mol. Gen. Genet. 262, 239–249. [DOI] [PubMed] [Google Scholar]
- Allen, D.J., Makhov, A., Grilley, M., Taylor, J., Thresher, R., Modrich, P., and Griffith, J.D. (1997). MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 16, 4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M., et al. (1998). Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488. [DOI] [PubMed] [Google Scholar]
- Chen, C., Merrill, B.J., Lau, P.J., Holm, C., and Kolodner, R.D. (1999). Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 19, 7801–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, N.-W., and Kolodner, R.D. (1994). Purification and characterization of MSH1, a yeast mitochondrial protein that binds to DNA mismatches. J. Biol. Chem. 269, 29984–29992. [PubMed] [Google Scholar]
- Culligan, K.M., and Hays, J.B. (1997). DNA mismatch repair in plants. An Arabidopsis thaliana gene that predicts a protein belonging to the MSH2 subfamily of eukaryotic MutS homologs. Plant Physiol. 115, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K.M., Meyer-Gauen, G., Lyons-Weiler, J., and Hays, J.B. (2000). Evolutionary origin, diversification, and specialization of eukaryotic MutS-homolog mismatch-repair proteins. Nucleic Acids Res. 28, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, S.S., Baart, E.B., Dekker, M., Siezen, A., de Rooij, D.G., de Boer, P., and te Riele, H. (1999). Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, C.K., and Hays, J.B. (2000). Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J., in press. [DOI] [PubMed]
- Drummond, J.T., Anthony, A., Brown, R., and Modrich, P. (1996). Cisplatin and adriamycin resistance are associated with MutLα and mismatch repair deficiency in an ovarian tumor cell line. J. Biol. Chem. 271, 19645–19648. [DOI] [PubMed] [Google Scholar]
- Drummond, J.T., Genschel, J., Wolf, E., and Modrich, P. (1997). DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSα/hMutSβ ratio and reduces the efficiency of base–base mismatch repair. Proc. Natl. Acad. Sci. USA 94, 10144–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett, D.R., Drummond, J.T., Murchie, A.I.H., Reardon, J., Sancar, A., Lilley, D.M.J., and Modrich, P. (1996). Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. USA 93, 6443–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, W.H., and Modrich, P. (1993). Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J. Biol. Chem. 268, 11838–11844. [PubMed] [Google Scholar]
- Gradia, S., Acharya, S., and Fishel, R. (1997). The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91, 995–1005. [DOI] [PubMed] [Google Scholar]
- Gradia, S., Subramanian, D., Wilson, T., Acharya, S., Makhov, A., Griffith, J., and Fishel, R. (1999). hMSH2–hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. (1981). Sequence specificity of methylation in higher plant DNA. Nature 292, 860–862. [DOI] [PubMed] [Google Scholar]
- Habraken, Y., Sung, P., Prakash, L., and Prakash, S. (1998). ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2–MSH6 and MLH1–PMS1 protein complexes. J. Biol. Chem. 273, 9837–9841. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N.M., Ponte, L., and Halsey, C. (1995). MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9, 1728–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, N., Chambers, S.R., Louis, E.J., and Borts, R.H. (1996). The mismatch-repair system contributes to meiotic sterility in an interspecies yeast hybrid. EMBO J. 15, 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.E., Kovvali, G.K., Guzder, S.N., Amin, N.S., Holm, C., Habraken, Y., Sung, P., Prakash, L., and Prakash, S. (1996). Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 271, 27987–27990. [DOI] [PubMed] [Google Scholar]
- Kat, A., Thilly, W.G., Fang, W.-H., Longley, M.J., Li, G.-M., and Modrich, P. (1993). An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. USA 90, 6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kolodner, R., and Marsischky, G.T. (1999). Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9, 89–96. [DOI] [PubMed] [Google Scholar]
- Kornberg, A., and Baker, T.A. (1992). Replication mechanisms and operations. In DNA Replication, A. Kornberg and T.A. Baker, eds (New York: W.H. Freeman and Co.), pp. 497–498.
- Leadon, S.A., and Avrutskaya, A.V. (1997). Differential involvement of the human mismatch repair proteins, hMLH1 and hMSH2, in transcription-coupled repair. Cancer Res. 57, 3784–3791. [PubMed] [Google Scholar]
- Leadon, S.A., and Avrutskaya, A.V. (1998). Requirement for DNA mismatch repair proteins in the transcription-coupled repair of thymine glycols in Saccharomyces cerevisiae. Mutat. Res. 407, 177–187. [DOI] [PubMed] [Google Scholar]
- Le Page, F., Kwoh, E.E., Avrutskaya, A., Gentil, A., Leadon, S.A., Sarasin, A., and Cooper, P.K. (2000). Transcription-coupled repair and mutation avoidance at 8-oxoguanine: Requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101, 159–171. [DOI] [PubMed] [Google Scholar]
- Li, G.-M., and Modrich, P. (1995). Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl. Acad. Sci. USA 92, 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G.-M., Wang, H., and Romano, L.J. (1996). Human MutSα specifically binds to DNA containing aminofluorene and acetylaminofluorene adducts. J. Biol. Chem. 271, 24084–24088. [PubMed] [Google Scholar]
- Liu, H., Hewitt, S., and Hays, J.B. (2000). Antagonism of ultraviolet-light mutagenesis by the MutHLS mismatch repair system of Escherichia coli. Genetics 154, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkov, V.A., Biswas, I., Camerini-Otero, R.D., and Hsieh, P. (1998). Photocross-linking of the NH2-terminal region of Taq MutS protein to the major groove of a heteroduplex DNA. J. Biol. Chem. 272, 23811–23817. [DOI] [PubMed] [Google Scholar]
- Marra, G., Iaccarino, I., Lettieri, T., Roscilli, G., Delmastro, P., and Jiricny, J. (1998). Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl. Acad. Sci. USA 95, 8568–8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsischky, G.T., Filosi, N., Kane, M.F., and Kolodner, R. (1996). Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10, 407–420. [DOI] [PubMed] [Google Scholar]
- Modrich, P., and Lahue, R. (1996). Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65, 101–133. [DOI] [PubMed] [Google Scholar]
- Mu, D., Tursun, M., Duckett, D.R., Drummond, J.T., Modrich, P., and Sancar, A. (1997). Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell. Biol. 17, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides, N.C., Palombo, F., Kinzler, K.W., Vogelstein, B., and Jiricny, J. (1996). Molecular cloning of the N-terminus of GTBP. Genomics 31, 395–397. [DOI] [PubMed] [Google Scholar]
- Otterlei, M., Warbrick, E., Nagelhus, T.A., Haug, T., Slupphaug, G., Akbari, M., Aas, P.A., Steinsbekk, K., Bakke, O., and Krokan, H.E. (1999). Post-replicative base excision repair in replication foci. EMBO J. 18, 3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, M.-A., Dimpfl, J., Radman, M., and Echols, E. (1991). Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics 129, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla, T.A., Pang, Q., Alani, E., Kolodner, R.D., and Liskay, R.M. (1994). MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265, 1091–1093. [DOI] [PubMed] [Google Scholar]
- Rayssiguier, C., Thaler, D.S., and Radman, M. (1989). The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342, 396–401. [DOI] [PubMed] [Google Scholar]
- Reenan, R.A., and Kolodner, R.D. (1992). Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: Evidence for separate mitochondrial and nuclear functions. Genetics 132, 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.J. (1997). DNA methylation and plant development. Trends Genet. 13, 319–323. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald, P., and Roeder, G.S. (1994). Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Umar, A., Buermeyer, A.B., Simon, J.A., Thomas, D.C., Clark, A.B., Liskay, R.M., and Kunkel, T.A. (1996). Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87, 65–73. [DOI] [PubMed] [Google Scholar]
- Umar, A., Risinger, J.I., Glaab, W.E., Tindall, K.R., Barrett, J.C., and Kunkel, T.A.. (1998). Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics 148, 1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Lawrence, C.W., Li, G.-M., and Hays, J.B. (1999). Specific binding of human MSH2•MSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J. Biol. Chem. 274, 16894–16900. [DOI] [PubMed] [Google Scholar]