Abstract
Fumonisin B1 (FB1), a programmed cell death–eliciting toxin produced by the necrotrophic fungal plant pathogen Fusarium moniliforme, was used to simulate pathogen infection in Arabidopsis. Plants infiltrated with 10 μM FB1 and seedlings transferred to agar media containing 1 μM FB1 develop lesions reminiscent of the hypersensitive response, including generation of reactive oxygen intermediates, deposition of phenolic compounds and callose, accumulation of phytoalexin, and expression of pathogenesis-related (PR) genes. Arabidopsis FB1-resistant (fbr) mutants were selected directly by sowing seeds on agar containing 1 μM FB1, on which wild-type seedlings fail to develop. Two mutants chosen for further analyses, fbr1 and fbr2, had altered PR gene expression in response to FB1. fbr1 and fbr2 do not exhibit differential resistance to the avirulent bacterial pathogen Pseudomonas syringae pv maculicola (ES4326) expressing the avirulence gene avrRpt2 but do display enhanced resistance to a virulent isogenic strain that lacks the avirulence gene. Our results demonstrate the utility of FB1 for high-throughput isolation of Arabidopsis defense-related mutants and suggest that pathogen-elicited programmed cell death of host cells may be an important feature of compatible plant–pathogen interactions.
INTRODUCTION
Programmed cell death (PCD) is an integral component of plant growth, development, and plant–pathogen interactions (Fukuda, 1996; Beers, 1997; Morel and Dangl, 1997; Richberg et al., 1998). A common but not pervasive feature of host plant resistance to pathogen attack involves rapid, localized cell death termed the hypersensitive response (HR), which is activated by gene-for-gene interactions between plant resistance (R) genes and pathogen avirulence (avr) genes (Flor, 1947; Hammond-Kosack and Jones, 1997; Richberg et al., 1998). Although accumulating evidence has shown that the HR is a form of PCD that requires host cell functions (Greenberg, 1997; Morel and Dangl, 1997; Gilchrist, 1998), the underlying molecular mechanisms of the HR and its significance in conferring resistance remain unclear. Even less is known about the host cell death that occurs during so-called compatible interactions, in which gene-for-gene interactions are either absent or weak and disease develops (Greenberg, 1997; Richael and Gilchrist, 1999). In contrast to cell death associated with the HR, cell death in compatible interactions is generally thought to be the result of pathogen-mediated necrosis rather than host-induced PCD. However, increasing evidence suggests that cell death associated with the HR and disease symptom development in some compatible interactions share common features (Greenberg, 1997; Morel and Dangl, 1997; Gilchrist, 1998; Richberg et al., 1998).
Although relatively little is known about the mechanistic details of PCD in plants, some aspects of the molecular machinery might be conserved between plants and animals (Aravind et al., 1999; Lam et al., 1999). For example, some R genes resemble regions of the CED-4 and Apaf-1 genes (van der Biezen and Jones, 1998) that participate in PCD in Caenorhabditis elegans and humans, respectively. A homolog of an inhibitor of the proapoptotic BCL-2 family member Bax has been identified in Arabidopsis (Xu and Reed, 1998), but its involvement in plant PCD has not been demonstrated. However, heterologous expression of human Bax leads to HR-like cell death in tobacco (Lacomme and Santa Cruz, 1999), and expression of prosurvival human Bcl-XL in tobacco suppresses HR cell death (Mitsuhara et al., 1999). Furthermore, cysteine aspartate protease (caspase)–like activity, a critical mediator of PCD in animal cells, has been detected in plants undergoing pathogen attack, and inhibition of caspase activity suppresses the HR (del Pozo and Lam, 1998; D'Silva et al., 1998). Interestingly, however, no plant genes encoding proteins similar to animal caspases have been identified, even though >90% of the Arabidopsis genome has been sequenced.
The cell death response in plants is under strict genetic control, as evidenced by the existence of mutants that spontaneously form HR-like lesions (lesion mimic mutants) in many plant species (Johal et al., 1994; Delaney, 1997; Greenberg, 1997; Morel and Dangl, 1997; Rate et al., 1999; Takahashi et al., 1999). PCD in plants associated with pathogen infection or spontaneously manifested in so-called lesion mimic mutants is associated with the induction of other components of the plants' defense arsenal, including accumulation of reactive oxygen intermediates (ROIs), expression of pathogenesis-related (PR) genes, production of phytoalexin, and reinforcement of cell walls (Greenberg and Ausubel, 1993; Dietrich et al., 1994; Rate et al., 1999). Many lesion mimic mutants that display constitutive resistance to pathogens also exhibit constitutive expression of these HR-related characteristics (Greenberg, 1997). On the other hand, some mutants that exhibit constitutive resistance to pathogens lack a spontaneous cell death phenotype, suggesting that PCD is not necessarily required for pathogen resistance (Bowling et al., 1994; Ryals et al., 1996; Clarke et al., 1998). Host cell death can also be caused by pathogen-produced phytotoxic compounds that function as key virulence determinants. Necrotrophic phytopathogenic fungi synthesize a wide range of phytotoxic compounds, including the sphinganine analog mycotoxins, which are produced by at least two unrelated groups of fungi, Alternaria and Fusarium spp. Fumonisin B1 (FB1) is one of several related sphinganine analog mycotoxins produced by some Fusarium spp, including F. moniliforme, that may play a role in virulence (Desjardins et al., 1995; Gilchrist et al., 1995; Dutton, 1996; Gilchrist, 1997; Jardine and Leslie, 1999). FB1 elicits an apoptotic form of PCD in both plants and animal tissue culture cells (Tolleson et al., 1996; Wang et al., 1996a, 1996b; Yoo et al., 1996; Gilchrist, 1997), most probably through competitive inhibition of ceramide synthase, a key enzyme in sphingolipid biosynthesis (Wang et al., 1990; Abbas et al., 1994; Gilchrist et al., 1995; Yoo et al., 1996). Sphingolipids play diverse roles in many cellular processes, functioning both as anchors for membrane proteins (Futerman, 1995) and as second messengers regulating various cellular functions, including differentiation, growth, and apoptosis (Spiegel and Merrill, 1996). The sphingolipid ceramide is a key component of the mammalian stress response pathway, activating several stress-activated protein kinases and phosphatases (Nickels and Broach, 1996; Zhang et al., 1997).
Among the plant PCD elicitors that have been studied are avirulent pathogens (Morel and Dangl, 1997), preparations from pathogen culture fluids (He et al., 1993), cell wall preparations (Delledonne et al., 1998), abiotic elicitors such as H2O2 (Levine et al., 1996), and FB1 (Wang et al., 1996b; Gilchrist, 1997). FB1 is experimentally attractive as an elicitor of PCD because it can be used to simulate cell death in response to pathogen attack and because highly purified FB1 is commercially available. In this article, we report striking similarities between lesions formed on Arabidopsis plants in response to FB1 and the HR mounted in response to avirulent pathogens. These similarities include accumulation of ROIs, deposition of phenolic compounds and callose, production of the phytoalexin camalexin, and induction of defense-related gene expression. We also show that FB1 can be used to select directly for FB1-resistant (fbr) mutants, some of which are more resistant to pathogen attack.
RESULTS
FB1 Induces Lesion Formation in Arabidopsis
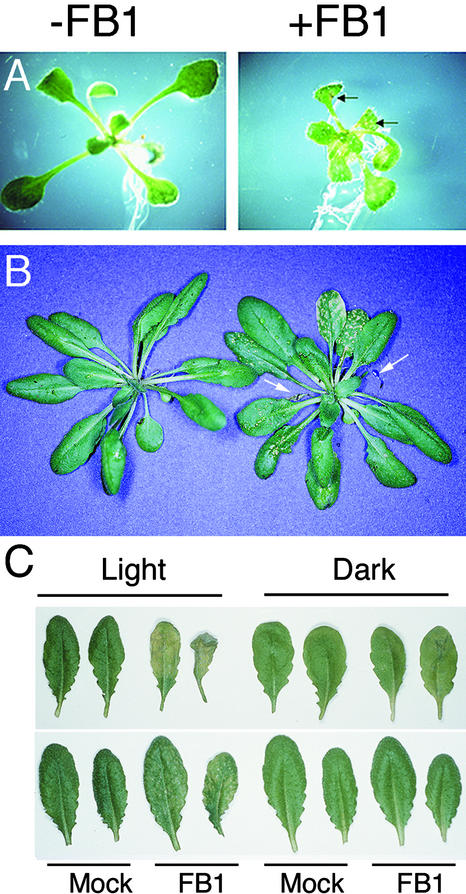
FB1 elicits both necrotic lesion formation in detached tomato leaves and a characteristic PCD response in susceptible tomato leaflets and protoplasts (Gilchrist et al., 1992; Wang et al., 1996b). To determine whether Arabidopsis responds to FB1 in a similar fashion, Arabidopsis ecotype Columbia (Col-0) seedlings grown on Murashige and Skoog (1962) (MS) agar medium for 10 days were transferred to MS medium containing 0, 10 nM, 0.1 μM, 1 μM, or 10 μM FB1. As illustrated in Figure 1A, macroscopic lesions formed on the leaves exposed for 4 days to 1 μM FB1. Lesion formation was dose dependent and was evident at concentrations of ⩾0.1 μM FB1. In addition, FB1 treatment resulted in a general growth arrest of the shoots and leaves, consistent with its effect on tomato and maize seedlings (Lamprecht et al., 1994). Root growth, however, was largely unaffected.
Figure 1.
FB1 Induces Lesion Formation in Arabidopsis.
(A) Ten-day-old Col-0 Arabidopsis seedlings were transferred to MS plates (−FB1) or MS plates supplemented with 1 μM FB1 (+FB1) and photographed 4 days later. Necrotic lesions on the FB1-treated seedling are indicated by arrows.
(B) Two lower leaves of 4-week-old Arabidopsis plants were infiltrated with 10 mM MgSO4 (left) or 10 μM FB1 in 10 mM MgSO4 (right) and photographed 7 days later. Dead FB1-infiltrated leaves are indicated with arrows.
(C) Lower leaves of 4-week-old Arabidopsis plants were infiltrated with 10 mM MgSO4 (Mock) or 10 μM FB1 in 10 mM MgSO4 (FB1). Plants were exposed to a 12-hr-light photoperiod (Light) or covered with aluminum foil (Dark) for 3 days. Infiltrated leaves are shown above; systemic, noninfiltrated leaves are shown below.
When a 10 μM FB1 solution was infiltrated into two lower leaves of 4-week-old Arabidopsis plants grown in soil in a greenhouse, macroscopic lesions formed on the infiltrated leaves within 1 to 2 days. As shown in Figure 1B, after 1 week the infiltrated leaves were completely dead, and smaller punctate lesions had formed on upper leaves that had not been infiltrated. The formation of lesions distant from the site of infiltration suggests either that FB1 is being transported to these upper leaves through the vasculature or that it induces a systemic signal that causes lesions throughout the plant.
Light is required for lesion formation in response to various pathogens (Peever and Higgins, 1989; Guo et al., 1995). Light is also required for lesion formation in some lesion mimic mutants and in transgenic plants that form spontaneous HR-like lesions (Elkind et al., 1990; Johal et al., 1994; Chamnongpol et al., 1996; Morel and Dangl, 1997; Genoud et al., 1998). The same is true for FB1; as shown in Figure 1C, FB1-elicited lesion formation in Arabidopsis leaves was greatly reduced in the dark.
FB1-Induced Lesions Share Many Features with HR Lesions
During the HR, a cell death program is triggered in host cells at or around the site of pathogen infection, resulting in cellular collapse and the formation of brown (necrotic) lesions (Morel and Dangl, 1997). HR lesions are also characterized by accumulation of ROIs (Jabs et al., 1996; Yang et al., 1997), deposition of callose and phenolic derivatives (Hahlbrock and Scheel, 1989; Dixon and Lamb, 1990), and biosynthesis of the low molecular mass antimicrobials, phytoalexins (Tsuji et al., 1992; Glazebrook and Ausubel, 1994).
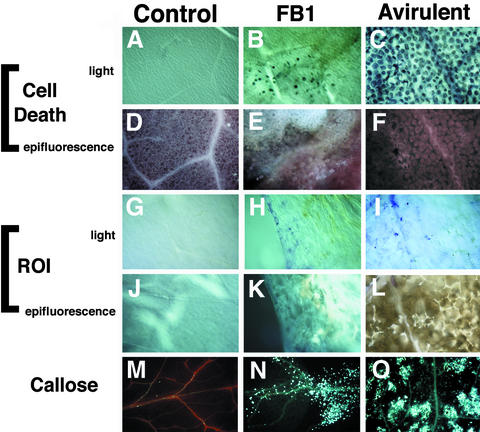
To determine whether FB1-induced lesions exhibit features characteristic of HR lesions, FB1-elicited lesions on the leaves of Arabidopsis seedlings were examined for cell death (trypan blue staining), accumulation of phenolic compounds (autofluorescence), production of ROIs (nitroblue tetrazolium staining), and deposition of callose (aniline blue staining). For comparison, HR lesions were elicited on 4-week-old Arabidopsis leaves by infiltration with the avirulent bacterial pathogen Pseudomonas syringae pv maculicola strain ES4326 expressing the avrRpt2 gene. As a negative control, 4-week-old leaves were infiltrated with MgSO4.
Figure 2B shows that FB1-treated leaves exhibited many dying cells that stained darkly with lactophenol–trypan blue at the periphery of a necrotic lesion. The lesion exhibits a light brown color, which can be seen at the top of Figure 2B. Figure 2E shows that the lesion was strongly autofluorescent. For comparison, Figures 2C and 2F show that an HR lesion elicited by P. s. maculicola ES4326 (avrRpt2) contained weakly autofluorescent dying cells. In contrast, as shown in Figures 2A and 2D, mock-inoculated control leaves lacked trypan blue staining, and autofluorescence was restricted to the vascular tissue. Our laboratory has also shown that leaves infiltrated with P. s. maculicola lacking avrRpt2 accumulate low levels of autofluorescent compounds (Yu et al., 1993; J.E. Heard, J.M. Stone, and F.M. Ausubel, unpublished results). Figures 2G to 2I show that nitroblue tetrazolium staining to detect ROI accumulation was absent in the control leaves (Figure 2G) but was visible in both FB1-treated (Figure 2H) and P. s. maculicola ES4326 (avrRpt2)–infected leaves (Figure 2I). Figures 2K and 2L show that areas of necrosis in the FB1-treated and P. syringae–infected leaves were highly autofluorescent. As was the case with trypan blue staining, the necrotic lesions themselves lacked nitroblue tetrazolium staining, which suggests that ROI accumulation occurred at the periphery of the lesions or was associated with initiation of lesion formation. Figures 2M to 2O show that callose deposition was barely visible and was restricted to vascular cells in control leaves (Figure 2M); in FB1-treated leaves, however, high amounts of callose were present in both vascular and nonvascular tissues (Figure 2N), similar to the response to infection with P. s. maculicola ES4326 (avrRpt2) (Figure 2O).
Figure 2.
FB1-Induced Lesions Resemble HR Lesions.
Transfer of 10-day-old seedlings to plates containing 1 μM FB1 or inoculation of 4-week-old leaves with P. s. maculicola 4326 avrRpt2 induced formation of macroscopic lesions. Comparisons were made by light or epifluorescence microscopic examination of stained leaves. See Methods for details.
(A) to (F) Leaves stained with lactophenol–trypan blue, revealing cell death.
(G) to (L) Leaves stained with nitroblue tetrazolium, revealing ROIs.
(M) to (O) Leaves stained with aniline blue, revealing callose deposition.
Control, MgSO4-treated leaves; FB1, FB1-treated leaves; Avirulent, P. s. maculicola ES4326 (avrRpt2)–infected leaves.
Camalexin is the only phytoalexin known to be produced in any substantial quantity in Arabidopsis in response to infection with pathogenic bacteria, fungi, and abiotic elicitors (Tsuji et al., 1992; Glazebrook and Ausubel, 1994; Zhao and Last, 1996; Glazebrook et al., 1997; Zhou et al., 1998; Thomma et al., 1999). Excised Arabidopsis leaves were placed on MS agar medium containing 1 μM FB1, and camalexin was extracted and measured fluorometrically. The concentrations of camalexin detected in two experiments (control/FB1-treated; 0.3/2.5 and 1.7/30 μg of camalexin g−1 fresh weight) were similar to those induced by bacterial and fungal inoculation (Zhao and Last, 1996; Thomma et al., 1999).
In summary, the data presented in this section demonstrate that FB1-induced lesions on Arabidopsis leaves share some of the features of the HR lesions elicited by avirulent pathogens.
FB1 Induces Expression of Defense-Related Genes
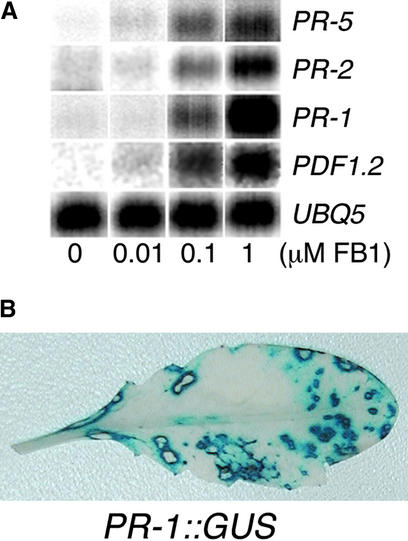
The HR is also characterized by the transcriptional activation of a set of PR genes (Stintzi et al., 1993; van Loon and van Strien, 1999). Figure 3A shows that FB1-induced lesion formation was accompanied by a dose-dependent accumulation of PR gene mRNAs. Expression of PR-5, PR-2, PR-1, and PDF1.2 (this last encoding the low molecular mass defensin polypeptide) was induced after 4 days of treatment with FB1 at 10 nM or more. To determine the pattern of PR gene expression, transgenic plants harboring the PR-1 promoter fused to the β-glucuronidase (GUS) reporter gene were infiltrated with FB1 and analyzed histochemically for GUS activity (Jefferson, 1987). As shown in Figure 3B, PR-1 expression in noninfiltrated leaves was restricted to the cells surrounding the punctate lesions, suggesting that PR-1 expression might depend on local, short-distance signals emanating from cells undergoing cell death.
Figure 3.
Treatment of Arabidopsis with FB1 Induces Expression of Defense-Related Genes.
(A) Defense gene activation in response to increasing concentrations of FB1. RNA was isolated from seedlings treated for 4 days on agar media with various concentrations of FB1 and analyzed by RNA gel blot analysis. UBQ5 was used as a loading control.
(B) Two lower leaves of transgenic Arabidopsis harboring a PR-1 promoter::GUS reporter gene fusion were infiltrated with 10 μM FB1 solution. After 1 week, the plants were histochemically stained for GUS activity. A noninfiltrated leaf that developed lesions is shown. Plants infiltrated with 10 mM MgSO4 showed no detectable GUS activity.
Selection of fbr Arabidopsis Mutants and Genetic Analyses
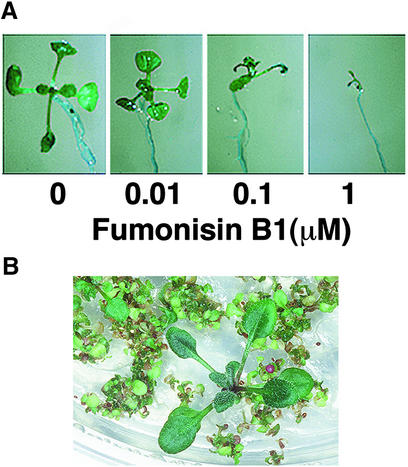
As shown in Figure 1A, FB1 treatment was associated with a general growth arrest of Arabidopsis seedlings (Figure 1A). This observation was substantiated by the experiment shown in Figure 4A in which Arabidopsis seeds were germinated directly on MS media containing FB1. The strong, dose-dependent inhibition of growth observed provides a powerful selection for fbr mutants. Among ∼55,000 ethyl methanesulfonic acid–mutagenized Arabidopsis Col-0 seeds subjected to selection on 1 μM FB1, five fbr mutants were identified; of these, two mutants that exhibited the most penetrant phenotypes, fbr1 and fbr2, were characterized in further detail. fbr1 and fbr2 were backcrossed to the wild-type parent Col-0, the resulting F1 plants were allowed to self-fertilize, and the F2 progeny were scored for resistance to FB1. The progeny segregated in a 3:1 susceptible/resistant ratio (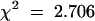 for fbr1, and
for fbr1, and 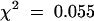 for fbr2), as would be expected if fbr phenotypes are conferred by single recessive mutations. Resistant F2 progeny from the first backcross were also crossed to each other (fbr1 × fbr2) to determine whether the two mutants are allelic. Analysis of the F2 progeny from this cross revealed that the phenotypes of fbr1 and fbr2 resulted from mutations at distinct genetic loci. In addition, fbr1 and fbr2 were crossed to Arabidopsis ecotype Landsberg erecta, and resistant F2 progeny were analyzed by using cleaved amplified polymorphisms and simple sequence-linked polymorphic markers to obtain rough map positions (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). fbr1 maps to the lower arm of chromosome I between markers F22K20 and PT1, whereas fbr2 maps to the upper arm of chromosome V between markers nga225 and nga249.
for fbr2), as would be expected if fbr phenotypes are conferred by single recessive mutations. Resistant F2 progeny from the first backcross were also crossed to each other (fbr1 × fbr2) to determine whether the two mutants are allelic. Analysis of the F2 progeny from this cross revealed that the phenotypes of fbr1 and fbr2 resulted from mutations at distinct genetic loci. In addition, fbr1 and fbr2 were crossed to Arabidopsis ecotype Landsberg erecta, and resistant F2 progeny were analyzed by using cleaved amplified polymorphisms and simple sequence-linked polymorphic markers to obtain rough map positions (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). fbr1 maps to the lower arm of chromosome I between markers F22K20 and PT1, whereas fbr2 maps to the upper arm of chromosome V between markers nga225 and nga249.
Figure 4.
FB1 Inhibits Seedling Germination.
(A) Arabidopsis Col-0 seeds were germinated on MS agar media supplemented with various concentrations of FB1 as indicated and photographed 10 days later.
(B) Mutagenized seeds were sowed directly on MS agar media supplemented with 0.5 μM FB1. An example of selection of a putative fbr mutant is shown.
fbr Mutants Are Affected in Defense-Related Gene Expression
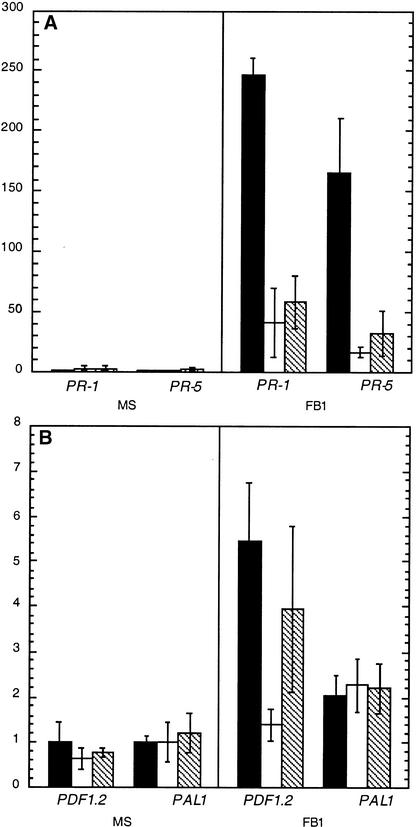
Because FB1 induces PR gene expression in wild-type Arabidopsis (Figure 3), we tested whether fbr1 and fbr2 exhibited aberrant expression of defense genes in response to FB1. Ten-day-old seedlings were transferred to MS agar medium or to MS agar medium supplemented with 1 μM FB1 and were analyzed by RNA gel blot analysis. As shown in Figure 5A, FB1-mediated elicitation of PR-1 and PR-5 mRNA accumulation in fbr1 and fbr2 seedlings was ∼20% of that in wild-type Col-0 seedlings. Although the decrease was less dramatic, FB1 also elicited less PDF1.2 mRNA in fbr1 (and most likely in fbr2 as well) than it did in wild-type plants (Figure 5B). In contrast to PR-1, PR-5, and PDF1.2, however, the mutations in fbr1 and fbr2 appeared to have no effect on the FB1-mediated induction of PAL1 mRNA. These results demonstrate that the fbr phenotype is associated with aberrant profiles of defense gene induction in response to FB1 but apparently not with a global defect in defense gene activation in these mutants.
Figure 5.
fbr Mutants Display Aberrant Defense Gene Induction in Response to FB1.
Wild-type Col-0, fbr1, and fbr2 seedlings were grown axenically for 10 days, then transferred to MS plates (MS) or MS plates supplemented with 1 μM FB1 (FB1). After 4 days of treatment, RNA was isolated and analyzed by RNA gel blot analysis as described in Methods. A representative experiment with triplicate RNA samples is shown. Experiments were repeated four times with similar results, except that PDF1.2 expression in fbr2 seedlings was generally found to be similar to that in fbr1. Data are presented as means ±sem. The relative expression of wild-type Col-0 is shown by black bars, fbr1 by white bars, and fbr2 by hatched bars.
(A) RNA gel blot analysis for SA-dependent and systemic acquired resistance–associated pathogenesis-related genes PR-1 and PR-5.
(B) RNA gel blot analysis for SA-independent genes PDF1.2 and PAL1.
fbr Mutants Are Differentially Responsive to Plant Pathogens
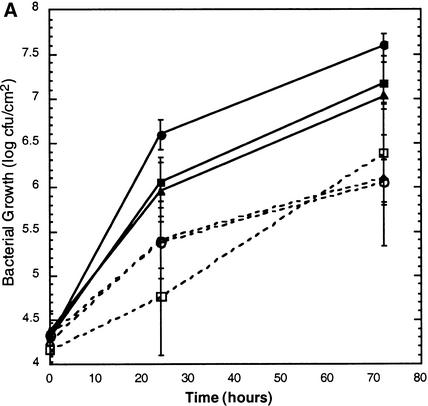
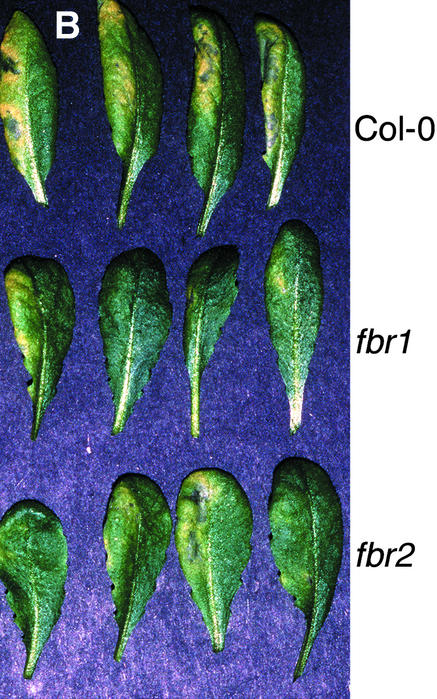
To determine whether fbr1 or fbr2 displays altered responses to pathogen attack, 4-week-old plants grown in soil were infected with virulent and avirulent strains of P. s. maculicola and with the obligate biotrophic fungal pathogen Erysiphe orontii. No differential response to E. orontii was observed for fbr1 or fbr2 in comparison with the response of wild-type Col-0 (data not shown). As shown in Figure 6A, growth of the avirulent bacterial pathogen P. s. maculicola ES4326 (avrRpt2) (Figure 6A) was also unaffected in fbr1 and fbr2, and the ability of the pathogen to elicit the HR did not appear to be diminished. Surprisingly, however, the isogenic virulent P. s. maculicola ES4326 strain lacking avrRpt2 consistently grew to a 10-fold lower titer in both fbr1 and fbr2 leaves than in wild-type plants (Figure 6A) and elicited less-pronounced disease symptoms (Figure 6B).
Figure 6.
fbr Mutants Display Enhanced Resistance to a Virulent Bacterial Pathogen.
(A) Growth of P. s. maculicola ES4326 in wild-type Col-0 (circles), fbr1 (squares), and fbr2 (triangles) plants. Leaves were infiltrated with virulent P. s. maculicola ES4326 (closed symbols) or with the avirulent isogenic strain expressing the avrRpt2 avirulence gene (open symbols). Leaf disks were harvested at 0, 24, and 72 hr after infiltration, and bacterial counts were determined by serial dilution. The data shown are means for six leaves ±sem from a representative experiment; these experiments were repeated three times with fbr2 and five times with fbr1 with similar results. cfu, colony-forming units.
(B) Disease symptoms elicited by inoculation with P. s. maculicola ES4326. The left half of each leaf was infiltrated with bacterial suspension (104 cfu/mL), and the leaves were photographed 2 days later.
DISCUSSION
Although cell death is a central feature of both compatible and incompatible plant–pathogen interactions, its respective roles in resistance and susceptibility are largely unknown. In this article, we describe a relatively simple pathogen-free system in Arabidopsis involving the fungal toxin FB1 that can be exploited to study the signal transduction events involved in pathogen-elicited cell death.
FB1-induced lesions in Arabidopsis are similar to pathogen-induced lesions in many respects, including deposition of phenolic compounds and callose, production of ROIs, accumulation of camalexin, and expression of PR genes. FB1 elicits expression of the PR-1, PR-2, and PR-5 genes, which are also induced after infection by various pathogens and by treatment with salicylic acid (SA) or its analogs (Yang et al., 1997). The pattern of PR-1 expression in response to FB1 in transgenic plants harboring a PR-1 promoter::GUS reporter suggests that PR-1 expression may occur in response to signals emanating from dead or dying cells rather than to signals within the cells undergoing cell death. Other examples of diffusible signals eliciting PR gene expression in cells neighboring HR-like lesions have been noted (Samac and Shah, 1991; Levine et al., 1994; Chappell et al., 1997). The expression of PR-1 in the cells surrounding FB1-elicited lesions may be related to our finding that ROI accumulation occurred at the margins of lesions, similar to the observation that superoxide production is detected at the margins of lesions in the Arabidopsis lsd1 mutant (Jabs et al., 1996). On the other hand, we cannot rule out the possibility that expression of PR-1 is a precursor to cell death rather than a consequence of a diffusible cell death–generated signal.
If PR-1 gene expression in Arabidopsis is activated by a diffusible signal from dying cells, then probably expression of PR-1 is neither necessary nor sufficient for host cell death. This conclusion is supported by several reports in the literature. First, exogenous application of SA induced expression of several PR genes but did not in itself induce cell death (Shirasu et al., 1997). Second, suppression of tobacco mosaic virus–induced HR by low oxygen pressure or caspase inhibition only marginally affected PR-1a and PR-2 expression in tobacco (Mittler et al., 1996; del Pozo and Lam, 1998). Third, when avirulent bacterial pathogen–induced cell death was suppressed by scavengers of NO in soybean cell suspensions, expression of the gene encoding glutathione S-transferase was not substantially affected (Delledonne et al., 1998).
In addition to systemic acquired resistance–associated and SA-dependent PR genes (e.g., PR-1, PR-2, and PR-5), FB1 induces expression of PDF1.2. PDF1.2 encodes a small, cysteine-rich secreted protein related to insect defensins, which is induced by necrotizing fungal pathogens and abiotic elicitors such as jasmonic acid and ethylene (Penninckx et al., 1996, 1998). In the companion paper to this article (Asai et al., 2000, in this issue), we demonstrate that signal transduction pathways dependent on SA, jasmonic acid, and ethylene are required for FB1-induced PCD in Arabidopsis protoplasts. Thus, FB1 appears to activate directly or indirectly a variety of signaling pathways associated with the defense response to pathogen attack.
Another similarity between FB1-induced lesion formation and HR lesions is their dependence on light. A similar light dependence for lesion formation has also been observed in tomato treated with AAL toxin, which is structurally and functionally related to FB1 (Moussatos et al., 1993). The reason for light-dependent augmentation of lesion formation is poorly understood. Light can enhance the oxidative burst (Allen et al., 1999), but it is also possible that phytochrome signaling (Genoud et al., 1998) or photosynthesis (Gray et al., 1997) is required for some lesion phenotypes. Under some circumstances, lesion formation has been shown to be dependent on SA accumulation (Weymann et al., 1995; Rate et al., 1999; Shah et al., 1999), and interestingly, as shown in the companion paper, FB1-induced cell death in Arabidopsis protoplasts is correlated with the light-dependent accumulation of SA (Asai et al., 2000, in this issue).
The cellular target or targets of FB1 in Arabidopsis are not clear. FB1 is known to be a competitive inhibitor of ceramide synthase in other species (Wang et al., 1990; Abbas et al., 1994; Yoo et al., 1996; Norred et al., 1997), and some FB1-induced effects can be suppressed by exogenous ceramide (Harel and Futerman, 1993; Wakita et al., 1996; Tonnetti et al., 1999). However, several other mechanisms of toxicity have also been noted, including membrane perturbation (Yin et al., 1996) and direct effects on signaling molecules such as protein phosphatases (Fukuda et al., 1996). We have been unable to rescue FB1-induced cell death with cell-permeable C2-ceramide (T. Asai, J.M. Stone, and F.M. Ausubel, unpublished data), which suggests that FB1-induced cell death in Arabidopsis cannot be attributed simply to ceramide depletion. Interestingly, however, ceramide was recently shown to suppress cell death in tomato induced by AAL toxin, which is, as mentioned above, structurally related to FB1 (Brandwagt et al., 2000).
The fact that FB1 inhibits germination in Arabidopsis provides a powerful genetic screen to identify mutants. fbr mutants were readily obtained by directly sowing mutagenized seeds on FB1-containing agar. The recessively inherited fbr phenotypes of fbr1 and fbr2 could indicate an inability to incorporate or transport FB1, the absence or alteration of FB1 targets or signaling cascade components, or the inability to undergo PCD. The two mutants characterized in this study, fbr1 and fbr2, form lesions on FB1-containing plates (data not shown), which suggests that they remain capable of responding to FB1. Moreover, protoplasts derived from fbr1 and fbr2 are not markedly different from wild-type Col-0 in their susceptibility to FB1-induced cell death (T. Asai and F.M. Ausubel, unpublished results). On the other hand, both fbr1 and fbr2 express less PR-1, PR-5, and PDF1.2 transcripts than do wild-type plants in response to FB1. Therefore, we think it unlikely that fbr1 and fbr2 are blocked in a primary FB1 receptor or in the ability to perform PCD; instead, they may have a lesion or lesions in an FB1-mediated signaling pathway that leads to activation of PCD and expression of defense genes.
Because of the similarities between FB1- and pathogen-induced lesions and the altered PR gene expression in fbr1 and fbr2, we reasoned that fbr1 and fbr2 might be affected in response to pathogens. Most plant–pathogen interactions result in some extent of host cell death. Although the HR is considered one of the hallmarks of incompatible (or resistant) interactions, its actual contribution to the restriction of pathogen growth is unclear. The Arabidopsis dnd1 (for defense, no death) mutant, which is defective in undergoing the HR yet still capable of hindering growth of avirulent pathogens (Yu et al., 1998), negates the supposition that the HR is strictly required for disease resistance. In another study, however, blocking the P. s. maculicola avrRpm1–elicited HR in Arabidopsis with inhibitors of NO synthase led to increased growth of the avirulent pathogen (Delledonne et al., 1998), suggesting, at least in this case, that the HR contributes to restriction of pathogen growth.
Unexpectedly, we found that both fbr1 and fbr2 exhibited no difference in response to the avirulent P. s. maculicola pathogen expressing avrRpt2, but they were markedly more resistant than wild-type plants to the isogenic virulent bacterial pathogen. One interpretation of this result is that virulent bacterial pathogens such as P. syringae intentionally trigger PCD to obtain nutrients from host tissues, but P. syringae is not capable of fully activating this PCD pathway in fbr1 and fbr2. The dnd1 mutant, which is defective in mounting an HR, is also resistant to growth of virulent bacterial pathogens, further supporting a role for host cell death in compatible interactions. However, the dnd1 mutant, like many other mutants displaying enhanced resistance to virulent pathogens, has increased concentrations of SA and a small stature (Yu et al., 1998). In contrast, fbr1 and fbr2 are normal in size and do not constitutively express PR genes, which would be indicative of high SA concentrations. Recently, ihr (for intermediate HR) mutants have been described as having normal stature and increased resistance to virulent pathogens (Yu et al., 2000). Some of these latter mutants may be allelic to the fbr mutants.
Why fbr1 and fbr2 mutants are more resistant than wild-type plants to P. s. maculicola is not clear. PR genes are induced in the fbr1 and fbr2 mutants in response to both virulent and avirulent P. s. maculicola ES4326 (J.M. Stone and F.M. Ausubel, unpublished data). However, we observed no important differences from wild-type plants regarding PR gene expression that could account for the resistance to the virulent pathogen. One problem in interpreting this result is that the set of inducible defense-related genes used to analyze defense-related mutants is limited. Transcription profiling analysis using DNA array technology will, we hope, increase the number of target responses that can be monitored during the defense response, which will help correlate the phenotypes of the mutants with gene expression patterns.
In summary, the importance of PCD in plant–pathogen interactions, coupled with the evident complexity of cell death–mediated defense responses, has led us to develop a simple model system to study the role of cell death in plant pathogenesis. We demonstrate the utility of FB1 for high-throughput isolation of Arabidopsis mutants altered in cell death–associated processes. Cloning the genes corresponding to these altered phenotypes promises to expand our understanding of the mechanisms of PCD and its role in plant defense responses.
METHODS
Plant Growth Conditions and Treatment with FB1
Arabidopsis thaliana seedlings were germinated axenically on Murashige and Skoog (MS; Murashige and Skoog, 1962) medium containing 2% sucrose and 0.6% Phytagar (Gibco BRL) and maintained in a temperature-controlled growth room (20 ± 2°C) under continuous illumination (50 μE m–2 sec–1). Arabidopsis seedlings were grown for 10 days before being transferred to fresh MS plates or to fresh MS plates supplemented with various concentrations of FB1 (Sigma). Seedlings were returned to the growth room for 4 days, at which time samples were taken for RNA gel blot analysis or staining and preparation for microscopy. For camalexin quantitation, leaves were removed from 10-day-old seedlings and transferred to MS plates or to plates of MS supplemented with 1 μM FB1 for 5 days before extraction and quantification, as described previously (Glazebrook et al., 1997). For some experiments, Arabidopsis plants were grown in Metromix 2000 (W.R. Grace, Ontario, Canada) in a temperature-controlled greenhouse (20 ± 2°C) on a 12-hr-light/12-hr-dark cycle (100 μE m–2 sec–1). A 10 μM FB1 solution in 10 mM MgSO4 was infiltrated into two lower leaves with a 1-mL syringe. For the dark experiment, plants were covered with aluminum foil and the leaves were photographed after 3 days. For selection of fbr mutants, ethyl methanesulfonic acid–mutagenized seeds (Lehle Seeds, Tucson, AZ) were germinated axenically on MS plates supplemented with 1 μM FB1.
Microscopy
Microscopy was performed with a Zeiss universal light microscope. Cell death was visualized in whole seedlings after staining with lactophenol–trypan blue, as described previously (Dietrich et al., 1994; Bowling et al., 1997). Autofluorescence was observed by epifluorescence microscopy (excitation at 460 nm, emission at >478 nm). Before visualization, tissue was fixed in autofluorescence-fixing solution (Bowling et al., 1997). For aniline blue staining of callose, plant samples were vacuum-infiltrated with ethanol/lactophenol (2:1 [v/v]) and incubated at 60°C for 30 min. Samples were then rinsed with water to remove the lactophenol and stained overnight with aniline blue (0.01% aniline blue powder in 150 mM K2PO4, pH 9.5). Before samples were mounted, they were equilibrated in 50% glycerol. Aniline blue staining was visualized by epifluorescence microscopy. Nitroblue tetrazolium staining was performed as described previously (Jabs et al., 1996).
RNA Gel Blot Analyses
Total RNA was isolated and purified from control and FB1-treated seedlings (Reuber and Ausubel, 1996). Five to 10 μg of total RNA per treatment was separated on formaldehyde–agarose gels, blotted to GeneScreen membrane (New England Nuclear, Boston, MA), and hybridized with various 32P-labeled probes (described below) overnight at 42°C. Blots were washed twice with 1% SDS in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 65°C for 45 min each. Quantitation was performed with a PhosphorImager (Molecular Dynamics), the ratio of the signal for each probe to that from UBQ5 was calculated to compensate for loading differences, and the values were normalized to a relative gene expression level of 1 for ecotype Columbia (Col-0) on MS.
Single-stranded 32P-labeled probes were synthesized from a linear double-stranded DNA template by polymerase chain reaction (Greenberg et al., 1994). Gene-specific probes for PR-1, PR-5, PR-2, PAL1, and UBQ5 were synthesized by using templates and primers described previously (Rogers and Ausubel, 1997). The PDF1.2 template and primers were also based on a previous report (Penninckx et al., 1996).
Histochemical Staining for β-Glucuronidase Activity
Four-week-old transgenic plants harboring the PR-1 promoter::GUS reporter construct (kindly provided by Allan Shapiro, University of Delaware, Wilmington) were treated by infiltrating two lower leaves with 10 μM FB1. After 1 week, leaves were excised and vacuum-infiltrated with a modified stain solution containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl glucuronide, incubated at 37°C overnight, and cleared with 70% ethanol (Jefferson, 1987) before photography.
Pathogen Infections and Analyses
Four-week-old Arabidopsis plants were inoculated with Erysiphe orontii and scored for susceptibility, as described previously (Reuber et al., 1998). Bacterial strains were Pseudomonas syringae pv maculicola ES4326 carrying pLAFR3 (virulent) or pLH12 (avirulent; avrRpt2) plasmids (Whalen et al., 1991). For growth determinations and microscopic analyses, fully expanded leaves were infiltrated with a bacterial suspension (104 colony-forming units [cfu]/mL) in 10 mM MgSO4. At various times, leaves were excised and prepared for microscopy or two leaf discs were ground in 10 mM MgSO4, serially diluted, and plated on King's B-agar plates supplemented with 100 μg/L streptomycin and 10 μg/L tetracycline.
Acknowledgments
We thank Drs. Mary Wildermuth, Peter Yorgey, Julia Dewdney, Andrew Diener, and other members of the Ausubel laboratory for critical comments on the manuscript and useful discussions. We also acknowledge Allan Shapiro (University of Delaware) for the transgenic line harboring the PR-1 promoter::GUS reporter construct. This work was supported by a National Science Foundation postdoctoral fellowship (No. DBI-9750297) award to J.M.S.; a Life Sciences Research Foundation postdoctoral fellowship award to J.E.H.; fellowships from the Toyobo Biotechnology Foundation and the Uehara Memorial Foundation awarded to T.A.; and a National Institutes of Health grant (No. R01-GM48707) awarded to F.M.A.
References
- Abbas, H.K., Tanaka, T., Duke, S.O., Porter, J.K., Wray, E.M., Hodges, L., Sessions, A.E., Wang, E., Merrill, A.H., and Riley, R.T. (1994). Fumonisin- and AAL-toxin–induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 106, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, L.J., MacGregor, K.B., Koop, R.S., Bruce, D.H., and Karner, J., and Bown, A.W. (1999). The relationship between photosynthesis and a mastoparan-induced hypersensitive response in isolated mesophyll cells. Plant Physiol. 119, 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Dixit, V.M., and Koonin, E.V. (1999). The domains of death: Evolution of the apoptosis machinery. Trends Biochem. Sci. 24, 47–53. [DOI] [PubMed] [Google Scholar]
- Asai, T., Stone, J.M., Heard, J.E., Kovtun, Y., Yorgey, P., Sheen, J., and Ausubel, F.M. (2000). Fumonisin B1–induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12, in press. [DOI] [PMC free article] [PubMed]
- Beers, E.P. (1997). Cell death during plant growth and development. Cell Death Differ. 4, 649–661. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandwagt, B.F., Mesbah, L.A., Takken, F.L.W., Laurent, P.L., Kneppers, T.J.A., Hille, J., and Nijkamp, H.J.J. (2000). A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl. Acad. Sci. USA 97, 4961–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamnongpol, S., Willekens, H., Langebartels, C., Van Montagu, M., Inzé, D., and Van Camp, W. (1996). Transgenic tobacco with a reduced catalase activity develops necrotic lesions and induces pathogenesis-related expression under high light. Plant J. 10, 491–504. [Google Scholar]
- Chappell, J., Levine, A., Tenhaken, R., Lusso, M., and Lamb, C. (1997). Characterization of a diffusible signal capable of inducing defense gene expression in tobacco. Plant Physiol. 113, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6–1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P. (1997). Genetic dissection of acquired resistance to disease. Plant Physiol. 113, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne, M., Xia, Y., Dixon, R.A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. [DOI] [PubMed] [Google Scholar]
- del Pozo, O., and Lam, E. (1998). Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol. 8, 1129–1132. [DOI] [PubMed] [Google Scholar]
- Desjardins, A.E., Plattner, R.D., Nelsen, T.C., and Leslie, J.F. (1995). Genetic analysis of fumonisin production and virulence of Gibberella fujikuroi mating population A (Fusarium moniforme) on maize (Zea mays) seedlings. Appl. Environ. Microbiol. 61, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., and Lamb, C.J. (1990). Molecular communication in interactions between plants and microbial pathogens. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 339–367. [Google Scholar]
- D'Silva, I., Poirer, G.G., and Heath, M.C. (1998). Activation of cysteine protease in cowpea plants during the hypersensitive response—A form of programmed cell death. Exp. Cell. Res. 245, 389–399. [DOI] [PubMed] [Google Scholar]
- Dutton, M.F. (1996). Fumonisins, mycotoxins of increasing importance: Their nature and their effects. Pharmacol. Ther. 70, 137–161. [DOI] [PubMed] [Google Scholar]
- Elkind, Y., Edwards, R., Mavandad, M., Hedrick, S.A., Ribak, O., Dixon, R.A., and Lamb, C.J. (1990). Abnormal plant development and downregulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc. Natl. Acad. Sci. USA 87, 9057–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1947). Host–parasite interactions in flax rust—Its genetics and other implications. Phytopathology 45, 680–685. [Google Scholar]
- Fukuda, H. (1996). Xylogenesis: Initiation, progression, and cell death. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 299–325. [DOI] [PubMed] [Google Scholar]
- Fukuda, H., Shima, H., Vesonder, R.F., Tokuda, H., Nishino, H., Katoh, S., Tamura, S., Sugimura, T., and Nagao, M. (1996). Inhibition of protein serine/threonine phosphatases by fumonisin B1, a mycotoxin. Biochem. Biophys. Res. Commun. 220, 160–165. [DOI] [PubMed] [Google Scholar]
- Futerman, A.H. (1995). Inhibition of sphingolipid synthesis—Effects on glycosphingolipid-GPI–anchored protein microdomains. Trends Cell Biol. 5, 377–380. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Millar, A.J., Nishizawa, N., Kay, S.A., Schafer, E., Nagatani, A., and Chua, N.-H. (1998). An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell 10, 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, D.G. (1997). Mycotoxins reveal connections between plants and animals in apoptosis and ceramide signaling. Cell Death Differ. 4, 689–698. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D.G. (1998). Programmed cell death in plant disease: The purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 36, 393–414. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D.G., Ward, B., Moussatos, V., and Mirocha, C.J. (1992). Genetic and physiological response to fumonisins and AAL-toxin by intact tissue of a higher plant. Mycopathologia 117, 57–64. [Google Scholar]
- Gilchrist, D.G., Wang, H., and Bostock, R.M. (1995). Sphingosine-related mycotoxins in plant and animal diseases. Can. J. Bot. 73 (suppl. 1), 459.–467. [Google Scholar]
- Glazebrook, J., and Ausubel, F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91, 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J., Close, P.S., Briggs, S.P., and Johal, G.S. (1997). A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89, 25–32. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997). Programmed cell death in plant–pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Ausubel, F.M. (1993). Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4, 327–341. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Guo, A., Reimers, P.J., and Leach, J.E. (1995). Effect of light on incompatible interactions between Xanthomonas oryzae pv. oryzae and rice. Physiol. Mol. Plant Pathol. 42, 413–425. [Google Scholar]
- Hahlbrock, K., and Scheel, D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 347–369. [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Harel, R., and Futerman, A.H. (1993). Inhibition of sphingolipid synthesis affects axonal outgrowth in cultured hippocampal neurons. J. Biol. Chem. 268, 14476–14481. [PubMed] [Google Scholar]
- He, S.Y., Huang, H.-C., and Collmer, A. (1993). Pseudomonas syringae pv. syringae HarpinPss: A protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Jabs, T., Dietrich, R.A., and Dangl, J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. [DOI] [PubMed] [Google Scholar]
- Jardine, D.J., and Leslie, J.F. (1999). Aggressiveness to mature maize plants of Fusarium strains differing in ability to produce fumonisin. Plant Dis. 83, 690–693. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Johal, G.S., Hulbert, S.H., and Briggs, S.P. (1994). Disease lesion mimics of maize: A model for cell death in plants. Bioessays 17, 685–692. [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 96, 7956–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Pontier, D., and del Pozo, O. (1999). Die and let live—Programmed cell death in plants. Curr. Opin. Plant Biol. 2, 502–507. [DOI] [PubMed] [Google Scholar]
- Lamprecht, S.C., Marasas, W.F.O., Alberts, J.F., Cawood, M.E., Gelderblom, W.C.A., Shepherd, G.S., Thiel, P.G., and Calitz, F.J. (1994). Phytotoxicity of fumonisins and TA-toxin to corn and tomato. Phytopathology 84, 383–391. [Google Scholar]
- Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Levine, A., Pennell, R.I., Alvarez, M.E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6, 427–437. [DOI] [PubMed] [Google Scholar]
- Mitsuhara, I., Malik, K.A., Miura, M., and Ohashi, Y. (1999). Animal cell-death suppressors Bcl-XL and Ced-9 inhibit cell death in tobacco plants. Curr. Biol. 9, 775–778. [DOI] [PubMed] [Google Scholar]
- Mittler, R., Shulaev, V., Seskar, M., and Lam, E. (1996). Inhibition of programmed cell death in tobacco plants during a pathogen-induced hypersensitive response at low oxygen pressure. Plant Cell 8, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.-B., and Dangl, J.L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Moussatos, V., Witsenboer, H., Hille, J., and Gilchrist, D. (1993). Behaviour of the disease resistance gene Asc in protoplasts of Lycopersicon esculentum mill. Physiol. Mol. Plant Pathol. 43, 255–263. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nickels, J.T., and Broach, J.R. (1996). A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10, 382–394. [DOI] [PubMed] [Google Scholar]
- Norred, W.P., Plattner, R.D., Dombrink-Kurtzman, M.A., Meredith, F.I., and Riley, R.T. (1997). Mycotoxin-induced elevation of free sphingoid bases in precision-cut rat liver slices: Specificity of the response and structure–activity relationships. Toxicol. Appl. Pharmacol. 147, 63–70. [DOI] [PubMed] [Google Scholar]
- Peever, T.L., and Higgins, V.J. (1989). Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and non-specific elicitors from Cladosporium fulvum. Plant Physiol. 90, 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defense, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., and Ausubel, F.M. (1996). Isolation of Arabidopsis genes that differentiate between disease resistance responses mediated by RPS2 and RPM1 disease resistance genes. Plant Cell 8, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Richael, C., and Gilchrist, D. (1999). The hypersensitive response: A case of hold or fold? Physiol. Mol. Plant Pathol. 55, 5–12. [Google Scholar]
- Richberg, M.H., Aviv, D.H., and Dangl, J.L. (1998). Dead cells do tell tales. Curr. Opin. Plant Biol. 1, 480–485. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E., and Ausubel, F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac, D.A., and Shah, D.M. (1991). Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expession salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, N., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C.J. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel, S., and Merrill, A.H. (1996). Sphingolipid metabolism and cell growth regulation. FASEB J. 10, 1388–1397. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., Heitz, T., Prasad, V., Wiedemann-Merdinoglu, S., Kauffmann, S., Geoffroy, P., Legrand, M., and Frittig, B. (1993). Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie 75, 687–706. [DOI] [PubMed] [Google Scholar]
- Takahashi, A., Kawasaki, T., Henmi, K., Shii, K., Kodama, O., Satoh, H., and Shimamoto, K. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17, 535–545. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Nelissen, I., Eggermont, K., and Broekaert, W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- Tolleson, W.H., Melchior, W.B., Jr., Morris, S.M., McGarrity, L.J., Domon, O.E., Muskhelishvili, L., James, S.J., and Howard, P.C. (1996). Apoptotic and anti-proliferative effects of fumonisin B1 in human keratinocytes, fibroblasts, asophageal epithelial cells and hepatoma cells. Carcinogenesis 17, 239–249. [DOI] [PubMed] [Google Scholar]
- Tonnetti, L., Veri, M.-C., Bonvini, E., and D'Adamio, L. (1999). A role for neutral sphingomyelinase-mediated ceramide production in T cell receptor–induced apoptosis and mitogen-activated protein kinase–mediated signal transduction. J. Exp. Med. 189, 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, J., Jackson, E.P., Gage, D.A., Hammerschmidt, R., and Somerville, S.C. (1992). Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv. syringae. Plant Physiol. 98, 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D.G. (1998). Homologies between plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R227. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C., and van Strien, E.A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Wakita, H., Nishimura, K., Tokura, Y., Furukawa, F., and Takigawa, M. (1996). Inhibitors of sphingolipid synthesis modulate interferon (IFN)-γ–induced intercellular adhesion molecular (ICAM)-1 and human leukocyte antigen (HLA)–DR expression on cultured normal human keratinocytes—Possible involvement of ceramide in biologic action of IFN-γ. J. Invest. Dermatol. 107, 336–342. [DOI] [PubMed] [Google Scholar]
- Wang, E., Norred, W.P., Bacon, C.W., Riley, R.T., and Merrill, A.H. (1990). Inhibition of sphingolipid biosynthesis by fumonisins: Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266, 14486–14490. [PubMed] [Google Scholar]
- Wang, H., Jones, C., Ciacci-Zanella, J., Holt, T., Gilchrist, D.G., and Dickman, M.B. (1996. a). Fumonisins and Alternaria alternata lycopersici toxins: Sphinganine analog mycotoxins induce apoptosis in monkey kidney cells. Proc. Natl. Acad. Sci. USA 93, 3461–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Li, J., Bostock, R.M., and Gilchrist, D.G. (1996. b). Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8, 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann, K., Hunt, M., Uknes, S., Neuenschwander, U., Lawton, K., Steiner, H.Y., and Ryals, J. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., and Reed, J.C. (1998). Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell 1, 337–346. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]
- Yin, J.-J., Smith, M.J., Eppley, R.M., Troy, A.L., Page, S.W., and Sphon, A. (1996). Effects of fumonisin B1 and (hydrolyzed) fumonisin backbone AP1 on membranes: A spin-label study. Arch. Biochem. Biophys. 335, 13–22. [DOI] [PubMed] [Google Scholar]
- Yoo, H.-S., Norred, W.P., Showker, J., and Riley, R.T. (1996). Elevated sphingoid bases and complex sphingolipid depletion as contributing factors in fumonisin-induced toxicity. Toxicol. Appl. Pharmacol. 138, 211–218. [DOI] [PubMed] [Google Scholar]
- Yu, G.-L., Katagiri, F., and Ausubel, F.M. (1993). Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol. Plant-Microbe Interact. 6, 434–443. [DOI] [PubMed] [Google Scholar]
- Yu, I.-C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, I.-C., Fengler, K.A., Clough, S.J., and Bent, A.F. (2000). Identification of Arabidopsis mutants exhibiting an altered hypersensitive response in gene-for-gene disease resistance. Mol. Plant-Microbe Interact. 13, 277–286. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Yao, B., Delikat, S., Bayoumy, S., Lin, X.-H., Basu, S., McGinley, M., Chan-Hui, P.-Y., Lichenstein, H., and Kolesnick, R. (1997). Kinase suppressor of Ras is ceramide-activated protein kinase. Cell 89, 63–72. [DOI] [PubMed] [Google Scholar]
- Zhao, J., and Last, R.L. (1996). Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell 8, 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream of salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]