Abstract
We have examined the behavior of pre-replication complex (pre-RC) proteins in relation to key cell cycle transitions in Chinese Hamster Ovary (CHO) cells. ORC1, ORC4 and Cdc6 were stable (T1/2 >2 h) and associated with a chromatin-containing fraction throughout the cell cycle. Green fluorescent protein-tagged ORC1 associated with chromatin throughout mitosis in living cells and co-localized with ORC4 in metaphase spreads. Association of Mcm proteins with chromatin took place during telophase, ∼30 min after the destruction of geminin and cyclins A and B, and was coincident with the licensing of chromatin to replicate in geminin-supplemented Xenopus egg extracts. Neither Mcm recruitment nor licensing required protein synthesis throughout mitosis. Moreover, licensing could be uncoupled from origin specification in geminin-supplemented extracts; site-specific initiation within the dihydrofolate reductase locus required nuclei from cells that had passed through the origin decision point (ODP). These results demonstrate that mammalian pre-RC assembly takes place during telophase, mediated by post-translational modifications of pre-existing proteins, and is not sufficient to select specific origin sites. A subsequent, as yet undefined, step selects which pre-RCs will function as replication origins.
Keywords: cell cycle/DNA replication/origin decision point/origin recognition complex/pre-replication complex
Introduction
Eukaryotic DNA replication is tightly regulated to achieve duplication of the genome exactly once per cell cycle. Central to this regulation is the assembly of pre-replication complex (pre-RC) proteins at origins of replication during telophase (Takisawa et al., 2000), a process often referred to as replication ‘licensing’. Pre-RC proteins are largely conserved in budding yeast, fission yeast, Drosophila, Xenopus and mammalian cells, and a universal model for the mechanism of pre-RC assembly in all eukaryotes has emerged (Leatherwood, 1998; Takisawa et al., 2000, and references therein). In budding yeast, the hetero-hexameric origin recognition complex (ORC) is bound to the consensus sequence of replication origins throughout the cell cycle. After the destruction of mitotic cyclins during anaphase, resulting in the inactivation of Cdc28 kinase activity, ORC recruits Cdc6, which in turn serves as a loading factor for the hetero-hexameric minichromosome maintenance (Mcm2-7) proteins. Pre-RCs can assemble at sites of ORC binding until the point during G1 phase at which Cdc28 activity begins to rise as a result of the induction of G1 cyclins, preventing further pre-RC assembly. Shortly thereafter, a combination of Cdc28 and Cdc7 kinase activity triggers the initiation of replication, which converts the pre-RC to an active replication complex. Since Cdc28 activity continues to rise during S phase, pre-RCs can not re-assemble until the following anaphase, ensuring that no segment of DNA can replicate more than once per cell cycle.
In metazoa, some aspects of pre-RC assembly have been difficult to study, primarily due to the fact that specific origin DNA sequences have yet to be defined (Gilbert, 2001). For example, it has not been possible to address whether sites of ORC residence and pre-RC assembly are coincident with the sites of once-per-cell cycle initiation of replication (albeit, Drosophila ORC has been shown to bind near origins of programmed gene amplification; Austin et al., 1999). Nonetheless, biochemical studies in Xenopus egg extracts have defined the order of events that must take place for replication to initiate in vitro. These studies have shown that the physical association of pre-RC proteins with detergent-washed chromatin reflects the assembly of these proteins into functional pre-RCs and that the ordered assembly of pre-RC proteins, ORC– Cdc6–Mcm2-7, is essential for subsequent initiation of replication (Takisawa et al., 2000, and references therein). These results have prompted similar studies of the physical association of pre-RC proteins with detergent-washed chromatin in mammalian cells (Dimitrova et al., 1999; Mendez and Stillman, 2000), largely confirming this model of ordered assembly. However, some important differences with the budding yeast model have been identified. First, in addition to the S phase and mitotic cyclin-dependent kinase (Cdk) activity, multi-cellular organisms have a specific inhibitory protein called geminin (McGarry and Kirschner, 1998). Geminin binds and inactivates Cdt1 (Wohlschlegel et al., 2000; Tada et al., 2001), which is required along with Cdc6 to load the Mcm2-7 proteins (Maiorano et al., 2000; Nishitani et al., 2000). Geminin is an extremely potent inhibitor of pre-RC assembly, accounting for most of the inhibition of pre-RC assembly during mitosis in Xenopus egg extracts (Tada et al., 2001). The mammalian homolog to geminin is also a Cdt1 binding protein and a potent inhibitor of DNA synthesis (Wohlschlegel et al., 2000).
Other aspects of pre-RC assembly in higher eukaryotes are not yet clear. In particular, the precise behavior of ORC and Cdc6 has been something of an enigma. In both Saccharomyces cerevisiae (Aparicio et al., 1997; Tanaka et al., 1997) and Schizosaccharomyces pombe (Ogawa et al., 1999), it is clear that ORC is bound to origins of replication throughout the cell cycle, including mitosis. However, in metazoa, some investigators find ORC to be removed from chromatin during mitosis (Romanowski et al., 1996; Loupart et al., 2000), while others do not (Pak et al., 1997; Tatsumi et al., 2000). In mammalian cells, results from different laboratories have not produced a consistent picture, even with identical cell lines. Some have reported that Orc1 and Orc6 fail to co-immunoprecipitate with a complex containing Orc2-5 (Dhar and Dutta, 2000), while other laboratories have successfully co-immunoprecipitated ORC1 with other ORC subunits (Gavin et al., 1995; Kreitz et al., 2001; Vashee et al., 2001). Some reports have concluded that ORC1 (but not ORC2) is selectively removed from chromatin during mitosis (Natale et al., 2000), while others have cross-linked ORC1 to DNA during mitosis (Tatsumi et al., 2000), and still others find both ORC1 and ORC2 to be removed from chromatin during S phase (Kreitz et al., 2001).
The behavior of Cdc6 is also not entirely clear. In Xenopus, some experiments indicate that Cdc6 remains bound to chromatin during S phase (Coleman et al., 1996), while others observe removal of Cdc6 during S phase (Hua and Newport, 1998). In mammalian cells, a fraction of Cdc6 remains chromatin bound throughout S phase and mitosis, while any unbound Cdc6 is phosphorylated by Cyclin A–Cdk2 and actively exported at the onset of S phase (Saha et al., 1998; Fujita et al., 1999; Jiang et al., 1999; Petersen et al., 1999; Coverley et al., 2000; Mendez and Stillman, 2000; Pelizon et al., 2000). Furthermore, some investigators have detected constant steady-state levels of Cdc6 throughout the cell cycle (Saha et al., 1998; Fujita et al., 1999; Jiang et al., 1999), while others have found Cdc6 levels to be considerably lower in early G1, resulting from ubiquitin-mediated proteolysis elicited by the anaphase-promoting complex (APC) (Mendez and Stillman, 2000; Petersen et al., 2000).
We have used Chinese Hamster Ovary (CHO) cells to examine events taking place during early G1 phase that establish a temporal and spatial program for DNA replication (Wu et al., 1998; Dimitrova and Gilbert, 1999, 2000; Dimitrova et al., 1999). CHO cells are ideal for studying early G1-phase events as they are readily synchronized in mitosis by selective detachment and then enter G1 phase rapidly and synchronously. In addition, initiation sites within the amplified dihydrofolate reductase (DHFR) locus can be readily mapped when G1-phase CHO nuclei are introduced into cell-free extracts made from Xenopus eggs (Gilbert et al., 1995). With nuclei isolated during the first few hours of G1 phase, Xenopus egg extracts initiate replication at sites dispersed throughout the DHFR locus. With nuclei isolated after a distinct point during G1 phase (origin decision point; ODP), replication initiates at the same sites that are utilized in vivo.
Recently, it was reported that the ORC1 subunit of ORC selectively dissociates from chromatin during mitosis and re-associates with chromatin at the ODP (Natale et al., 2000). This finding prompted a model (Natale et al., 2000; Cimbora and Groudine, 2001) in which the release and re-association of ORC1 in metazoa selects origin sites, providing a mechanism to reprogram replication during development. This model, while attractive, is difficult to reconcile with our previous findings that Xenopus egg extracts lacking ORC efficiently initiate replication at dispersed sites within pre-ODP nuclei (Yu et al., 1998), suggesting that ORC is assembled into a functional complex within pre-ODP nuclei. Furthermore, we have shown that Mcm proteins associate with chromatin during telophase, several hours prior to the ODP (Dimitrova and Gilbert, 1999; Dimitrova et al., 1999).
Here, we have directly related the behavior of pre-RC proteins during the CHO cell cycle to the assembly of functional pre-RCs and the timing of the ODP. Our findings indicate that ORC1, ORC4 and Cdc6 associate with chromatin throughout the proliferating cell cycle. We provide evidence that apparent contradictions in results from different laboratories with respect to ORC and Cdc6 during the cell cycle may be due to different methods of fixation and cellular extraction and/or to the overexpression of these proteins in certain cell lines. We show that, like ORC1, Cdc6 is sensitive to proteolysis during extraction specifically during mitosis. However, both proteins are stable in living cells and functional pre-RCs can be assembled without the need for protein synthesis. Finally, the transient sensitivity of ORC1 and Cdc6 to extraction and the assembly of functional pre-RCs is completed during telophase, and can be uncoupled from the DHFR ODP.
Results
ORC1 associates with chromatin during mitosis
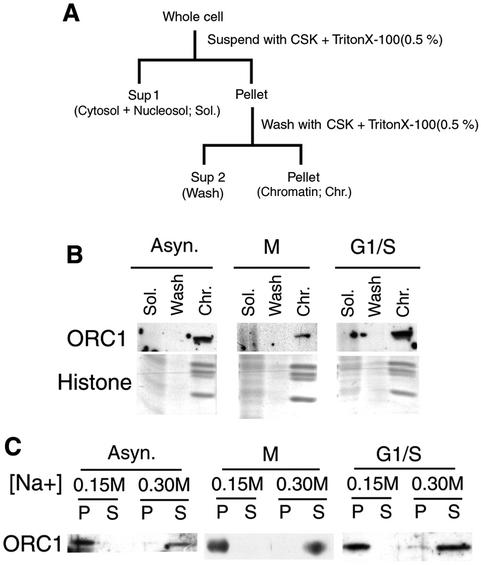
To determine whether we could detect any changes in the affinity of ORC1 for chromatin during the cell cycle, CHO cells were synchronized in metaphase by a brief treatment with nocodozole followed by mitotic shake-off. Cells were then either collected as metaphase populations, or released into the cell cycle and accumulated at the G1–S border by incubation in aphidicolin prior to extraction. Cells were then lysed in a Triton X-100-containing buffer. Soluble proteins were separated from the insoluble pellet by centrifugation, the insoluble pellet was washed with the same buffer and all three fractions (Sup1, cytosol and nucleosol, 70–80% of total cellular protein; Sup2, wash, ∼10% total protein; pellet, chromatin 15–20% total protein) were analyzed by immunoblotting with affinity-purified polyclonal antibodies raised against full-length ORC1 protein (Figure 1). As shown in Figure 1B, all detectable ORC1 was found in the chromatin fraction in cells synchronized in either M phase or G1–S, as well as in asynchronous cells. Next, the pellet fraction from Figure 1B was further incubated with increasing concentrations of salt, and the solubilized proteins were separated from the chromatin fraction by centrifugation. As shown in Figure 1C, with all three preparations, ORC1 remained in the chromatin fraction at 0.15 M salt but was completely removed from chromatin fraction by increasing the salt concentration to 0.3 M. Intermediate salt concentrations gave an intermediate degree of extraction, and no consistent differences in salt lability were observed between the three cell preparations at intermediate salt concentrations (not shown). Hence, ORC1 associates with Triton-washed chromatin throughout most of the cell cycle, including metaphase.
Fig. 1. Salt extractability of ORC1 during the cell cycle. (A) Protocol for cellular extraction. (B) ORC1 is detected exclusively in the chromatin-containing fraction (Chr.). Either asynchronously growing cells (Asyn.), cells arrested in metaphase with nocodozole and collected by mitotic shake-off (M), or metaphase cells that were released into G1 phase and arrested at the G1–S border with aphidicolin (G1/S) were extracted as in (A) and fractions were analyzed by immunoblotting with affinity-purified polyclonal anti-ORC1 antibodies. Protein from equal numbers of cells was loaded in each lane. Aliquots of each fraction were independently analyzed for the presence of histones (Coomassie Blue staining) and DNA (not shown). In all experiments shown in this report, no signal for any pre-RC protein was detected in the wash fraction and all histones and DNA were found exclusively in the chromatin fraction. (C) Salt extractability of ORC1 during the cell cycle. The chromatin fraction from (B) was further incubated for 30 min with CSK buffer containing the indicated salt concentrations, separated into soluble (S) and insoluble (P) fractions, and the two fractions were subjected to immunoblotting as in (B).
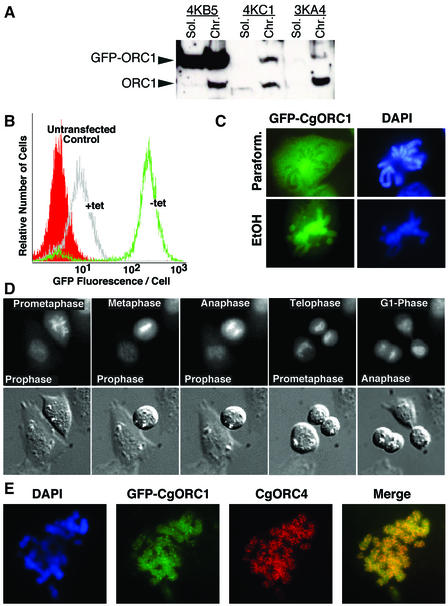
To verify by an independent method that ORC1 associates with chromatin throughout mitosis, we constructed stable CHO cell lines expressing a green fluorescent protein (GFP)-tagged ORC1 protein. We have previously defined conditions to establish homogeneous, tetracycline-regulatable gene expression in CHO cells (Izumi and Gilbert, 1999). Central to this expression system is a cell line (CHOC 400-Hyg16) that allows direct continuous selection for the tetracycline transactivator (tTA) protein. CHOC 400-Hyg16 was transfected with a vector containing GFP–ORC1 under the control of the tetracycline-repressible promoter Ptet, selection was made for the linked neomycin marker in the presence of tetracyline to prevent ORC1 expression, and G418-resistant colonies were scored for the ability to express GFP–ORC1 after the removal of tetracycline. Figure 2A shows immunoblots of fractionated cell extracts made from three of these cell lines and probed with anti-ORC1 antibody, which recognizes both the endogenous ORC1 migrating at 100 kDa and the GFP-tagged ORC1 migrating at 130 kDa. Each of these cell lines expresses GFP–ORC1 to different levels relative to endogenous ORC1. Cell line 3KA4 was used for all further experiments because the levels of exogenous GFP–ORC1 were significantly lower than the endogenous pool of ORC1 molecules, minimizing the possibility of artifacts caused by ORC1 overexpression. Flow cytometry analysis (Figure 2B) indicated that GFP–ORC1 was readily detectable and induced to homogenous levels in nearly all cells in the 3KA4 population. The low basal level of GFP fluorescence detected by flow cytometry in the presence of tetracycline was below the level of detection by fluorescence microscopy.
Fig. 2. Live cell imaging of GFP–ORC1 during M phase. (A) Cell line 3KA4 expresses a low level of GFP–ORC1. Three stable GFP–ORC1-expressing cell lines were cultured in the absence of tetracycline to fully induce GFP–ORC1. Cells were extracted and fractions subjected to immunoblotting as in Figure 1A. Immunoblots were probed with anti-ORC1 antibody, which detects both the ∼100 kDa endogenous ORC1 and the ∼130 kDa GFP–ORC1. (B) 3KA4 cells express homogeneous amounts of GFP–ORC1. Live 3KA4 cells cultured in either the presence or absence of tetracycline (induced for 24 h), as well as untransfected CHOC 400 cells, were analyzed by flow cytometry to evaluate the amount of GFP fluorescence per cell. (C) Fixation conditions for immunofluorescence influence the apparent localization of ORC1 during mitosis. 3KA4 cells were induced for GFP–ORC1 expression and then fixed with either 4% PFA (Paraform.) or 70% ethanol (EtOH). DNA was stained with DAPI, and cells were observed by fluorescence microscopy. (D) In living cells, GFP–ORC1 is associated with chromatin throughout mitosis. Differential interference contrast microscopy (DIC) and GFP fluorescence images of two cells undergoing mitosis are shown. (E) ORC1 and ORC4 co-localize during metaphase. Induced 3KA4 cells were synchronized in mitosis and attached to coverslips in a cytocentrifuge. ORC4 was detected by indirect immunofluorescence with an anti-ORC4 antibody and an Alexa 594-labeled anti-mouse secondary antibody. Co-localization with GFP–ORC1 was evaluated directly through a dual FITC/Rhodamine filter without computer merging.
We first examined the metaphase localization of GFP–ORC1 in fixed cells, under conditions typically employed for immunofluorescence. These experiments revealed results that depended upon the conditions of fixation. Using paraformaldehyde (PFA) as a fixative, GFP–ORC1 was dispersed throughout the cell, specific ally excluded from the sites of metaphase chromatin (Figure 2C). However, when these same cells were fixed with ethanol, GFP–ORC1 was localized almost exclusively to metaphase chromatin (Figure 2C). These results can potentially explain discrepancies between previous reports of ORC localization during mitosis (Romanowski et al., 1996; Pak et al., 1997).
The ability to examine GFP-tagged ORC1 in living cells provided a means to determine which of these fixation techniques preserved the physiological localization of ORC1. Observation of living cells revealed that GFP– ORC1 could be detected within 5 h after the removal of tetracycline, and was found concentrated in the nucleus and bound to chromatin as early as it could be detected. Figure 2D shows an example of time-lapsed video microscopy performed on CHOC 400 3KA4. GFP– ORC1 was unambiguously found to be chromatin-associated throughout mitosis. Measurements of the amount of GFP fluorescence per cell detected no fluctuations throughout mitosis and early G1 phase (not shown), demonstrating that there is no change in the steady state pool of GFP–ORC1 molecules during this period of time. To examine whether another ORC subunit is also associated with chromatin during metaphase, 3KA4 metaphase spreads were extracted with Triton X-100, fixed with ethanol, and stained with anti-ORC4 antibody. As shown in Figure 2E, ORC4 is also associated with metaphase chromatin under these conditions, co-localizing with ORC1. In fact, when asynchronously growing populations of cells were first extracted with Triton X-100 and then stained with anti-ORC4 antibody, nearly 100% of cells were positive for both ORC1 and ORC4, indicating that these proteins associate with chromatin at all times during the cell cycle. Taken together, we conclude that ORC1 and ORC4 associate with chromatin throughout mitosis.
Pre-RC assembly takes place between anaphase and telophase and does not require protein synthesis
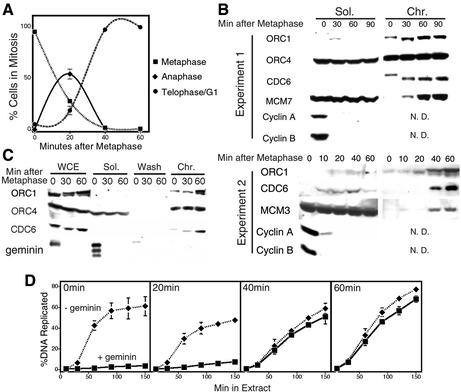
In budding yeast, B-type cyclin–cdc28 activity prevents the formation of pre-RCs during mitosis (Noton and Diffley, 2000). Destruction of B-type cyclin activity by the APC during anaphase allows Cdc6p to accumulate, associate with chromatin and recruit the binding of Mcm2-7 family of proteins to form the pre-RC. Metazoa have an additional inhibitor of pre-RC assembly, geminin, which is also degraded by the APC (McGarry and Kirschner, 1998; Wohlschlegel et al., 2000; Tada et al., 2001). CHO cells are ideal for examining events during this period of the cell cycle as nearly 100% of cells enter telophase within 40 min after release from a nocodozole block (Figure 3A). We have previously shown that Mcm2 associates with chromatin in CHO cells during telophase, roughly coincident with the formation of a nuclear envelope (Dimitrova and Gilbert, 1999; Dimitrova et al., 1999). Since that time, additional antibodies, capable of recognizing other pre-RC proteins in CHO cells, have become available. To examine the association of pre-RC proteins with chromatin as cells enter G1 phase, CHOC 400 cells were synchronized in mitosis, released into G1 phase, and collected at various time points thereafter. Figure 3B shows examples of experiments in which cells were collected at either 30 or 10–20 min intervals after metaphase. Cells were then extracted as in Figure 1 and the soluble and chromatin-containing fractions were subjected to western blotting. Cyclins A and B, as well as geminin (Figure 3C), were detected only in the soluble fraction and were almost completely destroyed within 10 min of release from the nocodozole block. ORC1 and Cdc6 were found almost exclusively in the chromatin fraction throughout mitosis, including metaphase. Low mobility forms of both ORC1 and Cdc6 during mitosis, previously shown to be due to phosphorylation by mitotic Cdk activity (Carpenter and Dunphy, 1998; Tatsumi et al., 2000), were rapidly converted to higher mobility forms after release from the nocodozole block. ORC4 was found in both the chromatin and soluble fractions at constant levels throughout this time period. By contrast, Mcm2 (not shown), -3 and -7 associated with chromatin late in mitosis, 20–30 min after the destruction of cyclins A and B (Figure 3B) and geminin (Figure 3C).
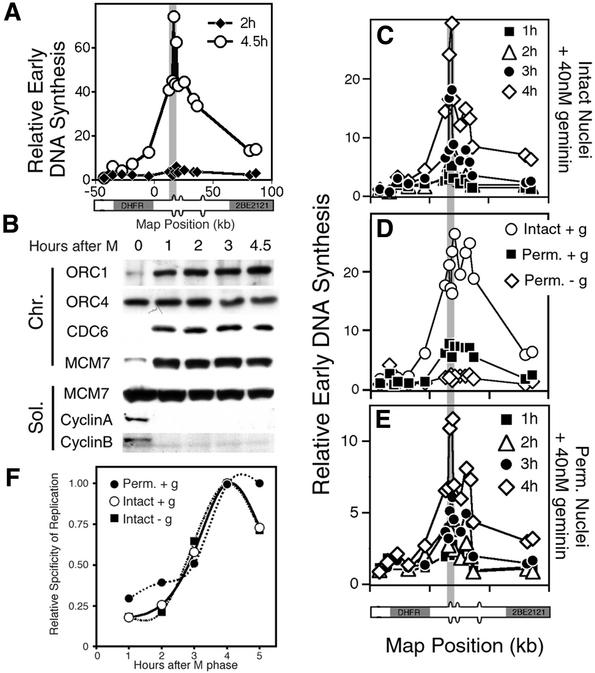
Fig. 3. Association of pre-RC proteins with chromatin during telophase. (A) Rapid and synchronous release of CHO cells from metaphase. Cells were synchronized in metaphase, collected at the indicated time points, and the percentage of cells in different stages of mitosis were determined microscopically after staining DNA with DAPI and monitoring nuclear envelope closure by the exclusion of fluorescent IgG (Wu et al., 1997). Cells were scored as metaphase if chromosomes were largely at the metaphase plate, anaphase when chromosomes were clearly separating, and telophase/G1 as soon as nuclear membrane formation was completed. Shown are the mean values for more than five experiments ± SEM (when >5). (B) ORC and Cdc6 associate with chromatin throughout the M–G1 transition, while Mcm proteins associate during telophase. At the indicated time points, cells were fractionated as in Figure 1A and the fractions examined by immunoblotting with the indicated antibodies. Two experiments are shown to illustrate the variable loss of ORC1 and Cdc6 signals during metaphase. N.D. = not done. (C) Variable degradation of ORC1 and Cdc6 takes place during extraction. Cells synchronized as in (A) and (B) were either lysed directly to make a whole-cell extract (WCE) or extracted as in Figure 1 and immunoblots of each sample were probed with the indicated antibodies. (D) Association of Mcm proteins with chromatin during telophase renders CHO chromatin independent of Xenopus Mcm proteins. Cells were permeabilized at the indicated time points after metaphase, and incubated in Xenopus egg extract supplemented with [α-32P]dATP in the presence (squares) or absence (diamonds) of geminin, and the percentage of input DNA replicated was determined by acid precipitation. Shown are the mean values for three independent experiments ± SEM (when >3.5).
In both budding yeast (Donovan et al., 1997) and Xenopus embryos (Prokhorova and Blow, 2000), Triton-resistant association of pre-RC proteins with chromatin is correlated with the assembly of functional pre-RCs. One way to evaluate the functionality of pre-RCs in mammalian cells is to introduce nuclei from synchronized cells into Xenopus egg extracts that have been supplemented with a non-degradable form of geminin (McGarry and Kirschner, 1998), a potent inhibitor of the Mcm loading reaction. Under these conditions, geminin completely prevents the association of Xenopus Mcm proteins with CHO chromatin and completely inhibits the replication of metaphase, but not G1-phase chromatin (D.S.Dimitrova, T.A.Prokhorova, J.J.Blow, I.Todorov and D.M.Gilbert, submitted). To evaluate precisely when functional pre-RCs are assembled during mitosis, the replication of CHO chromatin in geminin-supplemented extract was examined at 20 min intervals after metaphase (Figure 3D). Replication of both metaphase chromatin and chromatin from cells 20 min after metaphase (consisting mostly of anaphase cells; Figure 3A) was almost completely inhibited by geminin, whereas chromatin from cells isolated 40 or 60 min after metaphase was almost completely independent of the presence of geminin. Hence, the association of Mcm proteins with chromatin during telophase represents the assembly of functional pre-RCs, rendering CHO chromatin independent of the pre-RC assembly activities of the extract.
In some experiments, both ORC1 and Cdc6 were difficult to detect in either the soluble or chromatin fractions during metaphase and the first 20 min after release (Figure 3B, Experiment 2). A similar observation has previously been reported for ORC1 (Natale et al., 2000), leading these investigators to conclude that ORC1 was easily eluted from chromatin during mitosis and early G1 phase, but became stably bound during mid-G1 phase. In our hands, all detectable ORC1 (and Cdc6) was found in the chromatin fraction (Figures 1 and 3), and the amount of both Cdc6 and ORC1 detected during metaphase was highly variable (compare Experiments 1 and 2 in Figure 3B). In many experiments there was only a modest reduction in chromatin-bound ORC1 and Cdc6 (Figure 3B, Experiment 1). This suggested that instability arises during extraction, not within the living cells. To investigate this possibility directly, cells synchronized during metaphase and released for 30 and 60 min were divided into two aliquots. One aliquot was lysed directly to make a whole-cell extract, while the other was subjected to Triton extraction as before. As shown in Figure 3C, no signal for Cdc6 or ORC1 was found in the Triton-soluble or in the wash fraction at any time, while the chromatin fraction showed a relative increase of detectable Cdc6 and ORC1 at 30 and 60 min after metaphase. By contrast, there was no detectable fluctuation in the amounts of these proteins in whole-cell extracts. Although we can not rule out the possibility that subtle changes in the association of ORC1 and Cdc6 with chromatin during metaphase accompany the instability, ORC1 and Cdc6 were not detected free in the cytosol. These results suggest that both ORC1 and Cdc6 experience a conformational change during mitosis that renders them susceptible to proteolysis during extraction, but these proteins are stable in intact cells.
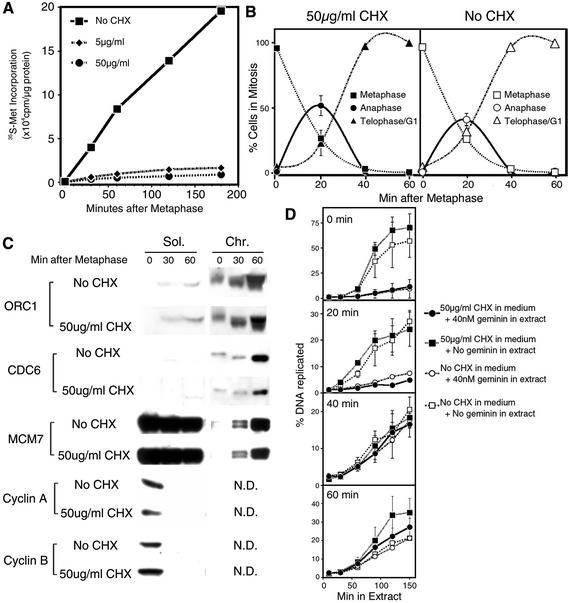
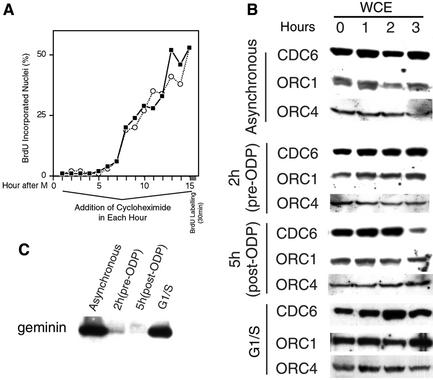
The finding that Cdc6 was stable during mitosis was somewhat surprising, since pre-RC assembly in budding yeast requires de novo Cdc6 synthesis during mitosis (Cocker et al., 1996). To confirm that ORC1 and Cdc6 are not degraded during mitosis and re-synthesized before telophase, we treated cells with the protein synthesis inhibitor cycloheximide 30 min prior to mitotic selection and then released the synchronized cells into fresh medium in the continued presence of cycloheximide. We reasoned that, if the diminished amounts of detectable ORC1 and Cdc6 were a result of their degradation during mitosis, chronic inhibition of protein synthesis would prevent the re-appearance of these proteins after mitosis. Furthermore, since the assembly of pre-RCs requires ORC1 and Cdc6, these proteins would necessarily have to be re-synthesized to load Mcm proteins onto chromatin. To verify the successful inhibition of protein synthesis in these experiments, aliquots of these cell populations were labeled with [35S]methionine and the amount of protein synthesis was measured by acid precipitation relative to parallel populations of synchronized cells that were not treated with cycloheximide. As shown in Figure 4A, cycloheximide treatment rapidly inhibited protein synthesis. However, cycloheximide treatment had no effect on the progression of cells from metaphase into G1 phase (Figure 4B). Aliquots of these same cells were then collected at various times during cycloheximide treatment, extracted and subjected to immunoblotting. Figure 4C shows that the apparent re-appearance of ORC1 and Cdc6 on chromatin following exit from metaphase is not significantly inhibited in the presence of cycloheximide. Therefore, the reduction in detectable ORC1 and Cdc6 during metaphase must be an artifact caused by their proteolysis following cell lysis. Consistent with Figures 1 and 3, virtually all of the ORC1 and Cdc6 that was detectable in these experiments was associated with chromatin from metaphase to G1 phase. Figure 4C also shows that the association of Mcm7 with chromatin still occurs in extracts treated with cycloheximide. To determine whether the association of Mcm proteins with chromatin represents the assembly of functional pre-RCs (‘licensing’), the replication of chromatin from cycloheximide-treated cells after introduction into geminin-supplemented extracts was evaluated as in Figure 3. As shown in Figure 4D, the licensing of chromatin took place at the same time, regardless of the presence of cycloheximide. We conclude that, in mammalian cells, both ORC1 and Cdc6 are stable proteins that associate with chromatin throughout mitosis and early G1 phase and do not need to be newly synthesized to promote the assembly of functional pre-RCs.
Fig. 4. Pre-RC formation is independent of protein synthesis. (A) Cycloheximide inhibition of protein synthesis during mitosis. Cells were treated with or without either 5 or 50 µg/ml of cycloheximide during the final 30 min of nocodozole treatment; metaphase cells were collected and then continuously incubated with cycloheximide and [35S]methionine during release from mitosis. Aliquots of cells were collected at the indicated times and protein synthesis was measured by incorporation of [35S]methionine into acid-precipitable protein. Rates of protein synthesis (slope of the respective curves) in 5 or 50 µg/ml cycloheximide were 8 and 4% of control, respectively. (B) Cycloheximide does not affect the synchronous release of CHO cells from metaphase. Cells treated with or without 50 µg/ml cycloheximide as in (A) were collected at the indicated time points, and the percentage of cells in different stages of mitosis was determined microscopically. Shown are the mean values for two or three experiments ± SEM (when >4.5). (C) Association of pre-RC proteins with chromatin in the presence of cycloheximide. Cells treated with or without 50 µg/ml of cycloheximide as in (A) were collected at the indicated time points, extracted as in Figure 1, and fractions were analyzed by immunoblotting. N.D. = not done. (D) Licensing of chromatin does not require protein synthesis. Cells treated with or without 50 µg/ml of cycloheximide as in (A) were collected at the indicated time points, and incubated in Xenopus egg extract supplemented with [α-32P]dATP in the presence or absence of geminin. The percentage of input DNA replicated was determined by acid precipitation as in Figure 3. Shown are the mean values for two independent experiments ± SEM.
Stabilization of ORC1 and Cdc6 to extraction, and the functional assembly of pre-RCs, are completed prior to the ODP
A previous report concluded that specification of the DHFR origin at the DHFR ODP coincides with stabilization of ORC1 and the binding of Mcm3 to chromatin (Natale et al., 2000), giving rise to the model that ORC1 selects sites of initiation (Natale et al., 2000; Cimbora and Groudine, 2001). We were puzzled by this conclusion, since our data (Figures 3 and 4) indicate that the stabilization of ORC1 and the association of Mcm proteins with chromatin take place during telophase, while specification of the DHFR origin does not take place until 3–4 h after mitosis (Wu and Gilbert, 1996). Since the previous report measured ORC1 stabilization and origin specification in separate experiments, it remained possible that some variability in cell synchrony may have been experienced. Hence, we examined these two events simultaneously with the same synchronized cell populations. CHO cells were synchronized in metaphase and collected at hourly intervals that encompass the ODP. Nuclei from cells collected at 2 and 4.5 h after metaphase were introduced into Xenopus egg extracts and the specificity of initiation of replication at the DHFR locus was evaluated by the early labeled fragment hybridization (ELFH) assay. In this assay, replication forks are arrested close to their sites of initiation by supplementing extracts with aphidicolin, which allows the initiation of replication and the synthesis of short (∼500 bp) nascent strands, but prevents the processive elongation of those forks. After washing away the aphidicolin, short nascent DNA strands are then labeled with [32P]dATP, and the 32P-labeled DNA strands are isolated and hybridized to a panel of probes spanning 120 kb of the DHFR locus. Results (Figure 5A) revealed that nuclei passed through a transition between 2 and 4.5 h after mitosis that allowed Xenopus egg cytosol to specifically recognize the DHFR origin locus, a transition that we have previously termed the ODP (Wu and Gilbert, 1996). In parallel, aliquots of these same synchronized cells were collected at hourly intervals, extracted with Triton, and analyzed by immunoblotting (Figure 5B). As in Figures 3 and 4, both ORC1 and Cdc6 were stable to extraction within 60 min after metaphase. Importantly, no detectable changes in the amounts of ORC, Cdc6 and Mcm proteins associated with chromatin were observed between 3 and 4.5 h after metaphase. Hence, specification of the DHFR origin takes place several hours after the stabilization of ORC1 and Cdc6 and the association of Mcm proteins with chromatin.
Fig. 5. The ODP takes place after the association of Mcm proteins with chromatin. Cells were synchronized in metaphase and released as in Figure 3. (A) Aliquots of cells were collected at 2 or 4.5 h after metaphase, and intact nuclei from these cells were introduced into a Xenopus egg extract supplemented with aphidicolin to arrest replication forks close to their sites of initiation. After a 60 min incubation period, nuclei were washed free of aphidicolin, nascent DNA strands were labeled with [32P]dATP, and the distribution of nascent DNA was evaluated by hybridizing radiolabeled nascent DNA to a panel of probes spanning the DHFR locus. The horizontal axis includes a diagram of the genomic region encompassed by these probes, including the positions of the DHFR and 2BE2121 genes. The vertical shaded line shows the positions of probes encompassing the major initiation site, ori-β (probes B–E; Wu et al., 1997). (B) Aliquots of these same synchronized cell populations were collected at the indicated times, extracted and the fractions were analyzed by immunoblotting as in Figure 1. (C) At various times (1 h, filled square; 2 h, open triangle; 3 h, filled circle; 4 h, open diamond) after mitosis, intact nuclei were introduced into Xenopus egg extracts supplemented with geminin. Sites of initiation of replication were evaluated with the ELFH assay as in (A). Similar results were obtained in three independent experiments. (D) CHOC 400 cells were synchronized in the post-ODP stages of G1 phase (5 h after metaphase), and either intact nuclei or nuclei that were permeabilized by exposure to higher concentrations of digitonin were introduced into Xenopus egg extracts in the presence or absence of geminin. Sites of initiation of replication were evaluated with the ELFH assay. Similar results were obtained in three independent experiments. (E) Permeabilized nuclei prepared from cells at various times (1 h, filled square; 2 h, open triangle; 3 h, filled circle; 4 h, open diamond) after mitosis were introduced into Xenopus egg extracts supplemented with geminin. Sites of initiation of replication were evaluated with the ELFH assay. (F) The relative specificity of initiation was defined as the average values for probes in the region of peak initiation activity [highlighted by the gray line in (A) and (C–E)] and then normalized to a value of 1 to facilitate a comparison between intact and permeabilized nuclei. Results with intact nuclei in geminin-free extract (closed squares), or either intact (open circles) or permeabilized (closed circles) nuclei in geminin-supplemented extracts was plotted as a function of the time after metaphase. Five hour time points [omitted in (C–E) for presentation] illustrate the plateau in specificity by 4 h post-metaphase. No specificity was observed at any time point with permeabilized nuclei introduced into extracts without geminin.
To verify that the G1-phase schedule for the appearance of site-specific initiation within the DHFR locus was independent of the pre-RC assembly activities of the extract, the sites of initiation of replication were evaluated directly in geminin-supplemented extracts using nuclei prepared at various times after mitosis. As shown in Figure 5C, site-specific initiation of replication did not appear until 3–4 h after mitosis. We next wanted to determine whether the change in specification of origins at the ODP is mediated by the population of Mcm molecules associated with chromatin, by evaluating origin specificity with nuclei in which the soluble pool of Mcm proteins had been removed. Unfortunately, Triton-washed nuclei make extremely poor substrates for replication in Xenopus egg extracts, likely due to irreparable damage to the nuclear envelope (Dimitrova and Gilbert, 1998). By contrast, digitonin permeabilization results in nuclei that have been stripped of soluble Mcm proteins (Dimitrova et al., 1999), but are efficiently replicated in Xenopus egg extract (Dimitrova and Gilbert, 1998). However, in complete extracts, digitonin permeabilization of post-ODP nuclei results in a complete loss of DHFR origin specificity (Dimitrova and Gilbert, 1998; Figure 5D). This loss of origin specificity is likely to be due to the assembly of additional pre-RCs by Xenopus proteins, as a significant degree of origin specificity is retained when permeabilized nuclei are introduced into geminin-supplemented extracts (Figure 5D; D.S.Dimitrova, T.A.Prokhorova, J.J.Blow, I.Todorov and D.M.Gilbert, submitted). The nuclear envelope is known to provide a barrier to activities within Xenopus egg extracts that license chromatin, although the activity that is prevented by the envelope has not been identified (Blow, 2001). Nevertheless, the ability to examine origin specification with nuclei that had been washed free of soluble Mcm proteins provided the opportunity to examine the specification of the DHFR origin mediated solely by the population of Mcm proteins bound tightly to CHO chromatin. Nuclei prepared from cells at various times after mitosis were permeabilized and washed to remove soluble Mcm proteins. They were then introduced into geminin-supplemented extracts and the sites of initiation of replication within the DHFR locus were mapped as before (Figure 5E). Although the relative specificity was not as robust as with intact nuclei, it is clear that initiation was not maximally focused to within the DHFR origin region until 4 h after mitosis.
Figure 5F summarizes the origin mapping results in geminin-supplemented and unsupplemented extracts. The relative specificity of initiation was defined as the average relative early DNA synthesis values for four probes within the region of peak initiation activity (ori-β), and plotted as a function of the time after metaphase. Clearly, the presence of geminin had no influence on the schedule for specification of origin sites within the DHFR locus. Importantly, the total amount of radioactive replication intermediates hybridizing to probes within the DHFR locus was the same in all experiments, demonstrating that the capacity to initiate replication within the DHFR locus was similar, regardless of the time during G1 phase at which nuclei were prepared or whether the pre-RC assembly activity of the extracts was inhibited with geminin. These results provide a direct demonstration that the assembly of functional pre-RCs within CHO nuclei during telophase is not sufficient to specify the DHFR replication origin. Additional events, taking place 3–4 h after mitosis, are required to select which of many potential sites can function as an origin when exposed to S phase-promoting factors.
Cdc6 and ORC1 are stable and geminin is absent throughout pre-restriction point G1 phase
Although the steady-state amounts of ORC4, ORC1 and Cdc6 do not change during G1 phase, the possibility remained that their turnover rates might fluctuate during G1. In yeast, Cdc6 is targeted for degradation by the SCF ubiquitin ligase, constituting a ‘point of no return’ for the formation of pre-RCs (Piatti et al., 1996). In fact, in human HeLa cells, microinjected GFP-tagged Cdc6 is degraded ∼3–4 h after mitosis and the half-life of endogenous Cdc6 is shortened during G1 phase (Petersen et al., 2000). Potentially, the pre-ODP stage of G1 phase could represent a period of dynamic pre-RC formation, while an increase in the turnover rate of either ORC1 or Cdc6 (or the re-synthesis of geminin) could constitute a point of no return for the assembly of pre-RCs, allowing only specific pre-RCs to be stabilized, contributing to the ODP. To examine directly the stability of these proteins in pre- and post-ODP cells, CHOC 400 cells were synchronized in mitosis and released into G1 phase. Cycloheximide was added at either 2 or 5 h after mitosis, whole-cell extracts were prepared from aliquots of these cells at hourly intervals thereafter, and immunoblots of these extracts were probed with Cdc6 and ORC1 antibodies. As shown in Figure 6B, both ORC1 and Cdc6 were found to be stable throughout the pre- and post-ODP stages of G1 phase. In fact, no degradation of either protein was detectable from 2 to 5 h, the period during which cells pass through the ODP (cells pass through the ODP on schedule in the continued presence of cycloheximide; S.M.Keezer and D.M.Gilbert, submitted). Abrupt degradation of Cdc6 (but not ORC1) could be observed, however, upon prolonged incubation of cells in the presence of cycloheximide. This treatment drives cells into G0, which is known to result in the elimination of all detectable Cdc6 (Stoeber et al., 1998; Petersen et al., 1999). To verify that cells did not enter S phase under these conditions, aliquots of cells treated with cycloheximide at various times during G1 phase were pulse labeled with 5-bromo-2-deoxyuridine (BrdU) 15 h after mitosis. Cells treated with cycloheximide throughout the first 5 h of G1 phase were completely prevented from entering S phase (Figure 6A). Hence, the abrupt degradation of Cdc6 (Figure 6B, 5 h) is not due to the entry of cells into S phase. Furthermore, when cells were first allowed to enter early S phase and then incubated with cycloheximide, Cdc6 (as well as ORC1 and ORC4) was stable for at least 3 h during early S phase (Figure 6B). The simplest interpretation of these results is that, under conditions that promote cellular proliferation, both Cdc6 and ORC1 are stable proteins throughout G1 phase. However, when cells are prevented from passing through the R-point and begin to enter quiescence, Cdc6 (but not ORC1) is rapidly degraded.
Fig. 6. ORC1, ORC4 and Cdc6 are stable and geminin is absent throughout the pre-restriction point stages of G1 phase. (A) The execution point for cell cycle arrest by cycloheximide is just before entering S phase. Cycloheximide (50 mg/ml) was added to aliquots of cells at hourly intervals after metaphase, all cultures were labeled for 30 min with BrdU at 15 h after metaphase, and the percentage of cells labeled with BrdU was determined by immunofluorescence with anti-BrdU antibodies (filled squares). To evaluate the time of entry into S phase, parallel cultures were labeled with BrdU for 30 min at each hourly interval and similarly stained (open circles). (B) Stability of ORC1, ORC4 and Cdc6 during G1 and early S phase. Cycloheximide was added to either asynchronously growing cells (Asynchronous), cells synchronized in the pre-ODP (2 h after metaphase) or post-ODP (5 h after metaphase) stages of G1 phase, or cells arrested at the G1–S border with aphidicolin. Cells were either collected immediately (0 h) or at 1, 2 or 3 h thereafter, and whole-cell extracts were prepared and immunoblotted with antibodies to Cdc6, ORC1 and ORC4. (C) Geminin is not re-synthesized until after the ODP. Whole-cell extracts were prepared either from asynchronously growing cells (Asynchronous) or from cells synchronized in metaphase and released for either 2 or 5 h or arrested at the G1–S border in aphidicolin. Immunoblots were then probed with anti-geminin antibody.
To determine whether geminin is re-synthesized at the ODP, whole-cell extracts were prepared from cells at 2 and 5 h after mitosis, while parallel cultures were allowed to accumulate at the G1–S border in the presence of aphidicolin. As shown in Figure 6C, no geminin was detected in either 2 or 5 h G1 phase extracts, whereas geminin was readily detected at the G1–S border, demonstrating that the re-synthesis of geminin takes place after the ODP. Together, these results raise the possibility that cells remain in a state favorable for pre-RC formation throughout the pre-restriction point stages of G1 phase.
Cdc6 and ORC1 are associated with chromatin throughout S phase
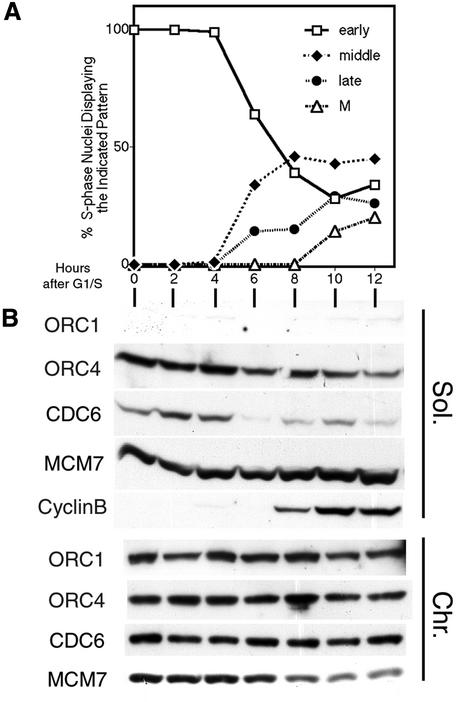
To examine the association of ORC1 and Cdc6 during passage of CHO cells through S phase, CHOC 400 cells were synchronized in mitosis and then released into medium containing aphidicolin for a period of time sufficient to accumulate cells at the G1–S boundary. Cells were then transferred to aphidicolin-free medium and aliquots were pulse labeled with BrdU at various times thereafter. In addition, nocodozole was added to prevent cells from passing through mitosis and entering the following cell cycle. The progression of cells through S phase was monitored by calculating the percentage of BrdU-labeled cells, as well as the fraction of these labeled cells displaying each of the spatial patterns of DNA synthesis representative of early, middle and late S phase (Dimitrova and Gilbert, 1999). In addition, the appearance of mitotic cells, the synthesis of mitotic Cyclin B, and the removal of Mcm proteins from chromatin were monitored. In parallel, populations of cells collected at each of these times were extracted as in Figure 1, and the soluble and chromatin-containing fractions were subjected to immunoblotting with Cdc6 and ORC1 antibodies. As shown in Figure 7, all detectable ORC1 and nearly all Cdc6 were found in the chromatin-containing fraction, and no significant changes were detected in the amounts of either of these proteins in the chromatin fraction throughout S phase and into G2 phase. We conclude that, in CHO cells, ORC1 and Cdc6 are stable proteins that associate with chromatin throughout the cell cycle.
Fig. 7. ORC1, ORC4 and Cdc6 associate with chromatin throughout S phase. CHOC 400 cells were synchronized in metaphase and released into medium containing aphidicolin for 12 h to arrest cells at the G1–S boundary. Cells were then transferred to aphidicolin-free medium containing nocodazole (to prevent cells from entering the following G1 phase) and collected at the indicated times thereafter. (A) To monitor progression through S phase, aliquots of cells at each time point were pulse labeled with BrdU and the percentage of BrdU-positive cells, as well as the spatial pattern for DNA synthesis (early, middle or late) at each time point, was scored. Separate aliquots of these same cells were fixed in ethanol and stained with DAPI to evaluate the percentage of cells in mitosis (M). (B) Aliquots of these same cells were extracted as in Figure 1A and the fractions subjected to immunoblotting with the indicated antibodies.
Discussion
Here, we demonstrate that the majority of detectable Cdc6 and ORC1 associates with chromatin throughout the CHO cell cycle, including mitosis. Although both Cdc6 and ORC1 were unstable to extraction during mitosis, we found no evidence for in vivo degradation of either protein. Both Cdc6 and ORC1 exhibited a mobility shift during mitosis, consistent with previous results demonstrating phosphorylation of these proteins during mitosis (Carpenter and Dunphy, 1998; Tatsumi et al., 2000). It is therefore possible that phosphorylation of either or both of these proteins during mitosis may render them unstable to extraction. However, it is clear that neither protein is degraded in the cell, since the inhibition of protein synthesis during mitosis did not inhibit pre-RC formation or significantly reduce the amounts of these proteins in the following G1 phase. The high degree of synchrony achieved in these experiments allowed us to examine the timing of pre-RC assembly with greater precision than previously reported. The ORC1 and Cdc6 mobility shift is rapidly reversed as cells exit metaphase, coincident with the degradation of Cyclins A and B, and geminin. Approximately 20 min thereafter, Mcm proteins are assembled into functional pre-RCs that can serve as efficient substrates for initiation of replication in geminin-supplemented Xenopus egg extracts. This process of licensing chromatin is completed in a brief period between anaphase and telophase. Surprisingly, this entire process, including the functional licensing of chromatin, did not require protein synthesis. This differs from the situation in budding yeast, where cells that exit mitosis without de novo Cdc6 protein synthesis fail to assemble pre-RCs (Cocker et al., 1996). Hence, pre-RC formation in mammals differs from that in yeast in that it is regulated post-translationally. An interesting possibility is that phosphorylation of ORC1 and Cdc6 by mitotic Cdk activity acts in conjunction with geminin to inhibit pre- RC formation. This would explain why ORC-depleted Xenopus egg extracts do not assemble pre-RCs on CHO metaphase chromatin despite the presence of hamster ORC on the chromatin (Yu et al., 1998). Rapid de-phosphorylation during anaphase may then allow pre-RCs to assemble.
We find that mammalian geminin is degraded very shortly after metaphase and is not synthesized again until late G1 phase. Since Cdk activity is also not induced until after the restriction point, our results suggest the possibility that licensing can take place continually throughout the pre-restriction point period of G1 phase. Since the interaction of many proteins with chromatin is highly dynamic (Misteli, 2001), pre-RCs may assemble and disassemble during early G1 phase. If this is true, then changes in chromatin structure taking place at any time during early G1 phase could influence the sites of pre-RC assembly.
Sub-cellular localization of Cdc6 and ORC1
Our findings for Cdc6 raise some apparent discrepancies with those presented by previous investigators, but also offer a possible means to reconcile the results from various groups. First, several groups have reported that a significant fraction of human Cdc6 is exported from nuclei upon entry of cells into S phase (Fujita et al., 1999; Jiang et al., 1999; Coverley et al., 2000; Mendez and Stillman, 2000), whereas we find most detectable Cdc6 to be localized to chromatin throughout the cell cycle. Since human cells also contain a substantial chromatin-bound fraction of Cdc6 during S phase (Fujita et al., 1999; Coverley et al., 2000; Mendez and Stillman, 2000), this discrepancy could be explained if Cdc6 is limiting in CHO cells, leaving them with a negligible, soluble Cdc6 pool. In fact, stable CHO cell lines expressing high levels of HA-tagged human Cdc6 export a substantial portion of HA-Cdc6 to the cytosol during S phase (A.J.McNairn, unpublished observations). A second discrepancy is that, in transformed human cells, Cdc6 is degraded by the APC during early G1 phase and is re-synthesized at the onset of S phase (Mendez and Stillman, 2000; Petersen et al., 2000). By contrast, we found no evidence for rapid turnover of CHO Cdc6 throughout mitosis and early G1 phase, while geminin and Cyclins A and B, all targets of the APC, were degraded within 10 min after metaphase. In fact, we did not identify any point during the cell cycle at which the half-life of Cdc6 (or ORC1) in whole-cell extracts was <2 h, including S phase. The only point at which Cdc6 became unstable was after prolonged incubation with cycloheximide prior to the restriction point, which causes cells to enter a quiescent state. This apparent discrepancy also could be explained if the majority of the degradation observed in transformed human cells is accounted for by an excess soluble pool of Cdc6. In fact, one of these reports (Mendez and Stillman, 2000) fractionated cells into soluble and chromatin-bound forms, revealing very little change in the amount of Cdc6 protein bound to chromatin throughout the cell cycle, consistent with our results. Since the majority of human Cdc6 detected in this previous study was found to be in a soluble form, the cell cycle-regulated changes observed by these investigators may be entirely accounted for by the soluble fraction. Of course we can not rule out the possibility that there are fundamental differences in the regulation of Cdc6 in human versus CHO cells. In summary, a universal model for Cdc6 can be offered in which a fraction of Cdc6 remains stable and associates with chromatin throughout the cell cycle, while excess Cdc6 is either degraded by the APC during early G1 phase or phosphorylated and exported from the nucleus during S phase.
With respect to ORC1, our results challenge the popular model (Natale et al., 2000; Cimbora and Groudine, 2001) that ORC1 is released from chromatin during mitosis and re-associates during early G1 phase. Consistent with our results, a recent report has shown that ORC1 in HeLa cells is associated with chromatin throughout the cell cycle (Tatsumi et al., 2000). In fact, these investigators successfully UV cross-linked ORC1 to chromatin during mitosis. UV induces covalent coupling between thymine bases and amino acids that contact DNA directly (Biggin, 1999). By contrast, two other studies concluded that ORC1 dissociates from chromatin during S phase and/or mitosis (Natale et al., 2000; Kreitz et al., 2001). However, no re-distribution of ORC1 to the soluble fraction was detected in these prior studies. In fact, all detectable ORC1 signal was still in the chromatin fraction, consistent with our data presented here. Furthermore, we show that the loss of ORC1 during metaphase is seen only in fractionated cells and not in whole-cell extracts, suggesting that proteolysis occurring during cell fractionation is responsible for the reduction of detectable ORC1 in the chromatin fraction. We observed no variation in chromatin association or detectability of ORC1 when the conditions of extraction were varied, including the presence or absence of ATP, phosphatase inhibitors, and the concentration of Triton X-100 (not shown). We can not rule out subtle changes in the interaction of ORC1 with chromatin during mitosis. For example, Tatsumi et al. (2000) find that 200 mM NaCl dissociates ORC1 from metaphase chromatin but that 250 mM NaCl was required to dissociate ORC1 from chromatin isolated at other stages of the cell cycle. In our hands, there was more variation from experiment to experiment at these salt concentrations than we could observe between metaphase and other stages of the cell cycle (not shown). In addition, we have shown that PFA fixation of cells disperses ORC1 from chromatin, unlike ethanol fixation, which could explain previous discrepancies in the immunolocalization of ORC subunits during mitosis (Romanowski et al., 1996; Pak et al., 1997; Loupart et al., 2000). Finally, and perhaps most compelling, we show that GFP-tagged ORC1 is chromatin-associated throughout mitosis in living cells. We conclude that ORC1 associates with chromatin throughout mitosis but that, during metaphase and anaphase, ORC1 (and Cdc6) is susceptible to degradation during the process of cellular extraction.
Interestingly, our results demonstrate that ORC1 and Cdc6 behave similarly throughout the cell cycle, including their differential sensitivity to extraction. Both proteins recover from this state between anaphase and telophase, and both are stable throughout the remainder of the proliferating cell cycle. These results suggest that there is a common modification to both proteins. ORC1 and Cdc6 share significant homology in the C-terminal domain region that includes a series of Cdk phosphorylation sites (Gavin et al., 1995). Furthermore, the fact that this common modification is limited to mitosis suggests that it may be mediated by mitotic Cdk activity. Also, ORC1 and Cdc6 are known to interact physically (Liang et al., 1995; Saha et al., 1998). It will be interesting to determine whether ORC1 and Cdc6 form a complex throughout the cell cycle, and whether their common modification during mitosis has a functional significance, for example, to prevent pre-RC formation during nuclear envelope breakdown. The behavior of these two proteins parts when cells enter late G1 phase under conditions that prevent passage through the R-point. Under these conditions, ORC1 remains stable while Cdc6 is rapidly degraded (Figure 6).
Pre-RC formation is not sufficient to specify the DHFR replication origin
Our data also provide the clearest demonstration to date that de-phosphorylation of ORC1 and Cdc6, stabilization of ORC1 and Cdc6 to the conditions of cellular extraction, association of Mcm proteins with chromatin, and the assembly of functional pre-RCs all take place prior to, and can be uncoupled from the ODP. Thus, the assembly of functional pre-RCs within CHO nuclei during telophase is not sufficient to specify the DHFR replication origin. Previously, we have shown that replication initiates at sites dispersed throughout the DHFR locus when intact pre-ODP nuclei are introduced into ORC-depleted (Yu et al., 1998), Mcm-depleted or geminin-supplemented (D.S.Dimitrova, T.A.Prokhorova, J.J.Blow, I.Todorov and D.M.Gilbert, submitted) Xenopus egg extracts. Here, we have extended those findings to show that the ability of CHO chromatin to be replicated in Mcm-deficient extracts coincides precisely with the detergent-resistant association of Mcm proteins with chromatin during telophase, which occurs at least 2 h before the ODP. Furthermore, when nuclei isolated at various times during G1 phase were first permeabilized to remove unbound Mcm proteins and then introduced into geminin-supplemented Xenopus egg extracts, site-specific initiation within the DHFR locus was still not observed until 3–4 h after mitosis. In the absence of geminin, these same extracts initiated at dispersed sites within permeabilized nuclei prepared at all G1-phase time points, confirming that geminin effectively prevented the assembly of Xenopus pre-RCs under the conditions described in these experiments. Hence, dispersed initiation within pre-ODP nuclei is mediated by mammalian pre-RCs that are likely to form at many more sites than are actually utilized during S phase. Additional events, taking place at the ODP, dictate which of these potential sites will function as the preferred initiation sites during S phase.
Our results differ from those of Natale et al. (2000), who reported that the ODP coincides temporally with the stabilization of ORC1 and the ability of CHO chromatin to be replicated in ORC- or Mcm-depleted egg extracts. In our hands, the formation of pre-RCs and stabilization of ORC1 (and Cdc6) is completed within 1 h after metaphase, while the ODP takes place several hours later. A critical difference in protocol is that Natale et al. correlated the timing of ORC1 ‘stabilization’ and the ODP from measurements made in separate experiments, with separately synchronized populations of cells. By contrast, we have evaluated origin specification directly in Mcm-deficient extracts, and with the same synchronized populations of cells that were utilized for evaluating pre-RC formation. Our results clearly demonstrate that dispersed initiation within pre-ODP nuclei is mediated by functional pre-RCs assembled within the CHO cell prior to the preparation of nuclei. Hence, pre-RC formation can be uncoupled from the ODP and is not sufficient to specify DHFR origin sites. Furthermore, this conclusion is consistent with our previous studies demonstrating that SV40-transformed CHO cells treated with protein kinase inhibitors prior to the ODP (but not after) will enter S phase and initiate replication in vivo at sites dispersed throughout the DHFR locus (Wu et al., 1998), demonstrating that the capacity to initiate (replication licensing) can be uncoupled from origin specification. The implication is that, as has been shown in S.cerevisiae (Greenfeder and Newlon, 1992; Santocanale and Diffley, 1996), mammalian cells assemble many more pre-RCs than are actually utilized. The mechanism that selects which pre-RCs will be activated is not known. However, in budding yeast, the assembly of silent chromatin (Stevenson and Gottschling, 1999) and the positioning of nucleosomes near origins can directly influence the efficiency with which origins fire (Lipford and Bell, 2001). It is possible that chromatin structure changes taking place as nuclei mature after mitosis lead to the potentiation and/or repression of certain pre-RCs. Alternatively, additional steps in the preparation of G1-phase nuclei for entry into S phase may directly modify certain pre-RCs, making them better or worse substrates for initiation.
The ODP is a distinct G1-phase event that takes place after pre-RC formation and prior to the restriction point. The ODP is also independent of growth signaling mechanisms (mitogens, PKA, PKG, focal adhesion kinase and MAP kinase activity), and does not require protein synthesis (S.M.Keezer and D.M.Gilbert, submitted). The formidable challenge now is to identify molecular events that regulate the origin decision process. Naturally, it will be important to examine the G1-phase behavior of other proteins involved in the initiation process. However, it is possible that the ODP represents an epigenetic alteration that establishes favorable initiation sites within the context of chromatin. It seems that additional descriptive information will be necessary to eventually understand the nature of the ODP. Importantly, we have characterized the ODP for only one replication origin locus. An important question is whether the ODP is a global event that specifies all replication origins, only a subset of origins, or whether different origins are specified at different times during G1 phase. This information will be critical for us to understand whether we are searching for a global molecular transition during G1 phase, or a series of local, locus-specific events.
Materials and methods
Cell culture
CHOC 400 cells, which contain ∼1000 copies of a 243 kb segment of DHFR gene region (Hamlin et al., 1994), are maintained in Dulbecco’s modified Eagle’s medium + 5% fetal bovine serum (FBS) with nonessential amino acids. The pattern of initiation within the amplified locus is indistinguishable from that within the parental CHO cell line (Dijkwel and Hamlin, 1995). Metaphase synchronization was performed by mitotic shake-off after a brief nocodozole treatment, as described (Wu and Gilbert, 1997). Since nearly 60 min are required for metaphase cells to re-attach to a solid substratum, events taking place within 60 min of metaphase cells were examined by releasing cells into medium containing 50 mM HEPES–KOH pH 7.6 and incubated in a shaking water bath at 37°C. Nuclear envelope assembly, the association of Mcm proteins with chromatin, and the passage of cells through the ODP all take place at the same time whether cells are plated or released in suspension (Dimitrova and Gilbert, 1999; Dimitrova et al., 1999; and data not shown). However, this method allows one to collect homogeneous populations of cells exiting mitosis with a high degree of synchrony at many time intervals (e.g. Figure 3A). Release from mitosis was monitored by permeabilizing cells with digitonin and mixing with Texas-red-conjugated anti-rat IgG and 0.1 mg/ml of 4,6-diamino-2-phenylindole (DAPI) as described for examining intact nuclei (Wu et al., 1997). Cells were synchronized at the G1–S boundary with 5 µg/ml aphidicolin as described (Dimitrova et al., 1999). The percentage of BrdU-labeled cells was determined as described (Wu et al., 1997). [35S]methionine incorporation was monitored by TCA precipitation (Ausubel et al., 1994) and normalized to the total protein in each sample.
Protein expression in Escherichia coli
To construct the expression vector for full-length ORC1 (pET9hORC1), a 512 bp fragment from the start codon to a ScaI site in the ORC1 open reading frame (Natale et al., 2000) was generated by PCR (primers: 5′-TTCGGCGTGACATATGCCATCGTATCACA-3′, 5′-CTGGCACTTAGGGCTATC-3′), and digested with NdeI and ScaI. This PCR fragment was inserted, together with a ScaI–KpnI C-terminal fragment from the ORC1 cDNA, into NdeI and BamHI sites of pET9, with a linker oligonucleotide (5′-CATGCTAG-3′) to combine the KpnI and BamHI sites. pET9hORC1 was transformed into E.coli BL21 and induced with 1 mM isopropyl-β-d-galactopyranoside. Inclusion bodies containing the ORC1 protein were purified, dissolved into SDS–PAGE loading buffer, and electro-eluted from SDS–PAGE gels (Rosenberg, 1998). Purification of the DEL mutant of Xenopus geminin-H was performed as described (McGarry and Kirschner, 1998), and dialyzed with 10 mM HEPES pH 7.3, 300 mM NaCl and 50% glycerol for storage in liquid nitrogen. All buffers used for purification of geminin-H contain 1 mM phenylmethylsulfonyl fluoride (PMSF), except the outer solution for dialysis. Transport buffer (20 mM HEPES pH 7.3, 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 1 mM EGTA; Wu et al., 1997) was used for diluting geminin just before adding to egg extract.
Antibodies and western blotting
Rabbit polyclonal antibodies were raised against full-length ORC1. For affinity purification, purified ORC1 protein expressed in E.coli was subjected to SDS–PAGE and transferred to polyvinylene difluoride (PVDF) membranes (Millipore). PVDF membrane strips were incubated overnight at 4°C with anti-ORC1 sera diluted with an equal volume of Tris-buffered saline (TBS). The antibody-absorbed membrane was washed three times with TBS, and the antibody was eluted in 100 mM glycine pH 3.0 for 2 min and immediately neutralized by adding a one-tenth volume of 1 M Tris–HCl pH 8.0. The eluted IgG fraction was concentrated with a Protein A affinity column (Hi-trap column; Amersham-Pharmacia Biotech), according to the manufacturers instructions. In western blots with CHO cell extracts, affinity-purified antibodies recognized a 100 kDa band that co-migrated with bacterially expressed ORC1. Semi-quantitative comparison of the amount of ORC1 in these extracts relative to known quantities of bacterially expressed ORC1 revealed an amount equivalent to ∼1 × 105 ORC1 molecules per cell. The other primary antibodies used in this study were as follows: anti-Xenopus MCM3 antibody (Prokhorova and Blow, 2000), anti-human Mcm7 (Santa Cruz), anti-human ORC4 (BD Transduction Labs), anti-human CDC6 (Santa Cruz), anti-Cyclin A (Santa Cruz), anti-Cyclin B (Sigma) and anti-geminin (Wohlschlegel et al., 2000). For immunoblotting, all antibodies except ORC4 were incubated in 10 mM Tris–HCl pH 8.0, 137 mM NaCl, 0.5% Tween 20 with 1% bovine serum albumin and 0.3% skim milk. The ORC4 antibody was incubated in 10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5% Tween 20.
Preparation of whole-cell extract and chromatin isolation
Whole-cell extracts were prepared as described (Wu and Gilbert, 1997), with the following modifications. Cells were collected by trypsinization, washed twice in cold phosphate-buffered saline (PBS), and resuspended with lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 1 mM Na vanadate, 50 mM NaF, 1 mM PMSF, 1 µg/ml pepstatin, 1 µg/ml aprotinin, 1 µg/ml leupeptin) at 1.67 × 104 cells/µl. Samples were briefly sonicated to break DNA and a one-half volume of 3× SDS-loading buffer was added. Chromatin fractionation with Triton X-100 was performed as described (Reyes et al., 1997), with modifications. In brief, exponentially growing or synchronized CHOC 400 cells were collected by trypsinization and washed twice with ice-cold PBS. Cells (5 × 106) were resuspended in 300 µl of CSK buffer [10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 1 mM PMSF, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 µg/ml aprotinin, 50 mM NaF, 0.1 mM Na vanadate] containing 0.5% Triton X-100 and incubated for 3 min at 0°C. Soluble (‘Cytosol + Nucleosol’) and insoluble fractions were separated in a microfuge (7500 r.p.m., 3 min) and the pellet was washed for 3 min at 0°C with the same buffer. The insoluble fraction was briefly sonicated before adding SDS–PAGE loading buffer. DNA from 10 µl of each fraction was purified by sequential incubation with RNase A at 37°C and proteinase K at 65°C, and then precipitated with ethanol after phenol–chloroform extraction. Localization of core histones was detected by running 22% (T:C = 60:0.5) SDS–PAGE gels and staining with Coomassie Blue. Protein from equivalent numbers of cells was loaded on each lane (3 × 105cells/lane, except geminin which was 2 × 105 cells/lane).
Construction of stable GFP–ORC1 cell lines
To construct pEGFP-ORC1, a 5′ piece of the ORC1 cDNA was PCR amplified (primers: 5′-AATTGGGCCCCCATCGTATCTCACAAGG-3′ and 5′-ATTAGGGCCCTCGAGCCACTGTACTCGAGCAC-3′) using Pfu polymerase (Stratagene) to incorporate a Bsp120I site into the 5′ end. The PCR product was then digested with Bsp120I and ligated to the (Klenow-filled) EcoRI site of ptetEGFP-ins1/1 (Izumi and Gilbert, 1999) to form ptetEGFP ins1/1ORC15. The remaining portion of the ORC1 cDNA was digested with ScaI and SalI and inserted into the SacI–SalI sites of ptetEGFP ins1/1ORC15′ to form pEGFP-ORC1. One microgram of pGFP-ORC1 was linearized with MluI and transfected into Hyg16 (Izumi and Gilbert, 1999) using Lipofectamine (Gibco-BRL). Trans fected cells were selected with 1 mg/ml G418 (Gibco-BRL) and maintained in the presence of 2 µg tetracycline (Sigma) and 0.5 mg/ml Hygromycin B (Calbiochem). To screen colonies, cells were plated onto coverslips, GFP–ORC1 expression was induced by removal of tetyracycline, and GFP fluorescence was monitored with an inverted fluorescence microscope at 24 h intervals. Six out of 60 colonies displayed GFP expression in >75% of cells. FACS analysis (with live cells) and western blotting were then used to determine the level and homogeneity of GFP–ORC1 expression.
Fluorescence microscopy
For immunofluorescence analysis of fixed cells, 3KA4 cells were plated onto coverslips and induced for 72 h. The coverslips were then washed with PBS and fixed with either 4% PFA or 70% ethanol. Mitotic cells were identified by DAPI staining and photographed. For co-localization of GFP–ORC1 with ORC4, mitotic 3KA4 cells were re-suspended at 1 × 106 cells/ml in hypotonic buffer (10 mM Tris pH 7.4, 10 mM NaCl, 5 mM MgCl2) and swollen for 15 min at 37°C. One hundred microliters of cells were then cytospun onto coverslips (2 min at 1500 r.p.m., using a Shandon Cytospin 2), extracted with CSK buffer containing 0.5% Triton X-100 on ice for 3 min, and then fixed with 70% ethanol. Endogenous ORC4 was detected with monoclonal anti-HsORC4 (BD Transduction Labs) and goat anti-mouse Alexa 594 (Molecular Probes). Images were captured using a SPOT CCD camera mounted on a Nikon Labophot-2 microscope and processed using Adobe Photoshop 5.0.
Time-lapse imaging and analysis
For time-lapse imaging, 3KA4 cells were plated onto ΔT 0.15 mm dishes (Bioptechs), incubated in CO2-independent medium lacking phenol red and containing 5% FBS and nonessential amino acids (Gibco-BRL), overlaid with mineral oil, and maintained at 37°C using the ΔT system (Bioptechs). Fluorescence and DIC images were acquired as described (Karlsson and Pines, 1998). Cells were defined as being in prophase when chromosomes were seen to condense; prometaphase when nuclear envelope breakdown was complete; metaphase when chromosomes were aligned at the cell equator; anaphase when sister chromatids began separating; telophase when chromosomes began decondensing; and G1 phase when the nuclear envelope reformed and chromosomes were no longer visible by DIC.
Replication in Xenopus egg extract
Extract preparation, handling, TCA precipitation and ELFH assay were performed as described (Chong et al., 1997; Wu et al., 1997). Preparation of intact and permeabilized nuclei by digitonin treatment was performed as described (Dimitrova and Gilbert, 1998). Generally, CHO nuclei were introduced at a concentration of 10 000/µl of extract, previously shown to be optimal for origin specification (Dimitrova and Gilbert, 1998). In some experiments, permeabilized mitotic cells were incubated at 1000/µl extract to ensure nuclear membrane formation. Where appropriate, calculation of the DNA content per cell was adjusted to reflect the ratio of metaphase/anaphase cells (13.2 pg DNA/cell) and telophase/G1 cells (6.6 pg DNA/cell). Geminin was added at a concentration of 40 nM.
Acknowledgments
Acknowledgements
We thank J.Chen for assistance with flow cytometry; D.Natale and M.DePamphilis for providing ORC1 cDNA; G.Yu for help constructing the ORC1 pET vector; T.McGarry for providing the geminin pET vector; J.Blow for anti-Mcm3 antibody; A.Dutta for anti-geminin antibodies; and J.Blow and K.Helin for helpful criticism of the manuscript. This work was supported by American Cancer Society grant #RPG-97-098-04-CCG and NIH grant #GM57233-01 to D.M.G.
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Austin R.J., Orr-Weaver,T.L. and Bell,S.P. (1999) Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev., 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Biggin M.D. (1999) Ultraviolet cross-linking assay to measure sequence-specific DNA binding in vivo. Methods Enzymol., 304, 496–515. [DOI] [PubMed] [Google Scholar]
- Blow J.J. (2001) Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J., 20, 3293–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter P.B. and Dunphy,W.G. (1998) Identification of a novel 81-kDa component of the Xenopus origin recognition complex. J. Biol. Chem., 273, 24891–24897. [DOI] [PubMed] [Google Scholar]
- Chong J., Thommes,P., Rowles,A., Mahbubani,H. and Blow,J. (1997) Characterisation of the Xenopus replication licensing system. Methods Enzymol., 283, 549–564. [DOI] [PubMed] [Google Scholar]
- Cimbora D.M. and Groudine,M. (2001) The control of mammalian DNA replication: a brief history of space and timing. Cell, 104, 643–646. [PubMed] [Google Scholar]
- Cocker J.H., Piatti,S., Santocanale,C., Nasmyth,K. and Diffley,J.F. (1996) An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature, 379, 180–182. [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- Coverley D., Pelizon,C., Trewick,S. and Laskey,R.A. (2000) Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A–cdk2 dependent process. J. Cell Sci., 113, 1929–1938. [DOI] [PubMed] [Google Scholar]
- Dhar S.K. and Dutta,A. (2000) Identification and characterization of the human ORC6 homolog. J. Biol. Chem., 275, 34983–34988. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.A. and Hamlin,J.L. (1995) The chinese hamster dihidrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol., 15, 3023–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D. and Gilbert,D. (1998) Regulation of mammalian replication origin usage in Xenopus egg extracts. J. Cell Sci., 111, 2989–2998. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S. and Gilbert,D.M. (1999) The spatial position and replication timing of chromosomal domains are both established in early G1-phase. Mol. Cell, 4, 983–993. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S. and Gilbert,D.M. (2000) Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nature Cell Biol., 2, 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D.S., Todorov,I.T., Melendy,T. and Gilbert,D.M. (1999) Mcm2, but not RPA, is a component of the mammalian early G1-phase pre-replication complex. J. Cell Biol., 146, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Yamada,C., Goto,H., Yokoyama,N., Kuzushima,K., Inagaki,M. and Tsurumi,T. (1999) Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human mcm complex and CDC2 kinase-mediated hyperphosphorylation. J. Biol. Chem., 274, 25927–25932. [DOI] [PubMed] [Google Scholar]
- Gavin K., Hidaka,M. and Stillman,B. (1995) Conserved initiator proteins in eukaryotes. Science, 270, 1667–1671. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M. (2001) Making sense of eukaryotic DNA replication origins. Science, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.M., Miyazawa,H. and DePamphilis,M.L. (1995) Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol., 15, 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S.A. and Newlon,C.S. (1992) A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell, 3, 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin J.L., Mosca,P.J. and Levenson,V.V. (1994) Defining origins of replication in mammalian cells. Biochim. Biophys. Acta, 1198, 85–111. [DOI] [PubMed] [Google Scholar]
- Hua X. and Newport,J. (1998) Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol., 140, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M. and Gilbert,D.M. (1999) Homogeneous tetracycline-regulatable gene expression in mammalian fibroblasts. J. Cell. Biochem., 76, 280–289. [DOI] [PubMed] [Google Scholar]
- Jiang W., Wells,N.J. and Hunter,T. (1999) Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl Acad. Sci. USA, 96, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson C. and Pines,J. (1998) Green fluorescent protein. In Celis,J. (ed.), Cell Biology: A Laboratory Handbook. Vol. 4. Academic Press, San Diego, CA, pp. 246–252.
- Kreitz S., Ritzi,M., Baack,M. and Knippers,R. (2001) The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem., 276, 6337–6342. [DOI] [PubMed] [Google Scholar]
- Leatherwood J. (1998) Emerging mechanisms of eukaryotic DNA replication initiation. Curr. Opin. Cell Biol., 10, 742–748. [DOI] [PubMed] [Google Scholar]
- Liang C., Weinreich,M. and Stillman,B. (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell, 81, 667–676. [DOI] [PubMed] [Google Scholar]
- Lipford J.R. and Bell,S.P. (2001) Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell, 7, 21–30. [DOI] [PubMed] [Google Scholar]
- Loupart M., Krause,S. and Heck,M.S. (2000) Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr. Biol., 10, 1547–1556. [DOI] [PubMed] [Google Scholar]
- Maiorano D., Moreau,J. and Méchali,M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- McGarry T.J. and Kirschner,M.W. (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell, 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Mendez J. and Stillman,B. (2000) Chromatin association of human origin recognition complex, cdc6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol., 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (2001) Protein dynamics: implications for nuclear architecture and gene expression. Science, 291, 843–847. [DOI] [PubMed] [Google Scholar]
- Natale D.A., Li,C.J., Sun,W.H. and DePamphilis,M.L. (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M–G1 transition in mammals. EMBO J., 19, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Noton E. and Diffley,J.F. (2000) CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell, 5, 85–95. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Takahashi,T. and Masukata,H. (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol., 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak D., Pflumm,M., Chesnokov,I., Huang,D., Kellum,R., Marr,J., Romanowski,P. and Botchan,M. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Pelizon C., Madine,M.A., Romanowski,P. and Laskey,R.A. (2000) Unphosphorylatable mutants of cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev., 14, 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O., Lukas,J., Sorensen,C.S., Bartek,J. and Helin,K. (1999) Phosphorylation of mammalian CDC6 by cyclin A–CDK2 regulates its subcellular localization. EMBO J., 18, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O. et al. (2000) Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev., 14, 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm,T., Cocker,J., Diffley,J. and Nasmyth,K. (1996) Activation of S-phase-promoting CDKs in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev., 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Prokhorova T.A. and Blow,J.J. (2000) Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem., 275, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.C., Muchardt,C. and Yaniv,M. (1997) Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol., 137, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Rosenberg A.H. (1998) Producing proteins in a T7 expression system. In Spector,D. (ed.), Cells: A Laboratory Manual. Vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 64.61–64.24.
- Saha P., Chen,J., Thome,K.C., Lawlis,S.J., Hou,Z.H., Hendricks,M., Parvin,J.D. and Dutta,A. (1998) Human CDC6/Cdc18 associates with Orc1 and cyclin–cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J. (1996) ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J., 15, 6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Stevenson J.B. and Gottschling,D.E. (1999) Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev., 13, 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber K., Mills,A.D., Kubota,Y., Krude,T., Romanowski,P., Marheineke,K., Laskey,R.A. and Williams,G.H. (1998) Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J., 17, 7219–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S., Li,A., Maiorano,D., Méchali,M. and Blow,J.J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nature Cell Biol., 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takisawa H., Mimura,S. and Kubota,Y. (2000) Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr. Opin. Cell Biol., 12, 690–696. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y., Tsurimoto,T., Shirahige,K., Yoshikawa,H. and Obuse,C. (2000) Association of human origin recognition complex 1 with chromatin DNA and nuclease-resistant nuclear structures. J. Biol. Chem., 275, 5904–5910. [DOI] [PubMed] [Google Scholar]
- Vashee S., Simancek,P., Challberg,M.D. and Kelly,T.J. (2001) Assembly of the human origin recognition complex. J. Biol. Chem., 276, 26666–26673. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer,B.T., Dhar,S.K., Cvetic,C., Walter,J.C. and Dutta,A. (2000) Inhibition of eukaryotic DNA replication by geminin binding to cdt1. Science, 290, 2309–2312. [DOI] [PubMed] [Google Scholar]
- Wu J.-R. and Gilbert,D.M. (1996) A distinct G1 step required to specify the chinese hamster DHFR replication origin. Science, 271, 1270–1272. [DOI] [PubMed] [Google Scholar]
- Wu J.-R. and Gilbert,D.M. (1997) The replication origin decision point is a mitogen independent, 2-aminopurine sensitive, G1-phase step that precedes restriction point control. Mol. Cell. Biol., 17, 4312–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-R., Keezer,S. and Gilbert,D. (1998) Transformation abrogates an early G1-phase arrest point required for specification of the Chinese hamster DHFR replication origin. EMBO J., 17, 1810–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-W., Yu,G. and Gilbert,D.M. (1997) Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods, 13, 313–324. [DOI] [PubMed] [Google Scholar]
- Yu G., Wu,J.-R. and Gilbert,D.M. (1998) Analysis of mammalian origin specification in ORC-depleted Xenopus egg extracts. Genes Cells, 3, 709–721. [DOI] [PubMed] [Google Scholar]