Abstract
TEL is a transcriptional repressor that is a frequent target of chromosomal translocations in a large number of hematalogical malignancies. These rearrangements fuse a potent oligomerization module, the SAM domain of TEL, to a variety of tyrosine kinases or transcriptional regulatory proteins. The self-associating property of TEL–SAM is essential for cell transformation in many, if not all of these diseases. Here we show that the TEL–SAM domain forms a helical, head-to-tail polymeric structure held together by strong intermolecular contacts, providing the first clear demonstration that SAM domains can polymerize. Our results also suggest a mechanism by which SAM domains could mediate the spreading of transcriptional repression complexes along the chromosome.
Keywords: chromatin silencing/leukemia/oncogene/Pointed domain/polycomb group proteins
Introduction
SAM domains (also known as SPM, HLH or Pointed domains) have been found in >250 regulatory proteins including receptor tyrosine kinases, serine/threonine kinases, adapter proteins and transcription factors (Ponting, 1995; Schultz et al., 1997; Kyba and Brock, 1998). Some SAM domains are known to form homotypic or heterotypic oligomers and could thereby serve to organize protein complexes in the cell (Carroll et al., 1996; Golub et al., 1996; Jousset et al., 1997; Peterson et al., 1997; Tu et al., 1997; Kwiatkowski et al., 1998; Poirel et al., 2000; Potter et al., 2000; Baker et al., 2001). Structures have been reported of SAM domains from the Eph receptors EphA4 (Stapleton et al., 1999) and EphB2 (Smalla et al., 1999; Thanos and Bowie, 1999; Thanos et al., 1999b), from the tumor suppressor p53 homolog p73 (Chi et al., 1999) and from the Ets transcription factor Ets1 (Slupsky et al., 1998). In spite of the large number of structures, the mechanism of SAM domain association remains unclear since none of these SAM domain structures describes oligomers that are clearly biologically relevant. In both the EphA4 and EphB2 SAM domain structures, possible complexes were observed between monomers in the crystals that could represent oligomeric SAM structures that form upon receptor clustering. A possible dimeric structure was proposed for EphA4 and a possible polymeric structure was observed in the crystal of EphB2 (Stapleton et al., 1999; Thanos et al., 1999b). In both cases, however, the Eph receptor SAM domains were found to associate very weakly in solution (Kd > 1 mM), thus the biological relevance of these models remains speculative. Here we report structural information on a SAM domain that strongly self-associates—the SAM domain from TEL (translocation Ets leukemia). The structure reveals that TEL–SAM self-associates as a polymeric structure that is completely different to any of the oligomers previously reported.
TEL is a transciptional repressor that contains an N-terminal SAM domain, a central co-repressor binding domain and a C-terminal Ets DNA binding domain. The central domain is required for the recruitment of the co-repressors mSin3A, SMRT and N-CoR, which in turn can recruit histone deacetylases (Chakrabarti and Nucifora, 1999; Fenrick et al., 1999; Guidez et al., 2000). TEL was first discovered as the target of a chromosomal rearrangement in a chronic myelomonocytic leukemia patient (Golub et al., 1994), and chromosomal translocations involving TEL have since been found in a large number of additional hematological malignancies (Rubnitz et al., 1999). These genetic rearrangements result in a variety of oncogenic TEL chimeras, most of which fuse the SAM domain of TEL to a variety of tyrosine kinase domains such as PDGFRβ, ABL, JAK2 and NTRK3 (Golub et al., 1994, 1996; Papadopoulos et al., 1995; Peeters et al., 1997; Knezevich et al., 1998; Eguchi et al., 1999; Lacronique et al., 2000), or to transcription factors such as AML1 and ARNT (Golub et al., 1995; Romana et al., 1995; Salomon-Nguyen et al., 2000). In the tyrosine kinase fusions, TEL–SAM-mediated oligomerization leads to the constitutive activation of the enzyme and is essential for cell transformation (Carroll et al., 1996; Golub et al., 1996; Jousset et al., 1997; Lacronique et al., 1997). In the TEL–AML1 fusions both the SAM domain and central domain of TEL become fused to AML1 and convert the transcriptional activator AML1 into a transcriptional repressor. In various studies of TEL–AML1, the SAM domain has been shown to be essential for transcriptional repression (Hiebert et al., 1996; Fears et al., 1997; Fenrick et al., 1999; Uchida et al., 1999). Because of the common involvement of the SAM domain in all the chimeric oncoproteins, therapeutic strategies that abrogate TEL–SAM oligomerization could have a significant impact on a variety of different hematological diseases.
Results
Generation of a soluble form of TEL–SAM
When the TEL–SAM domain was independently expressed in Escherichia coli, the protein was found in inclusion bodies and we were unable to refold the protein into a soluble form. Although proteins commonly cannot be recovered by refolding from inclusion body preparations, additional experiments discussed below suggest that our inability to obtain soluble TEL–SAM was due to its intrinsic polymerization activity.
To overcome the solubility problems that precluded detailed biochemical or structural characterization of TEL–SAM, we screened for soluble variants of the domain using the method of Waldo et al. (1999). The TEL–SAM domain was fused to a variant of green fluorescent protein called cyan fluorescent protein (CFP) (Tsien, 1998). Since the insoluble TEL–SAM domain also renders the fused CFP protein insoluble, cells expressing the fusion protein are colorless. We randomly mutagenized the TEL–SAM domain and screened for cyan-colored colonies, which would indicate the expression of a soluble, colored fusion protein. After screening 15 000 colonies, six independent soluble clones were obtained. Although most contained multiple mutations, four of the six bore the same Val to Glu mutation at position 80. The independently expressed single mutant, V80E, was found in the soluble fraction in E.coli extracts.
The V80E mutant was purified to homogeneity and found to be monomeric at high concentrations above pH 7, but precipitated reversibly at low pH. Although many scenarios could explain this behavior, we demonstrate below that the V80E mutation places a charged residue in the natural polymer interface that blocks polymerization at high pH, but permits polymerization upon protonation at low pH.
Crystal structure of the polymeric TEL–SAM domain
High quality crystals were obtained of the V80E mutant containing selenomethionine and the structure was solved by the multiwavelength anomolous diffraction (MAD) phasing method. The structure was refined to an Rfree of 0.195 at 1.45 Å resolution. Details of the structure determination and refinement are given in Table I. The three TEL–SAM monomers in the asymmetric unit of the crystal each adopt the same global fold as the previously determined SAM domain structures. The root mean square deviation (r.m.s.d.) of Cα atoms is 2.8 Å between the TEL–SAM and Ets1–SAM (Slupsky et al., 1998) domain structures (also see Discussion).
Table I. Crystallographic data.
| Space group Cell | C2 | ||||||
|---|---|---|---|---|---|---|---|
| a = 132.865 | b = 52.678 | c = 58.701 | α = 90° | β = 113.898° | γ = 90 | ||
| λ (Å) | Resolution (Å) | Observed reflections | Unique reflections | Completeness (%) | <I>/<σi> | Rsym (%) | |
| Anomalous peak | 0.97880 | 1.40 | 244 968 | 139 887 | 97.4 (97.3) | 9.9 (1.8) | 5.7 (38.3) |
| Inflection point | 0.97902 | 1.40 | 216 872 | 139 471 | 97.1 (97.1) | 8.3 (2.0) | 5.7 (24.6) |
| High energy remote | 0.97135 | 1.40 | 269 457 | 140 500 | 98.5 (98.3) | 8.7 (2.3) | 5.1 (26.5) |
| R factor | |||||||
| Rcryst | 18.4% | ||||||
| Rfree | 19.5% | ||||||
| R.m.s.d. from ideality | |||||||
| bond lengths | 0.0056 Å | ||||||
| bond angles | 1.17° | ||||||
| Ramachandran analysis | |||||||
| most favored | 91.6% | ||||||
| additionally favored | 8.4% | ||||||
| Non-protein molecules | |||||||
| sulfates | 5 | ||||||
| waters | 242 | ||||||
| B factors | |||||||
| overall | 17.72 Å2 | ||||||
| sulfates | 31.19 Å2 | ||||||
| waters | 29.55 Å2 | ||||||
Values in parentheses are for the highest resolution bin: 1.45–1.40 Å.
Rsym = ∑|I – <I>|/∑<I>, where I is the observed intensity and <I> is the average intensity from observations of symmetry-related reflections. Rcryst = ∑|Fobs – Fcalc|/∑Fobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated for a set of reflections (10%) that were not included in atomic refinement. Both Rcryst and Rfree are calculated from 500.0 to 1.45 Å resolution, after bulk solvent correction and with no reflection intensity cut-off.
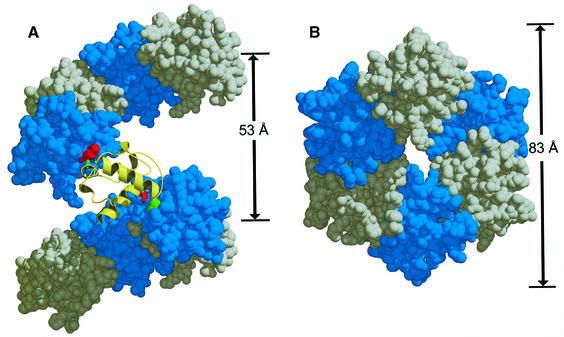
Each TEL–SAM monomer forms a large interface with other monomers in the crystal. A consequence of this interface is the formation of a polymeric structure shown in Figure 1. The molecules in this polymer are arranged in head-to-tail fashion with approximate 65 screw symmetry, and a repeat distance of 53 Å that corresponds to the y-axis of the crystal unit cell. Both the N- and C-termini point outward from the polymeric helix.
Fig. 1. TEL–SAM polymer structure. (A) The V80E polymer structure viewed with the helix axis in the plane of the page, pointing up. The figure shows nine subunits of the polymer. The central subunit is shown in gold ribbon. V80E mutation on the EH surface is shown in red and ML surface apolar residue Ala61 is shown in green. The red and green coloring scheme representing the EH and ML surfaces, respectively, will be consistent for the other figures. (B) The V80E polymer structure viewed down the helix axis of the polymer.
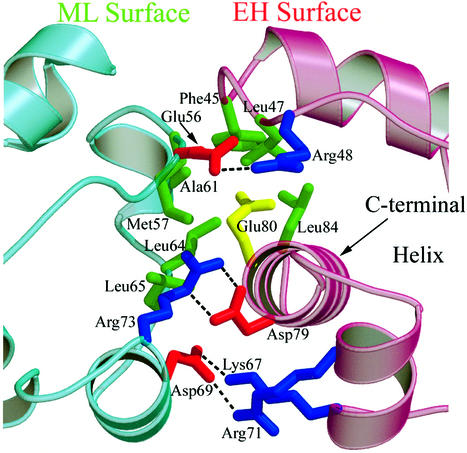
The interface between monomers of the polymer is constructed from two patches of residues, which we will refer to as the mid-loop (ML) surface because important apolar residues involved in the interaction are on the loop portions near the center of the peptide and the end-helix (EH) surface because the C-terminal helix provides important residues involved in the interaction (see Figures 1 and 2). The solvent-accessible surface area buried in the interface ranges from 1070 to 1250 Å2 in the different molecules of the crystal asymmetric unit. As shown in Figure 2, the core of the ML surface is comprised of Met57, Ala61, Leu64 and Leu65, and the core of the EH surface is comprised of residues Phe45, Leu47, Glu80 (Val80 in the wild-type protein) and Leu84. All these residues are hydrophobic with one exception—the Glu80 mutant side-chain that renders the protein soluble at high pH. The position of this mutation in the center of what would otherwise be a completely hydrophobic interface is consistent with the idea that the V80E mutation solubilizes TEL–SAM by preventing polymerization. In addition to the hydrophobic interior of the binding interface, a number of salt-bridges are found on the external surface (Figure 2). Although there are some differences in electrostatic interactions between the three different interfaces in the crystal, a number of salt-bridges are always observed: Glu56 to Arg48, Asp69 to Lys 67, Asp69 to Arg71 and Arg73 to Asp79. In addition, Lys60 to Glu44 and Glu68 to Arg71 salt-bridges are observed in two of the three interfaces.
Fig. 2. Detailed view of the TEL–SAM V80E polymer interface. Hydrophobic residues that make up the core of the interface are shown in green. The V80E mutation that renders the protein soluble is shown in yellow. In the wild-type sequence a hydrophobic Val side-chain would occupy this position. Salt-bridges in the interface that surround the core residues are also shown. Negatively charged residues are shown in red and positively charged residues are shown in blue.
Native TEL–SAM interface
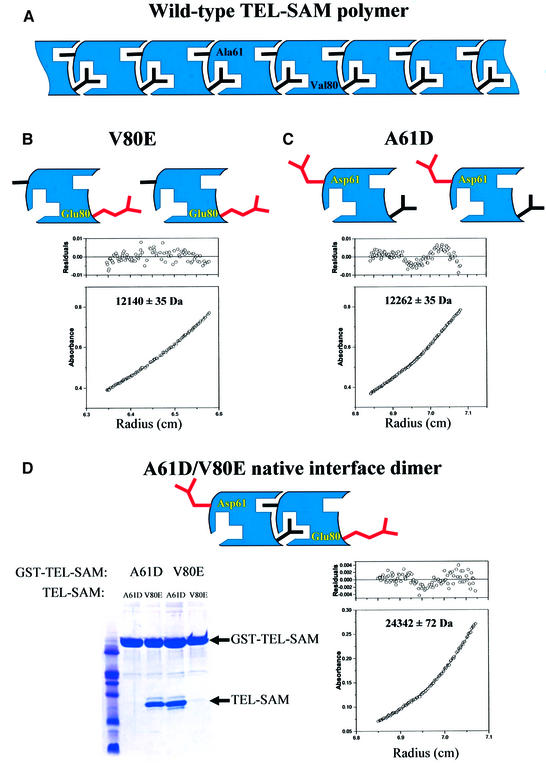
Although the interface observed in Figure 2 is large and resembles a typical oligomeric interface, it remained possible that the observed polymer was simply an artifact of crystallization. We therefore designed a set of experiments, outlined in Figure 3, to test the polymer model and measure the affinity of the wild-type interface in the polymer.
Fig. 3. Test of the polymer model. (A) Schematic illustration of the putative wild-type polymeric structure highlighting two key residues, A61 and V80, on the different binding surfaces, ML and EH, respectively. (B) Equilibrium sedimentation results for the V80E mutant. The observed molecular weight was found to be 12 140 ± 35 Da and the calculated molecular weight for the monomer is 12 079 Da. (C) Equilibrium sedimentation results for the A61D mutant. The observed molecular weight was found to be 12 262 ± 35 Da and the calculated molecular weight for the monomer is 12 093 Da. (D) Testing mixed V80E–A61D dimer formation. The ability of V80E or A61D to bind to themselves or to each other was tested in GST pull-down experiments. As shown in the gel, A61D bound to V80E but not to itself, and V80E bound to A61D but not to itself. Equilibrium sedimentation results for an equimolar mixture of V80E and A61D are also shown. The observed molecular weight was found to be 24 342 ± 72 Da and the calculated molecular weight for the dimer is 24 172 Da.
First, we hypothesized that the V80E mutation renders the TEL–SAM domain soluble at high pH by destroying the EH surface in the polymer. Based on our polymer structure, we would expect that the V80E mutant should be monomeric and, as shown in Figure 3B, equilibrium sedimentation results verify this prediction.
Secondly, from the polymer structure, we expected that a mutation on the ML surface could also block polymerization and produce a different soluble, monomeric variant of TEL–SAM. We constructed such a mutant, A61D, placing a charged residue at the ML surface. The A61D mutant was indeed found in the soluble fraction in E.coli extracts and equilibrium sedimentation, shown in Figure 3C, verifying that the protein is monomeric. Unlike the V80E mutant, the A61D mutant remained soluble even at lower pH. Thus, we were able to rationally design a monomeric variant of TEL–SAM, based on the interface seen in the crystal structure. These results suggest that the reason for wild-type TEL–SAM insolubility is indeed its propensity to polymerize and that the structure of the polymer is represented in the crystal structure.
Finally, the polymer model also suggests that the two monomeric mutants described above would form a stable mixed dimer because the A61D mutation on the ML surface still contains a wild-type EH surface, and the V80E mutation on the EH surface still contains a wild-type ML surface. The results shown in Figure 3D verify this hypothesis. First, in glutathione S-transferase (GST) pull-down experiments we found that a GST–V80E fusion bound to the A61D mutant, but not the V80E mutant. In the complementary experiment, a GST–A61D fusion bound to the V80E mutant but not the isolated A61D mutant. As shown in Figure 3D, equilibrium sedimentation experiments demonstrate that an equimolar mixture of V80E and A61D has the molecular weight expected for a dimer.
To determine the strength of the V80E–A61D heterodimer interface, we turned to surface plasmon resonance experiments. We found that the dimer is very stable, with a dissociation constant of 1.7 nM. This dissociation constant reflects the affinity of the wild-type interface in the polymer. Our ability to design this dimeric TEL–SAM and its strong association strongly suggests that the TEL–SAM polymer is not simply an artifact of crystallization and is likely to form in the cell.
Visualization of the wild-type TEL–SAM polymer
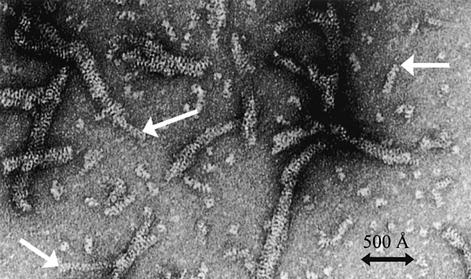
Because the TEL–SAM crystal structure is a mutant, we sought to visualize the wild-type polymer by electron microscopy. Wild-type TEL–SAM formed large aggregates, however, and we were unable to observe any individual polymer fibers. To reduce the size and solubilize smaller polymer units, the A61D and V80E mutants were mixed with the wild-type protein to cap the wild-type polymers. This procedure led to soluble filaments that have similar width dimensions to the TEL–SAM helical polymer seen in the crystal structure (Figure 4). No filaments were observed for the mutants alone or for a mixture of the two mutants. Because the polymer structures observed in Figure 4 reflect the structure of the wild-type TEL–SAM, it further suggests that the helical polymer structure observed in the crystal structure accurately reflects the true state of the TEL–SAM polymer. The filaments also exhibit ordered striations on their surface perhaps reflecting the repeating helical structure of the TEL–SAM polymer.
Fig. 4. Electron microscopy of TEL–SAM. Electron microscopy photograph of the TEL–SAM polymer. Alhough most filaments appear tangled, isolated single filaments, indicated by arrows, have width dimensions similar to the width of the helical polymer observed in the crystal structure.
Discussion
Until now, as with various other protein modules involved in signal transduction, the oligomeric state of TEL–SAM has only been discussed in terms of a closed oligomer. In this work, however, we have presented clear evidence that TEL–SAM actually forms an open-ended polymeric structure. This finding not only clarifies the mechanism of transformation described for the tyrosine kinase fusion oncogenes (Golub et al., 1996; Jousset et al., 1997; Lacronique et al., 1997), in which kinase domain oligomerization leads to constitutive tyrosine kinase activation, but also raises questions concerning the role of TEL–SAM polymerization in transcriptional repression. We suggest that by linking many DNA binding elements together, TEL–SAM polymerization may provide a mechanism for spreading repression over a large segment of chromatin.
A possible model for spreading of transcriptional repression
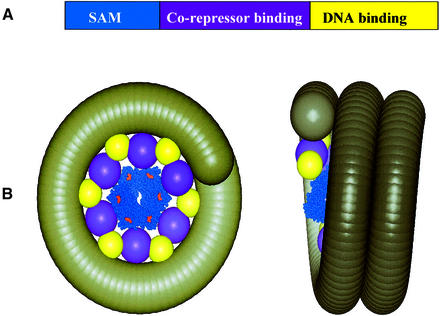
To illustrate how polymerization could lead to spreading of transcriptional repression, we constructed a possible model of a TEL-mediated repression complex shown in Figure 5. In the structure of the TEL–SAM polymer, the C-termini point outward, away from the polymer axis. In the full-length TEL protein, the C-terminal regulatory and DNA binding domains must therefore be displayed on the outside of the polymer. The DNA binding domains could in turn recruit chromatin, leading to a toroidal wrapping of the chromatin around the polymer. It is notable that the 53 Å repeat distance of the polymer almost exactly matches the width of the nucleosome core particle, which would lead to close packing of the nucleosome core particles along the polymer axis (Luger et al., 1997). It is easy to envisage that such a compact structure would block access to transcriptional machinery, leading to a repressed region of the chromosome. Although the structural details are highly speculative and many other architectures are possible, the key feature of this model is polymerization of a DNA binding element engaging a large chromosomal region.
Fig. 5. Model of one possible TEL repression complex. (A) The domain structure of TEL. (B) A possible complex of TEL with chromatin. The TEL–SAM polymer structure from this work is shown as a space-filling model in blue. The C-termini are colored red and orient away from the axis of the polymer. This is the only known structure in the model shown. Spheres representing the co-repressor and DNA binding domains of TEL are placed on the outside of the polymer with the same helical pitch as the TEL–SAM polymer. The sphere volumes were determined assuming a partial specific volume of 0.7 ml/g. A coil representing chromatin is wrapped around the polymer, interacting with the DNA binding domains. The thickness of the coil is the same as the width of a nucleosome core particle (54 Å) (Luger et al., 1997). We note that this is a highly speculative model and serves only to illustrate one way that polymerization could lead to spreading of transcriptional repression. Many other models are possible.
Once nucleated at a specific locus on DNA, only weak DNA binding affinity would be required for each additional polymer subunit to bind DNA productively since much of the entropic cost of uniting protein and DNA would be paid by the strong SAM–SAM interactions. Ets DNA binding domains, including TEL, can bind with reasonable affinity to DNA elements with core sequences of GGA, a sequence that occurs frequently in DNA (Szymczyna and Arrowsmith, 2000). A commonly occurring recognition motif may be exactly what is required for organizing chromatin in this fashion since the DNA must be bound at many sites in the putative repression complex.
Our polymeric repression model could also suggest a mechanism of repression in artificial fusions of TEL–SAM to the GAL4 DNA binding domain. In GAL4–TEL–SAM fusions, transcriptional repression was observed even when the GAL4 binding site was located 600 bp upstream of the promoter site (Fenrick et al., 1999). Although it has been suggested that TEL–SAM may be involved in recruiting co-repressors (Fenrick et al., 1999; Guidez et al., 2000), another report disputes this finding (Chakrabarti and Nucifora, 1999). Given the polymeric nature of TEL–SAM, an alternative mechanism of repression can be envisaged. The binding of the GAL4–TEL–SAM fusion to the GAL4 binding site could act to nucleate a repression complex similar to the one shown in Figure 5. Due to TEL–SAM polymerization, non-specific binding to DNA via the GAL4 DNA binding domain would likely become energetically favorable. Thus, in the case of the TEL–SAM–GAL4 fusions, the polymeric structure could extend out from the high affinity binding site by non-specific DNA binding, thereby blocking the access of transcriptional machinery. A similar mode of repression can be envisaged for the TEL–AML1 chimeric oncoprotein that would utilize the DNA binding activity of the Runt homology domain to nucleate a repression complex.
Polycomb group (PcG) proteins are known to spread over large regions of the chromosome. Two members of the PcG proteins, Scm and ph, contain SAM domains that are known to self-associate (Peterson et al., 1997). It is tempting to speculate that a similar mechanism of spreading, mediated by SAM domains, could occur in these repression complexes. In the case of the PcG complexes, however, no specific DNA binding domains have been identified, so interactions with the chromatin may involve binding to chromatin-associated proteins (Pirrotta, 1998).
Polymerization via SAM domains presents an attractive model for the extension of repression complexes over long distances. While the extension provided by oligomeric SAM domains and the resultant quaternary structural changes could play an important role in repression, it is also part of a more complex system that includes the recruitment of co-repressors in the establishment of repression. For example, using an artificial promoter system, Lopez et al. (1999) showed that TEL-mediated repression still occurs when the SAM domain is replaced by a leucine-zipper dimerization motif. The results of Fenrick et al. (2000) and Buijs et al. (2000) suggest that oligomerization may not even be required in some cases. It is therefore possible that SAM domain polymerization only plays a role in spreading or maintaining transcriptional repression, but not in establishing the repressed state. Future work will further our understanding of the role SAM domain polymerization plays in the wild-type protein and how polymerization is regulated.
Do other SAM domains polymerize?
While the results reported here indicate that at least one function of SAM domains is to form polymers, it remains unclear whether it is a general property of SAM domains. Not all SAM domains have been shown to form oligomers and many are monomeric even at high concentrations (Slupsky et al., 1998; Chi et al., 1999; Thanos et al., 1999a). While these SAM domains may provide a different function, it is also possible that in many cases SAM domain polymerization is regulated, requiring an appropriate trigger. For example, in the case of the Eph receptor SAM domains, we have proposed that SAM domain polymer formation could be triggered by receptor clustering (Thanos et al., 1999b).
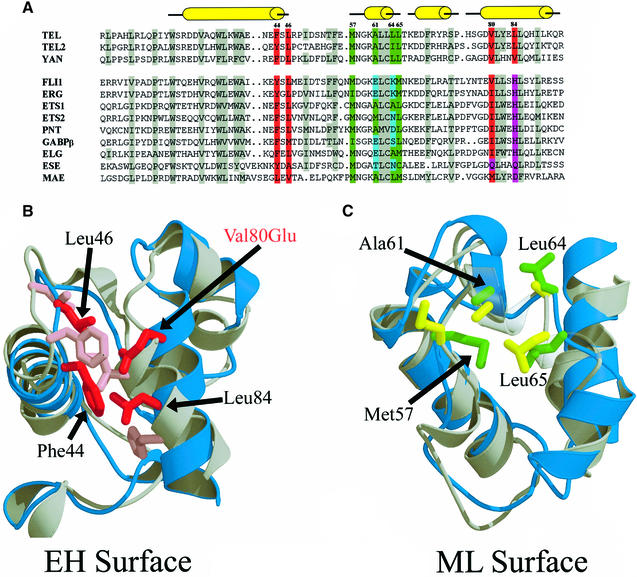
Based on our structure of the TEL–SAM polymer, it is possible to define a subset of Ets family SAM domains that may also polymerize in the same fashion. Figure 6A shows a sequence alignment of a set of ETS family SAM domains. Residues that comprise the hydrophobic core of the ML binding surface are colored in green and residues that comprise the hydrophobic core of the EH binding surface are colored red. Sequences that maintain hydrophobic residues in both interfaces (TEL, TEL2 and YAN) are grouped separately from the rest. Of this group, TEL and TEL2 are already known to self-associate (Carroll et al., 1996; Golub et al., 1996; Jousset et al., 1997; Lacronique et al., 1997; Kwiatkowski et al., 1998; Poirel et al., 2000; Potter et al., 2000). It seems entirely possible that TEL and TEL2 could even form a co-polymer.
Fig. 6. Sequence and structural alignment of the Ets family SAM domains. (A) The sequence of wild-type TEL–SAM aligned with SAM domains from members of the Ets family of transcription factors. The residues highlighted in red and green represent the EH and ML binding surfaces, respectively. Amino acids that have >75% of the residue buried in the core of the TEL–SAM structure and conserved apolar residues at that position are shaded in gray. The numbering scheme above the TEL–SAM sequence is of the construct used in this study. (B) Structural alignment of TEL–SAM with Ets1–SAM (Slupsky et al., 1998) showing the EH binding surface. TEL–SAM is shaded in blue while Ets1–SAM is shaded in gray. The apolar residues and the Val80Glu mutation that form the EH binding surface of TEL–SAM are colored red. The analogous Ets1–SAM residues from the sequence alignment in Figure 6A are colored pink. (C) The ML binding surface. The TEL–SAM hydrophobic residues that make up the core of the ML surface are green while the equivalent Ets1–SAM residues are colored yellow.
In the group of ETS family members that do not maintain a hydrophobic core, the SAM domain of Ets1 is known to be monomeric (Slupsky et al., 1998) and we have found that the isolated Fli1–SAM is also monomeric (data not shown). In the case of Fli1–SAM, two charged residues occur in the ML surface that could easily block polymerization. The reason for the lack of Ets1 polymerization is less clear from the sequence, however. Although the sequence contains a His residue in one of the EH positions, it is not inconceivable that such a change would be tolerated. A structure alignment of TEL–SAM and Ets1–SAM shown in Figure 6B and C, however, reveals a second factor that could block polymerization of the Ets1 SAM domain. Although the ML binding surfaces are quite similar, the EH binding surface structures are substantially altered, possibly preventing productive interactions. It remains possible that a regulated conformational change in Ets1–SAM could lead to a polymerization-competent form.
The ETS family SAM domains that appear incompatible with a TEL-like polymeric structure may form different homopolymers, form a defined and closed oligomeric state, or form heteropolymers with other ETS family SAM domains. They may also play a role in regulating polymerization of other SAM domains, for example, by capping SAM polymers in a similar fashion to the TEL–SAM mutants we have described in this paper. Alternatively, these SAM domains may not polymerize but provide alternative functions such as binding to other proteins.
If a common function of SAM domains is polymerization, it seems likely that different polymeric structures must exist. For example, Eph receptors contain SAM domains, but the helical polymer created by TEL–SAM would be incompatible with a protein that is inserted into the bilayer. For a protein inserted into the membrane, any protein domains attached to the SAM domain would need to point toward the membrane, but in a helical polymer they would radiate out in all directions. Thus, if the Eph receptor SAM domain polymers are indeed made, it would require conservation of function, but not structure over the course of evolution. Indeed, the proposed structure of EphB2–SAM polymer is completely different from the polymer observed for TEL–SAM. This is not unprecedented. For example, the T1 domain of potassium channels and the BTB/POZ domain are structurally similar but oligomerize in distinct ways (Ahmad et al., 1998; Kreusch et al., 1998), and members of the cytokine receptor family exhibit diverse receptor binding modes in spite of their common structure and function (Deller and Jones, 2000). It seems likely that SAM domains will also exhibit diverse protein–protein interaction modes that remain to be uncovered.
Materials and methods
TEL–SAM–CFP screen for soluble mutants
The wild-type TEL–SAM amino acid sequence from residues 38 to 124 was cloned into a modified pET3c vector (Novagen) as an N-terminal fusion to CFP. In so doing, a leader sequence of MEKTR was incorporated. The end of the TEL–SAM sequence (wild-type residue 124) was fused to the initiator Met of CFP with an eight amino acid linker sequence, DHHHHHHS. The PCR-mutagenized TEL–SAM sequence was cloned into the CFP fusion vector and then transformed into BL21(DE3) pLysS cells. The resulting colonies were lifted from the agar using filter paper and induced by placing the paper on top of another soaked in LB media containing 100 µg/ml ampicillin and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After incubation at 37°C for 1–3 h, colony fluorescence was observed by viewing the lifted colonies under UV light. Colonies that were brighter than the background colonies were picked from the original plate and re-plated several times, each time re-checking for fluorescence with the colony lift assay. Starting from 15 000 original colonies, ∼30 colonies, which remained fluorescent, were screened in small cultures for the production of cyan-colored lysate. Potential positive clones were grown in 2 ml of LB media containing 100 µg/ml ampicillin to an OD600 of 1.0 and fusion protein production induced by adding IPTG to a final concentration of 1 mM. A 1.5 ml portion of the culture was pelleted in a microfuge, resuspended in 100 µl of 50 mM Tris pH 8.0, 200 mM NaCl buffer with 5 mg/ml lysozyme and 4 U of DNase I, lysed by three freeze–thaw cycles and insoluble material removed by centrifugation. Six cultures were observed to have cyan-colored soluble lysate. We obtained readable sequence from five of the six clones. Although most were multiple mutants, four of the five bore the same V80E mutation. The only single mutant we obtained was V80E.
Preparation of TEL–SAM mutants
The V80E mutant was PCR amplified using the single mutant CFP fusion protein as the template and cloned into the modifed pET3c vector described above. The expressed protein sequence includes an N-terminal MEKTR leader sequence, residues 38–124 of the TEL protein and a C-terminal DHHHHHH sequence that provides a purification tag. In our numbering scheme, residue 6 corresponds to residue 38 of the wild-type TEL protein sequence. The V80E construct was transformed into BL21(DE3) pLysS cells and grown to an approximate OD600 of 0.8 and induced with 1 mM IPTG for 5 h. Cells from a 1 l culture were resuspended in 10 ml of 50 mM Tris pH 8.0, 200 mM NaCl, 30 mM imidazole pH 7.8, and lysed by sonication. The protein in the soluble extract was applied to a 1.5 ml column volume of Ni-NTA agarose and washed extensively in the same buffer. The bound protein was then eluted with 15 ml of 50 mM Tris pH 8.0, 200 mM NaCl and 300 mM imidazole pH 7.8. The elution was dialyzed against 1 l of 25 mM bis tris propane pH 6.4, 50 mM NaCl. Upon removing precipitated protein from the dialysis, the remaining soluble protein solution was applied to a 1 ml HiTrap SP column (Pharmacia), washed with 25 mM bis tris propane pH 6.4, and eluted with a gradient from 50 mM to 1 M NaCl. The purified protein was precipitated by adding (NH4)2SO4 to 60% saturation then suspended and dialyzed in 10 mM bis tris propane pH 8.5, 200 mM NaCl to remove the (NH4)2SO4. The TEL–SAM A61D mutant was cloned into the same vector using Stratagene’s QuikChange Mutagenesis Kit. The same purification protocol used for the V80E mutant was used for the A61D mutant.
Crystal structure determination
Fully incorporated Se-Met protein samples were prepared according to the protocol of Van Duyne et al. (1993). Crystals were grown by hanging drop vapor diffusion, mixing 2–3 µl of ∼7 mg/ml protein in 10 mM Tris pH 8.5, 200 mM NaCl with an equal volume of well buffer: 50 mM Tris pH 8.0, 1.6 M Li2SO4. Crystals grew over a 4 week period at room temperature. Data were collected under a liquid nitrogen stream at the National Synchrotron Light Source at the Brookhaven National Laboratory on Beamline X8-C. The data were processed with DENZO/SCALEPACK (Otwinowski and Minor, 1997) and data sets collected at the three wavelengths were input directly into SOLVE (Terwilliger and Berendzen, 1999). SOLVE located three Se sites from which phases were calculated. Solvent flattening was carried out using DM in the CCP4 suite of programs (CCP4, 1994). Electron density maps calculated from these phases were readily interpreted and an initial model was built using the program O (Jones et al., 1991). Refinement was carried out using CNS (Brunger et al., 1998) with the MLHL target function. The PDB accession code is 1JI7.
Equilibrium sedimentation
A61D, V80E and the equimolar combination of the two were all prepared in 10 mM Tris pH 8.5, 200 mM NaCl at a total protein concentration of 0.3 mg/ml. Lower concentrations, 0.071, 0.038 and 0.014 mg/ml, of the equimolar mixture were also prepared. Sedimentation equilibrium runs were performed at 20°C in a Beckman Optima XL-A analytical ultracentrifuge at speeds of 17 000, 25 000 and 30 000 r.p.m. using a 12 mm pathlength six-sector cell and absorption optics at 280 nm. For the lower concentration protein samples, 228 nm was used. The data were fitted with a non-linear least-squares exponential fit for a single ideal species using the Beckman Origin-based software (version 3.01). A partial specific volume of 0.715 calculated from the amino acid composition and corrected to 20°C was used. No dissociation of the A61D + V80E complex was observed at the lowest concentration tested.
Electron microscopy
Approximately 0.2 mg of insoluble protein pellet of wild-type TEL–SAM were resuspended with 200 µl of a 0.2 mg/ml solution of an equimolar mixture of TEL–SAM V80E and A61D mutants in 25 mM Tris pH 8.0, 200 mM NaCl. The three-protein mixture was allowed to incubate for several hours at room temperature prior to mounting. Carbon-coated parlodion support films mounted on copper grids were made hydrophilic immediately before use by high voltage, alternating current glow-discharge. Samples were applied directly onto grids and allowed to adhere for 2 min. Grids were rinsed with distilled water and negatively stained with 1% uranyl acetate. Specimens were examined in a Hitachi H-7000 electron microscope at an accelerating voltage of 75 kV.
GST fusion protein binding assay
Forty microliters of the supplied slurry of glutathione Sepharose 4B beads (Pharmacia) were equilibrated in assay buffer (typically 50 mM bis tris propane pH 7.5, 150 mM NaCl and 10 mM βME). Four hundred microliters of ∼1 mg/ml GST fusion proteins were incubated with the beads for 1 h at 4°C. The beads were washed twice with 500 µl of the assay buffer and then incubated with 300 µl of ∼1 mg/ml of the TEL–SAM mutants in assay buffer for 1 h at 4°C. The beads were washed three times with 500 µl of the assay buffer, followed by elution of the bound proteins with 60–80 µl of 20 mM reduced glutathione in the assay buffer. Equivalent amounts of the elution were separated using 15% Tris–tricine SDS–PAGE.
Surface plasmon resonance
The surface plasmon resonance experiments for the TEL–SAM mutants were performed in 25 mM Tris pH 8.5, 200 mM NaCl and 0.005% Surfactant P20. The TEL–SAM A61D mutant was immobilized on a Biacore Pioneer B1 sensor chip. Various concentrations of the TEL–SAM V80E mutant were injected onto the chip and the resulting binding data analyzed with the BIAevaluation 3.0 software. The apparent dissociation constant was found to be 1.7 ± 0.5 nM. The association rate constant was 7.9 ± 1.3 × 105 M–1 s–1 and the dissociation rate constant was 1.3 ± 0.1 × 10–3 s–1.
Acknowledgments
Acknowledgements
The authors would like to thank Al Courey, Aaron Chamberlain, Sarah Yohannan and Ranjini Ramachander for helpful comments on the manuscript, Michael Sawaya and Duilio Cascio for help with synchrotron data collection, and Owen Witte for providing a clone of TEL–ABL. This work was supported by a grant from the Margaret E. Early Medical Research Trust and NIH grant R01 CA81000.
References
- Ahmad K.F., Engel,C.K. and Prive,G.G. (1998) Crystal structure of the BTB domain from PLZF. Proc. Natl Acad. Sci. USA, 95, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.A., Mille-Baker,B., Wainwright,S.M., Ish-Horowicz,D. and Dibb,N.J. (2001) Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature, 411, 330–334. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buijs A. et al. (2000) The MN1-TEL fusion protein, encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity. Mol. Cell. Biol., 20, 9281–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M., Tomasson,M.H., Barker,G.F., Golub,T.R. and Gilliland,D.G. (1996) The TEL/platelet-derived growth factor β receptor (PDGF β R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF β R kinase-dependent signaling pathways. Proc. Natl Acad. Sci. USA, 93, 14845–14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S.R. and Nucifora,G. (1999) The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem. Biophys. Res. Commun., 264, 871–877. [DOI] [PubMed] [Google Scholar]
- Chi S.W., Ayed,A. and Arrowsmith,C.H. (1999) Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J., 18, 4438–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) CCP4 programs. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Deller M.C. and Jones,E.Y. (2000) Cell surface receptors. Curr. Opin. Struct. Biol., 10, 213–219. [DOI] [PubMed] [Google Scholar]
- Eguchi M. et al. (1999) Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood, 93, 1355–1363. [PubMed] [Google Scholar]
- Fears S., Gavin,M., Zhang,D.E., Hetherington,C., Ben-David,Y., Rowley,J.D. and Nucifora,G. (1997) Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter. Proc. Natl Acad. Sci. USA, 94, 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenrick R., Amann,J.M., Lutterbach,B., Wang,L., Westendorf,J.J., Downing,J.R. and Hiebert,S.W. (1999) Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol. Cell. Biol., 19, 6566–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenrick R. et al. (2000) TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stromelysin-1. Mol. Cell. Biol., 20, 5828–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub T.R., Barker,G.F., Lovett,M. and Gilliland,D.G. (1994) Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell, 77, 307–316. [DOI] [PubMed] [Google Scholar]
- Golub T.R. et al. (1995) Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA, 92, 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub T.R., Goga,A., Barker,G.F., Afar,D.E., McLaughlin,J., Bohlander,S.K., Rowley,J.D., Witte,O.N. and Gilliland,D.G. (1996) Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol. Cell. Biol., 16, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidez F. et al. (2000) Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood, 96, 2557–2561. [PubMed] [Google Scholar]
- Hiebert S.W. et al. (1996) The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol. Cell. Biol., 16, 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jousset C. et al. (1997) A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFR β oncoprotein. EMBO J., 16, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevich S.R., McFadden,D.E., Tao,W., Lim,J.F. and Sorensen,P.H. (1998) A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nature Genet., 18, 184–187. [DOI] [PubMed] [Google Scholar]
- Kreusch A., Pfaffinger,P.J., Stevens,C.F. and Choe,S. (1998) Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature, 392, 945–948. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski B.A., Bastian,L.S., Bauer,T.R.,Jr, Tsai,S., Zielinska-Kwiatkowska,A.G. and Hickstein,D.D. (1998) The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J. Biol. Chem., 273, 17525–17530. [DOI] [PubMed] [Google Scholar]
- Kyba M. and Brock,H.W. (1998) The SAM domain of polyhomeotic, RAE28, and scm mediates specific interactions through conserved residues. Dev. Genet., 22, 74–84. [DOI] [PubMed] [Google Scholar]
- Lacronique V. et al. (1997) A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science, 278, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Lacronique V. et al. (2000) Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood, 95, 2076–2083. [PubMed] [Google Scholar]
- Lopez R.G., Carron,C., Oury,C., Gardellin,P., Bernard,O. and Ghysdael,J. (1999) TEL is a sequence-specific transcriptional repressor. J. Biol. Chem., 274, 30132–30138. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307. [DOI] [PubMed] [Google Scholar]
- Papadopoulos P., Ridge,S.A., Boucher,C.A., Stocking,C. and Wiedemann,L.M. (1995) The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res., 55, 34–38. [PubMed] [Google Scholar]
- Peeters P. et al. (1997) Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood, 90, 2535–2540. [PubMed] [Google Scholar]
- Peterson A.J., Kyba,M., Bornemann,D., Morgan,K., Brock,H.W. and Simon,J. (1997) A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol., 17, 6683–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. (1998) Polycombing the genome: PcG, trxG, and chromatin silencing. Cell, 93, 333–336. [DOI] [PubMed] [Google Scholar]
- Poirel H., Lopez,R.G., Lacronique,V., Della Valle,V., Mauchauffé,M., Berger,R., Ghysdael,J. and Bernard,O.A. (2000) Characterization of a novel ETS gene, TELB, encoding a protein structurally and functionally related to TEL. Oncogene, 19, 4802–4806. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. (1995) SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci., 4, 1928–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M.D., Buijs,A., Kreider,B., van Rompaey,L. and Grosveld,G.C. (2000) Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood, 95, 3341–3348. [PubMed] [Google Scholar]
- Romana S.P., Mauchauffe,M., Le Coniat,M., Chumakov,I., Le Paslier, D., Berger,R. and Bernard,O.A. (1995) The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood, 85, 3662–3670. [PubMed] [Google Scholar]
- Rubnitz J.E., Pui,C.H. and Downing,J.R. (1999) The role of TEL fusion genes in pediatric leukemias. Leukemia, 13, 6–13. [DOI] [PubMed] [Google Scholar]
- Salomon-Nguyen F., Della-Valle,V., Mauchauffe,M., Busson- Le Coniat,M., Ghysdael,J., Berger,R. and Bernard,O.A. (2000) The t(1;12)(q21;p13) translocation of human acute myeloblastic leukemia results in a TEL-ARNT fusion. Proc. Natl Acad. Sci. USA, 97, 6757–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Ponting,C.P., Hofmann,K. and Bork,P. (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci., 6, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupsky C.M., Gentile,L.N., Donaldson,L.W., Mackereth,C.D., Seidel,J.J., Graves,B.J. and McIntosh,L.P. (1998) Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc. Natl Acad. Sci. USA, 95, 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla M., Schmieder,P., Kelly,M., Ter Laak,A., Krause,G., Ball,L., Wahl,M., Bork,P. and Oschkinat,H. (1999) Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci., 8, 1954–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton D., Balan,I., Pawson,T. and Sicheri,F. (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nature Struct. Biol., 6, 44–49. [DOI] [PubMed] [Google Scholar]
- Szymczyna B.R. and Arrowsmith,C.H. (2000) DNA-binding specificity studies of 4 ETS proteins support an ‘indirect read-out’ mechanism of protein–DNA recognition. J. Biol. Chem., 275, 28363–28370. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos C.D. and Bowie,J.U. (1999) p53 family members p63 and p73 are SAM domain-containing proteins. Protein Sci., 8, 1708–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos C.D., Faham,S., Goodwill,K.E., Cascio,D., Phillips,M. and Bowie,J.U. (1999a) Monomeric structure of the human EphB2 sterile α motif domain. J. Biol. Chem., 274, 37301–37306. [DOI] [PubMed] [Google Scholar]
- Thanos C.D., Goodwill,K.E. and Bowie,J.U. (1999b) Oligomeric structure of the human EphB2 receptor SAM domain. Science, 283, 833–836. [DOI] [PubMed] [Google Scholar]
- Tsien R.Y. (1998) The green fluorescent protein. Annu. Rev. Biochem., 67, 509–544. [DOI] [PubMed] [Google Scholar]
- Tu H., Barr,M., Dong,D.L. and Wigler,M. (1997) Multiple regulatory domains on the Byr2 protein kinase. Mol. Cell. Biol., 17, 5876–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H., Downing,J.R., Miyazaki,Y., Frank,R., Zhang,J. and Nimer,S.D. (1999) Three distinct domains in TEL-AML1 are required for transcriptional repression of the IL-3 promoter. Oncogene, 18, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Waldo G.S., Standish,B.M., Berendzen,J. and Terwilliger,T.C. (1999) Rapid protein-folding assay using green fluorescent protein. Nature Biotechnol., 17, 691–695. [DOI] [PubMed] [Google Scholar]