Abstract
To understand better the control of DNA methylation, we cloned and characterized the dim-2 gene of Neurospora crassa, the only eukaryotic gene currently known in which mutations appear to eliminate DNA methylation. The dim-2 gene is responsible for methylation in both symmetrical and asymmetrical sites. We mapped dim-2 between wc-1 and un-10 on linkage group (LG) VIIR and identified the gene by RFLP mapping and genetic complementation. Dim-2 encodes a 1454 amino acid protein including a C-terminal domain homologous to known DNA methyltransferases (MTases) and a novel N-terminal domain. Neither a deletion that removed the first 186 amino acids of the protein nor a mutation in a putative nucleotide binding site abolished function, but a single amino acid substitution in the predicted catalytic site did. Tests for repeat-induced point mutation (RIP) indicated that dim-2 does not play a role in this process, i.e. duplicated sequences are mutated in dim-2 strains, as usual, but the mutated sequences are not methylated, unlike the situation in dim-2+ strains. We conclude that dim-2 encodes an MTase that is responsible for all DNA methylation in vegetative tissues of Neurospora.
Keywords: dim-2/DNA methyltransferase/methylation mutant/Neurospora/repeat-induced point mutation
Introduction
DNA methylation is common in organisms as diverse as bacteria, fungi, higher plants and mammals. In eukaryotes, methylation is almost exclusively found at the C5 position of selected cytosines; and in many organisms, nearly all 5mC is found in symmetrical sites, such as the sequence 5′CpG/3′GpC. This is thought to reflect the operation of a ‘maintenance DNA methyltransferase (MTase)’ that preferentially recognizes and methylates hemimethylated sites (Holliday and Pugh, 1975; Riggs, 1975). Methylation patterns are consistently maintained, but it is common to find cytosines that are ‘partially’ methylated, i.e. methylated in some fraction of a cell population. The status of any given site presumably reflects a combination of de novo methylation, maintenance methylation and demethylation, e.g. by failure to methylate DNA after replication. Remarkably little is known about what determines whether a particular sequence or site is subject to these processes in eukaryotes. This has motivated us to study control of methylation in a model system, the filamentous fungus Neurospora crassa.
The function of DNA methylation is somewhat better understood than its control. It is known that DNA methylation can indirectly interfere with transcription initiation (Kass et al., 1997; Schubeler et al., 2000) or transcription elongation (Barry et al., 1993; Rountree and Selker, 1997), and methylation is involved in a range of epigenetic phenomena, such as X chromosome inactivation (Beard et al., 1995; Panning and Jaenisch, 1996; Heard et al., 1997), genomic imprinting (Li et al., 1993; Peterson and Sapienza, 1993; Bartolomei and Tilghman, 1997) and silencing of selfish DNA (Bestor, 1996; Selker, 1997). DNA methylation is required for normal development in both mammals and plants. Methylation deficiencies in mice homozygous for mutations in either of two MTase genes, Dnmt1 (Li et al., 1992) or Dnmt3b (Okano et al., 1999), lead to lethality during embryonic development. Mutations in a third MTase known to be active in mouse, Dnmt3a, cause mice to become runted and to die at 4 weeks of age (Okano et al., 1999). In humans, mutations in DNMT3B are associated with an autosomal recessive disorder, ICF syndrome (Hansen et al., 1999; Okano et al., 1999; Xu et al., 1999). In the plant Arabidopsis thaliana, reduction in DNA methylation, caused by expression of a MTase (MET1) antisense construct or by a mutation in a modifier gene (ddm1), results in flowering abnormalities, including homeotic transformations of floral organs and partial female sterility (Finnegan, 1996; Kakutani et al., 1996; Ronemus et al., 1996). The various consequences of reduced DNA methylation in animals and plants may result from expression of genes normally silenced by methylation.
One approach to elucidating the control and function of DNA methylation is to identify and characterize the components of the methylation machinery, starting with the MTases. The first eukaryotic DNA MTase to be characterized, DNMT1, was isolated from mouse (Bestor et al., 1988). Although DNMT1 appears responsible for the maintenance of most DNA methylation in mammals, mutant cells that produce no DNMT1 retain a low level of methylation and are capable of de novo methylation (Li et al., 1992; Lei et al., 1996). Three other potential DNA MTase genes were found in mouse and human databases: Dnmt2 (Okano et al., 1998; Yoder and Bestor, 1998), and Dnmt3a and Dnmt3b (Okano et al., 1998). Biochemical studies demonstrated that Dnmt3a and Dnmt3b are bona fide DNA MTases and genetic studies suggested that these two enzymes are responsible for de novo methylation in embryonic cells (Hsieh, 1999; Okano et al., 1999; Robertson et al., 1999). Sequence conservation among DNA MTases facilitated isolation of putative MTase genes from several other model eukaryotes, including A.thaliana (Finnegan and Dennis, 1993; Scheidt et al., 1994; Henikoff and Comai, 1998; Genger et al., 1999), Xenopus laevis (Kimura et al., 1996), Ascobolus immersus (Chernov et al., 1997; Malagnac et al., 1997, 1999), Aspergillus nidulans (D.W.Lee, M.Freitag, E.U.Selker and R.Aramayo, personal communication), the fission yeast Schizosaccharomyces pombe (Wilkinson et al., 1995) and Drosophila melanogaster (Hung et al., 1999; Tweedie et al., 1999; Gowher et al., 2000; Lyko et al., 2000). Some of these organisms lack obvious DNA methylation, suggesting that their MTase-like sequences may represent pseudogenes. Some putative DNA MTase genes from organisms with obvious methylation may also be inactive or relatively insignificant. For example, disruption of the known Ascobolus DNA MTase-like genes (masc1 and masc2), singly or together, does not noticeably affect methylation in this fungus, and attempts to detect MTase activity in vitro from Masc1 have failed (Malagnac et al., 1997).
Genetic approaches to identifying the components of the methylation machinery offer notable advantages over molecular approaches. First, no prior knowledge about the identity of the ‘players’ is required. Thus, a mutant hunt for genes that affect DNA methylation may uncover novel MTases as well as unexpected components of the methylation machinery. Secondly, by definition, genetic approaches reveal genes that have a recognizable phenotype. This makes it possible to distinguish between genes that are major players and genes with minor roles (and pseudogenes). To date, DNA methylation mutants have been successfully isolated in A.thaliana (Vongs et al., 1993) and the filamentous fungus N.crassa (Foss et al., 1993, 1995, 1998). In the case of Arabidopis, this approach yielded mutants in two genes, a previously identified DNA MTase, MET1 (E.Richards, personal communication), and DDM1 (decrease in DNA methylation; Kakutani et al., 1995, 1996, 1999; Jeddeloh et al., 1999), a protein related to the yeast SWI/SNF2 family of chromatin-remodeling proteins. Mutant hunts in Neurospora have revealed a greater number of genes involved in DNA methylation—five so far—but with the exception of the gene described here, their identities are not yet known. One possible reason why genetic studies have revealed more methylation genes in Neurospora than in plants or animals is that DNA methylation appears to be non-essential in this organism: mutations in dim-2 (defective in methylation) appear to eliminate all DNA methylation without causing growth defects (Foss et al., 1993). This is consistent with the idea that the primary function of DNA methylation in Neurospora is to control ‘selfish’ DNA. Although a sizeable fraction of the cytidines in the wild-type genome of Neurospora is methylated (∼1.5%; Foss et al., 1993), all known functional genes are unmethylated. Methylation appears concentrated in relics of RIP (repeat induced point mutation), a genome defense system that operates specifically in the haploid nuclei of dikaryotic cells formed by fertilization (Selker et al., 1987a; Grayburn and Selker, 1989; Selker, 1990; Kinsey et al., 1994; Perkins et al., 1997; Margolin et al., 1998). RIP searches for linked and unlinked sequence duplications, such as those that result from transposons, and then introduces numerous G:C to A:T transitions in both copies of the duplicated sequences (Selker and Garrett, 1988; Cambareri et al., 1989). All duplicated sequences longer than ∼1 kb are efficiently inactivated by the mutations and/or by methylation, which is typically found on remaining cytosines in sequences heavily mutated by RIP. The relationship between RIP and DNA methylation is not well understood. On the one hand, it has been shown that RIP frequently creates ‘signals’ for de novo cytosine methylation that cause the mutated sequences, and some surrounding sequences, to be methylated in vegetative tissues (Selker et al., 1993; Singer et al., 1995b; Irelan and Selker, 1997; Miao et al., 2000). On the other hand, it has been suspected that methylation of cytosines may also be a part of the RIP mutagenic mechanism in the sexual dikaryotic cells (Selker, 1990). Consistent with this possibility, an analogous process called MIP (methylation induced premeiotically), which operates in the fungus A.immersus, results in methylation of cytosines without concomitant mutations in duplicated sequences (Rhounim et al., 1992; Rossignol and Faugeron, 1994). Interestingly, inactivation of the Ascobolus DNA MTase-like gene masc1 interferes with MIP, but, as noted above, does not appear to affect DNA methylation in vegetative tissues (Malagnac et al., 1997).
As a step towards a better understanding of the control of DNA methylation in eukaryotes, we cloned and characterized the dim-2 gene of N.crassa, currently the only gene known in which mutations appear to abolish all DNA methylation. Loss of all methylation is particularly striking in Neurospora because this organism normally has 5mC in both symmetrical and asymmetrical sites (Selker and Stevens, 1985; Selker et al., 1993). We wished to determine whether dim-2 encodes a key regulator of the methylation machinery and/or encodes a novel methyltransferase. We also addressed the role of Dim-2 in the RIP process. We report that dim-2 encodes a novel MTase responsible for all DNA methylation that has been detected in N.crassa, and that the dim-2 MTase plays no role in mutagenesis by RIP.
Results
Previous work has shown that the original mutation in dim-2 eliminated methylation and allowed transcription of genes that were methylated after the action of RIP (Foss et al., 1993; Rountree and Selker, 1997). Nevertheless, neither the original dim-2 mutation identified in a heavy chemical mutagenesis (Foss et al., 1993), nor an allele identified in a subsequent screen for spontaneous mutants (J.Irelan, A.Tharp, P.Paine and E.U.Selker, unpublished), display a strong phenotype that would allow for selection of transformants complementing the mutation. We therefore decided to clone the dim-2 gene by positional cloning using cosmid, lambda and YAC libraries and to test candidate clones for complementation of the methylation defect of a dim-2 strain. As the first step, we built dim-2/dim-2+ heterokaryons to verify that the original mutation could be complemented (Figure 1). To test for induction of methylation in dim-2 nuclei, the mtrRIP8 allele, which is methylated in dim-2+ strains (Figure 1B, lanes 1 and 2), was included in the dim-2 partner, and an mtr deletion was included in the dim-2+ partner (Figure 1A). Heterokaryons with various ratios of dim-2+/dim-2 nuclei (1:1 to 1:200) were constructed and tested for methylation of mtrRIP8 by Southern hybridization. Substantial, but incomplete methylation was detected in all of the heterokaryons, indicating that the dim-2 allele can be complemented and that the product of the dim-2 gene is not nucleus limited (Figure 1B, lanes 5–12). This result suggested that dim-2+ nuclei generated by transformation should be detectable even if most of the nuclei in the dim-2 strain were untransformed.
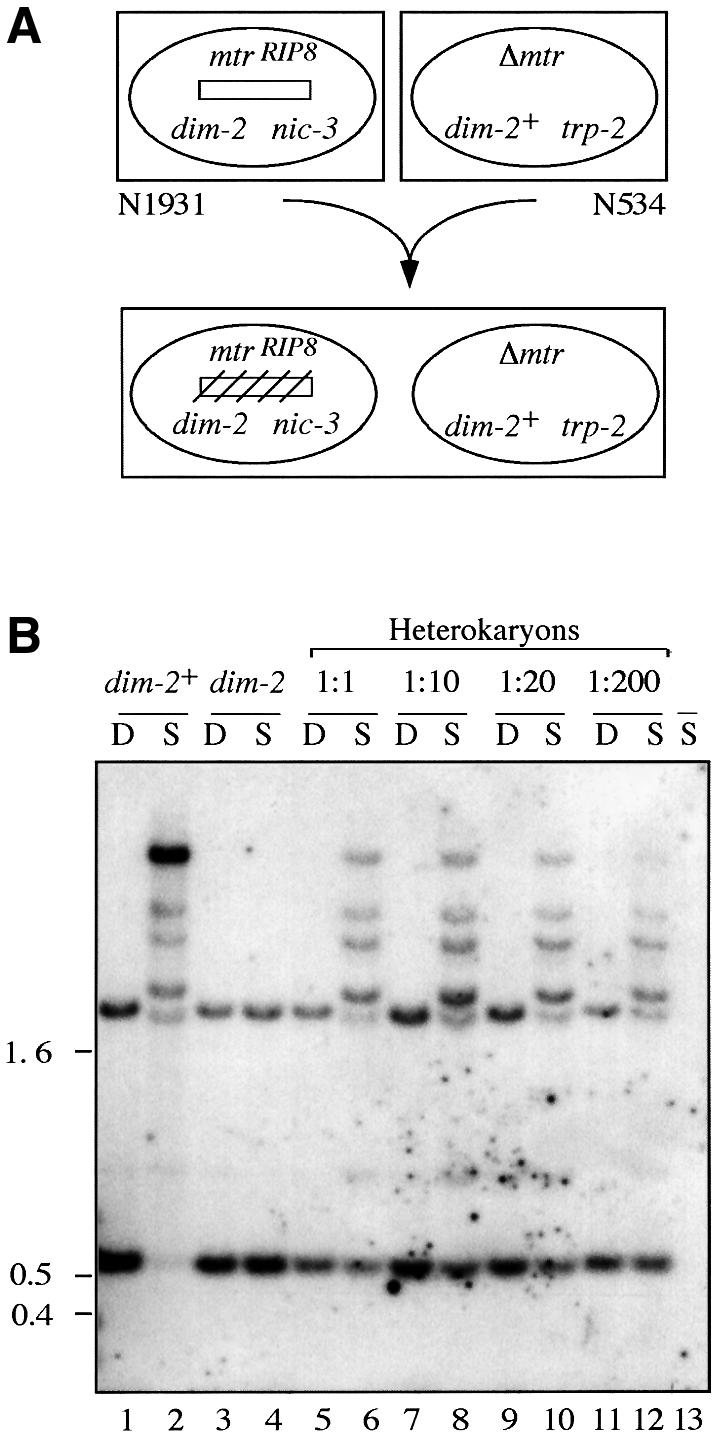
Fig. 1. The dim-21 mutation is complemented in heterokaryons. (A) Schematic diagram of the experiment. Two compatible auxo trophic N.crassa strains (N1931 and N534) were forced to form a heterokaryotic strain on minimal medium. The initially unmethylated mtrRIP8 allele (open rectangle) of the dim-2 nucleus became methylated (crossed-hatched rectangle) in the heterokaryon. (B) Southern analysis of methylation at mtrRIP8 in the dim-2; mtrRIP8/Δmtr heterokaryon. Heterokaryons were formed by mixing N534 (col-4 mtrSR62; trp-2) and N1931 (mtrRIP8; nic-3 wc-1 dim-21) in liquid minimal medium in the indicated ratios. Genomic DNA was purified, digested with either DpnII (D) or Sau3AI (S), fractionated, blotted and hybridized to an mtr probe. Lanes 1 and 2, DNA sample from N1930 (a control for methylation at mtrRIP8 in dim-2+ background); lanes 3 and 4, DNA sample from N1931; lanes 5–12, DNA sample from heterokaryons; lane 13, DNA sample from N534. All the bands except for the two that also show up in DpnI digests (e.g. lane 1) reflect methylation. The position of size standards (kb) is indicated on the left.
Mapping and positional cloning of the dim-2 gene
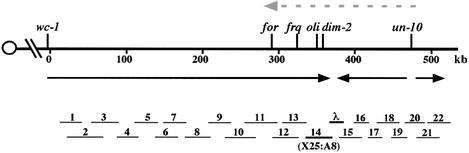
To isolate the dim-2 gene by positional cloning, we first mapped it genetically. Linkage tests with various tester strains revealed that dim-2 is linked to csp-2 on linkage group (LG) VII (Kouzminova, 2000). Results of a three-point cross with wc-1 and arg-10 mapped dim-2 between these markers on the right arm of LG VII (∼10 cM to the right of wc-1 and ∼6 cM to the left of arg-10); other crosses confirmed that dim-2 is between wc-1 and un-10. Based on published genetic maps and contigs established in previous walks in the region (Cabibbo et al., 1991; Ballario et al., 1996), we initiated what should have been converging chromosome walks from wc-1 and un-10. We discovered, however, that the previously published genetic map has an inversion between un-10 and for (Kouzminova and Selker, 1999). This led us to reverse the orientation of our chromosome walk from un-10 (Figure 2). We covered the 550 kb interval between wc-1 and un-10 and confirmed that the previously published genetic map is incorrect—the correct order of the genes in the region being wc-1, for, frq, oli, un-10. Before attempting to locate dim-2 by complementation, we assigned the gene to an individual cosmid within the contig by RFLP mapping as described in Materials and methods. Our results indicated that dim-2 must be either at the very right end of cosmid X25:A8 or on lambda clone λ25:A8 (Figure 2, step 14 and λ).
Fig. 2. Chromosome walk to the dim-2 gene. The physical map of LG VIIR is shown with the centromere indicated by the open circle. The inverted order of the for, frq, oli, dim-2 and un-10 loci, relative to a previous map, is indicated by the dashed gray arrow above the map. Arrows below the physical map show the extent and orientation of our chromosomal walks from wc-1 and un-10. Line segments below the map indicate positions of the key cosmid (1–22) and λ clones identified in the dim-2 chromosome walk. The identities of all clones are listed in Materials and methods.
Identification of the dim-2 gene by complementation
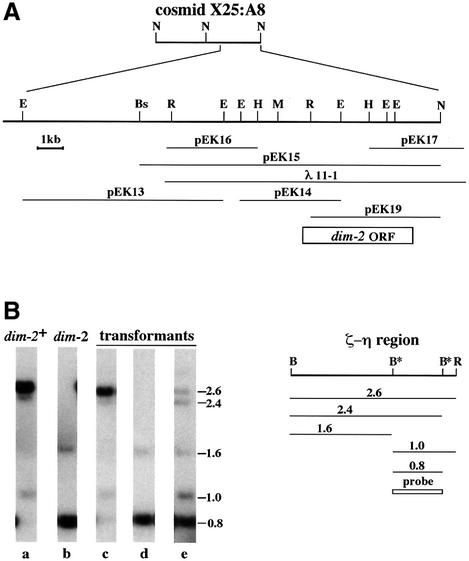
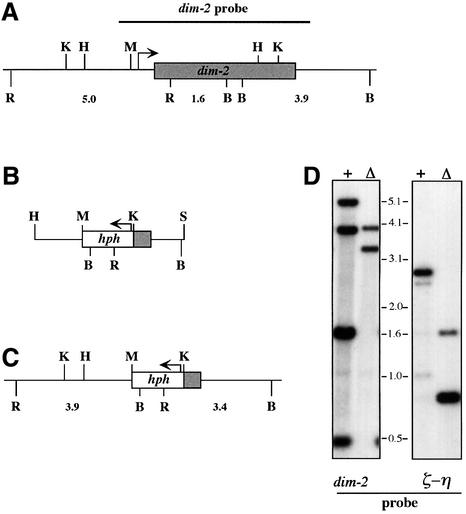
We transformed a dim-2 strain (N1257) with cosmid X25:A8 and analyzed 20 transformants by Southern hybridization to test for methylation at two BamHI sites that are normally methylated in the zeta-eta (ζ-η) region (Selker et al., 1987b) (Figure 3B). Two transformants showed partial restoration of methylation at both BamH1 sites in the region, confirming that this cosmid contains the dim-2 gene (data not shown). To map further dim-2, we divided X25:A8 into two 18–20 kb NotI fragments (Figure 3A) and introduced them separately into the dim-2 strain. Two of 12 transformants generated with the larger NotI fragment showed unambiguous complementation of the methylation defect. The complementing fragment was further subcloned to yield plasmids pEK13, pEK14, pEK15 and pEK19 (Figure 3A). We also tested for complementation with lambda clones λ25:A8 and λ11-1. The latter gave complementation and was subcloned to generate plasmids pEK16 and pEK17. Full complementation was achieved in transformants with either pEK15 or λ11-1 (Figure 3B, compare lanes a and c), while partial complementation, revealed as weak 2.6 and 2.4 kb bands, was found with pEK19 (Figure 3B, compare a and e).
Fig. 3. Complementation of dim-2 mutation. (A) Physical map of cosmid X25:A8 and related DNA segments. The positions of the dim-2 ORF (open box) and of restriction sites for NotI (N), EagI (E), Bsp120 I (Bs), EcoRI (R), HindIII (H) and MluI (M) are shown. (B) Southern analysis of methylation in the ζ-η region. The positions of EcoRI (R) and BamHI (B) restriction sites in the region are shown, and the size (kb) and origin of fragments that can be detected with the indicated probe are represented below the map. Sites that are sometimes methylated, and thus not digested, are marked with asterisks. EcoRI/BamHI digests of DNA from the following strains were probed to assess methylation at the ζ-η BamHI sites: a, wild-type (N150); b, dim-21 (N1257); c–e, N1257 transformed with pEK15, pEK14 and pEK19, respectively.
The dim-2 gene encodes a putative DNA (cytosine 5) methyltransferase
We sequenced the 5 kb insert in pEK19, and the adjacent 4 kb of DNA in pEK15, and found an open reading frame (ORF) with a region of strong similarity to the catalytic domains of known eukaryotic and prokaryotic MTases (Figure 4). The sequence of the dim-2 gene suggested that it contains a single 64 nucleotide intron. The 5′- and 3′- splice junctions, as well as the internal sites of the predicted intron, fit the consensus for N.crassa introns (Bruchez et al., 1993a). Using a cDNA library made from conidial RNA (Nelson et al., 1997), we identified and sequenced cDNA clones for the gene, confirming that the ORF is transcribed and that the predicted intron is indeed absent from the polyadenylated mRNA (data not shown). Translation of the dim-2 transcript probably initiates at the first ATG of the ORF. An in-frame stop codon is located 18 nucleotides before this codon, and the sequence surrounding it is consistent with the conserved context of the known initiator codons in N.crassa (Bruchez et al., 1993b). Three other candidate translation initiator codons are found near the beginning of the ORF (underlined in Figure 4).
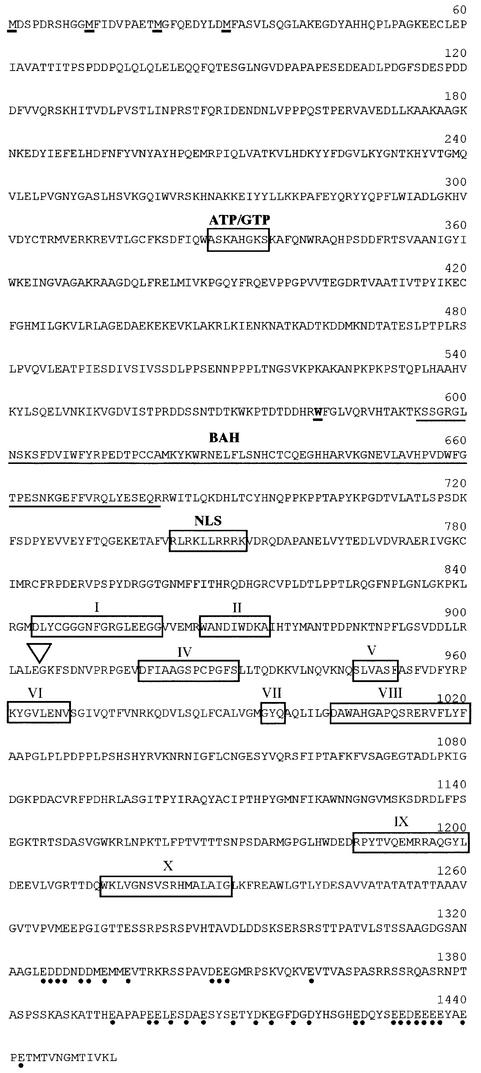
Fig. 4. The predicted amino acid sequence of the Dim-2 polypeptide. Four potential initiation methionines (M) are underlined. The boxed regions labeled with Roman numerals indicate motifs characteristic of DNA MTases (Kumar et al., 1994). A putative nuclear localization signal (NLS) and a nucleotide (ATP/GTP) binding motif are also boxed, and a bromo adjacent homology (BAH) domain (Callebaut et al., 1999) predicted by PFAM and SMART protein prediction web servers is underlined. The position of the only intron in the gene is indicated by the inverted triangle. The tryptophan (W) at amino acid position 581 that is mutated to a stop codon in the original dim-21 allele is shown in bold and underlined. The acidic amino acids in the C-terminus are indicated by dots.
The presumptive Dim-2 protein contains 1454 amino acids (160 kDa), including a C-terminal 600 amino acid segment that appears to correspond to the catalytic domain of known MTases (Figure 5). The six most conserved motifs (I, IV, VI, VIII, IX, X) of the 10 that characterize DNA MTases, as first defined with prokaryotic MTases (Posfai et al., 1989; Kumar et al., 1994), are prominent in the C-terminal domain (Figures 4 and 5). Sequence comparisons of the presumptive catalytic domains of known MTases, carried out using the BLAST 2 program (Tatusova and Madden, 1999), suggest that Dim-2 (amino acids 840–1454) is most similar to known and presumed MTases from plants, mammals and the fungus A.immersus. In the conserved regions of the C-terminal domain, Dim-2 shows 32–34% identity and 48–52% similarity, relative to the corresponding regions of Ascobolus Masc2, Arabidopsis MET1 and mouse Dnmt1. Interestingly, these three MTases show 49–64% identity and 63–80% similarity among themselves (Figure 5). The N-terminal domains of known eukaryotic MTases are much less conserved than the C-terminal domains, but some MTases show similarities (Colot and Rossignol, 1999). The large N-terminal region of the predicted Dim-2 protein does not show significant similarity to any known protein in current databases, although it does show a few sequence features, including a bromo adjacent homology (BAH) domain (Callebaut et al., 1999) (amino acids 594–681), a putative nuclear localization signal (NLS), RLRKLLRRRK (amino acids 742–751) and a ‘Walker A’ ATP/GTP-binding motif (Walker et al., 1982) (amino acids 327–334; Figure 4).
Fig. 5. Sequence alignment of the C-terminal domains of Mus musculus Dnmt1 (DDBJ/EMBL/GenBank accession No. P13864), Ascobulus immersus Masc2 (accession No. CAB09661), Arabidopsis thaliana MET1 (accession No. P34881) and Neurospora crassa Dim-2 (accession No. AF348971). Green boxes indicate amino acid identity in all four sequences, yellow boxes indicate amino acid identity in three out of four sequences, blue boxes indicate conservative changes, and dashes indicate gaps introduced by the CLUSTAL_W algorithm to maximize alignment (Thompson et al., 1994). The numbers on the left correspond to amino acid positions in the protein sequences. Six of the 10 conserved motifs defined previously based on prokaryotic MTases (Kumar et al., 1994) are indicated by brackets and Roman numerals above the aligned sequences. Pairwise BLAST 2 comparisons of the underlined regions demonstrated that Dim-2 is the most divergent MTase in the group. The expected values obtained from comparisons of Dim-2 to Masc2, Dnmt1 and MET1 are 3e–45, 1e–42 and 3e–44, respectively; expected values from comparisons of Masc2 to Dnmt1 and MET1 are 6e–84 and 5e–75, respectively; the expected value from the comparison of Dnmt1 to MET1 is 1e–110.
Mapping the dim-2 transcript
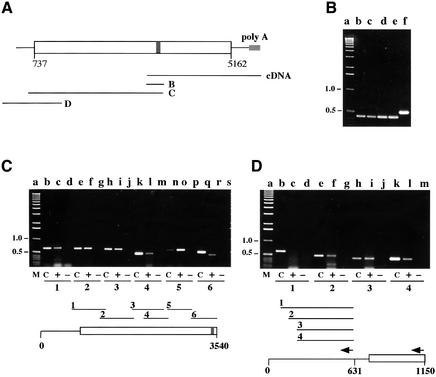
We had a clue from our complementation tests that the N-terminal 186 amino acids of Dim-2 are not essential: pEK19 transformants with only 3.9 of the 4.4 kb Dim-2 ORF showed partial complementation of the dim-2 mutation (sample transformant shown in Figure 3B). The partial complementation by pEK19 could reflect truncation of the Dim2 protein and/or absence of regulatory sequences from the transforming DNA. The pEK19 transformant shown in Figure 3 is one of two with the strongest complementation. No complementation was achieved when the same dim-2 segment was targeted to the his-3 locus (data not shown), consistent with the idea that transcription of the pEK19 sequences was driven by promoters that by chance were near the arbitrary genomic integration sites. As a step to determine whether the full dim-2 ORF is expressed in wild-type cells, we investigated the length of the dim-2 transcript. No signals were obtained for dim-2 in northern hybridizations using as much as 20 µg of polyA-RNA prepared from a wild-type strain (N150) grown for either 7 or 24 h from conidia, whereas control probings for the oli and am genes produced strong hybridization signals (data not shown), suggesting that the dim-2 transcript is rare. Further evidence that dim-2 is not highly expressed came from screening cDNA libraries; only two dim-2 clones, with 1.1 and 2.5 kb inserts, were found among 40 000 conidial cDNA clones (Nelson et al., 1997) screened, and only one clone (NMB5-9; 0.9 kb insert) was identified in N.crassa EST databases (see http://molbio.ahpcc.unm.edu/search/ngp.html). To determine whether the dim-2 transcript is long enough to encode the full predicted dim-2 ORF, we performed RT–PCR using polyA-RNA from a wild-type strain (N150). The dim-2 transcript was detected in non-germinated conidia (Figure 6B, lane b) as well as in growing vegetative cells (at least up to 24 h of growth; Figure 6B, lane e). Using random primers for the RT reaction, and a variety of PCR primers, we confirmed that the transcript includes the first ATG of the dim-2 ORF (Figure 6C). Thus, the transcript is long enough to be translated into the longest predicted Dim-2 protein. To map the 5′ end of the message, we employed specific primers for RT reactions (Figure 6D). The RT–PCR analysis also confirmed the absence of additional introns in the ORF and leader of the dim-2 transcript. The total length of the dim-2 transcript is at least 5.3 kb and appears to include an ∼700 nucleotide 5′ untranslated sequence and a 276 nucleotide 3′ untranslated sequence.
Fig. 6. Analysis of dim-2 transcription by RT–PCR. PCR products were fractionated on 1.1% agarose gels. In each panel, the numbering of the sets of lanes indicates the gene segments tested, as illustrated below. In each set, ‘C’ (control) indicates that PCR was performed on genomic DNA, ‘+’ indicates that RT was included in the RT reaction, while ‘–’ indicates that RT was omitted. The primers are described in Materials and methods. (A) Diagram of the dim-2 gene showing the ORF (starting at nucleotide 737 relative to the MluI site; open rectangle) and intron (nucleotides 3449–3512 relative to the MluI site; filled rectangle). Lines B, C and D represent regions tested by RT–PCR in the corresponding panels. The line marked ‘cDNA’ represents the longest identified dim-2 cDNA. (B) Expression of dim-2 in dormant conidia (lane b), and in conidia germinated for 3 (lane c), 7 (lane d) or 24 h (lane e). The RT reaction was performed with random hexamer primers using polyA-RNA from strain N150. The subsequent PCR was for the intron-containing region B, using primers #715 and #716. Lane f shows the PCR product from genomic DNA and lane a contains size markers. (C) Mapping the upstream portion of the dim-2 transcript. RT reactions were performed with random hexamer primers. The following primer pairs were used for PCR products 1–6, respectively: #717/#692; #691/#728; #690/#684; #720/# 721; #693/#686; and #715/#716. (D) Mapping the 5′ end of the dim-2 transcript. The RT reaction was performed with primer #713 (indicated by arrow left of position 631) and subsequent PCR reactions to generate products 1–3 were performed with primer pairs #713/#765, #713/#768 and #713/#712, respectively. Product 4 was obtained using primer #714 (indicated by arrow left of position 1150) in the RT reaction and primer pair #713/#712 for the subsequent PCR.
Characterization of the dim-21 allele and construction of a deletion allele
The unique, predicted large N-terminal domain of Dim-2 might confer a function distinct from the presumed MTase activity of the protein. This possibility prompted us to consider that the original mutation, dim-21, may not have eliminated all Dim-2 function(s) even though it appears to prevent all cytosine methylation. To address this possibility we isolated and sequenced the dim-21 allele as described in Materials and methods. Analysis of nucleotides 431–5009 revealed a single, G→A mutation at nucleotide position 2449, ∼40% into the ORF. The mutation changed the tryptophan codon at amino acid position 581 into a stop codon (Figure 4). Thus, the dim-21 mutant should produce a truncated protein lacking the predicted catalytic domain and the putative NLS.
As a step to investigate further the function of the N-terminal segment of the Dim-2 protein, which is potentially expressed in the dim-21 mutant, we constructed a dim-2 null allele by gene replacement (Figure 7). To do so, we transformed Neurospora with a construct in which the N-terminal region, and most of the C-terminal domain of dim-2, was replaced with the bacterial hph gene driven by the A.nidulans trpC promoter (Cullen et al., 1987). We then screened 105 hygromycin resistant transformants by Southern hybridization and found several in which homologous recombination resulted in replacement of the native dim-2 gene. The autoradiogram in the left panel of Figure 7D verifies the structure of one such strain (allele designated Δdim-22), and the autoradiogram in the right panel demonstrates the absence of methylation in the ζ-η region. Examination of several other loci that are normally methylated [e.g. ψ63 (Margolin et al., 1998) and rDNA (Russell et al., 1985)], and also of total genomic DNA, by comparison of ethidium bromide-stained Sau3AI and DpnII digests, confirmed the absence of methylation in this strain (data not shown). The Δdim-22 allele, like the dim-21 allele, confers no obvious additional phenotypes in either the vegetative or sexual phase of the life cycle (see also ‘RIP in the dim-2 mutants’ below). This suggests that the original dim-21 mutant and the Δdim-22 mutant have equivalent defects.
Fig. 7. Construction and characterization of the Δdim-22 allele. The gray and open bars represent the dim-2 and hph ORFs, respectively, and the arrows indicate the direction of transcription. (A) Restriction map of the dim-2+ allele showing sites for BamHI (B), HindIII (H), KpnI (K), MluI (M) and EcoRI (R), and various fragment lengths below (kb). (B) Diagram of the transforming SphI (S)–HindIII fragment of pEK22 with most of dim-2 replaced by the hph gene. (C) Structure of the resulting Δdim-22 allele. (D) Southern analysis of dim-2+ and ζ-η regions in wild-type (N150; +) and Δdim-2 (N1877; Δ) strains using EcoRI and BamHI (see Figure 3).
Construction and analysis of Dim-2 variants
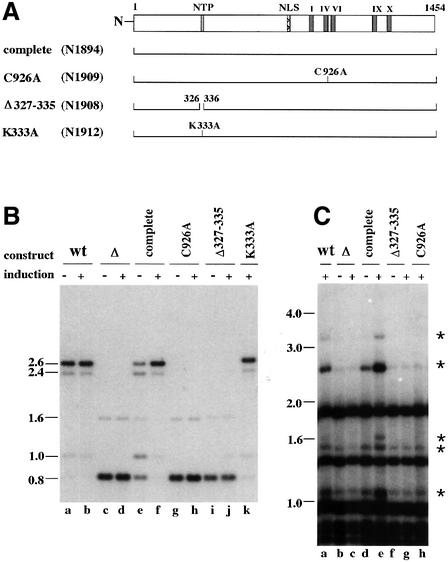
The striking similarity between the C-terminal 600 amino acid region of Dim-2 and known eukaryotic DNA MTases strongly suggests that Dim-2 is a DNA MTase, consistent with the phenotype of dim-2 strains. It remained possible, however, that Dim-2 worked in a complex with an unknown DNA MTase that was directly responsible for the methylation defect of dim-2 mutants. Attempts to detect the Dim-2 MTase using antibodies prepared against a segment of the protein produced in E.coli, or using epitope-tagged Dim-2, failed, perhaps because Dim-2 is present in very low levels in the cell, consistent with the low level of its mRNA. This hampered the testing of variant forms of the protein. The best we could do was to test all constructs as single copies, integrated by homologous recombination at a common chromosomal site under the regulation of a common promoter. We were able to verify that all constructs were expressed identically at the transcriptional level (data not shown). To test specifically that Dim-2 was a DNA MTase rather than a regulator of another MTase, we created and examined the effect of a single amino acid substitution in the putative catalytic site. The cysteine in the PC doublet of MTase motif IV serves as the catalytic nucleophile (Santi et al., 1983), as demonstrated, for example, by high-resolution structural data showing this cysteine linked to C6 of the cytosine subject to methylation (Klimasauskas et al., 1994). We therefore replaced cysteine 926 in the conserved PC with an alanine residue (construct C926A) and compared this strain (N1909) with a control strain (N1894) that was identical except that it lacked the amino acid substitution (Figure 8A). No methylation was observed in the mutant strain, while full methylation was restored in the control strain (Figure 8B, compare lanes e and g with lane a, and f and h with lane b; Figure 8C, compare lanes e and h with lane a). A similar construct, bearing a different single amino acid substitution, also complemented the dim-2 mutation, unlike the construct with the change in the presumed catalytic site (see below). This demonstration that substitution of a single amino acid in the putative catalytic site prevents methylation renders the idea that Dim-2 is a regulator of another MTase extremely unlikely.
Fig. 8. DNA methylation in Δdim-2 strains expressing various dim-2 constructs. All mutant constructs were driven by the N.crassa qa-2 promoter, which is inducible by quinic acid (Geever et al., 1989), and they were targeted to the his-3 locus (Margolin et al., 1997) of strain N1877 (Δdim-22) as single copies. All constructs contain 94 nucleotides upstream and 1 kb downstream of the dim-2 ORF sequence. Strains were grown for 41 h in minimal medium with 1.5% sucrose in the presence (+) or absence (–) of 0.4% quinic acid as an inducer. Expression of all constructs was confirmed by RT–PCR (data not shown). (A) Structure of dim-2 mutant constructs. The diagram at the top shows the position of an ATP/GTP binding motif (NTP), a putative nuclear localization signal (NLS) and the conserved MTase motifs of the Dim-2 protein (I, IV, VI, IX and X). The following Dim-2 constructs are shown: complete, full dim-2 ORF; C926A, substitution of cysteine 926 with alanine in the conserved PC dipeptide of motif IV; Δ327–335, nine amino acid deletion removing the NTP binding motif, and K333A, substitution of lysine 333 with alanine. The numbering refers to amino acid positions relative to the predicted N-terminus. (B) Southern analysis of DNA methylation at the ζ-η locus in mutant constructs introduced into a Δdim-2 host strain. DNA of wild-type control (N150; lanes a and b), the Δdim-2 host strain (N1877; lanes c and d), a transformant with the complete gene (N1894; lanes e and f), a transformant with construct C926A (N1909; lanes g and h), a transformant with NTP binding motif deletion (N1908; lanes i and j) and a transformant with construct K333A (N1912; lane k) were digested with EcoRI and BamHI and probed with the 0.8 kb BamHI fragment from the ζ-η region (see Figure 3). (C) Southern analysis of rDNA using Sau3AI and a 9 kb KpnI fragment carrying the entire rDNA unit as the probe. Lanes: a, wild-type strain (N150); b and c, Δdim-2 host strain (N1877); d and e, transformant (N1894) with the complete construct; f and g, transformant (N1908) with a deletion of the NTP binding motif; and h, transformant (N1909) with C926A construct. The membrane was initially probed with a fragment of the am gene (Rountree and Selker, 1997) to verify complete digestion of all DNA samples with Sau3AI. Asterisks indicate bands that become more prominent due to methylation in strain N1894 under inducing conditions.
To explore the possible importance of the ATP/GTP binding motif found in Dim-2, we first constructed a mutant (N1908, Δ327–335; Figure 8A) with a nine amino acid deletion including the motif (ASKAHGKS). Unlike control strains grown under identical conditions, the Δ327–335 construct did not rescue the methylation deficiency of the dim-2 mutant (Figure 8B, compare lanes j and f; Figure 8C, compare lanes g and e), suggesting that the nine amino acid segment is essential for the activity, stability or conformation of Dim-2. To test more specifically the possible role of the nucleotide binding motif, we replaced the lysine in the GKS sequence with an alanine, generating construct K333A. Mutation analyses of NTP-binding domains in various proteins has revealed that this lysine is important for the coordination of the nucleotide (Hung et al., 1998; Iaccarino et al., 1998). The K333A construct restored methylation (Figure 8B, lane k), suggesting that NTP binding is not necessary for Dim-2 activity.
Dim-2 is not involved in the mechanism of RIP
As discussed in the Introduction, the relationship between RIP and DNA methylation is not well understood. It is commonly assumed that the mutations by RIP result from methylation of cytosines followed by deamination, and it has been proposed that the deamination itself might be catalyzed by a MTase or a MTase-like enzyme (Selker, 1990). The Dim-2 MTase was an obvious candidate for one or both hypothetical activities. We therefore examined the possible involvement of Dim-2 in RIP by looking at inactivation of the mtr gene in crosses of dim-2+ and dim-2– strains (N1255, N1258 and N1259) bearing an mtr duplication. The duplication was created by crossing an mtr+ strain with a strain bearing a deletion of the native mtr gene but having an ectopic copy of mtr. The four possible crosses between strains carrying the dim-2+ or dim-21 alleles were performed (Table I). Because the frequency of RIP is known to be low in ascospores produced early (Singer et al., 1995a), we collected and analyzed spores produced at both 11 days (‘early’) and 17 days (‘late’) after fertilization. Duplications of mtr were inactivated by RIP at a frequency of 18–34% among early ascospores, compared with 77–87% among late ascospores (Table I). No effect of the dim-21 allele on the frequency of RIP was detected at either time point, even in crosses homozygous for dim-21. Since the dim-21 allele can not encode a functional MTase, it seems unlikely that the Dim-2 MTase participates in either of the postulated steps of RIP.
Table I. Testing effect of the dim21 allele on RIP.
| Cross | Relevant genotypea | RIP frequency (%)b |
|
|---|---|---|---|
| 11 daysc | 17 days | ||
| N1259 × N1257 | dim-2, mtr+/mtr+ × dim2 | 18.9 ± 0.4 (135) | 86.9 ± 5.4 (256) |
| N1255 × N1150 | mtr+/mtr+ × (+) | 22.6 (47) | 76.9 (52) |
| N1258 × N1150 | dim-2, mtr+/mtr+ × (+) | 17.2 ± 5.7 (157) | 80.8 ± 5.8 (208) |
| N1255 × N1257 | mtr+/mtr+ × dim2 | 34.6 (52) | 80.8 (51) |
adim-2 designates the dim-21 allele and mtr+/mtr+ designates a duplication of mtr involving an ectopic copy of mtr plus the native allele. When not listed, the alleles of dim-2 and mtr are wild type (+). The female strains carried the duplication.
bRIP frequency was calculated as a ratio of Mtr– progeny to a half of the total progeny. For crosses that were repeated several times, the average and standard error are shown. The number of progeny analyzed in the crosses is indicated in parentheses.
cAscospores were harvested at the indicated time after fertilization.
To address the possibility that Dim-2 is involved in RIP through its N-terminal domain, which may be produced by the dim-21 allele, we constructed an unlinked duplication of am in strains that had either the wild-type or the Δdim-22 allele (strains N1879 and N1880). We crossed am non-duplication strains with strains carrying the am duplication in various combinations (Table II). RIP of the am duplication was assayed by loss of am function, and progeny from each of two crosses were tested for RFLPs resulting from RIP. In control crosses, in which both parents had the dim-2+ allele, the RIP frequency was 64 and 72% in ascospores produced 20 days post-fertilization. The frequency of RIP was 24–56% in crosses heterozygous for the Δdim-2 allele and 52–56% in crosses homozygous for this null allele (Table II). The observed variation in RIP frequency is expected from variations in the strain backgrounds (our unpublished results). The lack of a noticeable effect of the dim-2 deletion on RIP is consistent with the results from the mtr duplication and dim-21 allele. We conclude that Dim-2 is not involved in the mechanism of RIP.
Table II. Testing effect of the Δdim 22 allele on RIP.
| Cross | Relevant genotypea | RIP (%)b | hph (%)c |
|---|---|---|---|
| N1879 × N1851 | Δdim-2, am/am × Δdim-2 | 52 | |
| N1879 × N150 | Δdim-2, am/am × (+) | 48 | 42 |
| N1880 × N1851 | am/am × Δdim-2 | 56 | 58 |
| N1880 × N150d |
am/am × (+) |
72 |
|
| N1851 × N1879 | Δdim-2 × Δdim-2, am/am | 56 | |
| N150 × N1879 | (+) × Δdim-2, am/am | 24 | 38 |
| N1851 × N1880 | Δdim-2 × am/am | 40 | 44 |
| N150 × N1880d | (+) × am/am | 64 |
aΔdim-2 designates the Δdim-22 allele; am/am designates an am gene duplication resulting from an ectopic copy of am plus the native allele. When not listed, the alleles of dim-2 and am are wild type (+). The female strains are listed first.
bRIP was calculated in all crosses for 50 random progeny as the ratio of Am– progeny to one half of the number of total progeny.
cProgeny were tested for independent segregation of the hph marker (expectation: 50%), which was inserted at dim-2 in building the dim-2 deletion allele.
dProgeny from crosses with the strain N1880 were tested by Southern analysis for RFLP with BamH1 and EcoRI to verify the occurrence of RIP.
Discussion
Although the function of DNA methylation is not fully understood in any eukaryote, it is clearly responsible for gene silencing in a variety of organisms, including Neurospora (Cambareri et al., 1996; Irelan and Selker, 1997; Rountree and Selker, 1997; Zhou et al., 2001). The Neurospora dim-2 mutant is the only eukaryotic mutant presently known that appears to be completely devoid of DNA methylation (Foss et al., 1993). We cloned the dim-2 gene to identify its product, which was clearly a key component of the DNA methylation machinery. To do so, we first mapped the locus to the right arm of LG VII between wc-1 and un-10 and built a 550 kb contig spanning the wc-1 to un-10 interval. During the course of this work we uncovered an inversion of the published genetic map between un-10 and for. After localizing the dim-2 gene to a small region by RFLP mapping, we identified it unambiguously by complementation. Sequencing revealed a 1454 amino acid ORF ending in a 600 amino acid domain showing obvious homology to known prokaryotic and eukaryotic DNA MTases. Results from RT–PCR analyses indicated that the dim-2 transcript is at least 5.3 kb long, consistent with the long N-terminal region of the ORF. Other known eukaryotic DNA MTases also have substantial N-terminal domains that are not well conserved, unlike their C-terminal domains, e.g. mammalian DNMT1 MTases have an ∼1000 amino acid N-terminal domain (Colot and Rossignol, 1999). Work in the last few years has identified several proteins that interact with the N-terminal domain of mouse and/or human (Yen et al., 1992) DNMT1, including the replication processivity factor PCNA (proliferating cell nuclear antigen; Chuang et al., 1997), histone deacetylases HDAC1 (Fuks et al., 2000; Robertson et al., 2000) and HDAC2 (Rountree et al., 2000), the tumor suppressor protein Retinoblastoma (Rb; Robertson et al., 2000), the transcriptional activator E2F1 (Robertson et al., 2000), the transcriptional co-repressor DMAP1 (Rountree et al., 2000) and the methyl-binding domain protein MBD3 (Tatematsu et al., 2000). Curiously, the N-terminal part of the dim-2 ORF shows no significant similarity to any gene in current public databases. Analysis of mutant dim-2 alleles constructed in vitro, and targeted precisely to the his-3 locus, suggested that MTase function is abolished by an alanine to cysteine substitution in the presumptive catalytic domain but not by a single amino acid substitution at the conserved lysine in a putative NTP-binding motif. A deletion that would remove 186 amino acids from the N-terminus of the predicted protein did not abolish function. In contrast, deletions of the entire N- or C-terminal domains did not give detectable dim-2 function (our unpublished observations). Although we demonstrated equivalent expression of all constructs at the transcriptional level, we did not succeed in producing antibodies that could recognize the Dim-2 MTase, or derivatives thereof. Thus, it is possible that the segments of Dim-2 that failed to function in vivo were unstable or not properly localized. Nevertheless, it is interesting that our results are consistent with reports that the C-terminal domain of the Dnmt1 MTase, when expressed alone in COS-7 cells, lacks catalytic activity (Zimmermann et al., 1997; Margot et al., 2000). The bulk of the N-terminal domain of Dnmt1, except for the first 300 amino acids, seems to be required for methyltransferase activity, e.g. as assayed in vitro with a poly(dI–dC) template.
Notable sequence features of the Dim-2 MTase in addition to the predicted catalytic site of MTase motif IV and the ‘Walker A’ (NTP-binding) motif, include MTase motifs I, VI, IX and X, a putative nuclear localization signal (NLS) and a bromo-adjacent homology (BAH) region (Callebaut et al., 1999) in the N-terminal region. Except for the putative NTP-binding site, these motifs have been found in several other known, and presumed, DNA MTases (Colot and Rossignol, 1999). It is worth noting that the dim-2 BAH domain is a worse match to the consensus sequence (expected value: 9e–5) than those of mouse Dnmt-1 (expected value: 4.8e–37), Arabidopsis Met-1 (expected value: 1.5e–55) and Ascobolus Masc-2 (expected value: 1e–44 ). Dim-2 lacks other obvious motifs found in some eukaryotic MTases such as an PCNA-binding motif and dipeptide (e.g. GK) repeats. Unlike some MTases, Dim-2 has a 220 amino acid segment downstream of the conserved MTase motifs (Kumar et al., 1994; Colot and Rossignol, 1999). This segment shows no obvious homology to other known proteins, but, like the C-terminus of Masc2 of Ascobolus (Chernov et al., 1997), one of the closest relatives of Dim-2 currently known, this segment is rich in acidic amino acids (Figure 4).
Of the approximately two dozen eukaryotic DNA MTases and putative DNA MTases described to date, Dim-2 appears to be the most divergent. The length of the N-terminal region and the sequence of the C-terminal region make Dim-2 most similar to the Dnmt1 subfamily of DNA MTases, which are regarded as maintenance MTases (Colot and Rossignol, 1999). The Dim-2 N-terminus lacks significant sequence homology with N-termini of other described Dnmt1-like MTases, however. Furthermore, as mentioned above, Dim-2 lacks the dipeptide repeats that link the N- and C-terminal domains of all previously described Dnmt1-like MTases. Thus, the lack of homology in its N-terminal domain and its lack of repeats distinguishes Dim-2 from the MTases of the Dnmt1 subfamily. Moreover, pairwise comparisons of the conserved regions of Dim-2 and representatives of the most closely related DNA MTases (Arabidopsis MET1, mouse Dnmt1 and Ascobolus Masc2) using BLAST 2, revealed that Dim-2 is substantially less similar to these MTases than they are to each other (Figure 5). The expected values obtained from comparisons of Dim-2 to Masc2, Dnmt1 and MET1 are 3e–45, e–42 and 3e–44, respectively, whereas comparisons of Masc2 and Dnmt1, Masc2 and MET1, and Dnmt1 and MET1 gave expected values of 6e–84, 5e–75 and 1e–110, respectively. The observation that Dim-2 does not appear more similar to the fungal MTase (Masc2) than known plant MTases (especially that from carrot; DDBJ/EMBL/GenBank accession No. AF007807), suggests that Masc2 is not a true ortholog of dim-2. Consistent with this hypothesis is the observation that disruption of Masc2 does not affect methylation in Ascobolus (Malagnac et al., 1999).
Our finding that Dim-2 is responsible for all apparent DNA methylation in Neurospora raises the possibility that this single MTase is capable of both de novo and maintenance methylation. This contrasts with the situation in the mammals and plants that have been examined, which each sport several MTases, consistent with the idea that DNA methylation patterns in higher eukaryotes are established and propagated by separate enzymes. Although it is not certain that any eukaryotic DNA MTase is exclusively devoted to de novo or maintenance methylation, biochemical and genetic data have provided evidence that some MTases, such as Dnmt1 in mammals, are devoted primarily, or exclusively, to maintenance methylation, while others, such as Dnmt3a and Dnmt3b in mammals, are devoted to de novo methylation (Bestor, 1992; Okano et al., 1998; Hsieh, 1999; Lyko et al., 1999; Okano et al., 1999; Howell et al., 2001). It is generally assumed that maintenance methylation depends on the symmetry of methylated sites (e.g. CpGs in mammals and CpGs and CpXpGs in plants), and that methylation at non-symmetrical sites, such as is prevalent in Neurospora, is due to de novo methylation (Selker and Stevens, 1985; Selker et al., 1987b). This view is supported by a variety of observations, including the recent observation of non-CpG methylation by the candidate de novo MTase Dnmt3a (Ramsahoye et al., 2000). Nevertheless, there are clues that this view is overly simplistic. For example, the strict maintenance model does not easily account for the heterogeneity in methylation patterns commonly observed in plant, animal and fungal systems.
The fact that mutations in dim-2 promptly and completely eliminate DNA methylation, and that methylation is promptly restored after introduction of dim-2+ DNA into dim-2 strains, argues that Dim-2 is a de novo MTase. Nevertheless, Dim-2 may also be capable of maintenance methylation. Evidence of maintenance methylation in Neurospora has come from two studies. In a survey of am alleles generated by RIP, some exceptional, lightly mutated, methylated alleles were found that were not subject to de novo methylation after demethylation by either of two methods (Singer et al., 1995b). A similar observation was made with the bacterial antibiotic resistance gene hph located between copies of am that had been subjected to RIP (Irelan and Selker, 1997). Re-establishment of methylation (and silencing) was incomplete after treatments causing demethylation, unlike the situation with most methylated sequences in Neurospora, such as sequences heavily mutated by RIP. The implication was that some of the original methylation of hph was due to maintenance methylation. In both cases, the methylation was established in the sexual cycle and was stably maintained in vegetative cells, but could not be re-established in vegetative cells after demethylation. The methylation was not limited to symmetrical sites, implying that methylation can promote subsequent methylation in a different way than that predicted by the original maintenance methylation model (Holliday and Pugh, 1975; Riggs, 1975). The fact that residual methylation has not been observed in a dim-2 strain isolated from a Dim+ dim-2/dim-2+ heterokaryon (M.Freitag and E.U.Selker, unpublished observation) suggests that the Dim-2 MTase may be responsible for both maintenance and de novo methylation in a variety of sequence contexts. Biochemical studies on Dim-2 should help to define the substrate specificity of this enzyme and detailed genetic studies should help to identify functions of its long N-terminal domain and acidic C-terminal tail.
The relationship between cytosine methylation and RIP is intriguing but not fully understood. What is certain is that many sequences mutated by RIP serve as excellent substrates for de novo methylation in vivo. Evidence of methylation induced in the sexual phase of the Neurospora life cycle is consistent with the possibility of an additional connection between RIP and methylation, namely that the mechanism of RIP involves methylation of cytosines. Unfortunately, because of technical difficulties involved in analyzing the minuscule ascogenous tissue in which RIP takes place, it has not yet been possible to determine unequivocally whether or not methylation occurs coincidently with RIP. Curiously, preliminary analysis of DNA methylation in the ascogenous tissues indicates that at least some sites in the sequences that are methylated in vegetative tissues lose methylation during the sexual cycle (data not shown). Moreover, we presented genetic evidence indicating that the Dim-2 MTase does not participate in the RIP mechanism. As pointed out previously, based on chemical principles, it seems possible that RIP results from deamination of cytosines catalyzed by an MTase-like protein (Selker, 1990). Perhaps a sexual phase-specific MTase, or MTase-like protein, is responsible for RIP. Support for this possibility comes from the discovery of Masc1, a putative MTase in the fungus A.immersus that appears to play a role in the RIP-like process called MIP, but which does not appear to play a role in DNA methylation in vegetative cells (Malagnac et al., 1997). A Neurospora homolog of Masc1 has recently appeared in an N.crassa database (DDBJ/EMBL/GenBank accession No. AL442164.1) and experiments are under way to determine whether or not this gene plays a role in RIP. The Neurospora genome sequence is nearly complete, and no additional DNA MTase-related protein is found in the database (http://waldo.wi.mit.edu/annotation/fungi/neurospora), consistent with our conclusion that dim-2 is responsible for all known methylation in this organism.
Materials and methods
Strains, media, growth conditions and transformation procedures
Neurospora strains used in this study are described in Table III and were handled according to standard procedures (Davis, 2000). In general, liquid cultures of Neurospora were grown at 32°C for 2 days with vigorous shaking in Vogel’s medium (Davis, 2000) with 1.5% sucrose and required supplements. For quinic acid induction of Pqa-2dim-2 constructs, 30 ml liquid cultures were grown from an inoculum of 1 × 106 conidia/ml for 41 h at 32°C with shaking (130 rpm) in Vogel’s medium with 1.5% sucrose and 0.4% quinic acid pH 6. Hygromycin B (Calbiochem) was used at 200 µg/ml to select for the hph gene (Staben et al., 1989). Scoring for the following N.crassa genes was as described previously: wc-1, un-10 and arg-10 (Perkins et al., 1982); am (Kinsey et al., 1980); and mtr (Irelan et al., 1994).
Table III. Neurospora strains.
| Name | Genotype | Sourcea |
|---|---|---|
| N1 | a | FGSC 988 |
| N51 | A | FGSC 2225 |
| N150 | A | FGSC 2489 |
| N268 | am132 amecT–510 5.6 inl; a | Selker et al. (1988) |
| N534 | col-4 mtrSR62; trp-2; a | D.Stadler |
| N581 | col-4 mtrSR33; (mtr+/hph+)ecpAH33; trp-2; a | lab collection |
| N623 | his-3; A | FGSC 6103 |
| N1150 | lys-1; A | lab collection |
| N1255 | (mtr+/hph+)ecpAH33; a | this study |
| N1257 | dim-21; A | lab collection |
| N1258 | (mtr+/hph+)ecpAH33; dim-21; a | this study |
| N1259 | (mtr+/hph+)ecpAH33; dim-21; a | this study |
| N1847 | wc-1 frq10 dim-21 arg10; a | this study |
| N1850 | Δdim-22; a | this study |
| N1851 | Δdim-22; A | this study |
| N1877 | Δdim-22; his-3 a | this study |
| N1879 | amecT–510 5.6; Δdim-22; a | this study |
| N1880 | AmecT–510 5.6; a | this study |
| N1892 | Δdim-22; his-3+::dim-2pEK20–4 a | this study |
| N1894 | Δdim-22; his-3+::dim-2pEK24–1 a | this study |
| N1908 | Δdim-22; his-3+::dim-2pEK30–12 a | this study |
| N1909 | Δdim-22; his-3+::dim-2pEK32–2 a | this study |
| N1912 | Δdim-22; his-3+::dim-2pEK34 a | this study |
| N1930 | mtrRIP8; lys-1A | this study |
| N1931 | mtrRIP8; nic-3 wc-1 dim-21; a | this study |
aStrains with FGSC designations are from the Fungal Genetics Stock Center (Department of Microbiology, University of Kansas Medical Center, Kansas City, KS 66103).
Escherichia coli strains were normally grown in liquid or solidified Luria broth supplemented with ampicillin (200 µg/ml) as necessary. Plasmids were propagated in E.coli strain DH5αF′. Phage lambda was propagated in E.coli strain A585 (C600 recD1009 SuII SuIII; kindly provided by Frank Stahl, University of Oregon), grown in tryptone broth according to standard procedures (Ausubel et al., 1998).
Transformation of Neurospora strain N1257 was performed by electroporation (Margolin et al., 1997) using 1 µg of cosmid DNA or, for cotransformation experiments, a mixture of 0.25 µg of pCSN43 (Staben et al., 1989) and 1 µg of lambda DNA or 0.25 µg of pEK19. Transformants were isolated and propagated on minimal medium supplemented with hygromycin. To target DNA to the his-3 locus of strains N1877, plasmids were linearized with AflIII. Successful gene replacements were verified and analyzed by Southern analysis.
Neurospora crassa recombinant DNA libraries
Two N.crassa genomic DNA libraries were used primarily in the chromosome walk: the Orbach/Sachs cosmid library (Orbach, 1994) and a λJ1 library provided by the Fungal Genetic Stock Center (FGSC; University of Kansas Medical Center, Kansas City). These, as well as conidial (C1) and mycelial (M1) cDNA libraries (Nelson et al., 1997) were obtained from the FGSC. Cosmid libraries were replica-plated on LB plates covered with nylon membranes (Hybond-N; Amersham Pharmacia Biotech). Membranes with E.coli colonies or phage plaques were processed as recommended by the manufacturer. The cosmids and lambda clones identified in the dim-2 chromosome walk and described in Figure 2 are: 1, G7:G3; 2, G21:F8, G14:C1, 56:E7a; 3, X11:G8, X14:B3; 4, X3:B5, G11:G3; 5, G23:F12, G18:H18; 6, G12:D7, G3:C9, G5:C10; 7, G4:A3, G9:E9, X4:C5, X14:A5; 8, X9:G1, X3:A4, X6:G10, X12:C2; 9, X13:G3, X17:G9, X17:F12, G14:F11, G20:D9, G23:F9; 10, X5:G3, X15:D7, X19:H11, G6:G3; 11, G19:D1, G10:H7, X16:C9; 12, X15:E7; 13, G12:B8, G1:C1, G15:B2, G20:A9, G22:F2, G22:C11, X21:F1; 14, X25:A8; 15, cos2-10Ab; 16, G2:F8, X18:D6; 17, G1:H8, X20:E3; 18, G6:H11, G19:C12, X20:E8; 19, G17:D7, G24:F3, G2:F3; 20, cos10:12Eb, G17:C7, X20:C9; 21, G17:C9, X8:H8, 55:E3a; 22, X10:C4, X24:C10, G7:F11, G16:E5, 26:A8a, λ– λ25:A8, λ3–1, λ7–1, λ11–1. All cosmids were from the Orbach/Sachs library (Orbach, 1994), except those marked with a or b superscripts, which are from the CBM1 (Cabibbo et al., 1991) and Vollmer/Yanofsky (Vollmer and Yanofsky, 1986) libraries, respectively.
RFLP mapping
At each step in the chromosome walks, the cosmid chosen to be the next probe was RFLP mapped using 38 meiotic progeny from ordered asci from a cross between Oak Ridge (OR) and Mauriceville (M) strains (Metzenberg et al., 1985), to verify that it was specific for LG VIIR. DNA samples were digested with EcoRI, fractionated by gel electrophoresis and probed with radiolabeled total cosmid DNA. To assign a given cosmid to a particular chromosomal region, its pattern of inheritance was compared with that of previously mapped sequences on the seven chromosomes of Neurospora (Nelson and Perkins, 2000).
The position of dim-2 in the cosmid contig was determined by employing 45 recombinant strains in the frq to arg-10 interval from a cross between an OR lineage strain (N1847) and an M lineage strain (N51; Table III). DNA prepared from these strains was digested with EcoRI, BglII, HindIII or DpnII and analyzed by Southern hybridization with probes from cosmid or lambda clones in the region. Two recombinants proved to be the most informative: #12, which is wc-1 frq10 dim-2+arg-10+ and showed M-type RFLPs to the right of oli, indicating that the recombination occurred close to oli, and that the dim-2 gene is to the right of oli; #16, which is wc-1 frq10 dim-21 arg-10+ and showed M-type RFLPs to the right of step 15 of the walk, placing dim-2 to the left of step 15 (Figure 2).
Nucleic acid isolation, DNA hybridization and RT–PCR analysis
Plasmids and cosmids were purified by standard procedures such as the alkaline lysis procedure (Birnboim and Doly, 1979). Lambda DNA was isolated as described elsewhere (Kouzminova, 2000). Neurospora genomic DNA was isolated as described previously (Foss et al., 1993), as was total RNA (Luo et al., 1995). Poly(A)-RNA was prepared using the PolyATract mRNA Isolation System III (Promega).
Southern hybridization analyses were performed as described previously (Irelan and Selker, 1997) using DNA probes prepared by priming with random hexamers (Feinberg and Vogelstein, 1984).
For RT reactions, 1 µg of poly(A)-RNA or total RNA was first treated with DNAse I according to the protocol of the manufacturer (Gibco-BRL). First-strand cDNA synthesis was performed at 37°C for 60 min with MMLV RT according to the protocol of the supplier (Promega). PCR was carried out in 100 µl of 1× Promega Taq polymerase buffer supplemented with 1.5 mM MgCl2, 200 µM dATP, dCTP, dGTP and TTP, 0.4 µM of each primer, 2.5 U of Taq polymerase (Promega) and either 10 ng of plasmid DNA or 100 ng of genomic DNA. PCR were carried out using a Hybaid Omnigene thermocycler as follows: 94°C for 4 min, followed by 30 cycles of [94°C for 4 s, 50–54°C for 10 s, 72°C for 30 s per 1 kb], followed by 72°C for 5 min. Pfu polymerase was used according to the protocol of the manufacturer (NEB). PCR reactions described in Figure 6 were carried out with the following primer pairs: #717/#692, 5′-GCCATGGATTCGCCAGATCGC-3′/5′-CTTGGTCGC TACAAGCTG-3′; #691/#728, 5′-CATTGATCTCCTTCCAGA-3′/5′-TACCGCCTCCGCAGTCAAC-3′; #690/#684, 5′-GTTTGAACTGTC ATCTCG-3′/5′-TCTGGAAGGAGATCAATG-3′; #720/#721, 5′-TGG CAGGGGAGGACGCCG-3′/5′-CTGGCTCAACCAATGAGG-3′; #693/#686, 5′-CATAATGCACTTGCCTAC-3′/5′-GAGTACATACCGCCA AGA-3′; #715/#716, 5′-GCCTGGGCACGTTGTCGGAG-3′/5′-GTC TATACCGAAGATTTGGTG-3′; #713/#765, 5′-AGGTATTCAGGG ATGTCTTGG-3′/5′-CGAGGAAGTTCCGTTCGCTGC-3′; #713/#768, 5′-AGGTATTCAGGGATGTCTTGG-3′/5′-GTGGGCAAGGGAGGG GAAC-3′; #713/#712, 5′-AGGTATTCAGGGATGTCTTGG-3′/5′-ATT CCATCCGATCCAAGTTCG-3′). For the RT reaction described in Figure 6D the primer #714, 5′-GGTCGATACTGGTAGATCGAC-3′, was used.
Plasmid constructions
The largest NotI fragment of cosmid X25:A8 was cloned into the NotI site of pMOcosX (Orbach, 1994) to make pX25L. pEK13, pEK14 and pEK15 were constructed by cloning the 8 kb EagI–EagI fragment, the 4 kb EagI–EagI fragment or the 12 kb NotI–Bsp120I fragment from cosmid X25:A8, respectively, into NotI-digested pMOcosX. pEK16-1 and pEK17 were constructed by cloning the 4.3 and 4.8 kb HindIII fragments from λ11–1, respectively, into the HindIII site of pCSN43 (Staben et al., 1989). pEK19 was constructed by cloning the 4.9 kb NotI–EcoRI fragment from cosmid X25:A8 into pBluescriptII SK(+) (Stratagene, La Jolla, CA) digested with NotI and EcoRI. pEK21 was constructed by cloning the 3.1 kb BglII–HindIII fragment from pEK15 between the BamHI and HindIII sites of pBR322. To construct pEK22, a 2.2 kb PCR fragment generated from pEK19 [using primers #694 (5′-CTGCCTTCAAGTTCGTCA-3′) and #707 (5′-AACCTGCATGC TATGACCATGATTACGC-3′) and Pfu DNA polymerase] was digested with SphI and KpnI to yield a 1.7 kb fragment (containing the 3′ part of the dim-2 ORF) that was ligated with the 1.6 kb KpnI–MluI fragment of pCSN43 (containing the hph gene), and inserted into pEK21 digested with SphI and MluI. To build pEK24, the 0.9 kb EcoRI–XmaI fragment (containing the qa-2 promoter) from pMYX2 (Campbell et al., 1994) was ligated to the 0.9 kb BglII–NgoMIV fragment from pEK14 (starting 94 nucleotides upstream of the ATG codon in the dim-2 ORF) and the combined fragment was inserted into pEK20 digested with BglII and EcoRI. pEK20 was constructed by cloning the 4.9 kb NotI–EcoRI fragment from pEK19 into pBM61 (Margolin et al., 1997) digested with NotI and EcoRI. pEK27 was constructed by cloning the 2134 bp, dim-2-containing SphI–XmaI fragment from pEK19 into pMTL21 (Chambers et al., 1988) digested with SphI and XmaI. pEK28-33 was built from pEK27 by PCR with Pfu polymerase using primers #777 (5′-CCACTGGATGAAGTCGCTC-3′) and #778 (5′-AAGGCTTTC CAGAACTGGC-3′) and ligation. The product of the ligation reaction was digested with DpnI to destroy traces of the pEK27 template. A 2107 bp SphI–XmaI fragment of pEK28-33 was sequenced to verify the presence of the 27 bp deletion (corresponding to the ASKAHGKSK amino acid sequence of Dim-2) and the absence of mutations in the amplified sequence. pEK30–5 was constructed by cloning the 2107 bp SphI–XmaI fragment of pEK28–33 into pEK24 digested with SphI and XmaI, and the construct verified by DNA sequencing. pEK31 was constructed by PCR from pEK27 using Pfu polymerase and primers M13F (5′-GTAAAACGACGGCCAGT-3′) and #782 (5′-GTACCCGGG AGCAGGGCTACC-3′; the underlined sequence, which corresponds to the complement of an alanine codon, replaces a cysteine codon in the wild-type dim-2 sequence). The PCR product was digested with XmaI and SphI and inserted between sites for these enzymes in pMTL21. pEK32 was constructed by cloning the 2134 bp SphI–XmaI fragment from pEK31 into pEK24 digested with SphI and XmaI, and confirmed by sequencing. pEK34 was constructed by PCR overlap extension mutagenesis, using the primers #687 (5′-TCGAGGACGCCATACTTG-3′), #856 (5′-CGCATG GGGCGTCAAAGGCTTTCC-3′), #728 (5′-TACCGCCTCCGCAGT CAAC-3′) and #857 (5′-GGAAAGCCTTTGACGCCCCATGCG-3′), to replace the lysine at position 333 of Dim-2 with an alanine nine (the codon is underlined). The PCR fragments obtained with primer pairs #687/#856 and #728/#857 were used as templates in a PCR step using primers #728 and #687. The final PCR product was digested with XmaI and SphI and inserted between sites for these enzymes in pEK24. The 2.1 kb SphI–XmaI fragment of pEK32 and pEK34 was sequenced to verify the intended mutations and to rule out the occurrence of chance mutations in the amplified sequences.
DNA sequence analyses
The 4.8 kb dim-2 region (nucleotides 690–5500 in Figure 6) was sequenced on both strands from plasmids pEK15 and pEK19 on an ABI 377 automated sequencer at the University of Oregon Biotech Facility. The cDNA clone λ12-1-1 (Figure 6A) was also sequenced. To characterize the dim-21 allele of strain N1257, PCR fragments were amplified from genomic DNA and sequenced directly. Similarly, genomic DNA of strain N1912 was amplified (primers #691, #728) and sequenced to verify the presence of the alanine codon. To safeguard against mutations potentially introduced by PCR, the products of five independent 20 µl PCR reactions were pooled for each sequencing reaction. The PCR products were fractionated in 1% agarose and gel purified with a QIAquick Gel Extraction Kit (QIAGEN). Both strands of DNA were sequenced in the PCR fragment found to carry the mutation (primers: 5′-CATAATGCACTTGCCTAC-3′ and 5′-TCTGGAAGG AGATCAATG-3′). Computer-based sequence analyses were carried out using MacVector/AssemblyLIGN and GCG software (Oxford Molecular). The wild-type dim-2 sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. AF348971.
Acknowledgments
Acknowledgements
We thank M.Freitag for confirming that dim-2 strains do not maintain pre-existing methylation. We also thank M.Freitag, G.Kothe and A.Kuzminov for miscellaneous advice and comments on the manuscript, H.Foss for her enthusiasm for the project, K.Hicks for advice on positional cloning and G.Macino for providing clones to orient the cosmid walk. This study was supported by a grant from the National Institutes of Health (GM35690).
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1998) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY.
- Ballario P., Vittorioso,P., Magrelli,A., Talora,C., Cabibbo,A. and Macino,G. (1996) White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J., 15, 1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Barry C., Faugeron,G. and Rossignol,J.-L. (1993) Methylation induced premeiotically in Ascobolus: Coextension with DNA repeat lengths and effect on transcript elongation. Proc. Natl Acad. Sci. USA, 90, 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M.S. and Tilghman,S.M. (1997) Genomic imprinting in mammals. Annu. Rev. Genet., 31, 493–525. [DOI] [PubMed] [Google Scholar]
- Beard C., Li,E. and Jaenisch,R. (1995) Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev., 9, 2325–2334. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. (1992) Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J., 11, 2611–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T.H. (1996) DNA methyltransferases in mammalian development and genome defense. In Russo,V.E.A., Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 61–76.
- Bestor T., Laudano,A., Mattaliano,R. and Ingram,V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol., 203, 971–983. [DOI] [PubMed] [Google Scholar]
- Birnboim H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchez J.J., Eberle,P.J. and Russo,V.E.A. (1993a) Regulatory sequences in the transcription of Neurospora crassa gene: CAAT box, TATA box, introns, poly(A) tail formation sequences. Fungal Genetics Newsl., 40, 89–96. [Google Scholar]
- Bruchez J.J., Eberle,P.J. and Russo,V.E.A. (1993b) Regulatory sequences involved in the translation of Neurospora crassa mRNA: Kozak sequences and stop codons. Fungal Genetics Newsl., 40, 85–88. [Google Scholar]
- Cabibbo A., Sporeno,E., Macino,G. and Ballario,P. (1991) CBM1, a Neurospora crassa genomic library in pAC3 and its use for walking on the right arm of linkage group VII. Fungal Genetics Newsl., 38, 68–70. [Google Scholar]
- Callebaut I., Courvalin,J.C. and Mornon,J.P. (1999) The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett., 446, 189–193. [DOI] [PubMed] [Google Scholar]
- Cambareri E.B., Jensen,B.C., Schabtach,E. and Selker,E.U. (1989) Repeat-induced G-C to A-T mutations in Neurospora. Science, 244, 1571–1575. [DOI] [PubMed] [Google Scholar]
- Cambareri E.B., Foss,H.M., Rountree,M.R., Selker,E.U. and Kinsey,J.A. (1996) Epigenetic control of a transposon-inactivated gene in Neurospora is dependent on DNA methylation. Genetics, 143, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.W., Enderlin,C.S. and Selitrennikoff,C.P. (1994) Vectors for expression and modification of cDNA sequences in Neurospora crassa. Fungal Genetics Newsl., 41, 20–21. [Google Scholar]
- Chambers S.P., Prior,S.E., Barstow,D.A. and Minton,N.P. (1988) The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene, 68, 139–149. [DOI] [PubMed] [Google Scholar]
- Chernov A.V., Vollmayr,P., Walter,J. and Trautner,T.A. (1997) Masc2, a C5-DNA-methyltransferase from Ascobolus immersus with similarity to methyltransferases of higher organisms. Biol. Chem., 378, 1467–1473. [DOI] [PubMed] [Google Scholar]
- Chuang L.S., Ian,H.I., Koh,T.W., Ng,H.H., Xu,G. and Li,B.F. (1997) Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science, 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- Colot V. and Rossignol,J.L. (1999) Eukaryotic DNA methylation as an evolutionary device. BioEssays, 21, 402–411. [DOI] [PubMed] [Google Scholar]
- Cullen D., Leong,S.A., Wilson,L.J. and Henner,D.J. (1987) Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene, 57, 21–26. [DOI] [PubMed] [Google Scholar]
- Davis R.H. (2000) Neurospora: Contributions of a Model Organism. Oxford University Press, New York, NY.
- Feinberg A.P. and Vogelstein,B. (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity (II). Addendum. Anal. Biochem., 137, 266–267. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J. (1996) The role of DNA methylation in plant development. In Russo,V.E.A., Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 127–140.
- Finnegan E.J. and Dennis,E.S. (1993) Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res., 21, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H.M., Roberts,C.J., Claeys,K.M. and Selker,E.U. (1993) Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science, 262, 1737–1741. [DOI] [PubMed] [Google Scholar]
- Foss H.M., Roberts,C.J., Claeys,K.M. and Selker,E.U. (1995) Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation [correction]. Science, 267, 316. [DOI] [PubMed] [Google Scholar]
- Foss H.M., Roberts,C.J. and Selker,E.U. (1998) Reduced levels and altered patterns of DNA methylation caused by mutations in Neurospora crassa. Mol. Gen. Genet., 259, 60–71. [DOI] [PubMed] [Google Scholar]
- Fuks F., Burgers,W.A., Brehm,A., Hughes-Davies,L. and Kouzarides,T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet., 24, 88–91. [DOI] [PubMed] [Google Scholar]
- Geever R.F., Huiet,L., Baum,J.A., Tyler,B.M., Patel,V.B., Rutledge,B.J., Case,M.E. and Giles,N.H. (1989) DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J. Mol. Biol., 207, 15–34. [DOI] [PubMed] [Google Scholar]
- Genger R.K., Kovac,K.A., Dennis,E.S., Peacock,W.J. and Finnegan,E.J. (1999) Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol Biol., 41, 269–278. [DOI] [PubMed] [Google Scholar]
- Gowher H., Leismann,O. and Jeltsch,A. (2000) DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J., 19, 6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayburn W.S. and Selker,E.U. (1989) A natural case of RIP: degeneration of DNA sequence in an ancestral tandem duplication. Mol. Cell. Biol., 9, 4416–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R.S., Wijmenga,C., Luo,P., Stanek,A.M., Canfield,T.K., Weemaes,C.M. and Gartler,S.M. (1999) The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl Acad. Sci. USA, 96, 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Clerc,P. and Avner,P. (1997) X-chromosome inactivation in mammals. Annu. Rev. Genet., 31, 571–610. [DOI] [PubMed] [Google Scholar]
- Henikoff S. and Comai,L. (1998) A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics, 149, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. and Pugh,J.E. (1975) DNA modification mechanisms and gene activity during development. Science, 187, 226–232. [PubMed] [Google Scholar]
- Howell C.Y., Bestor,T.H., Ding,F., Latham,K.E., Mertineit,C., Trasler,J.M. and Chaillet,J.R. (2001) Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell, 104, 829–838. [DOI] [PubMed] [Google Scholar]
- Hsieh C.L. (1999) In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol., 19, 8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.W., Wang,I.X., Nikaido,K., Liu,P.Q., Ames,G.F. and Kim,S.H. (1998) Crystal structure of the ATP-binding subunit of an ABC transporter. Nature, 396, 703–707. [DOI] [PubMed] [Google Scholar]
- Hung M.S., Karthikeyan,N., Huang,B., Koo,H.C., Kiger,J. and Shen,C.J. (1999) Drosophila proteins related to vertebrate DNA (5-cytosine) methyltransferases. Proc. Natl Acad. Sci. USA, 96, 11940–11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino I., Marra,G., Palombo,F. and Jiricny,J. (1998) hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J., 17, 2677–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irelan J.T. and Selker,E.U. (1997) Cytosine methylation associated with repeat-induced point mutation causes epigenetic gene silencing in Neurospora crassa. Genetics, 146, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irelan J.T., Hagemann,A.T. and Selker,E.U. (1994) High frequency repeat-induced point mutation (RIP) is not associated with efficient recombination in Neurospora. Genetics, 138, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes,T.L. and Richards,E.J. (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet., 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh,J.A. and Richards,E.J. (1995) Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res., 23, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh,J.A., Flowers,S.K., Munakata,K. and Richards,E.J. (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl Acad. Sci. USA, 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Munakata,K., Richards,E.J. and Hirochika,H. (1999) Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics, 151, 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S.U., Landsberger,N. and Wolffe,A.P. (1997) DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol., 7, 157–165. [DOI] [PubMed] [Google Scholar]
- Kimura H., Ishihara,G. and Tajima,S. (1996) Isolation and expression of a Xenopus laevis DNA methyltransferase cDNA. J. Biochem. (Tokyo), 120, 1182–1189. [DOI] [PubMed] [Google Scholar]
- Kinsey J., Fincham,J., Siddig,M. and Keighren,M. (1980) New mutational variants of Neurospora NADP-specific glutamate dehydrogenase. Genetics, 95, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey J.A., Garrett-Engele,P.W., Cambareri,E.B. and Selker,E.U. (1994) The Neurospora transposon Tad is sensitive to repeat-induced point Mutation (RIP). Genetics, 138, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) Hhal methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- Kouzminova E.A. (2000) Isolation and characterization of a DNA-methyltransferase gene, dim-2, from Neurospora crassa. PhD thesis, Department of Biology, University of Oregon, Eugene, OR, p. 123.
- Kouzminova E.A. and Selker,E.U. (1999) Inversion in the published genetic map of linkage group VII. Fungal Genetics Newsl., 46, 14–15. [Google Scholar]
- Kumar S., Cheng,X., Klimasauskas,S., Mi,S., Posfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- Li E., Beard,C. and Jaenisch,R. (1993) Role for DNA methylation in genomic imprinting. Nature, 366, 362–365. [DOI] [PubMed] [Google Scholar]
- Luo Z., Freitag,M. and Sachs,M.S. (1995) Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol., 15, 5235–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F., Ramsahoye,B.H., Kashevsky,H., Tudor,M., Mastrangelo,M.A., Orr-Weaver,T.L. and Jaenisch,R. (1999) Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nature Genet., 23, 363–366. [DOI] [PubMed] [Google Scholar]
- Lyko F., Ramsahoye,B.H. and Jaenisch,R. (2000) DNA methylation in Drosophila melanogaster. Nature, 408, 538–540. [DOI] [PubMed] [Google Scholar]
- Malagnac F. et al. (1997) A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell, 91, 281–290. [DOI] [PubMed] [Google Scholar]
- Malagnac F., Gregoire,A., Goyon,C., Rossignol,J.L. and Faugeron,G. (1999) Masc2, a gene from Ascobolus encoding a protein with a DNA-methyltransferase activity in vitro, is dispensable for in vivo methylation. Mol. Microbiol., 31, 331–338. [DOI] [PubMed] [Google Scholar]
- Margolin B.S., Freitag,M. and Selker,E.U. (1997) Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genetics Newsl., 44, 34–36. [Google Scholar]
- Margolin B.S., Garrett-Engele,P.W., Stevens,J.N., Yen-Fritz,D., Garrett-Engele,C., Metzenberg,R.L. and Selker,E.U. (1998) A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by RIP. Genetics, 149, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margot J.B., Aguirre-Arteta,A.M., Di Giacco,B.V., Pradhan,S., Roberts,R.J., Cardoso,M.C. and Leonhardt,H. (2000) Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J. Mol. Biol., 297, 293–300. [DOI] [PubMed] [Google Scholar]
- Metzenberg R.L., Stevens,J.N., Selker,E.U. and Morzycka- Wroblewska,E. (1985) Identification and chromosomal distribution of 5S rRNA genes in Neurospora crassa. Proc. Natl Acad. Sci. USA, 82, 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao V.P., Freitag,M. and Selker,E.U. (2000) Short TpA-rich segments of the zeta-eta region induce DNA methylation in Neurospora crassa. J. Mol. Biol., 300, 249–273. [DOI] [PubMed] [Google Scholar]
- Nelson M. and Perkins,D.D. (2000) Restriction polymorphism maps of Neurospora crassa: 2000 update. Fungal Genetics Newsl., 47, 25–39. [Google Scholar]
- Nelson M.A. et al. (1997) Expressed sequences from conidial, mycelial, and sexual stages of Neurospora crassa. Fungal Genet. Biol., 21, 348–363. [DOI] [PubMed] [Google Scholar]
- Okano M., Xie,S. and Li,E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Orbach M.J. (1994) A cosmid with a HyR marker for fungal library construction and screening. Gene, 150, 159–162. [DOI] [PubMed] [Google Scholar]
- Panning B. and Jaenisch,R. (1996) DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev., 10, 1991–2002. [DOI] [PubMed] [Google Scholar]
- Perkins D.D., Radford,A., Newmeyer,D. and Björkman,M. (1982) Chromosomal loci of Neurospora crassa. Microbiol. Rev., 46, 426–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.D., Margolin,B.S., Selker,E.U. and Haedo,S.D. (1997) Occurrence of repeat induced point mutation in long segmental duplications of Neurospora. Genetics, 147, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. and Sapienza,C. (1993) Imprinting the genome: imprinted genes, imprinting genes, and hypothesis for their interaction. Annu. Rev. Genet., 27, 7–31. [DOI] [PubMed] [Google Scholar]
- Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye B.H., Biniszkiewicz,D., Lyko,F., Clark,V., Bird,A.P. and Jaenisch,R. (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA, 97, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhounim L., Rossignol,J.-L. and Faugeron,G. (1992) Epimutation of repeated genes in Ascobolus immersus. EMBO J., 11, 4451–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A.D. (1975) X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet., 14, 9–25. [DOI] [PubMed] [Google Scholar]
- Robertson K.D., Uzvolgyi,E., Liang,G., Talmadge,C., Sumegi,J., Gonzales,F.A. and Jones,P.A. (1999) The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res., 27, 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D., Ait-Si-Ali,S., Yokochi,T., Wade,P.A., Jones,P.L. and Wolffe,A.P. (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet., 25, 338–342. [DOI] [PubMed] [Google Scholar]
- Ronemus M.J., Galbiati,M., Ticknor,C., Chen,J. and Dellaporta,S.L. (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science, 273, 654–657. [DOI] [PubMed] [Google Scholar]
- Rossignol J.-L. and Faugeron,G. (1994) Gene inactivation triggered by recognition between DNA repeats. Experientia, 50, 307–317. [DOI] [PubMed] [Google Scholar]
- Rountree M.R. and Selker,E.U. (1997) DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev., 11, 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree M.R., Bachman,K.E. and Baylin,S.B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet., 25, 269–277. [DOI] [PubMed] [Google Scholar]
- Russell P.J., Rodland,K.D., Cutler,J.E., Rachlin,E.M. and McCloskey,J.A. (1985) DNA methylation in Neurospora: chromatographic and isoschizomer evidence for changes during development. In Timberlake,W. (ed.), Molecular Genetics of Filamentous Fungi. Alan Liss, Inc., pp. 321–332.
- Santi D.V., Garrett,C.E. and Barr,P.J. (1983) On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell, 33, 9–10. [DOI] [PubMed] [Google Scholar]
- Scheidt G., Weber,H., Graessmann,M. and Graessmann,A. (1994) Are there two DNA methyltransferase gene families in plant cells? A new potential methyltransferase gene isolated from an Arabidopsis thaliana genomic library. Nucleic Acids Res., 22, 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schubeler D., Lorincz,M.C., Cimbora,D.M., Telling,A., Feng,Y.Q., Bouhassira,E.E. and Groudine,M. (2000) Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol., 20, 9103–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E.U. (1990) Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet., 24, 579–613. [DOI] [PubMed] [Google Scholar]
- Selker E.U. (1997) Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion? Trends Genet., 13, 296–301. [DOI] [PubMed] [Google Scholar]
- Selker E.U. and Stevens,J.N. (1985) DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc. Natl Acad. Sci. USA, 82, 8114–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E.U. and Garrett,P.W. (1988) DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc. Natl Acad. Sci. USA, 85, 6870–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E.U., Cambareri,E.B., Jensen,B.C. and Haack,K.R. (1987a) Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell, 51, 741–752. [DOI] [PubMed] [Google Scholar]
- Selker E.U., Jensen,B.C. and Richardson,G.A. (1987b) A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science, 238, 48–53. [DOI] [PubMed] [Google Scholar]
- Selker E.U., Fritz,D.Y. and Singer,M.J. (1993) Dense non-symmetrical DNA methylation resulting from repeat-induced point mutation (RIP) in Neurospora. Science, 262, 1724–1728. [DOI] [PubMed] [Google Scholar]
- Singer M.J., Kuzminova,E.A., Tharp,A., Margolin,B.S. and Selker,E.U. (1995a) Different frequencies of RIP among early versus late ascospores of Neurospora crassa. Fungal Genetics Newsl., 42, 74–75. [Google Scholar]
- Singer M.J., Marcotte,B.A. and Selker,E.U. (1995b) DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol. Cell. Biol., 15, 5586–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staben C., Jensen,B., Singer,M., Pollock,J., Schectman,M., Kinsey,J. and Selker,E. (1989) Use of a bacterial Hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genetics Newsl., 36, 79–81. [Google Scholar]
- Tatematsu K.I., Yamazaki,T. and Ishikawa,F. (2000) MBD2–MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells, 5, 677–688. [DOI] [PubMed] [Google Scholar]
- Tatusova T.A. and Madden,T.L. (1999) BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences [published erratum appears in FEMS Microbiol. Lett., 1999, 177, 187–188]. FEMS Microbiol. Lett., 174, 247–250. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., Ng,H.H., Barlow,A.L., Turner,B.M., Hendrich,B. and Bird,A. (1999) Vestiges of a DNA methylation system in Drosophila melanogaster? Nature Genet., 23, 389–390. [DOI] [PubMed] [Google Scholar]
- Vollmer S.J. and Yanofsky,C. (1986) Efficient cloning of genes of Neurospora crassa. Proc. Natl Acad. Sci. USA, 83, 4869–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A., Kakutani,T., Martienssen,R.A. and Richards,E.J. (1993) Arabidopsis thaliana DNA methylation mutants. Science, 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C.R., Bartlett,R., Nurse,P. and Bird,A.P. (1995) The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res., 23, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.L. et al. (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 402, 187–191. [DOI] [PubMed] [Google Scholar]
- Yen R.-W.C. et al. (1992) Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res., 20, 2287–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.A. and Bestor,T.H. (1998) A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet., 7, 279–284. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Cambareri,E.B. and Kinsey,J.A. (2001) DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol. Gen. Genet., 265, 748–754. [DOI] [PubMed] [Google Scholar]
- Zimmermann C., Guhl,E. and Graessmann,A. (1997) Mouse DNA methyltransferase (MTase) deletion mutants that retain the catalytic domain display neither de novo nor maintenance methylation activity in vivo. Biol. Chem., 378, 393–405. [DOI] [PubMed] [Google Scholar]