Abstract
Numerous non-ribosomal trans-acting factors involved in pre-ribosomal RNA processing have been characterized, but none of them is specifically required for the last cytoplasmic steps of 18S rRNA maturation. Here we demonstrate that Rio1p/Rrp10p is such a factor. Previous studies showed that the RIO1 gene is essential for cell viability and conserved from archaebacteria to man. We isolated a RIO1 mutant in a screen for mutations synthetically lethal with a mutant allele of GAR1, an essential gene required for 18S rRNA production and rRNA pseudouridylation. We show that RIO1 encodes a cytoplasmic non-ribosomal protein, and that depletion of Rio1p blocks 18S rRNA production leading to 20S pre-rRNA accumulation. In situ hybridization reveals that, in Rio1p depleted cells, 20S pre-rRNA localizes in the cytoplasm, demonstrating that its accumulation is not due to an export defect. This strongly suggests that Rio1p is involved in the cytoplasmic cleavage of 20S pre-rRNA at site D, producing mature 18S rRNA. Thus, Rio1p has been renamed Rrp10p (ribosomal RNA processing #10). Rio1p/Rrp10p is the first non-ribosomal factor characterized specifically required for 20S pre-rRNA processing.
Keywords: pre-rRNA processing/ribosome/Saccharomyces cerevisiae/synthetic lethal screen
Introduction
In eukaryotic cells, most steps of ribosomal subunit biogenesis take place in a specialized nuclear structure, the nucleolus, which can be seen primarily, but not exclusively, as the ribosome factory. In the yeast Saccharomyces cerevisiae, transcription by RNA polymerase I of the repeated rDNA genes produces a primary transcript that not only contains the mature ribosomal RNAs (rRNAs), but also internal and external spacer sequences. The primary transcript is chemically modified and undergoes endo- and exonucleolytic reactions (see Figure 1) to lead ultimately to the mature rRNAs of 25S and 5.8S found in the large ribosomal subunit of 60S, and to the 18S rRNA found in the small ribosomal subunit of 40S. Concomitantly with the maturation process, numerous ribosomal and non-ribosomal proteins associate with the pre-rRNA intermediates to generate the pre-60S and 43S pre-ribosomal subunits, precursors to the 60S and 40S ribosomal subunits, respectively. These pre-ribosomal subunits are eventually exported to the cytoplasm, where the final assembly and maturation steps occur (for recent and thorough reviews see Kressler et al., 1999; Venema and Tollervey, 1999).
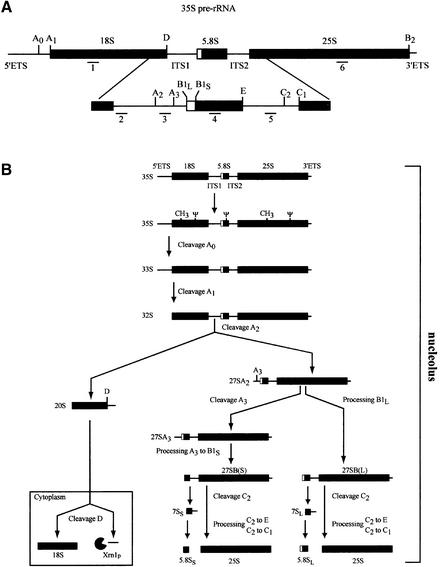
Fig. 1. Pre-rRNA processing in S.cerevisiae. (A) Structure of the 35S pre-rRNA. In the primary transcript the sequences of the mature 18S, 5.8S and 25S rRNAs are flanked by the external transcribed spacers (5′ and 3′ETS) and separated by the internal transcribed spacers (ITS1 and ITS2). Cleavage sites are indicated by uppercase letters A0–E, and oligonucleotide probes used in northern blot hybridizations by numbers 1–6. (B) Pre-rRNA processing pathway. Sequential cleavages of the 35S pre-rRNA at sites A0 and A1 generate the 33S and 32S pre-rRNAs. Cleavage of the 32S pre-rRNA at site A2 in ITS1 yields the 27SA2 and 20S pre-rRNAs, which are precursors to the RNA components of the large and small ribosomal subunits, respectively. The 27SA2 precursor is either processed at site A3 by RNase MRP, generating the 27SA3 pre-rRNA rapidly digested by the 5′–3′ exonucleases Rat1p and Xrn1p to site B1S yielding the 27SBS pre-rRNA. This constitutes the major pathway. Approximately 15% of the 27SA2 molecules are processed at site B1L leading to the 27SBL intermediate. Processing at site B2, the 3′ end of the 25S rRNA, occurs concomitantly with 27SB 5′-end formation. The 27SBS and 27SBL pre-rRNAs both follow the same processing pathway to 25S and 5.8SS/L through cleavage at site C2 in ITS2, followed by 3′–5′ exonucleolytic digestion of 7SS and 7SL from site C2 to E by the exosome complex, and 5′–3′ exonucleolytic digestion to the 5′ end of the 25S rRNA. The final maturation of the 20S pre-rRNA by an endonucleolytic cleavage at site D occurs in the cytoplasm and produces the mature 18S rRNA and a fragment D-A2 (5′ ITS1). The D-A2 fragment is then degraded by the 5′–3′ exonuclease Xrn1p.
Processing intermediates formed during 35S pre-rRNA maturation were characterized early in the course of rRNA processing studies. The identification and functional characterization of the trans-acting factors involved in this process constituted one of the ‘key questions’ in the early nineties (Woolford and Warner, 1991). During the past decade, thanks to the development of new biochemical, genetic and in silico technologies, dozens of these components have been identified in yeast. These are small nucleolar components of ribonucleoprotein particles (snoRNPs) or non-ribosomal nucleolar proteins: putative helicases, ribonucleases and other factors, the actual functions of which have not been defined yet (for reviews see Kressler et al., 1999; Venema and Tollervey, 1999). Although the processing cleavage sites along the 35S precursor have been precisely determined, the endoribonucleases acting on these sites have been identified for very few of them, namely the snoRNP MRP at site A3 and Rnt1p within the 3′ external transcribed spacer (3′ETS) (for review see Venema and Tollervey 1999).
Early cleavage steps of the 35S pre-rRNA at sites A0, A1 and A2 yield the 20S and 27SA pre-rRNAs, precursors to the rRNAs of the small ribosomal subunit and the large ribosomal subunit, respectively. In S.cerevisiae, mature 18S rRNA is then formed through an endoribonucleolytic cleavage of the 20S pre-rRNA at site D (Figure 1), which occurs in the cytoplasm (Udem and Warner, 1973; Trapman et al., 1975; Stevens et al., 1991). Impairment of this processing step results in 20S pre-rRNA accumulation and concomitant 18S rRNA depletion. This phenotype could stem from defects in 43S pre-subunit assembly and/or nucleocytoplasmic transport as well, as demonstrated for Ran regulators or nucleoporin mutants (Moy and Silver, 1999). While numerous factors involved in the early cleavages of the 35S pre-rRNA have been identified, very few factors specifically required for the processing of 20S pre-rRNA into 18S rRNA have been described. Deletion of DRS2 (Ripmaster et al., 1993) or of RPS31/UBI3, which encodes a ribosomal protein from the small subunit (Finley et al., 1989), results in decreased efficiency of the conversion of 20S pre-rRNA to 18S rRNA, which is still produced, but at slower rates. On the contrary, deletion of both RPS0A and RPS0B, two redundant genes encoding the rpS0 ribosomal proteins, leads to an arrest of 18S rRNA production correlated with 20S pre-rRNA accumulation (Ford et al., 1999).
Here we report the characterization of a new protein specifically required to process the 20S pre-rRNA to 18S rRNA. This factor, encoded by the essential RIO1/RRP10 gene (Angermayr and Bandlow, 1997), has homologues in all eukaryotes and archaebacterial organisms sequenced so far. We demonstrate that RIO1/RRP10 encodes a cytoplasmic non-ribosomal protein. Its inactivation results in drastic inhibition of cleavage at site D, leading to a strong accumulation of the 20S pre-rRNA in the cytoplasm and to the inhibition of 18S rRNA production. Rio1p/Rrp10p is the first non-ribosomal protein specifically required for 20S pre-rRNA processing in the cytoplasm to be character ized. We suggest that Rio1p/Rrp10p could catalyse, or be part of a complex catalysing, the endonucleolytic cleavage of 20S pre-rRNA at site D.
Results
A synthetic lethal screen with gar1-10 mutant identifies RIO1
Gar1p, one of the core proteins of H/ACA snoRNPs (Lubben et al., 1995; Ganot et al., 1997; Henras et al., 1998), is an essential protein required for the pseudouridylation of rRNAs and for 18S rRNA synthesis (Girard et al., 1992; Bousquet-Antonelli et al., 1997). In order to characterize factors involved in these processes, we carried out synthetic lethal (sl) screens with mutant alleles of GAR1. Strain YO89 is one of the three non-sectoring, 5′-fluoroorotic acid (5-FOA)-sensitive clones, isolated in a screen undertaken with the gar1-10 temperature-sensitive allele (see Materials and methods and Venema et al., 1997). This clone regained a sectoring phenotype and segregated FOA-resistant (FOAr) cells upon transformation with pJPG67 carrying the GAR1+ allele, but not upon transformation with a vector carrying the gar1-10 allele, demonstrating that it carries mutation(s) synthetically lethal with gar1-10. Strain YO89 was crossed with a MATa derivative of the parental strain YO126, the diploid clones obtained could segregate FOAr cells, and upon tetrad dissection, a 2:2 segregation of the sl character was observed, demonstrating that it is caused by a single recessive mutation.
To clone a wild-type allele of this sl mutation, strain YO89-13, a derivative of YO89 in which the LEU2::gar1-Δ disruption has been replaced by a TRP1::gar1-Δ allele, was transformed with a yeast multi-copy genomic library carried by the 2µ LEU2 vector pFL46S (Bonneaud et al., 1991). Eight leu+ sectoring colonies that could grow on 5-FOA plates were obtained (loss of pJPG203: ARS, CEN, ADE3, GAR1). Among these, seven had received from the library a GAR1-containing plasmid since they could segregate lysine-requiring cells (loss of pJPG225: ARS CEN, LYS2, gar1-10). The library plasmid was recovered from the remaining transformant and shown to complement the sl phenotype upon retransformation into strain YO89-13. This plasmid, pEV6, was partially sequenced and shown to carry a fragment from chromosome XV (from nucleotide 548 151 to nucleotide 552 896) containing three complete open reading frames (ORFs): (i) GCY1, which encodes an aldo/keto reductase of unknown substrate specificity (Angermayr and Bandlow, 1997); (ii) an essential gene RIO1 (Angermayr and Bandlow, 1997); and (iii) an uncharacterized ORF, YOR121C (included in GCY1 ORF). When a plasmid carrying the subcloned RIO1 wild-type allele (pEV21) was introduced in the sl strain YO89, red/white sectoring and growth on 5-FOA medium were restored, whereas this was not the case for plasmid pEV22 carrying the rio1-1 mutant allele derived from YO89 genomic DNA (see Materials and methods). Furthermore, this same plasmid pEV22 allows growth in glucose-containing medium of strain YO296, which carries a RIO1+ allele under the control of the GAL10 promoter, demonstrating that the mutation responsible for the colethality with gar1-10 indeed lies within the RIO1 gene.
This gene has previously been identified in S.cerevisiae, and was shown to be essential for cell viability (Angermayr and Bandlow, 1997). Occurrence of genes homologous to RIO1 in archaebacteria, Emericella nidulans (formerly Aspergillus nidulans), Caenorhabditis elegans and Homo sapiens has been reported (Anaya et al., 1998). Our search in databases identified homologues of Rio1p in all eukaryotic and archaebacterial sequenced genomes (for sequence alignments see Supplementary figure 1 in the Supplementary data, available at The EMBO Journal Online).
The predicted polypeptide has an unusual composition, containing 18% glutamic and aspartic acids and 11.5% serine and threonine. The only mutation found in the rio1-1 allele that causes lethality in gar1-10 strains is a missense mutation leading to the change of the universally conserved threonine 62 in the S.cerevisiae protein to isoleucine.
Since a mutant allele of RIO1 was retrieved as synthetically lethal with a mutant allele of the GAR1 gene, the product of which is clearly required for pre-rRNA processing and post-transcriptional modification (Girard et al., 1992; Bousquet-Antonelli et al., 1997), we investigated the possible involvement of Rio1p in the production of mature rRNAs.
As shown below, RIO1 appears to be involved in the processing of pre-rRNA and consequently it has been renamed RRP10 (ribosomal rNA processing #10).
Depletion of Rrp10p affects 18S rRNA production
Since RIO1/RRP10 has been shown to be an essential gene (Angermayr and Bandlow, 1997), a conditional RIO1/RRP10 allele was constructed by replacing its promoter by the inducible GAL10 promoter. The 5′ end of this construct was also tagged with two protein A epitopes (see Supplementary data for the description of this construction). Transcription driven by the GAL10 promoter allows high expression of the gene in culture media that contain galactose (YPG, YNB Gal), but is repressed in culture media containing glucose (YPD, YNB Glu), leading to the depletion of ProtA-Rrp10p (see Supplementary figure 2).
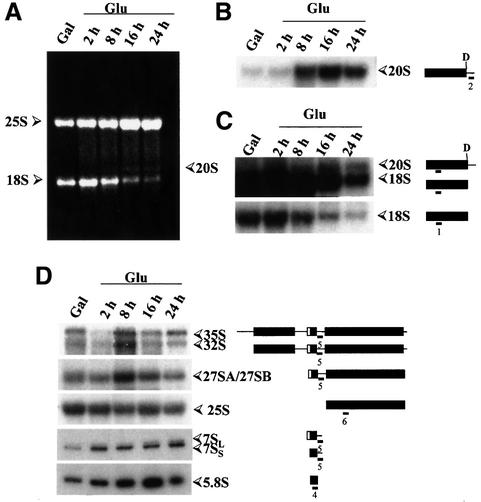
We made use of this conditional RRP10 allele to determine whether Rrp10p is necessary for pre-rRNA processing. We first assessed the steady-state levels of mature rRNAs by ethidium bromide staining and northern blotting of RNAs prepared from cells grown in permissive YPG medium or shifted to non-permissive YPD medium for 2–24 h. As shown in Figure 2A, depletion of Rrp10p has no effect on 25S rRNA accumulation but drastically affects the level of 18S rRNA. Furthermore, after 16 h culture in YPD medium, RNA molecules that migrate similarly to the 20S pre-rRNA accumulate at sufficient level to be detected by ethidium bromide staining (Figure 2A).
Fig. 2. Depletion of Rrp10p specifically affects the steady-state levels of mature 18S rRNA species and results in 20S pre-rRNA accumulation. Northern blot analysis of pre-rRNA processing: YO296 (GAL::PROTA-RRP10) cells were grown in YPG medium (Gal), or in YPD medium for up to 24 h (Glu). At the indicated time points, total RNA was extracted and separated in 1% agarose-formaldehyde gels to analyse 35S, 32S, 27SA2, 25S, 20S and 18S species, and in 6% polyacrylamide gels for 7S(L), 7S(S) and 5.8S species analysis. Equal amounts of total RNA (5 µg) were loaded in every lane. (A) Ethidium bromide staining of the gel. Northern blots of the gel were hybridized to: (B) probe 2 complementary to ITS1 upstream of site A2; (C) probe 1 complementary to 18S rRNA, a 7 h exposure (upper panel in C) and a 90 min exposure (lower panel in C) of the same blot are shown; (D) probe 4 complementary to 5.8S, probe 5 complementary to ITS2 and probe 6 complementary to 25S rRNA. Pre-rRNA and rRNA species are schematically represented on the right side; full rectangles represent the mature rRNAs and thin lines the transcribed spacers.
To determine whether this RNA species is related to the 20S pre-rRNA, precursor to the 18S rRNA, northern hybridizations using probe 1 complementary to the 18S rRNA, and probe 2 complementary to an ITS1 sequence between sites D and A2 (see Materials and methods and Figure 1), were performed. Results of these hybridizations are shown in Figure 2B and C. The RNA species that accumulates in Rrp10p depleted cells is detected with probes 1 and 2, and migrates to the same position as the normal 20S pre-rRNA intermediate (Figure 2B and C, lane Gal). Probing the same blot with probe 3 (complementary to ITS1 sequences between sites A2 and A3) did not reveal any pre-rRNA intermediate of this size or close to it (data not shown), indicating that the RNA species accumulating in Rrp10p depleted cells is the 20S pre-rRNA that extends from the 5′ end of 18S to site A2 in ITS1 (Figure 1). These data show that depletion of Rrp10p severely impairs 18S rRNA production and leads to accumulation of its 20S pre-rRNA precursor. Phosphorimager quantifications of the 20S and 18S bands in the northern blot shown in Figure 2C reveal a 40-fold reduction in the amount of 18S rRNA and a 20-fold increase of the 20S pre-rRNA levels, in cells grown for 16 h in YPD medium compared with the ones in cells grown in galactose medium. Considering the observed evolution of the 18S/25S ratio, and that the same amount of total cellular RNA has been loaded in every lane and the corresponding number of cells used to get this amount of RNA (given the dramatic inhibition of 18S rRNA production upon depletion of Rrp10p, more cells, collected from the later time point samples of the culture in YPD medium, were needed to get the same amount of total cellular RNA), the actual drop in 18S rRNA level is even more dramatic than 40-fold. Moreover, the small amount of 18S rRNA found in these cells (Figure 2A) probably stems from incomplete depletion of Rrp10p due to leaks in glucose transcriptional repression of the GAL10 promoter and/or survival of Rrp10p molecules produced before or early after the switch to glucose medium. Consequently we think that upon deletion of Rrp10p, production of 18S rRNA is completely inhibited. It also appears that the 20S pre-rRNA does not accumulate stoichiometrically with respect to 25S rRNA, although arising from the processing of the very same 35S pre-rRNA molecules. This suggests that the 20S pre-rRNA is intrinsically unstable if not converted to 18S rRNA and degraded by the exosome complex (Allmang et al., 2000), or that Rrp10p depletion leads to a less stable form of this precursor, possibly due to some defect in the assembly of the 43S pre-subunits, or both.
The high molecular weight blot shown in Figure 2B was also probed with probes 5 and 6, complementary to the ITS2 and the 25S rRNA, respectively (see Figure 1 and Materials and methods). A low molecular weight blot was probed with probes 4 (complementary to the 5.8S rRNA) and 5 (complementary to the ITS2). Apart from what was already observed for 18S and 20S rRNAs, neither changes in the levels of normal pre-rRNA processing intermediates, nor accumulation of aberrant species was detected upon depletion of Rrp10p in these experiments (Figure 2D).
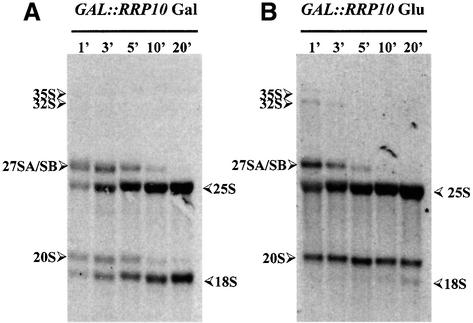
The kinetics of pre-rRNA processing and rRNA accumulation were analysed in (methyl-3H) methionine pulse– chase experiments (Figure 3). In a GAL::PROTA-RRP10 strain grown in the presence of galactose, most of the labelled 27S and 20S pre-rRNAs have disappeared after a 10 min chase. In contrast, after 20 min of chase, 20S pre-rRNA is still present and 18S rRNA is only faintly detectable in cells from the same GAL::PROTA-RRP10 strain grown for 14 h in glucose-containing medium. On the contrary, Rrp10p depletion does not affect the time course of the 27S pre-rRNA conversion to 25S rRNA. Moreover, no other pre-rRNA intermediate is observed during the chase experiment, indicating that the processing pathways leading to the 20S pre-rRNA and 25S rRNA are fully functional in Rrp10p depleted cells, while 18S rRNA formation is strongly inhibited in such conditions due to a defect in 20S pre-rRNA processing.
Fig. 3. Depletion of Rrp10p results in reduced synthesis of 18S rRNA. (A) YO296 (GAL::PROTA-RRP10) strain grown at 30°C on YNB Gal without methionine; (B) YO296 grown at 30°C in YPG medium (Gal) then shifted for 14 h to YPD medium (Glu), and finally grown for 9 h in YNB Glu without methionine. Cells were labelled for 4 min with (methyl-3H) methionine and chased with a large excess of unlabelled methionine for 1–20 min.
In order to determine whether Rrp10p depletion specifically affects the processing of 20S pre-rRNA, levels of small nuclear and nucleolar RNAs were also assessed in cells of strain YO296 shifted to YPD medium. All tested snoRNAs were found to be unaffected (data not shown). Likewise, levels of the RNase MRP RNA and of U1 snRNA were not altered even after 24 h of growth in YPD medium. Northern hybridizations with probes specific for ACT1 and rpS11 ribosomal protein mRNAs did not reveal any defect in pre-mRNA processing or accumulation (data not shown). Altogether these data show that depletion of Rrp10p strongly and specifically affects processing of the 20S pre-rRNA to 18S rRNA.
Cells depleted of Rrp10p accumulate 20S pre-rRNA in the cytoplasm
In yeast, it has been shown previously that the last steps of 18S rRNA maturation occur in the cytoplasm. After cleavage at site A2, the 20S pre-rRNA is exported from the nucleolus as part of a 43S pre-ribosomal particle and is subsequently processed at site D by an unknown endonuclease (Udem and Warner, 1973; Trapman et al., 1975), leading to 18S rRNA and a D-A2 5′-ITS1 fragment that is degraded by the cytoplasmic 5′–3′ exoribonuclease Xrn1p (Stevens et al., 1991). Thus, accumulation of the 20S pre-rRNA may be either due to nuclear retention of the 43S particles, precursors of the small ribosomal subunit, or to a defect in the cytoplasmic processing of 20S pre-rRNA. To sort out these two possibilities, the localization of the 20S pre-rRNA accumulating in Rrp10p depleted cells was determined by in situ hybridization with a probe complementary to the D-A2 fragment. Ultrastructural localization of the probe was analysed by electron microscopy (Figure 4). In wild-type cells (Figure 4A) and in YO296 cells grown in galactose medium (not shown), the labelling, which corresponds to the 35S, 32S or 20S precursors, is essentially restricted to the nucleolus. In contrast, Rrp10p depleted cells exhibit both a nucleolar and a cytoplasmic labelling (Figure 4C), a phenotype similar to the one observed in a Δxrn1 strain (Figure 4B). Thus, 20S pre-rRNA accumulation in Rrp10p depleted cells does not result from a defect in the nuclear export of the 43S particle, but rather from the impairment of the cytoplasmic endonucleolytic processing of 20S pre-rRNA.
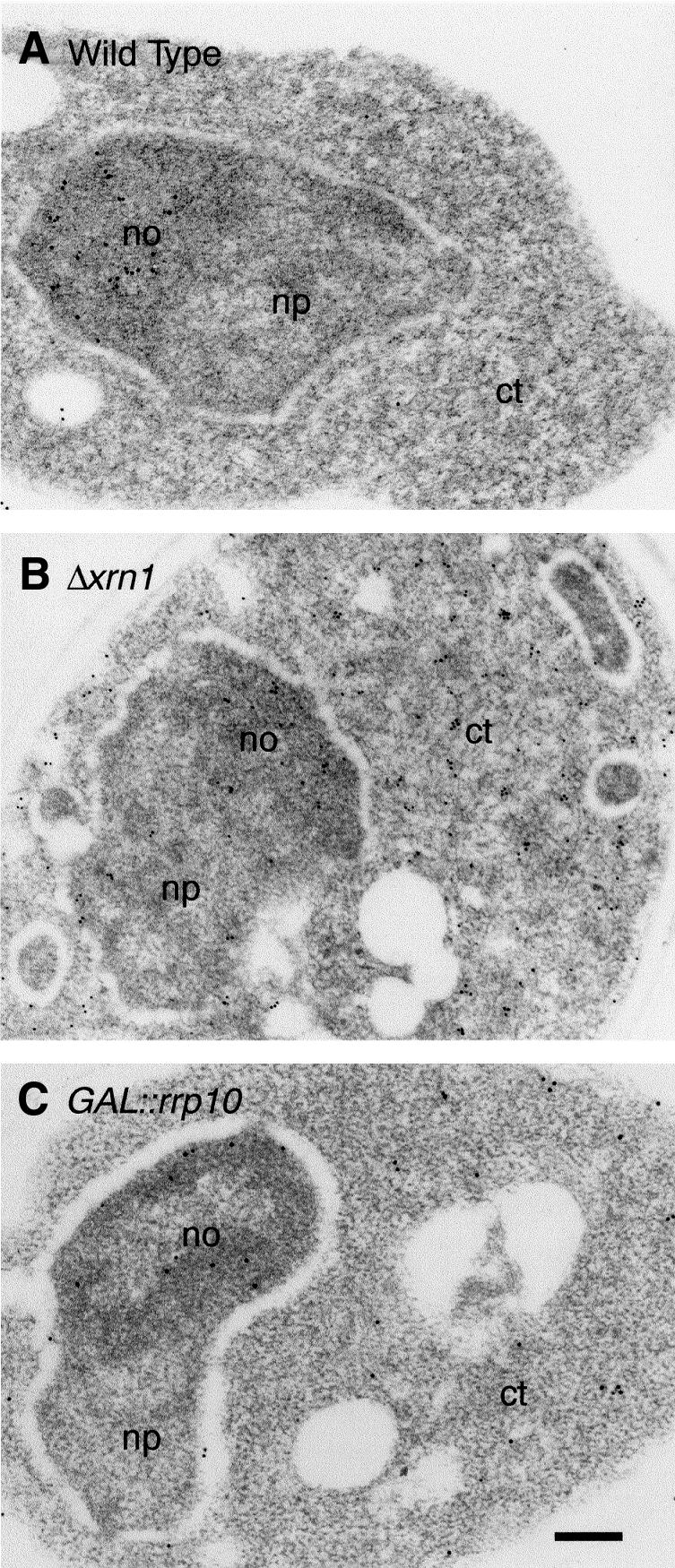
Fig. 4. Cells depleted of Rrp10p accumulate 20S pre-rRNA in the cytoplasm. Electron microscopic detection of the ITS1: (A) in wild-type yeast cells, the labelling is exclusively detected in the nucleolus (35S, 33S, 32S and 20S pre-rRNAs); (B) in Δxrn1 cells, the labelling is found in the nucleolus and in the cytoplasm (fragment D-A2 from the ITS1); (C) in GAL::PROTA-RRP10 cells grown for 16 h in YPD medium (Glu), the labelling is detected in the nucleolus and in the cytoplasm. np, nucleoplasm; no, nucleolus; ct, cytoplasm. Bar, 200 nm.
Rrp10p is a non-ribosomal cytoplasmic protein
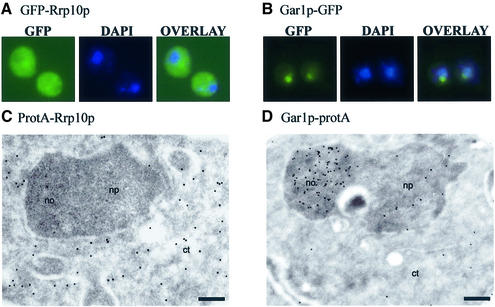
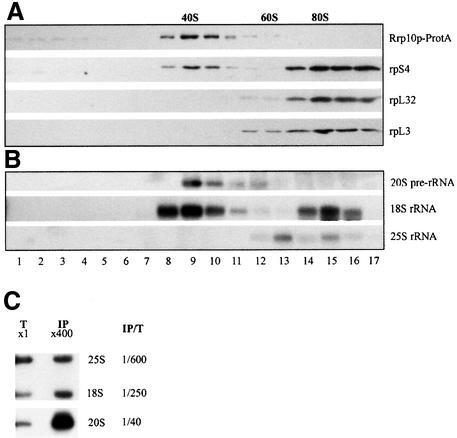
Subcellular localization of Rrp10p was investigated using a green fluorescent protein (GFP)-Rrp10p fusion protein. Strain YO296 (GAL::PROTA-RRP10) regained the ability to grow on YPD upon transformation with the pEV24 plasmid, which directs expression of the GFP-Rrp10p construct, showing that this fusion is functional. In strain JG540 (wild type) expressing a GFP-RRP10 fusion allele (pEV24), the green fluorescence is detected in the cytoplasm (Figure 5A). In the same strain, a fusion of the nucleolar protein Gar1p with the GFP localizes to the nucleolus, as expected (Girard et al., 1992) (Figure 5B). The cytoplasmic localization of Rrp10p is confirmed by immunodetection of a ProtA-Rrp10p fusion protein. Immunodetection of ProtA-Rrp10p and Gar1p-ProtA fusions by electron microscopy is shown in Figure 5C and D, respectively. Gar1p is found in the nucleolus, but Rrp10p exclusively localizes to the cytoplasm. Since Rrp10p is required for the maturation of the 20S pre-rRNA and is located in the cytoplasm, we assessed the possibility that Rrp10p is a ribosomal protein. In this analysis ProtA-Rrp10p could not be detected in either ribosomal subunit, whereas, as expected, the ribosomal proteins rpL3 and rpL32 are present in the 60S subunit, and rpS4 in the 40S subunit (see Supplementary figure 3). Moreover, Rrp10p is not found in the 80S ribosomes fractions of the gradient shown in Figure 6A (see below). These results show that although localized in the cytoplasm and required for cleavage at site D of the 20S pre-rRNA, Rrp10p is not a ribosomal protein. In agreement with these data, Rrp10p was not referenced in the recent compilation of ribosomal proteins (Planta and Mager, 1998).
Fig. 5. Rrp10p is a cytoplasmic protein. Subcellular localization using GFP-tagged proteins (A and B). (A) GFP-Rrp10p: cells transformed by pEV24 (GFP::RRP10) exhibit a cytoplasmic pattern of fluorescence. (B) Gar1p-GFP: in cells transformed with pZUT3 (GAR1::GFP) a punctate pattern characteristic of a nucleolar staining is observed. Positions of nuclei were determined by DAPI staining (blue). Overlay images are shown by superposition of the blue and green stainings. Immunolocalization by electron microscopy (C and D). (C) ProtA-Rrp10p; (D) Gar1p-ProtA. Tagged proteins were detected by treatment with anti-protein A antibodies followed by incubation with colloidal gold-conjugated protein A. no, nucleolus; np, nucleoplasm; ct, cytoplasm. Bars in C and D, 200 nm.
Fig. 6. Sedimentation profiles of ProtA-Rrp10p and 20S pre-rRNA in a glycerol gradient. A total extract was loaded on a 10–30% glycerol gradient and subjected to centrifugation. Fractions were collected, and proteins and RNA in each fraction were analysed. (A) Sedimentation profiles of ProtA-Rrp10p, rpS4p, rpL3p and rpL32p. Proteins pre cipitated with TCA and separated by SDS–PAGE were revealed by western blotting. (B) Sedimentation pattern of 18S and 25S rRNAs and of 20S pre-rRNA. Fractions containing the peaks of 40S, 60S ribosomal subunits and 80S ribosomes are indicated. (C) 20S pre-RNA selectively co-purifies with Rrp10-ProtAp. 1/400 of the clarified cell lysate (T) or the bulk of the immunoprecipitated RNA (IP) was probed with either a mix of probes 1 and 6 in Figure 1 (18S, 25S) or a D-A2 probe prepared by multiprime labelling (20S). The fraction of each rRNA or pre-rRNA recovered in the immunoprecipitate was determined by phosphoimager quantification (IP/T).
Rrp10p is associated with higher order structures but not found in translating ribosomes
To assess whether Rrp10p is present in the cytoplasm as free molecules or associated with higher order structures, a cellular extract from strain YO296 expressing ProtA-Rrp10p was fractionated by sedimentation through 10–30% glycerol gradients, and fractions from this gradient were analysed for their protein and RNA contents. The relative positions of 40S, 60S ribosomal subunits and 80S monosomes were determined by probing the RNA extracted from the fractions with 18S- and 25S-specific probes. Probing the same blot with a 20S pre-rRNA specific probe (fragment D-A2 in Figure 1) revealed that this precursor of 18S rRNA is exclusively found in particles that exhibit a sedimentation coefficient of ∼40S (Figure 6B). Likewise, immunodetection with PAP antibody showed that Rrp10p sediments in the very same fractions as 20S pre-rRNA (Figure 6A). Less than 5% of the ProtA-Rrp10p fusion protein is found in the first fractions of the gradient corresponding to soluble factors, and none is detected in the 80S fraction (Figure 6A). Thus, although Rrp10p is associated with higher order structures with a sedimentation coefficient of ∼40S, it is found neither in monosomes nor in polysomes, indicating that Rrp10p is not an integral component of translating ribosomes. Since Rrp10p depletion blocks 20S pre-rRNA processing, and Rrp10p is not found in monosomes or polysomes, the observed cosedimentation of Rrp10p and 20S pre-rRNA in the glycerol gradient suggests that Rrp10p might be associated, at least transiently, with the 43S particle, precursor of the 40S ribosomal subunit. In order to assess such an association, whole cell lysates from RRP10-protA or RRP10+ strains were incubated with IgG Sepharose beads at a salt concentration high enough to disrupt unspecific weak interactions (0.2 M KCl), and the RNA material retained onto the IgG–Sepharose beads in such conditions was extracted, separated in an agarose gel and probed with 18S-, 25S- or 20S-specific probes. As shown in Figure 6C, RNA immunoprecipitated from the lysate prepared from RRP10-protA cells was selectively enriched for 20S pre-rRNA. Such an effect was not observed when using an RRP10+ cell lysate (not shown), indicating that Rrp10p and 20S pre-rRNA are indeed found in the same molecular structure, namely the 43S precursor of the small ribosomal subunit. Another small RNA fragment that hybridizes to the D-A2 probe is also retrieved from the immunoprecipitated material (not shown). Such a small fragment is also observed in the fractions of the glycerol gradient containing the 20S pre-rRNA. This suggests that the slight enrichment in 18S rRNA compared with 25S rRNA (600/250 = 2.4) of the immunoprecipitated RNA is due to immunoprecipitation of precursor particles of the 40S ribosomal subunit still containing Rrp10p and the products of the processing of 20S pre-rRNA: 18S rRNA and the D-A2 fragment. This suggests that Rrp10p is directly involved in site D cleavage of this precursor of 18S rRNA.
Discussion
In this study, using an sl screen, we identified the first non-ribosomal factor specifically required for 20S pre-rRNA processing in yeast. sl screens developed in S.cerevisiae (Guarente, 1993) have proved very efficient in the analysis of large multi-molecular structures like the nuclear pore (Doye and Hurt, 1995) or complex pathways such as tRNA processing and export (Simos et al., 1996) or cell polarity and morphogenesis (Madden and Snyder, 1998). With respect to ribosome biogenesis, screening for mutations synthetically lethal with NOP1 mutant alleles led to the identification of NOP4/NOP77 (Berges et al., 1994), RRP5 (Venema and Tollervey, 1996), NOP56 and NOP58 (Gautier et al., 1997) as functional partners of NOP1. Using GAR1 mutant alleles as baits in sl screens, we already characterized mutations in ROK1, an essential gene encoding a putative RNA helicase required for 18S rRNA production (Venema et al., 1997), and in RRP8 that encodes a putative methyltransferase (Bousquet-Antonelli et al., 2000). As reported here, one of the sl mutations obtained in these screens resides in RIO1/RRP10, a constitutively expressed essential gene (Angermayr and Bandlow, 1997), which is conserved from yeasts to man and in archaebacteria (Anaya et al., 1998 and data herein), pointing to its possible involvement in an evolutionarily conserved biological process. So far no function had been ascribed to any of the Rrp10p homologues identified, either based on occurrence of putative functional motifs or on in vivo analyses. However, in E.nidulans, a cold-sensitive lethal mutation in the gene sudD, the RRP10 homologue, has been characterized as an extragenic suppressor of a thermosensitive mutation in bimD, a gene possibly involved in the control of chromosome segregation (Holt, 1996). At non-permissive temperatures, this sudD7 mutation itself leads to an abnormal nuclear morphology. Whether the phenotypes displayed by the sudD7 mutant reflect the direct involvement of this RRP10 homologue in chromosome segregation or are indirect consequences of the impairment of a more general pathway remains to be determined. In view of our results, an analysis of pre-rRNA processing in the sudD7 strain would be of interest and at the same time reveal whether the sudD/RRP10 structural homology is correlated with a functional one. In C.elegans, systematic investigation of loss of function phenotypes of predicted genes via RNA-mediated interference (RNAi), revealed that inhibition by RNAi of one of the RRP10 putative homologues (Ce2 in Supplementary figure 1) leads to post-embryonic growth defect and Sick phenotype (Fraser et al., 2000), but no molecular function has been ascribed to this gene. By specifically inhibiting the functions of selected genes, RNAi could provide a way to analyse the function(s) of the RRP10 homologues at the molecular level, especially during early embryogenesis in Drosophila, or in C.elegans.
The functional analysis of RRP10 described above shows that RRP10 encodes a cytoplasmic protein. It also demonstrates that Rrp10p-depleted cells exhibit a strong accumulation of the 20S pre-rRNA. Such an accumulation of 20S pre-rRNA might stem from: (i) defects in 43S pre-subunit assembly impairing either its export from the nucleus or its final processing or both; (ii) alteration of elements of the export machinery as exemplified for prp20-1, rna1-1 and yrb1-1 mutants (Moy and Silver, 1999); or (iii) direct inhibition of the endonucleolytic cleavage at site D.
To our knowledge, only a handful of the characterized S.cerevisiae mutants impaired in pre-rRNA processing specifically accumulate 20S pre-rRNA, amongst which are mutant alleles of genes encoding small subunit ribosomal proteins. Inactivation of the non-essential DRS2 gene leads to a delayed processing of 20S pre-rRNA into 18S rRNA and to impairment of 27SA pre-rRNA maturation, although to a lesser extent (Ripmaster et al., 1993). Drs2p localizes mostly in late Golgi membranes (Chen et al., 1999), and the deletion of DRS2 is lethal when coupled to thermosensitive mutations of CHC1, or deletion of ARF1. Since both CHC1 and ARF1 encode factors involved in the formation of coated transport vesicles, the 20S pre-rRNA processing defect observed in DRS2 mutants is probably indirect and reflects the coupling of ribosome biogenesis and membrane synthesis (Mizuta and Warner, 1994). The CLP-8 mutant described two decades ago (Carter and Cannon, 1980; Carter et al., 1980) exhibits decreased rates of 20S pre-rRNA export and cytoplasmic processing. Although this mutant has not been studied further, this phenotype suggests that the 43S pre-subunits produced are somehow altered in such a way that they are inefficiently recognized by both the export and the processing machineries. Deletion of RPS31/UBI3, which encodes one of the small ribosomal subunit proteins, specifically delays maturation of the 20S pre-rRNA but does not affect nuclear export of the 43S pre-ribosomal subunit. Enough mature 40S subunits are still synthesized to permit growth, although at a greatly decreased rate (Finley et al., 1989). Presumably pre-40S subunits devoid of rpS31 are poor substrates for the processing machinery. Finally, depletion of rpS0 results in a block in 18S rRNA synthesis, and pre-40S subunits containing 20S pre-rRNA accumulate. The inefficient incorporation of these rpS0 depleted pre-40S subunits in translating ribosomes leads to a shift to smaller polysomes, and eventually to translation inhibition (Ford et al., 1999). Clearly proper assembly of rpS0 with the pre-40S particles is required for their processing into mature 40S subunits.
Concerning the molecular role of Rrp10p, since 20S pre-rRNA is found in the cytoplasm of Rrp10p-depleted cells, its involvement in the pre-40S subunit nuclear export machinery itself is rather unlikely. Likewise, if Rrp10p inactivation led to improper assembly of the pre-40S ribosomal subunit, this would have no major consequence on its export from the nucleus. RRP10 is expressed constitutively (Angermayr and Bandlow, 1997) but at low level. According to transcriptome analysis (Holstege et al., 1998), on average one molecule of RRP10 mRNA is present per cell, to be compared with amounts in the range of 13–15 molecules per cell for mRNAs encoding snoRNP components Gar1p, Nhp2p or Nop1p, and of 30–50 molecules per cell for mRNAs encoding ribosomal proteins such as rpS0. In accordance with RRP10 low transcriptional expression level, its codon bias index of 0.098, calculated according to Bennetzen and Hall (Bennetzen and Hall, 1982; Coghlan and Wolfe, 2000), indicates that RRP10 mRNA is translated at low rate, suggesting that the cellular Rrp10p concentration should be low. Thus, Rrp10p is a low abundance protein that localizes to the cytoplasm, where it is found in structures with a sedimentation coefficient of ∼40S, and selectively interacts with 20S pre-rRNA-containing particles. Rrp10p inactivation rapidly leads to specific and total inhibition of cleavage at site D. These data suggest that Rrp10p is a factor transiently associated to the pre-40S subunit, and involved in a catalytic step of the cytoplasmic maturation of the 20S pre-rRNA. In support of our data it has been shown in a genome-wide two-hybrid analysis of S.cerevisiae (Ito et al., 2001) that Rrp10p and rpS1ap, a protein of the 40S ribosomal subunit, can physically interact. We propose that inhibition of maturation of 20S pre-rRNA arising from Rrp10p depletion is due to direct inhibition of the activity of the endonuclease or endonucleolytic complex, which cleaves this pre-rRNA at site D. The most tempting hypothesis is that Rrp10p itself carries this catalytic activity, or is a key component of the catalytic complex. Another possibility is that Rrp10p, transiently associated with the pre-40S particle, recruits the endonucleolytic factor(s). The development of an in vitro assay for site D processing, and the characterization of factors interacting with Rrp10p should allow us to define better its role in 18S rRNA synthesis.
Interestingly, very few factors involved in the maturation of the 3′ end of 18S rRNA in eukaryotic organisms, or of its 16S equivalent in bacteria and archaea, have been characterized. Implication of the snoRNAs U3, U22 and E2 in the processing of pre-rRNA at site 2 in Xenopus oocytes (the vertebrate equivalent of site D of S.cerevisiae pre-rRNA) has been reported (Tycowski et al., 1994; Mishra and Eliceiri, 1997; Borovjagin and Gerbi, 1999). Whether the observed impairment of site 2 cleavage is a direct or indirect consequence of the snoRNA depletions remains to be established. An endoribonucleolytic activity, partially purified from Ehrlich ascites tumor cells, which cleaves in vitro the 3′ region of in vitro transcribed pre-18S rRNA, has been reported (Shumard et al., 1990) but not further documented. It should be mentioned here that while maturation of the 3′ end of the 18S rRNA occurs in the cytoplasm in yeast cells, cleavage of the pre-rRNA at site 2 (3′ end of vertebrates 18S rRNA) is a nuclear event in vertebrate cells. Thus, if the vertebrate homologues of Rrp10p were functional ones, they should be nuclear proteins. Unfortunately, no information concerning the cellular localization of these proteins is presently available. Considering the occurrence of RRP10 homologues throughout the eukaryotic and archaebacterial genomes sequenced to date, the functional homology of the members of this protein family could be investigated by assaying for functional complementation of RRP10 S.cerevisiae mutants using cloned cDNAs encoding RRP10 homologues, or using RNAi approaches.
Materials and methods
Media, strains and plasmids
Yeast media were prepared as described previously (Bousquet-Antonelli et al., 2000). Escherichia coli DH5α was used for molecular cloning and propagation of plasmids. Yeast strains used and constructed in this study are listed in Table I. Constructions of vectors pJPG203 (CEN, ADE3, URA3, GAR1), pJPG225 (CEN, LYS2, gar1-10), pJPG219 (CEN, TRP1, gar1-10), pJPG67 (CEN, TRP1, GAR1) and pZUT3 (CEN, URA3, GAR1::GFP) have been described (Girard et al., 1994; Venema et al., 1997; Trumtel et al., 2000). Construction of pEV24, a plasmid directing the production of a GFP-Rrp10p fusion protein is described in the Supplementary data. For Rrp10p depletion, cells growing exponentially in YPG medium at 30°C were harvested by centrifugation, washed, and resuspended in YPD medium. During growth, cells were periodically diluted with pre-warmed medium in order to be kept in exponential growth phase. Yeast cells were transformed as previously described (Bousquet-Antonelli et al., 2000).
Table I. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| YO24 | MATa ade2 ade3 his4-260 leu2 lys2 trp1-1 ura3-52 can1-100 URA3::GAL::GAR1 | Henras et al. (1998) |
| YO89 | MATα ade2 ade3 his3 leu2 lys2 tyr7-1 trp1-1 ura3-52 can1-100 LEU2::gar1-Δ rrp10-1/pJPG203/pJPG225 | this study |
| YO89-13 | MATα ade2 ade3 his3 leu2 lys2 tyr7-1 trp1-1 ura3-52 can1-100 TRP1::gar1-Δ rrp10-1/pJPG203/pJPG225 | this study |
| YO126 | MATα ade2 ade3 his3 leu2 lys2 tyr7-1 trp1-1 ura3-52 can1-100 LEU2::gar1-Δ/pJPG203/pJPG225 | Venema et al. (1997) |
| YO296 | MATα leu2-Δtrp1 his3-Δ200 gal2 gal-Δ108 HIS3::GAL::PROTA-RRP10 | this study |
| JG540 | MATa ade2 ade3 his4-260 leu2 lys2 trp1-1 ura3-52 can1-100 | Bousquet-Antonelli et al. (2000) |
| R934 (Δxrn1) | MATa ade2-1 his3-11,15 trp1-1 ura3-52 URA3::xrn1-Δ | provided by S.Kearsey (Petfalski et al., 1998) |
| YDL402 | MATα leu2-Δtrp1 his3-Δ200 gal2 gal-Δ108 | Lafontaine and Tollervey (1996) |
Synthetic lethal screen
This screen, based on ADE2/ADE3 red/white sectoring coupled to fluoroorotate sensitivity/resistance tests (Kranz and Holm, 1990), has been previously described (Venema et al., 1997), and will be only briefly summarized here. Strain YO126, which carries an insertional disruption of the GAR1 gene, the wild-type GAR1 allele on a CEN, ADE3, URA3 plasmid (pJPG203), and the thermosensitive gar1-10 allele on a CEN, LYS2 plasmid (pJPG225), was UV mutagenized. Among 17 300 screened colonies, three failed to form white sectors on YPsec and did not grow on 5-FOA medium.
Cloning and sequencing of RRP10 and rrp10-1
Strain YO89-13, a derivative of YO89, was transformed with a multi-copy yeast genomic library cloned in pFL46S (2µS, LEU2) (Bonneaud et al., 1991), the leu+ transformants were replica plated onto YPSec to identify sectoring colonies, which were then streaked on 5-FOA-containing plates. Cloning of RRP10 ORFs from pEV6 (2µS, LEU2, insert) and from YO89 genomic DNA leading to plasmids pEV21 (CEN ARS LEU2 PROTA::RRP10) and pEV22 (CEN ARS LEU2 PROTA::rrp10-1), respectively, is described in the Supplementary data.
RNA analysis
RNA isolation and northern hybridizations were carried out as previously described (Henras et al., 1998). The following oligonucleotides were used for northern hybridizations (see Figure 1): 5′-CATGGCTTAATC TTTGAGAC-3′ (probe 1); 5′-TTAAGCGCAGGCCCGGCTGG-3′ (probe 2); 5′-GATTGCTCGAATGCCCAAAG-3′ (probe 3); 5′-TGC GTTCAAAGATTCGATG-3′ (probe 4); 5′-GGCCAGCAATTTCAAG TTA-3′ (probe 5); 5′-CTCACGACGGTCTAAACCC-3′ (probe 6). For analysis of the fractions from glycerol gradients and of the immunoprecipitated RNAs the 20S pre-rRNA probe used was prepared by multiprime labelling (Amersham) using a D-A2 DNA fragment as template.
Pulse–chase labelling of pre-rRNA
Metabolic labelling of RNA was performed as described previously (Bousquet-Antonelli et al., 2000). Strain YO296 (GAL::PROTA-RRP10) was grown at 30°C in YPG medium, shifted to YPD medium, grown in this medium for 14 h and then grown for an additional 9 h in YNB Glu medium without methionine (GAL::RRP10 Glu; Figure 3), or grown at 30°C in YPG medium and shifted to YNB Gal medium without methionine and grown for 9 h in this medium (GAL::RRP10 Gal; Figure 3). Labelling, chase and processing of the samples were carried out as previously described (Bousquet-Antonelli et al., 2000).
Immunoelectron microscopy
The ProtA-Rrp10p and Gar1p-ProtA expressing strains used were, respectively, strain YO296 (GAL::PROTA-RRP10) grown on galactose-containing medium, and strain YO24 transformed with pMCGZZ1 (CEN, TRP1, GAR1-PROTA) (Henras et al., 1998) grown on glucose-containing medium. Cell fixation, cutting and mounting of sections on nickel grids were performed as described in Leger-Silvestre et al. (1999). Grids were incubated for 2 h at room temperature with anti-protein A antibodies (Sigma) diluted to 1/1000 in phosphate saline buffer containing 2% bovine serum albumin. The sections were washed in the same buffer, and incubated for 1 h with colloidal gold-conjugated protein A (British Biolcell International) diluted to 1/20. Sections were contrasted with 5% aqueous uranyl acetate and imaged in a JEOL-1200 EX electron microscope operating at 80 kV.
In situ hybridization
To obtain a probe complementary to the 5′ region of the ITS1, the D-A2 rDNA fragment was PCR amplified and subcloned into pGEM4 (Promega). The resulting plasmid was linearized at a XbaI restriction site next to site D, and used for multiple cycles of DNA synthesis in a thermocycler using Goldstar DNA polymerase, primer 5′-CTCCACAGTGTGTTGTATTG-3′, which encompasses site A2, and a mix of dNTPs containing dUTP-biotin. The single-stranded DNA probe obtained was purified from free dUTP-biotin by precipitation and used for in situ hybridization on ultra-thin sections as described previously (Leger-Silvestre et al., 1999).
Glycerol gradients and protein and RNA analyses of the fractions
Sedimentation profiles on glycerol gradients were obtained as described by Bousquet-Antonelli et al. (2000).
Immunoprecipitations
Yeast cells were harvested from 0.5 l cultures in exponential growth phase (OD600 nm = 0.8), washed with the extraction buffer (20 mM HEPES pH 8.0, 1 mM MgCl2, 0.2 M KCl, 0.2% Triton X-100, 1 mM dithiothreitol, supplemented with a protease inhibitors cocktail), suspended in the same buffer and broken in ‘one shot cell disrupter’ (Henras et al., 1998). To 30 000 g supernatants of the broken cells, 0.1 ml of IgG– Sepharose beads were added, and these suspensions were incubated for 1 h at 6°C on a rocking table. The IgG–Sepharose beads were washed repeatedly with the same buffer, and bound RNAs were extracted as described previously (Ganot et al., 1997).
Western blotting
Proteins from total extracts were separated by SDS–PAGE. Fractions from sucrose gradients that contained the ribosomal subunits, or from glycerol gradients, were trichloroacetic acid precipitated and their protein content analysed by SDS–PAGE and western blotting. Samples were transferred onto nitrocellulose membranes (Hybond C-super; Amersham/Pharmacia). ProtA-Rrp10p was detected using the rabbit HRP-conjugated anti-protein A antibody (PAP; Dako, SA) diluted to 1/5000, Gar1p using a rabbit polyclonal anti-Gar1 antiboby (Girard et al., 1992) diluted to 1/200; ribosomal proteins rpL3, rpL32 and rpS4 were detected with a mouse monoclonal anti-rpL3 antiboby diluted to 1/5000, and a rabbit polyclonal anti-rpL32 antibody cross reacting with rpS4 (kindly provided by J.Warner) diluted to 1/2000, followed by ECL detection (Amersham/Pharmacia).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to J.Warner, Ed Hurt and D.Lafontaine for providing us with antibodies, plasmids and yeast strains. We are indebted to C.Normand who carried out the sl screen, and to R.Jahangir who participated in the 20S in situ localization. We also thank members of the Ferrer laboratory for helpful discussions and Y.Henry for critical reading of the manuscript. We would like to thank Y.de Préval (LBME, Toulouse, France) for the synthesis of oligonucleotides, D.Villa for the photographs and A.Rivals for technical support. Immunodetection by electron microscopy was performed in the laboratory of Professor N.Gas (LBME, Toulouse, France). This work was supported by the CNRS, the Université Paul Sabatier and grants from the Association pour la Recherche sur le Cancer (ARC) to P.-E.G., and from the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires du Ministère de l’Education Nationale de l’Enseignement Supérieur et de la Recherche (MENRT). E.V. was supported by grants from the MENRT, C.B.-A. was supported by grants from the MENRT and the ARC.
References
- Allmang C., Mitchell,P., Petfalski,E. and Tollervey,D. (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res., 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaya P., Evans,S.C., Dai,C., Lozano,G. and May,G.S. (1998) Isolation of the Aspergillus nidulans sudD gene and its human homologue. Gene, 211, 323–329. [DOI] [PubMed] [Google Scholar]
- Angermayr M. and Bandlow,W. (1997) The type of basal promoter determines the regulated or constitutive mode of transcription in the common control region of the yeast gene pair GCY1/RIO1. J. Biol. Chem., 272, 31630–31635. [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L. and Hall,B.D. (1982) Codon selection in yeast. J. Biol. Chem., 257, 3026–3031. [PubMed] [Google Scholar]
- Berges T., Petfalski,E., Tollervey,D. and Hurt,E.C. (1994) Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J., 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos,O., Li,G.Y., Labouesse,M., Minvielle-Sebastia,L. and Lacroute,F. (1991) A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/ E. coli shuttle vectors. Yeast, 7, 609–615. [DOI] [PubMed] [Google Scholar]
- Borovjagin A.V. and Gerbi,S.A. (1999) U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol., 286, 1347–1363. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Henry,Y., Gelugne J.P., Caizergues-Ferrer,M. and Kiss,T. (1997) A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J., 16, 4770–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Vanrobays,E., Gelugne,J.P., Caizergues-Ferrer,M. and Henry,Y. (2000) Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA, 6, 826–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C.J. and Cannon,M. (1980) Maturation of ribosomal precursor RNA in Saccharomyces cerevisiae. A mutant with a defect in both the transport and terminal processing of the 20 S species. J. Mol. Biol., 143, 179–199. [DOI] [PubMed] [Google Scholar]
- Carter C.J., Cannon,M. and Jimenez,A. (1980) A trichodermin-resistant mutant of Saccharomyces cerevisiae with an abnormal distribution of native ribosomal subunits. Eur. J. Biochem., 107, 173–183. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Ingram,M.F., Rosal,P.H. and Graham,T.R. (1999) Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol., 147, 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A. and Wolfe,K.H. (2000) Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast, 16, 1131–1145. [DOI] [PubMed] [Google Scholar]
- Doye V. and Hurt,E.C. (1995) Genetic approaches to nuclear pore structure and function [published erratum appears in Trends Genet., 1995, 11, 293]. Trends Genet., 11, 235–241. [DOI] [PubMed] [Google Scholar]
- Finley D., Bartel,B. and Varshavsky,A. (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature, 338, 394–401. [DOI] [PubMed] [Google Scholar]
- Ford C.L., Randal-Whitis,L. and Ellis,S.R. (1999) Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res., 59, 704–710. [PubMed] [Google Scholar]
- Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J.P., Lehtonen,H., Caizergues-Ferrer,M., Amalric,F., Tollervey,D. and Lapeyre,B. (1992) GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J., 11, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J.P., Bagni,C., Caizergues-Ferrer,M., Amalric,F. and Lapeyre,B. (1994) Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J. Biol. Chem., 269, 18499–18506. [PubMed] [Google Scholar]
- Guarente L. (1993) Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet., 9, 362–366. [DOI] [PubMed] [Google Scholar]
- Henras A., Henry,Y., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Gelugne,J.P. and Caizergues-Ferrer,M. (1998) Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J., 17, 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.L., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Holt C.L. and May,G.S. (1996) An extragenic suppressor of the mitosis-defective bim D6 mutation of Aspergillus nidulans codes for a chromosome scaffold protein. Genetics, 142, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz J.E. and Holm,C. (1990) Cloning by function: an alternative approach for identifying yeast homologues of genes from other organisms. Proc. Natl Acad. Sci. USA, 87, 6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and de La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger-Silvestre I., Trumtel,S., Noaillac-Depeyre,J. and Gas,N. (1999) Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae. Chromosoma, 108, 103–113. [DOI] [PubMed] [Google Scholar]
- Lubben B., Fabrizio,P., Kastner,B. and Luhrmann,R. (1995) Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem., 270, 11549–11554. [DOI] [PubMed] [Google Scholar]
- Madden K. and Snyder,M. (1998) Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol., 52, 687–744. [DOI] [PubMed] [Google Scholar]
- Mishra R.K. and Eliceiri,G.L. (1997) Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc. Natl Acad. Sci. USA, 94, 4972–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K. and Warner,J.R. (1994) Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol., 14, 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E., Dandekar,T., Henry,Y. and Tollervey,D. (1998) Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol., 18, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta R.J. and Mager,W.H. (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast, 14, 471–477. [DOI] [PubMed] [Google Scholar]
- Ripmaster T.L., Vaughn,G.P. and Woolford,J.L.,Jr (1993) DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7901–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumard C.M., Torres,C. and Eichler,D.C. (1990) In vitro processing at the 3′-terminal region of pre-18S rRNA by a nucleolar endoribonuclease. Mol. Cell. Biol., 10, 3868–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Tekotte,H., Grosjean,H., Segref,A., Sharma,K., Tollervey,D. and Hurt,E.C. (1996) Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J., 15, 2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Stevens A., Hsu,C.L., Isham,K.R. and Larimer,F.W. (1991) Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′–3′ exoribonuclease 1. J. Bacteriol., 173, 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J., Retel,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- Trumtel S., Leger-Silvestre,I., Gleizes,P.E., Teulieres,F. and Gas,N. (2000) Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell, 11, 2175–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Shu,M.D. and Steitz,J.A. (1994) Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science, 266, 1558–1561. [DOI] [PubMed] [Google Scholar]
- Udem S.A. and Warner,J.R. (1973) The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem., 248, 1412–1416. [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J., 15, 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Venema J., Bousquet-Antonelli,C., Gelugne,J.P., Caizergues-Ferrer,M. and Tollervey,D. (1997) Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol., 17, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J.L. Jr and Warner,J.R. (1991) The ribosome and its synthesis. In Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 587–626.