Abstract
The potential of expression analysis using cDNA microarrays to address complex problems in a wide variety of biological contexts is now being realised. A limiting factor in such analyses is often the amount of RNA required, usually tens of micrograms. To address this problem researchers have turned to methods of improving detection sensitivity, either through increasing fluorescent signal output per mRNA molecule or increasing the amount of target available for labelling by use of an amplification procedure. We present a novel DNA-based method in which an oligonucleotide is incorporated into the 3′ end of cDNA during second-strand cDNA synthesis. This sequence provides an annealing site for a single complementary heel primer that directs Taq DNA polymerase amplification of cDNA following multiple cycles of denaturation, annealing and extension. The utility of this technique for transcriptome-wide screening of relative expression levels was compared to two alternative methodologies for production of labelled cDNA target, namely incorporation of fluorescent nucleotides by reverse transcriptase or the Klenow fragment. Labelled targets from two distinct mouse tissues, adult liver and kidney, were compared by hybridisation to a set of cDNA microarrays containing 6500 mouse cDNA probes. Here we demonstrate, through a dilution series of cDNA derived from 10 µg of total RNA, that it is possible to produce datasets comparable to those produced with unamplified targets with the equivalent of 30 ng of total RNA. The utility of this technique for microarray analysis in cases where sample is limited is discussed.
INTRODUCTION
Expression analysis using DNA microarrays is rapidly becoming the method of choice for researchers wishing to identify differentially expressed transcripts in tissues of interest (for reviews see 1–3). However, for many researchers, such as developmental biologists, the requirement for large amounts of starting RNA, usually 10–100 µg of total RNA, frequently prohibits the analysis of specific structures or cells of interest. Several different approaches have been taken to address this issue. These fall primarily into two groups: either attempting to increase the fluorescent signal output per molecule, such as the use of the dendrimer technology (4), tyramide signal amplification (5) and amino-allyl labelling (6); or increasing the amount of target available for labelling through amplification, such as TPEA (7), RAGE (8), SMART-PCR (9) and in vitro transcription (IVT) (10–13).
Concerns arising from use of these technologies include reproducibility, reliability, conservation of differential expression, associated costs and ease of use. Here we describe a novel single primer amplification (SPA) method for use with spotted microarrays that addresses each of these concerns. The first steps in this protocol generate double-stranded cDNA, initially primed by a modified oligo(dT) primer. A primer equivalent to the heel of the modified oligo(dT) primer is then used to direct semi-linear Taq DNA polymerase amplification. The protocol is essentially a modified cycle sequencing reaction, familiar and accessible to the majority of molecular biologists.
MATERIALS AND METHODS
RNA extraction
Total RNA was extracted from adult female mouse liver and kidney (strain 3H1) using Trizol (Invitrogen) followed by a RNeasy midi kit (Qiagen) according to the manufacturer’s instructions. The quantity and quality of RNA was assessed using a Bioanalyzer 2100 (Agilent). RNA samples were pooled and aliquoted to ensure technical comparisons would not represent differences in RNA source or quality.
Target preparation
Preparation of labelled target cDNA for hybridisation to microarray slides was carried out using one of three methods.
Reverse transcriptase (RT) labelling. For the standard RT labelling method 100 µg of total RNA was heated for 10 min at 70°C in the presence of 4 µg oligo(dT15) (total volume 24 µl). Samples were cooled to 42°C and 26 µl labelling reaction mixture added [10 µl 5× RT buffer (BRL); 5 µl 0.1 M DTT (BRL); 25 mM dATP, dGTP and dTTP, 2.5 mM dCTP (Abgene); 1 mM Cy3 or Cy5-dCTP (APB); 2 µl RNase inhibitor; 400 U M-MLV II (Superscript II; BRL)]. Samples were incubated at 42°C for 4 h, with 200 U M-MLV II added each hour. Following first-strand cDNA synthesis, 20 µg RNase A was added to each sample and incubated at 37°C for 20 min. Reactions were stopped by the addition of 5 µl 0.5 M EDTA.
Klenow labelling. For labelling of unamplified RNA using Klenow fragment, 20 µg total RNA was employed in first- and second-strand synthesis reactions, employing the ‘cDNA synthesis system’ (Roche, catalogue no. 1117831). First-strand synthesis used oligo(dT) priming. Double-stranded cDNA samples were purified using a PCR purification kit (Qiagen) and eluted with 26 µl water. An aliquot of 5 µl was subjected to gel electrophoresis to confirm quantity and quality. Samples were fluorescently labelled using the genomic DNA protocol published online at http://cmgm.stanford.edu/pbrown/protocols/4_genomic.html. Briefly, a 21 µl sample was mixed with 20 µl random octamer oligonucleotides (Bioprime Kit; Invitrogen). Samples were heated to 90°C for 5 min and snap cooled on ice. An aliquot of 5 µl 10× dNTP mix was added to each sample (1.2 mM each dATP, dGTP and dTTP, and 0.6 mM dCTP), followed by 3 µl Cy5-dCTP or Cy3-dCTP (1 mM stocks; Amersham). An aliquot of 1 µl high concentration Klenow fragment (40–50 U/µl) was added and the samples incubated at 37°C for 2 h. Reactions were stopped by the addition of 5 µl 0.5 M EDTA pH 8.0.
Single primer amplification reactions. For production of SPA cDNA, 10 µg of total RNA was used in first- and second-strand synthesis reactions, again employing the ‘cDNA synthesis system’ (Roche, catalogue no. 1117831). However, a modified oligonucleotide replacing oligo(dT) (5′-AAACGACGGCCAGTGAATTGTAATACGACTCACTATAGGCGCTTTTTTTTTTTTTTTTV-3′) was used to prime first-strand synthesis. This oligonucleotide contains the T7 RNA polymerase promoter sequence. This was chosen as it represents a non-eukaryotic sequence. We also utilised the heel primer from the TPEA technique (7), with comparable results (data not shown).
After completion of second-strand synthesis, samples were purified using a PCR purification kit (Qiagen) and eluted with 100 µl water. This 100 µl sample was taken to represent 10 µg of starting RNA and dilutions representing 0.5–0.031 µg starting RNA were employed in the amplification step. The assumption of 100% recovery after cDNA synthesis and purification is likely to overestimate the amount of RNA represented in each sample. Amplification reactions were set up as follows. Aliquots of 10 µl 10× PCR buffer (including a final Mg2+ concentration of 1.5 mM), 10 µl dNTPs at 2 mM concentration, 100 pmol HPLC purified heel primer (5′-CGGCCAGTGAATTGTAATACGACTCACTA TAGGCG-3′), 12.5 U Taq DNA polymerase (Abgene) and cDNA template were combined in water to 100 µl total volume. Reactions were placed in a tetrad thermocycler (MJ Research) and incubated at 94°C for 3 min, then subjected to 40 cycles of 94°C (1 min), 59°C (1 min) and 72°C (2 min). Samples were again purified (Qiaquick PCR purification kit; Qiagen) and labelled using the method described in the Klenow labelling section above.
Microarray construction
Microarrays were manufactured using CMT-GAPS II slides (Corning) with a MicroTAS arrayer (BioRobotics). PCR probe elements were manufactured from plates 17–32 of the NIA mouse developmental set (14). PCR amplifications were performed incorporating 5 µl 10× PCR buffer, 5 µl 2 mM dNTPs, 2.5 µl 10 µM each primer (5′-CCAGTCACGACGT TGTAAAACGAC-3′ and 5′-NH2-GTGTGGAATTGTGAG CGGATAACAA-3′), 0.125 µl HotStart Taq DNA polymerase (Qiagen), 37.5 µl 1:10 glycerol stock and water to 150 µl. PCR samples were incubated at 95°C for 10 min, then subjected to 30 cycles of 95°C for 1 min, 60°C for 2 min and 72°C for 7 min. A final incubation step of 72°C for 10 min was carried out.
Amplification reactions were then purified using Multiscreen-PCR plates (Millipore) and eluted into 100 µl water. Two microlitres was subjected to agarose gel electrophoresis to determine quality and yield. Remaining sample was precipitated and resuspended in 25 µl arraying solution (150 mM sodium phosphate buffer pH 8.5, 0.001% sarkosyl). Samples were submitted for arraying if a single band was observed upon gel electrophoresis and a concentration between 250 and 500 ng/µl was obtained. Each EST probe was spotted on each slide in duplicate.
Microarray hybridisation
Following termination of the labelling reactions individual samples were mixed with the correctly labelled corresponding sample and passed through a nucleotide removal kit (Qiagen). Mouse Cot-1 DNA (10 µg) and 20 µg of T7-dT primer/oligo(dA)/oligo(dT) (amplified target/RT labelled target/Klenow labelled target, respectively) were then added to the mixed sample. Sample volume was reduced to 10 µl using a speed vac (Heto) and mixed with 30 µl hybridisation solution to give a final concentration of 50% formamide, 6× SSC, 0.2% SDS. Samples were heated to 80°C for 5 min then placed at 42°C for 30 min to allow repeat blocking. Samples were briefly subjected to centrifugation and pipetted beneath coverslips onto microarray slides incubated on a hot block at 42°C. Microarrays were placed into hybridisation chambers (Corning), the end wells were filled with 10 µl hybridisation buffer and the chambers sealed. Microarrays were placed in a waterbath at 42°C overnight.
Microarray washing
Microarrays were removed from the hybridisation chambers and washed in 2× SSC for 3 min; 0.1× SSC, 0.1% SDS for 3 min; 0.1× SSC for 3 min. Slides were dried by centrifugation for 5 min at 60 g and entered for scanning.
Data capture and analysis
Microarrays were scanned using an Affymetrix 428 scanner, producing a 16 bit tif file for each of the dyes used. The images were analysed and data quantified using ImaGene 4.1 (BioDiscovery). During image processing, an automatic flagging process was used to remove spots whose background-subtracted mean signal intensity was less than four times the standard deviation of the background pixel intensity. The cut-off level of four was chosen due to previous observations made with the hardware/software combination. This level was found to be most efficient at removing background level spots without flagging genuine signal. Manual flagging was also used to highlight spot irregularities such as dust, scratches and misaligned features. Data for each experiment were scaled for intensity using a least squares regression technique where each individual kidney data set was scaled against the mean of all kidney data sets and likewise for the liver data sets (Table 1) (15). Data were then normalised using the robust scatter plot smoothing technique, lowess, with print tip scaling from the ‘Statistical Microarray Analysis’ (16) library for statistical software package R (17).
Table 1. Experimental design.
| Microarray | Labelling method | Relative amount | Kidney label | Liver label | cDNA synthesis | Regression line between each kidney channel and mean of all kidney channels | Regression line between each liver channel and mean of all liver channels |
|---|---|---|---|---|---|---|---|
| 1 | RT | 100 | Cy 5 | Cy 3 | 5 | y = 0.80x + 0.35 | y = 1.09x – 0.76 |
| 2 | RT | 100 | Cy 3 | Cy 5 | 6 | y = 0.73x + 0.60 | y = 1.01x – 0.34 |
| 3 | RT | 100 | Cy 5 | Cy 3 | 7 | y = 0.95x – 0.46 | y = 0.79x + 0.44 |
| 4 | RT | 100 | Cy 3 | Cy 5 | 8 | y = 0.97x – 0.37 | y = 0.82x + 0.50 |
| 5 | Klenow | 20 | Cy 5 | Cy 3 | 3 | y = 1.01x – 0.12 | y = 0.66x + 1.05 |
| 6 | Klenow | 20 | Cy 3 | Cy 5 | 3 | y = 0.76x + 0.70 | y = 0.67x + 0.89 |
| 7 | Klenow | 20 | Cy 5 | Cy 3 | 4 | y = 0.89x + 0.28 | y = 0.74x + 0.80 |
| 8 | Klenow | 20 | Cy 3 | Cy 5 | 4 | y = 0.74x + 0.52 | y = 0.72x + 0.71 |
| 9 | SPA | 0.5 | Cy 5 | Cy 3 | 1 | y = 0.92x – 0.18 | y = 1.18x – 0.84 |
| 10 | SPA | 0.5 | Cy 3 | Cy 5 | 1 | y = 1.15x – 0.84 | y = 1.05x – 0.28 |
| 11 | SPA | 0.5 | Cy 5 | Cy 3 | 2 | y = 1.07x – 0.20 | y = 1.32x – 1.01 |
| 12 | SPA | 0.5 | Cy 3 | Cy 5 | 2 | y = 1.18x – 0.61 | y = 1.04x – 0.16 |
| 13 | SPA | 0.25 | Cy 5 | Cy 3 | 1 | y = 1.10x – 0.37 | y = 1.29x – 1.06 |
| 14 | SPA | 0.25 | Cy 3 | Cy 5 | 1 | y = 1.13x – 0.43 | y = 1.03x – 0.14 |
| 15 | SPA | 0.25 | Cy 5 | Cy 3 | 2 | y = 1.14x – 0.55 | y = 1.39x – 1.30 |
| 16 | SPA | 0.25 | Cy 3 | Cy 5 | 2 | y = 1.37x – 1.14 | y = 1.16x – 0.59 |
| 17 | SPA | 0.125 | Cy 5 | Cy 3 | 1 | y = 0.94x + 0.33 | y = 1.05x – 0.03 |
| 18 | SPA | 0.125 | Cy 3 | Cy 5 | 1 | y = 1.09x – 0.38 | y = 1.00x – 0.17 |
| 19 | SPA | 0.125 | Cy 5 | Cy 3 | 2 | y = 1.03x – 0.17 | y = 1.21x – 0.69 |
| 20 | SPA | 0.125 | Cy 3 | Cy 5 | 2 | y = 1.19x – 0.62 | y = 1.06x – 0.19 |
| 21 | SPA | 0.0625 | Cy 5 | Cy 3 | 1 | y = 0.92x + 0.08 | y = 1.09x – 0.43 |
| 22 | SPA | 0.0625 | Cy 3 | Cy 5 | 1 | y = 1.11x – 0.62 | y = 1.02x – 0.15 |
| 23 | SPA | 0.0625 | Cy 5 | Cy 3 | 2 | y = 1.06x – 0.23 | y = 1.23x – 0.73 |
| 24 | SPA | 0.0625 | Cy 3 | Cy 5 | 2 | y = 1.12x – 0.42 | y = 1.01x – 0.07 |
| 25 | SPA | 0.03125 | Cy 5 | Cy 3 | 1 | y = 0.97x + 0.26 | y = 1.07x – 0.19 |
| 26 | SPA | 0.03125 | Cy 3 | Cy 5 | 1 | y = 0.96x + 0.12 | y = 0.89x + 0.25 |
| 27 | SPA | 0.03125 | Cy 5 | Cy 3 | 2 | y = 1.00x – 0.04 | y = 1.16x – 0.52 |
| 28 | SPA | 0.03125 | Cy 3 | Cy 5 | 2 | y = 1.15x – 0.51 | y = 1.04x – 0.18 |
| 29 | SPA | 0.5 | Cy 5 & Cy 3 | 3 & 4 | y = 1.12x – 0.32, y = 0.96x + 0.27 | ||
| 30 | SPA | 0.5 | Cy 5 & Cy 3 | 4 & 3 | y = 1.25x – 0.90, y = 1.02x – 0.01 |
Dye incorporation methodology, dye orientation, relative amount of starting material and cDNA synthesis reaction number are shown for all 30 microarrays (29 and 30 are self–self hybridisations). Also shown are two equations for the linear regression lines used to scale each data set against the mean of all data sets for the kidney and liver channels, respectively.
On-slide duplicates were averaged and filtered to remove flagged data using GeneSpring (Silicon Genetics). Spots flagged in one or more arrays were excluded from further analysis. Data were exported to R where differential genes were selected from each slide using the single slide method described by Newton et al. (18). These lists of genes identified as significantly different on each slide were combined for the four slides within each target preparation methodology.
The reproducibility of the techniques was analysed by performing analysis of variance (ANOVA) on filtered datasets using SPSS.
RESULTS
Technique overview
SPA, as for IVT amplification, relies upon the use of a modified oligo(dT) primer to drive first-strand cDNA synthesis. The complement to this primer is incorporated into every transcript following second-strand synthesis. A primer complementary to the specific oligo in the oligo(dT) sequence then binds to the denatured second-strand cDNA and drives amplification by Taq DNA polymerase cycling extensions (similar to cycle sequencing).
Experimental design
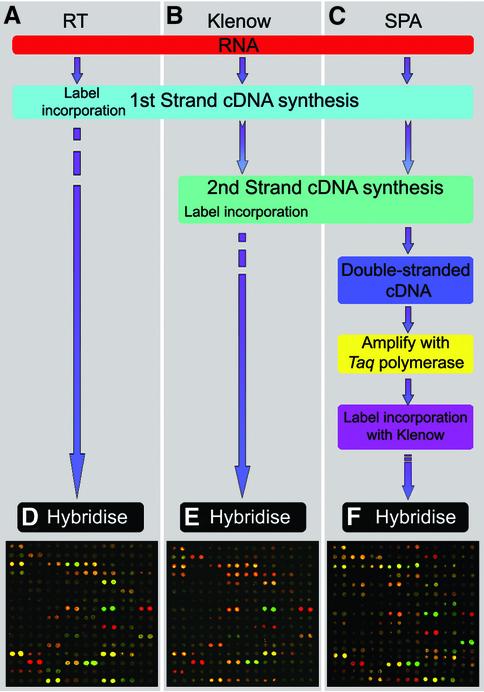
We performed 30 dual hybridisation microarray experiments using adult mouse kidney and liver RNA in order to determine the effect of single primer amplification on expression ratios obtained when compared to conventional labelling techniques (Table 1). A single source of liver and kidney total RNA was used for all experiments and either Cy3 or Cy5-dCTP was incorporated during labelling. We labelled four kidney and four liver RNA aliquots (100 µg) using direct incorporation by reverse transcriptase (henceforth called RT labelling; see Materials and Methods) and performed dual hybridisations in duplicate with both orientations of dye incorporation (dye swaps). Four equivalent hybridisations were performed using two kidney and two liver RNA samples (20 µg) and random primed labelling of first-strand cDNA with Klenow fragment (henceforth called Klenow labelling; see Materials and Methods), again incorporating dye-swap controls. Finally, two liver and two kidney samples were reverse transcribed with a modified oligo(dT) primer and converted to double-stranded cDNA. Each cDNA synthesis was diluted to provide five distinct starting concentrations equivalent to 0.5, 0.25, 0.125, 0.063 and 0.031 µg of total RNA. Each of these, totalling 20, was then amplified using the SPA method (see Materials and Methods). Amplified cDNA was labelled twice, once with Cy3 and once with Cy5 incorporation, by Klenow fragment using random priming (see Materials and Methods). Twenty dual hybridisations were then performed with reciprocal labelling reactions from distinct amplifications (Table 1). Figure 1 shows a schematic overview and representative microarray images for the three techniques.
Figure 1.
(A) A schematic overview of the three methods compared. In the RT experiments first-strand cDNA is fluorescently labelled through incorporation of Cy3-dCTP or Cy5-dCTP by reverse transcriptase. Samples are combined and hybridised to the microarray. (B) For the Klenow experiments cDNA is fluorescently labelled through random primed Klenow extension following first-strand cDNA synthesis. (C) In the SPA experiments double-stranded cDNA is entered for amplification and the resultant products labelled by random primed Klenow extension. (D–F) False colour overlay images of the same subsection of the array hybridised with cDNA derived from (D) 100 µg total RNA labelled with RT cDNA, (E) 20 µg of total RNA labelled with Klenow and (F) SPA cDNA equivalent to 0.031 µg total RNA.
Data analysis: reliability and utility
One of the primary concerns of researchers conducting amplification for microarray analysis is the reproducibility associated with the amplification step itself. To address this question self–self hybridisations derived from two independent SPA amplifications of both kidney and liver cDNA were characterised. The Pearson correlation coefficients for these are r = 0.985 and r = 0.986 for kidney and liver, respectively.
To determine whether target cDNA amplification had affected our ability to reliably profile gene transcription in the kidney and liver RNA samples, based on comparison with data produced with labelled target produced using conventional methods, we employed two complementary analyses of the 28 relevant datasets. First, we utilised a statistical analysis of variance (ANOVA) to ascertain the major determinants of differences across all data sets. Secondly, we identified differentially expressed genes observed in all data sets and determined the similarity between these sets of outliers across the three different target production and labelling techniques.
Analysis of variance (ANOVA). The Levene’s homogeneity of variance test (19) is utilised to examine the distribution of the individual data sets and test for equality of variances within the set, an assumption necessary for ANOVA. Application of this test to a random subset of genes from our data set was found to be not significant (throughout the ANOVA analysis significance was tested at the 95% level). To test for differences between methodologies we performed ANOVA (using SPSS) on the log-transformed ratios for each hybridisation. We examined the variation arising from methodology, replicates, amount of starting material, colour swaps and cDNA synthesis.
Variation between the datasets grouped by the three methodologies was found to be significant [Table 2(a)]. To ascertain the cause of this difference further analysis was undertaken. On examination of the variation within each methodology [Table 2(b)] there was a highly significant difference between the RT replicates, although no significant difference was observed between Klenow replicates and between SPA replicates, respectively. Comparing all data sets, variation between the two colour swap orientations is highly significant [Table 2(c)], however, when this is examined by methodology, the difference is only found in the RT group [Table 2(d)]. Perhaps surprisingly, the variation within amplification due to the amount of starting material used was not significant [Table 2(e)]. This was still not significant when comparing across all methodologies.
Table 2. Summary of ANOVA results for each source of variation.
| Source | d.f. | F | Significance | |
|---|---|---|---|---|
| (a) | Variation between RT, Klenow and SPA | 2 | 4.53 | 0.01076a |
| (b) | Variation within RT | 3 | 6.54 | 0.00021b |
| Variation within Klenow | 3 | 1.04 | 0.37375 | |
| Variation within SPA | 19 | 0.79 | 0.71678 | |
| (c) | Variation between colourswap | 1 | 15.61 | 0.00008b |
| (d) | Variation between colourswap within RT | 1 | 19.50 | 0.00001b |
| Variation between colourswap within Klenow | 1 | 2.48 | 0.11538 | |
| Variation between colourswap within SPA | 1 | 3.29 | 0.06988 | |
| (e) | Variation between amount of starting material (only within SPA) | 4 | 0.32 | 0.86389 |
| Variation between amount of starting material (across all experiments) | 6 | 1.70 | 0.11635 | |
| (f) | Variation between cDNA syntheses | 7 | 5.16 | 0.00001b,c |
(a) The variation between the three methodologies (RT incorporation, Klenow and SPA with Klenow labelling) is shown to be highly significant. (b) When examined within each of the three methodologies the variation is only significant within RT. (c) Differences between the two dye orientations are found to be highly significant across all data but when separated into methodologies (d) the differences are only significant within RT. (e) The amount of starting material used does not cause significant differences either within the SPA amplification or across all methodologies. (f) Examination of how the cDNA synthesis affects variation across all data shows highly significant results; to identify the cause of these differences a Bonferroni test was used to compare the individual cDNA synthesis to each other and identify which samples are different.
Significance is tested using the F distribution at the 95% confidence level.
aSignificant differences.
bHighly significant.
cA Bonferroni post hoc test was performed to identify the cause of the significant differences between cDNA synthesis and showed that there were differences between the following pairs of cDNA synthesis reactions: cDNA 1 versus 3; 1 versus 7; 1 versus 8; 3 versus 5; 5 versus 7; 5 versus 8; 6 versus 7, 6 versus 8.
Variation between cDNA syntheses [Table 2(f)] was highly significant and this accounted for the majority of variation observed across all data sets. A Bonferroni test indicated that several of the cDNA syntheses used are different but these are not confined to any one method consistently.
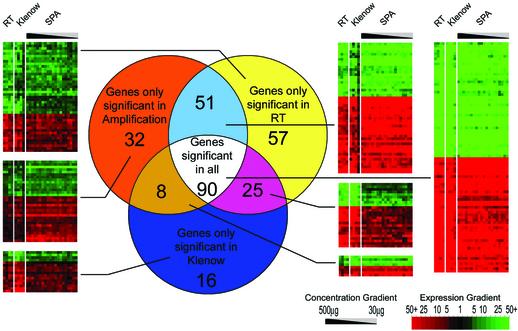
Identification of outliers. Lists of outliers [cDNA spots with differential expression above the set threshold (see Materials and Methods)] were generated to ascertain the ability of each methodology to reproducibly identify outliers. The lists generated from each of the methodologies were in turn set as the benchmark and the ability of other methodologies to reproducibly identify this list was then determined (Fig. 2). These data demonstrate that the SPA procedure allows identification of outliers as consistently as RT-based and Klenow-based labelling methods. A PubMed survey of known genes consistently identified as outliers confirmed relative expression differences highlighted by the microarray hybridisations (Fig. 2 and additional material).
Figure 2.
Reproducibility of outlier detection is demonstrated by comparison of genes selected as significant from each methodology [gene lists selected from individual slides using the methods described by Newton et al. (18) are combined if selected in all slides within each methodology to create the three segments of the Venn diagram]. The Venn diagram shows numbers of genes selected as significant in one, two or all three of the methodologies. Raster plots for the gene lists in each sector of the Venn diagram were created using single linkage clustering of each gene list in Cluster and Treeview (20). These plots demonstrate the variability of the ratio for a gene across each of the approaches used. Genes with relatively higher expression in kidney compared to liver are shown in red, while genes with relatively higher expression in liver compared to kidney are displayed in green. An expanded version of this figure listing the accession numbers of individual genes is available at www.mgu.har.mrc.ac.uk/microarray/amplification.
DISCUSSION
SPA is a reproducible and reliable method of target amplification for differential gene expression screens using cDNA microarrays. Not only does it identify outlier genes as efficiently as conventional methods, but also does so employing as little starting material as 32 ng total RNA.
The small level of disparity observed between SPA and the other techniques examined during this study can, for the most part, be attributed to the initial cDNA synthesis step, at present an unavoidable step for all microarray analysis. The amplification itself does not significantly increase the overall variability above that encountered during cDNA synthesis. Indeed, self–self hybridisation of independent amplifications demonstrates this point quite clearly.
Surprisingly, the amount of starting material was not found to be significant within the SPA hybridisations. A priori, we would have expected reproducibility to diminish as starting material was reduced. The observation that it does not suggests that 30 ng does not represent the lower limit of this technique. Indeed, recent experiments utilising this technique suggest that outlier identification may be possible using total RNA extracted from as few as 10 cells (Birgit Liss, University of Oxford, Oxford, UK, personal communication).
Our data also demonstrate that the ability to reproducibly identify outliers is undiminished in comparison to the other methods employed. The limitation for inclusion in the central grouping of Figure 2 (genes significant in all methods) is that the differential expression must have been observed in all 28 hybridisations. Omission from the central grouping is for the most part the result of failure of a single hybridisation to identify a differential greater than the threshold set. By visual inspection of the raster plots in Figure 2 it is clear to see that outliers identified by one technique are for the most part similarly coloured in the other two techniques, albeit below the 28 experiment detection threshold.
Although the efficacy of the SPA technique is apparent, the precise mechanism of amplification is unclear. The efficiency with which target can be produced from a small amount of starting material indicates that a model based on a merely linear reaction involving synthesis of one antisense cDNA copy per molecule at each round of extension is untenable. Rather, it is more likely that a combination of priming from the incorporated heel primer at the 3′ end of each cDNA molecule and mismatch priming at multiple sites transcriptome-wide results in levels of amplification significantly greater than linear, though falling short of the exponential levels associated with standard PCRs. Allied to this, Klenow labelling itself appears to possess a strand-switching function. First-strand cDNA labelled with Klenow recapitulates outlier identification when hybridised to slides arrayed with amino-linked sense strand cDNAs (data not shown). This was also observed when using sense oligonucleotide arrays. This suggests that further ‘amplification’ occurs during the labelling step that not only increases signal, but also allows hybridisation to both sense and antisense strands.
The data presented here also indicate the efficacy of incorporation of Cy dyes using Klenow fragment and random priming of first-strand cDNA. The signal intensity routinely obtained with 20 µg of total RNA is at least equivalent to that observed with conventional direct incorporation by reverse transcriptase of 100 µg of total RNA. The analyses performed also confirm that cDNA synthesis and labelling represent significant causes of variation between identical RNA samples and reaffirm the need for replicate hybridisations and, in the case of RT labelling, dye-swap replicates.
The experiments described here were performed on aliquots of total RNA that had previously been examined for high quality, e.g. absence of significant degradation. In real world examples of the use of limiting tissue samples such quality control measures may be impossible. We have recently used the SPA technique on total RNA extracted from individual male and female embryonic mouse gonads, equivalent to ∼200 ng, and have reliably detected appropriate expression of tissue-specific control genes and highlighted several novel genes not previously ascribed a sexually dimorphic expression pattern in the developing gonad (L.Sith and A.Greenfield, manuscript in preparation).
In conclusion, the Taq-based SPA protocol described here has certain advantages over previously published techniques. The simplicity of the protocol, requiring no modification, ligation or RNA amplification steps, will appeal to researchers wishing to rapidly characterise differential gene expression using DNA microarrays in cases of limiting tissue availability. This especially applies to those researchers with no previous experience in microarray sample preparation, as many will be familiar with the similar cycle sequencing protocol. Due to the stable DNA-based nature of the technique this methodology will also lend itself well to high throughput analysis of multiple small tissue samples. Our primary aim was also to develop a technique for microarray sample preparation that addressed the sometimes prohibitive costs associated with other methodologies. The technique itself uses off-the-shelf reagents rather than requiring expensive kits and thus can be considered an attractive alternative for researchers with limited funds.
Additional material is available from www.mgu.har.mrc.ac.uk/microarray/amplification. This includes the original tif image files generated from the 30 microarrays, the signal intensity results for each channel of each microarray and a detailed table of homologies associated with previously published expression data.
REFERENCES
- 1.Lockhart D. and Winzeler,E. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. [DOI] [PubMed] [Google Scholar]
- 2.Mills J.C., Roth,K.A., Cagan,R.L. and Gordon,J.I. (2001) DNA microarrays and beyond: completing the journey from tissue to cell. Nature Cell Biol., 3, E175–E178. [DOI] [PubMed] [Google Scholar]
- 3.Schulze A. and Downward,J. (2001) Navigating gene expression using microarrays – a technology review. Nature Cell Biol., 3, E190–E195. [DOI] [PubMed] [Google Scholar]
- 4.Stears R.L., Getts,R.C. and Gullans,S.R. (2000) A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiol. Genomics, 3, 93–99. [DOI] [PubMed] [Google Scholar]
- 5.Karsten S.L., Van Deerlan,V.M., Sabatti,C., Gill,L.H. and Geschwind,D.H. (2002) An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res., 30, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manduchi E., Scearce,L.M., Brestelli,J.E., Grant,G.R., Kaestner,K.H. and Stoeckert,C.J.,Jr (2002) Comparison of different labelling methods for two-channel high-density microarray experiments. Physiol. Genomics, 10, 169–179. [DOI] [PubMed] [Google Scholar]
- 7.Dixon A.K., Richardson,P.J., Lee,K., Carter,N.P. and Freeman,T. (1998) Expression profiling of single cells using 3 prime end amplification (TPEA) PCR. Nucleic Acids Res., 26, 4426–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang A., Pierce,A., Judson-Kremer,K., Gaddis,S., Aldaz,C.M., Johnson,D.G. and MacLeod,M.C. (1999) Rapid analysis of gene expression (RAGE) facilitates universal expression profiling. Nucleic Acids Res., 27, 4609–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livesey F.J., Furukawa,T., Steffen,M.A., Church,G.M. and Cepko,C.L. (2000) Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx.Curr. Biol., 10, 301–310. [DOI] [PubMed] [Google Scholar]
- 10.Van Gelder R.N., von Zastrow,M.E., Yool,A., Dement,W.C., Barchas,J.D. and Eberwine,J.H. (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl Acad. Sci. USA, 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahadevappa M. and Warrington,J. (1999) A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat. Biotechnol., 17, 1134–1136. [DOI] [PubMed] [Google Scholar]
- 12.Wang E., Miller,L.D., Ohnmacht,G.A., Liu,E.T. and Marincola,F.M. (2000) High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol., 18, 457–459. [DOI] [PubMed] [Google Scholar]
- 13.Baugh L.R., Hill,A.A., Brown,E.L. and Hunter,C.P. (2001) Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res., 29, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka T.S., Jaradat,S.A., Lim,M.K., Kargul,G.J., Wang,X., Grahovac,M.J., Pantano,S., Sano,Y., Piao,Y., Nagaraja,R. et al. (2000) Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray Proc. Natl Acad. Sci. USA, 97, 9127–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher G., Tibshirani,V.R. and Chu,G. (2001) Significance analysis of microarrays applied to the ionising radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y.H., Dudoit,S., Luu,P. and Speed,T. (2001) Normalization for cDNA microarray data. Available at: www.stat.berkeley.edu/users/terry/zarray/TechReport/589.pdf.
- 17.Ihaka R. and Gentleman,R. (1996) R: a language for data analysis and graphics. J. Comput. Graph. Stat., 5, 299–314. [Google Scholar]
- 18.Newton M.A., Kendziorski,C.M., Richmond,C.S., Blattner,F.R. and Tsui,K.W. (2001) On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J. Comput. Biol., 8, 37–52. [DOI] [PubMed] [Google Scholar]
- 19.Levene H. (1960) Robust tests for the equality of variance. In Olkin,I. (ed.), Contributions to Probability and Statistics. Stanford University Press, Palo Alto, CA, pp. 278–292.
- 20.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]