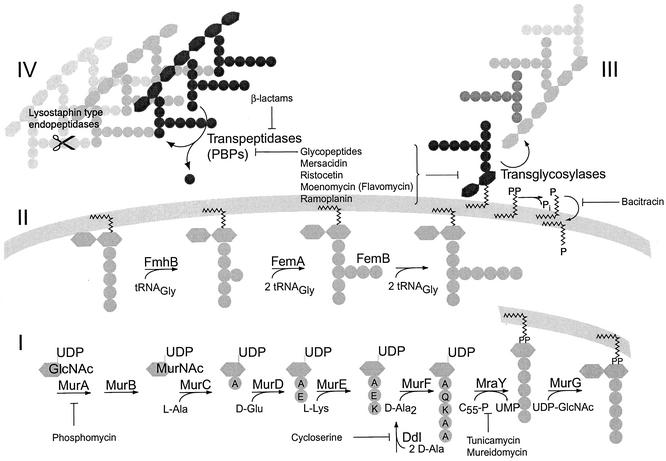
Peptidoglycan biosynthesis is one of the preferred targets for antibiotics, as peptidoglycan is unique to eubacteria and inhibition of its production is generally bactericidal (Fig. 1). Peptidoglycan, the major component of the bacterial cell wall, forms the murein sacculus, a giant macromolecule that surrounds the cell as a single, flexible meshwork and is intimately involved in cell division. Its structure determines cell shape and maintains cell integrity by protecting it against the high internal osmotic pressure. The most widely used class of antibiotics that target the cell wall are β-lactams and their derivatives. Their introduction into therapy has, however, invariably been followed by the development and spread of resistance in the bacterial community. Gram-positive bacteria have developed two main strategies against β-lactams: drug inactivation by β-lactamases and intrinsic resistance mediated by modified, low-affinity variants of the target enzymes, the so-called penicillin-binding proteins (PBPs). To reach high-level β-lactam resistance, the low-affinity PBPs of staphylococci and pneumococci, in particular, depend on correctly synthesized peptidoglycan precursors. Specifically, the interpeptide structure, which is characteristic of the peptidoglycan of many gram-positive organisms (Fig. 1; Table 1), has a great impact on the level of β-lactam resistance that is achieved, as will be discussed below.
FIG. 1.
Pathway and inhibitors of peptidoglycan synthesis in S. aureus (adapted in part from references 17 and 111). (I) The assembly of the lipid II precursor is universal for eubacteria; the MurA reaction is the first step committed to peptidoglycan synthesis (17, 111). (II) The interpeptide is synthesized on the membrane-bound precursor. (III and IV) Extracellular steps catalyzed by transglycosylases and transpeptidases (PBPs) lead to mature peptidoglycan.
TABLE 1.
All species of grampositive bacteria found to have a FemABX homolog also bear an interpeptide in their peptidoglycan
| Species | Interpeptide | Peptidyl transferase (FemABX homolog), accession no. | Reference(s) |
|---|---|---|---|
| Staphylococcus aureus | Gly5 | FemA, P14304; FemB, P14305; FmhB, AAD23961; FmhA, AAD23960; FmhC, AAD23963 | 10, 49, 84, 88, 109 |
| Staphylococcus epidermidis | Gly2-3-l-Ser1-2, l-Ala-Gly4 | FemA, JC5325; FemB, JC5326 | 1, 52, 88 |
| Staphylococcus spp. | Gly5, Gly2-3-l-Ser1-2, l-Ala-Gly4 | S. anaerobius FemA, AAD33940; S. capitis FemA, AAC69633; S. cohnii FemA, AAD45259; S. gallinarum FemA, AAD45258; S. haemolyticus FemA, AAC69631; S. hominis FemA, CAA73372; S. intermedius FemA, AAD33941; S. lugdunensis FemA, AAC69632; S. saprophyticus FemA, CAA73373; S. schleiferi FemA, AAC69636; S. sciuri FemA, AAC69635; S. xylosus FemA, AAC69634; S. warneri FemA, AAD33942; S. haemolyticus FemB, AAD22133; S. simulans Lif, AAB53784; S. capitis Epr, BAA21486; S. sciuri Epr, AAG16879; | |
| Streptococcus pneumoniae | l-Ala-l-Ser, l-Ala2 | FibA, CAB89120; FibB, CAB89121; MurMN, AJ250764 | 33, 88, 114 |
| Streptococcus mutans | l-Thr-l-Ala | 88 | |
| Streptococcus pyogenes | l-Ala2-3 | 88 | |
| Streptococcus thermophilus | l-Ala2-3 | AAK18829 | 88 |
| Streptococcus zooepidemicus | l-Ala2-3 | Zif, AAC46073 | 7, 88 |
| Streptococcus equi | l-Ala2-3 | ||
| Streptococcus milleri | l-Thr-Gly | MilF, AF243359 | 13, 88 |
| Enterococcus faecalis | l-Ala2-3 | 16, 88 | |
| Clostridium perfringens | Gly | BAB81796 | 88 |
| Streptomyces spp. | Gly | S. coelicolor,CAB45460; S. toyocaensis StaO, AAM80555 | 81, 88 |
| Weissella viridescens (Lactobacillus viridescens) | l-Ala-l-Ser | AAG21689 | 48, 88 |
| Deinococcus radiodurans | Gly2 | AAF10635 | 88 |
| Treponema pallidum | Gly | AAC65773 | 110 |
| Borrelia spp. | Gly | AAB91521 | 115 |
Mechanism of β-lactam action.
β-Lactams target the extracellular steps of peptidoglycan biosynthesis by inhibiting the PBPs that catalyze the transpeptidation reaction, the last step in peptidoglycan synthesis (Fig. 1). Their localization at the outer face of the cytoplasmic membrane makes PBPs a readily accessible target. β-Lactams accomplish inhibition of PBPs by their structural similarity to the extended conformation of the terminal d-Ala2 dipeptide of the peptide moiety of peptidoglycan (104). The active site of PBPs is rapidly acylated by the β-lactam, but the deacylation step is so slow that the proteins are terminally inactivated (37). Autolytic enzymes then trigger cell death under the influence of β-lactams (105). In Staphylococcus aureus, cell death occurs due to puncturing of the cell wall by the autolytic enzymes of the so-called splitting system (101), as the initiation of a new septum is inhibited, but the cell division cycle continues as scheduled. Bacteriolysis subsequently occurs when the cells are already dead (38).
PBPs belong to the protein superfamily of penicilloyl serine transferases, which also includes β-lactamases and β-lactam receptors such as BlaR1. All members of this protein family contain three common, conserved motifs, SXXK, (S/Y)X(N/C), and (K/H)(T/S)G, which together form the active site. Although the primary sequence can be highly divergent, β-lactam receptors and serine β-lactamases are considered to have evolved from PBPs, with β-lactamases having acquired the ability to rapidly hydrolyze the penicilloyl-enzyme intermediate, resulting in destruction of the β-lactam (37).
LOW-AFFINITY PENICILLIN-BINDING PROTEINS MEDIATING β-LACTAM RESISTANCE
Staphylococci.
Methicillin-resistant S. aureus (MRSA) isolates appeared shortly after the introduction of methicillin, the first penicillinase-resistant β-lactam to come into clinical use, which was in 1960 (5). Methicillin resistance is due to the acquisition of SCCmec (staphylococcal cassette chromosome mec [51], formerly termed the mec determinant). SCCmec is considered a novel type of mobile element that has been termed a “resistance island,” in analogy to pathogenicity islands (54, 58). SCCmec is thought to be of nonstaphylococcal origin, as it differs from the rest of the staphylococcal chromosome in its G+C content. However, its reservoir has not been clearly identified. Several SCCmec subtypes have been characterized (4, 51, 63) that differ in their sizes, genes, and the additional resistance determinants that they contain, as well as in their clonal distributions. The gene responsible for methicillin resistance, mecA, encodes an additional PBP, PBP2′ (synonym PBP2a), that has a lower affinity to methicillin and other β-lactam derivatives.
One of the characteristic features of methicillin resistance is its heterogeneity; in a given population of S. aureus, the majority of cells are often resistant to a relatively low concentration of methicillin, while a small proportion (10−8 to 10−2 of the cells) are able to grow at higher concentrations. This heterogeneity is the reason for the failure of β-lactam therapy in MRSA infections. It has been demonstrated that the level of resistance does not necessarily correlate with the amount of PBP2′ expressed (86), but there is no satisfactory genetic or biochemical explanation for this phenomenon, although many mutations leading from high-level resistance to heteroresistance in vitro have been identified (for a review, see reference 11).
In the absence of the mecA gene, staphylococci may acquire β-lactam resistance via mutations that lead to overexpression of endogenous PBP2 and/or PBP4 or mutations that reduce their penicillin affinities (43, 50). Clinical S. aureus isolates with such mutations have been identified and have been termed MODSA in reference to their modified PBPs (106). Furthermore, a decreased affinity of PBP3 for β-lactams was found to lead to low-level methicillin resistance in Staphylococcus epidermidis (78). Overexpression of a penicillinase in a specific genetic background, in the so-called borderline resistant S. aureus (BORSA), may also lead to low-level methicillin resistance (6). These two types of resistance mechanisms have less clinical impact than true methicillin resistance in S. aureus and were never observed to achieve more than borderline resistance in clinical isolates.
Pneumococci.
In Streptococcus pneumoniae, target alteration is the main mechanism of resistance to penicillin. In contrast to methicillin resistance in staphylococci, penicillin resistance arises by modification of endogenous PBPs. The first PBPs to be affected are PBP2b or the essential PBP, PBP2x, depending on the antibiotic used for selection (41, 59). Mutations in just three amino acids in the active-site region of PBP2x are sufficient to create low-affinity derivatives (55). Additional gene diversity is created by horizontal exchange of DNA fragments between strains or even with commensal streptococci, resulting in highly mosaic genes. Some very low affinity PBP2x variants from clinical isolates contain more than 100 amino acid substitutions spread over the entire sequence (47, 61, 83). Mutations in PBP2x and PBP2b are sufficient for midlevel resistance, but to achieve high-level penicillin resistance, all six endogenous PBPs can be modified. The combination of several highly resistant PBPs may raise the penicillin MIC 100- to 1,000-fold (45, 114). Similar to staphylococci, it was found that additional genes other than pbp genes are involved in pneumococcal β-lactam resistance, as transformation of susceptible strains with only the pbp genes from highly resistant strains was not sufficient to achieve the same level of resistance (46, 96).
Enterococci.
Enterococci are naturally less susceptible to β-lactams than pneumococci or staphylococci (35). Their intrinsic β-lactam resistance is due to endogenous low-affinity PBP5, which is not essential (91). Acquired resistance to higher antibiotic concentrations is associated with the overproduction of PBP5 and/or a decreased affinity of PBP5 for the β-lactam (34). However, the role of PBP5 in high-level resistance is likely to depend on additional factors. The expression of pbp5 alleles from highly resistant strains in a penicillin-susceptible pbp5 deletion mutant of Enterococcus faecium conferred only moderate, not high-level, resistance (91). As the sensitive PBPs are inactivated by β-lactams, PBP5 is able to synthesize muropeptide dimers and trimers in abundance, but higher oligomers are reduced (92). Inactivation of PBP5 of Enterococcus faecalis leads to hypersusceptibility to all β-lactams (93). Interestingly, the first low-affinity PBP found to be located on a plasmid is PBP3r from Enterococcus hirae, which is almost identical to PBP5 (80, 82).
STRUCTURAL CHARACTERISTICS OF NORMAL AND LOW-AFFINITY PBPS
Depending on the species, bacteria contain between four and eight PBPs, which are divided into several classes (39). Monofunctional PBPs contain a penicillin-binding (PB) domain and largely perform accessory functions in peptidoglycan synthesis. Class A multimodular, bifunctional PBPs contain a non-PB (n-PB) module that has transglycosylase activity, which has been demonstrated for PBP1a and PBP1b in Escherichia coli (71) and PBP2 in S. aureus (79). The PB domains of class A PBPs perform the transpeptidase function. Class A PBPs can therefore perform all necessary functions in peptidoglycan cross-linking.
Class B multimodular PBPs contain an n-PB domain which does not appear to have transglycosylase activity and a PB domain which has transpeptidase activity. The exact function of the n-PB module is unknown, but it may mediate interaction with other proteins of the cell wall synthesis machinery (53) and it is essential for folding (39). The low-affinity PBPs of enterococci and staphylococci are closely related and form PBP subclass B1, which stands apart from the unmodified class B PBPs and also from S. pneumoniae PBP2x (39). As an exception, Bacillus subtilis PBP3 groups in the B1 cluster but has not been linked to any low-affinity property (70). Class B PBPs must cooperate with either a monofunctional transglycosylase or the transglycosylase domain of a class A PBP to synthesize cross-linked peptidoglycan. In S. aureus, this assumption was recently supported by the observation that PBP2′ needs the intact transglycosylase module of the resident PBP2 in order to confer methicillin resistance (79). However, there are indications that PBP2′ is a very poor transpeptidase that only barely allows the cells to survive the action of methicillin. When highly methicillin resistant S. aureus strain COL is grown in the presence of methicillin concentrations sufficient to inactivate the endogenous PBPs, PBP2′ forms a poorly cross-linked cell wall containing mainly un-cross-linked muropeptides and only a few muropeptide dimers or trimers. The same strain grown in antibiotic-free medium produces highly cross-linked peptidoglycan (25).
Little is known about the structural changes that give rise to low-affinity PBPs. The recent determination of the structure of E. faecium PBP5fm, which belongs to the B1 group of PBPs, has provided some evidence into the mechanism that makes these proteins resistant to β-lactams. The active site of PBP5fm is surrounded by a number of amino acids that are strongly conserved in class B1 PBPs but not in PBPs of other classes and that may confer greater rigidity and a reduced affinity of β-lactams for the active site of this group of proteins (87).
In PBP2x from highly resistant strain SP328 (27), the helix that harbors the SXN motif is highly flexible, causing the serine to point away from the active site and leading to a decreased affinity for the antibiotic. Important amino acid exchanges are T338A in the STMK motif, which leads to a reduced rate of acylation by the β-lactam (27, 69). In a variant of PBP2x that has a higher affinity for penicillin (40), the active site lies in a deep groove of the protein. In contrast, the flexible architecture of PBP2x from resistant strain SP328 results in an “open” active-site pocket that may be able to accommodate branched peptidoglycan precursors (27), for which it apparently exhibits a substrate preference, as will be discussed below. Although kinetic studies of low-affinity PBP2x also demonstrated that it has a reduced affinity for a structural analog of the linear stem peptide (69, 117), studies with an analog of the branched stem peptide have not been done to confirm the substrate preference hypothesis.
LOW-AFFNITY PBPS AND VANCOMYCIN RESISTANCE
The recent first report of a vancomycin-resistant S. aureus isolate that has acquired the vanA gene from enterococci (19) raises a number of questions: does the incorporation of d-lactate into the peptidoglycan precursor affect the function of the cross-linking enzymes, particularly PBP2′? If yes, how does this affect resistance to agents other than vancomycin?
Some indications of the answers to this problem may be found in studies conducted with enterococci. It appears that the presence of d-lactate does not affect peptidoglycan cross-linking under unchallenged growth conditions (15, 21). However, vancomycin and β-lactams have been shown to exhibit a synergistic action in vancomycin-resistant enterococci, which indicates that their low-affinity PBPs do not cope well with the modified stem peptides (2, 42).
In a recent study with S. aureus (23), the addition of d-serine, d-threonine, and d-phenylalanine to the growth medium reduced the levels of cross-linking and methicillin resistance, as these amino acids were efficiently incorporated in the fifth position of the peptidoglycan stem peptide. The same effect has been observed when glycine was added to the growth medium (22). However, these experiments did not include d-lactate, and the effect on vancomycin resistance was not determined. Although the most recent of these studies concluded that vanA-mediated vancomycin resistance and mecA-mediated methicillin resistance may be mutually exclusive, this hypothesis does not hold true for the recently reported vancomycin-resistant S. aureus strain that also carries mecA and for which the oxacillin MIC is 16 μg/ml (19).
BIOSYNTHESIS OF BRANCHED CELL WALL PEPTIDES AND IMPACT ON β-LACTAM RESISTANCE
Cross-linking of peptidoglycan (Fig. 1) by transpeptidases (PBPs) always occurs via a diamino acid in the stem peptide, generally meso-diaminopimelic acid, l-Lys, or l-Orn. Cross-links occur either directly or via an interceding spacer (or interpeptide), which consists of one to five amino acids and which branches off the diamino acid. The interpeptide may contain amino acids in the l or d conformation (88).
In S. aureus, the pentaglycine interpeptide is synthesized in a sequential fashion on the cytoplasmic face of the membrane, with lipid II as a substrate (57, 66), where the first glycine is attached to the ɛ-amino group of l-lysine (56, 97). The glycine donor is glycyl-tRNA (65), and a fraction of tRNAs is exclusively used in interpeptide synthesis (18). Although tRNAs participate in interpeptide formation, the reaction is not inhibited by antibiotics that target protein translation, and thus, it was concluded that interpeptide synthesis is achieved by a nonribosomal mechanism (57). In an alternative pathway, in species such as E. faecium, whose interpeptide contains d-amino acids, the amino acids are incorporated from phosphate precursors (17, 88).
FemABX protein family.
The branched peptide chain is synthesized by a family of nonribosomal peptidyl transferases (Fig. 2) that use either the lipid II-linked peptidoglycan precursor or, alternatively, the soluble, UDP-MurNAc-linked stem peptide (cf. Fig. 1) as a substrate for the addition of one or several additional amino acids to the peptidoglycan stem peptide in a sequential fashion. The impact of these factors on cell wall composition and, in clinically relevant species, on antibiotic resistance is outlined below.
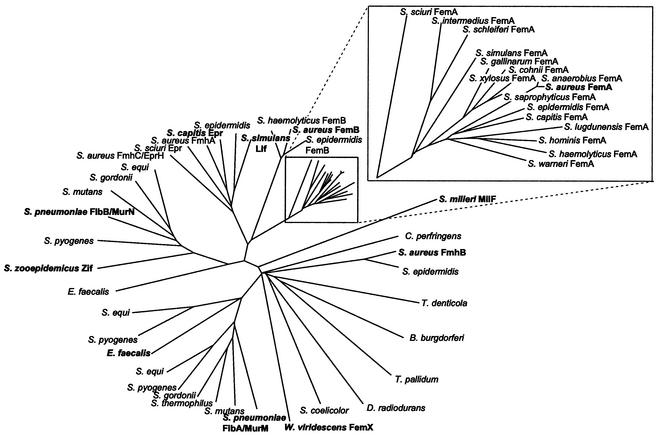
FIG. 2.
Dendrogram of the FemABX protein family. Branch lengths are proportional to phylogenetic distances. Experimentally confirmed family members are shown in boldface type. The number of sequenced FemA homologs of Staphylococcus spp. has become quite large, and they are shown in an enlarged inset. The GenBank database was searched with the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/) (3) by using S. aureus FmhB as the query sequence. In the case of Streptococcus equi and Streptococcus pyogenes, three FemABX homologs were found, two of which are related to FibA/MurM of S. pneumoniae and one of which is related to FibB/MurN of S. pneumoniae. Unfinished microbial genomes were searched by using the tblastn program (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). Preliminary sequence data from unfinished genomes were obtained from The Institute for Genomic Research website at http://www.tigr.org and from the Sanger Center at http://www.sanger.ac.uk. In the case of MurM, where multiple, more than 90% identical alleles are available, one representative sequence (that of strain R36A) was chosen. Protein sequences were aligned by using the ClustalW program (102), and the dendrogram was constructed by using the TreeView program (75).
Staphylococcus.
The cause of heterogeneity in methicillin resistance was initially investigated by transposon-mediated mutagenesis (9), which led to the identification of a number of loci whose inactivation lowered the resistance level, and the terms fem factors (for factors essential for methicillin resistance) and aux factors (for auxiliary) were coined (8, 12, 26). It was shown that all fem factors are housekeeping genes, most of which play a role in cell wall biosynthesis. Mutation of the femAB operon affects the glycine content in the cell wall (64). It was determined that FemA specifically adds glycines 2 and 3 of the pentaglycine interpeptide, while FemB, which shares 39% identity and 59% similarity with FemA on the amino acid level, adds glycines 4 and 5 (28). femAB mutants completely lose resistance to methicillin and become hypersusceptible to many unrelated antibiotics (62). femAB mutant cells are pseudomulticellular with thickened cell walls and abnormal septa due to impaired cell wall turnover and are resistant to the glycyl-glycine endopeptidase lysostaphin (49). It is clear from these observations that not only low-affinity PBP2′ but also the endogenous PBPs cope very badly with the shortened pentaglycine interpeptide. FemA and FemB homologs have been identified in all staphylococcal species tested (1, 112).
The factor (femX) that catalyzes the incorporation of the first glycine long remained elusive. Inactivation of femA or femB yielded viable cells, but it was demonstrated that they had acquired compensatory mutations in order to survive (62). It was deduced that the inactivation of femAB would otherwise be lethal and that the postulated femX was probably essential (60). The advent of whole-genome sequencing made it possible to search the complete S. aureus genome for additional homologs of femAB; and three sequences were identified and named fmhA, fmhB, and fmhC (for fem homolog) (109). fmhA and fmhC (alternative name, eprH [99]) were inactivated without a discernible phenotype, while inactivation was impossible for fmhB, making it a likely femX candidate (109). A promoter insertion strategy in which cells could be depleted of FmhB was used to demonstrate its role in the addition of glycine 1 of the interpeptide chain. FmhB depletion leads to the virtual disappearance of glycine-substituted muropeptide monomers and to a significant reduction in the degree of peptidoglycan cross-linking. The introduction of this construct into an MRSA background causes a complete loss of resistance to methicillin (84). It can be concluded from these observations that the staphylococcal cell can tolerate, if barely, the reduction of the interpeptide to one glycine, but the complete lack of glycine substitution is lethal. In addition, PBP2′, the low-affinity PBP responsible for methicillin resistance, is unable to perform its function in the absence of a five-membered interpeptide chain.
Pneumococcus.
In S. pneumoniae, the peptide side chain, consisting of Ala2 or Ser-Ala, is dispensable (90). In an early study of the pneumococcal cell wall, Garcia-Bustos and Tomasz (36) analyzed a number of penicillin-susceptible and -resistant clinical strains and observed branched as well as unbranched peptidoglycan stem peptides. While in resistant strains there was a large amount of Ala2- or Ser-Ala-substituted peptides, in susceptible strains, the situation was variable: in the clinical strains analyzed, the muropeptides were predominantly unsubstituted, whereas in laboratory strain R6 there was a significant fraction of branched muropeptides present. In both the susceptible and the resistant strains analyzed, the muropeptide monomer fractions contained a detectable amount of nonsubstituted muropeptides; but the dimer, trimer, and oligomer fractions appeared to contain almost exclusively substituted muropeptides. These findings led to the conclusion that the PBPs exhibit a substrate preference for substituted stem peptides; the preference appeared more marked in resistant strains, which have modified, low-affinity PBPs. In a penicillin-resistant transformant of strain R6, nonbranched muropeptide dimers were undetectable, leading Garcia-Bustos and Tomasz (36) to suggest that PBP2x may discriminate against unsubstituted stem peptides, a notion that has recently been supported by its crystal structure (discussed above).
The genes for the alanine and/or serine transferases of S. pneumoniae have recently been identified by two independent groups (33, 114) and named murMN and fibAB, respectively. Inactivation of these genes in penicillin-resistant strains results in greatly decreased levels of resistance to β-lactams (reductions of more than 32-fold), with a complete breakdown of resistance to cefoxitin, which predominantly targets PBP2x. The MIC is reduced twofold for penicillin-sensitive laboratory strain R6. In addition, the level of total cell wall cross-linking is reduced (114). As might be expected with such a weakened cell wall and in analogy to femAB mutants (62), murMN mutants are hypersusceptible to all other classes of cell wall synthesis inhibitors (32). Surprisingly, laboratory strain R36A contains a very small amount of alanine- or serine-substituted muropeptides compared to the amounts in strain R6, even with an intact murMN operon (33). The murM and murN genes of laboratory strains R6 and R36A, which have essentially the same genetic backgrounds, differ in only two and one amino acid positions, respectively. It is conceivable that variations in the transcriptional activity of murMN may cause this significant difference in their peptidoglycan compositions.
As is the case with many genes in pneumococci (44, 45), the murMN genes, like PBP2x, exhibit a mosaic structure which may explain the strain-dependent differences in branched muropeptide content. The variability of the murMN genes appears to be responsible for variations in the catalytic efficiencies of their products (30, 95). Again, in resistant strains, a larger amount of branched muropeptides indicates that these strains contain murM alleles that encode a highly efficient enzyme. The presence of a high-efficiency murM allele appears to be a prerequisite for the acquisition of high-level resistance to penicillin by remodeled, low-affinity PBPs, as only transformation by both the low-affinity PBPs and the murMN genes from a resistant donor strain results in high-level resistance in a susceptible recipient (95).
The correlation between branched peptidoglycan and low-affinity PBPs as the sole resistance mechanisms may be questionable due to a report that dissociated penicillin resistance from branched peptidoglycan peptides by transformation (89). When a susceptible, directly cross-linked strain was transformed with DNA from a resistant donor with low-affinity PBPs and a branched-chain peptidoglycan, the first round of transformation resulted in resistant strains that had acquired both the branched peptides and the low-affinity PBPs of the donor. However, in a second round of transformation, resistance was no longer coupled to the branched-chain peptidoglycan, as the strains were resistant to penicillin but had retained the recipient's peptidoglycan structure. Examination of the PBP profiles indicated that most of these second-round transformants had acquired changes, possibly point mutations, leading to PBPs with reduced β-lactam affinities. Conceivably, mutations in genes other than murMN or pbp were selected in these experiments; therefore, the study does not necessarily invalidate the link between resistance and peptidoglycan structure.
The description of MurMN in S. pneumoniae has given valuable insights into the specificity of the FemABX family of peptidyl transferases. Filipe et al. (31) have analyzed a set of murM alleles and pinpointed the region that determines which amino acid is incorporated into the peptidoglycan precursor. The exchange of codons 244 to 274 between murM alleles from two strains incorporating mainly seryl-alanine (strain KY17) or dialanine (strain DE1) determined the ratio of serine versus alanine that was incorporated. In particular, the amino acid in position 260 was determined as the major difference in these two strains. A single amino acid exchange (T260K) in murM of strain KY17, indeed, slightly increased the alanine content of the peptide side chain, but other residues in the region from codons 244 to 274 must be assumed to affect the specificity of murM as well. A Q27E/T mutation had no measurable effect, although it was suggested that this amino acid position may be involved in catalysis in FemX of Weissella viridescens (see below) (48). In addition, the deletion of as few as 50 amino acids at the N terminus or 10 amino acids at the C terminus inactivated MurM. This effect was also observed in the case of FemA and FemB of S. aureus, in which point mutations at the C terminus, which lead to a stop codon, completely inactivated the proteins (60).
Lactobacillus.
Hegde and Shrader (48) have recently identified FemX in W. viridescens (formerly Lactobacillus viridescens) and were able to reconstitute the addition of glycine to the soluble, UDP-linked peptidoglycan precursor from glycyl-tRNA in vitro. Their attempt to reconstitute the same reaction with S. aureus FmhB was unsuccessful, probably because this reaction occurs on lipid II, whereas in W. viridescens it occurs on the soluble precursor (cf. Fig. 1).
Enterococcus.
The catalytic activities of the FemABX homologs in E. faecalis have been analyzed (16). Two homologous open reading frames were found. Both orf1 (coding for a protein of 48.3 kDa) and orf2 (coding for a protein of 46 kDa) were recombinantly expressed and reconstituted in an in vitro assay. Only the product of orf2 has UDP-MurNAc-pentapeptide:l-alanine ligase activity and therefore corresponds to S. pneumoniae MurM, to which it is closely related (Fig. 2). It is conceivable that a coupled reaction with Orf1 and Orf2 would demonstrate the activity of Orf1, which groups with MurN in Fig. 2. The effect of mutations of orf1 and orf2 on β-lactam resistance has not yet been determined.
Endopeptidase resistance factors.
The family of FemABX-like proteins also includes immunity factors that protect producers of interpeptide-specific endopeptidases against their own products by substituting some of the amino acid positions in their interpeptides. epr (endopeptidase resistance) from Staphylococcus capitis (100) and lif (lysostaphin immunity factor) from Staphylococcus simulans bv. staphylolyticus (20, 103) incorporate serine into the interpeptide at specific positions and thereby make the cell wall resistant to glycyl-glycine endopeptidases such as lysostaphin (29, 108). Interestingly, the incorporation of serine into the peptide side chain due to expression of epr in an MRSA background does not appear to affect methicillin resistance (100). This finding indicates that although PBP2′ is dependent on a five-membered interpeptide bridge, it does not appear to discriminate between interpeptides consisting of glycine only or glycine and serine. Interestingly, the two additional FemABX homologs in S. aureus, FmhA and FmhC/EprH, are most closely related to Lif and Epr (Fig. 2). As mutation of these genes did not appear to cause a phenotype (99, 109), fmhA and fmhC may be silent or almost inactive remnants of genes encoding serine-incorporating enzymes in S. aureus. They may even be responsible for the somewhat elevated level of serine incorporation that was observed in a partial femAB mutant (24).
In Streptococcus milleri, the endopeptidase millericin B (milB) is coorganized with the immunity factor milF. MilF substitutes leucine in the interpeptide for the endogenous threonine (13). In Streptococcus zooepidemicus, zoocin (zooA) and its corresponding immunity factor, zif, have been described (94). However, it is unclear whether Zif substitutes any amino acids in the interpeptide (7).
ROLE OF BRANCHED-CHAIN PEPTIDES IN CELL DIVISION
In addition to their impact on β-lactam resistance, branched cell wall peptides play an important role in cell division. femAB mutants, which have a shortened interpeptide, exhibit thickened septa and are pseudomulticellular due to impaired cell separation (49). It has been shown that endogenous glycyl-glycine endopeptidases are involved in cell separation, and femAB mutants are largely resistant to the actions of such endopeptidases (98), which explains this defect in cell division. On the other hand, the major autolysin Atl, which is implicated in lytic death in the presence of penicillin (described above), is not impaired by the shortened interpeptide, as its mature domains have endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase activities (101), and is not affected by the interpeptide structure.
ANCHORING OF CELL SURFACE PROTEINS
In addition to their influence on resistance to β-lactams and their crucial role in cell wall integrity and cell division, branched-chain muropeptides also play a role in virulence. In S. aureus, important virulence factors are covalently attached to the cell wall interpeptide. Examples are protein A, fibrinogen-binding protein (clumping factor), and other so-called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) that enable adhesion to host tissues (73, 76). Branched side chains that do not participate in peptidoglycan cross-linking serve as anchoring points for such cell wall-linked proteins. Cell wall-localized proteins carry a characteristic C-terminal sorting signal with a conserved LPXTG sequence (72) that is recognized in S. aureus by the enzyme sortase (SrtA) (67). This enzyme tethers the proteins to the interpeptide in a transpeptidase reaction, which occurs on lipid II on the extracellular face of the membrane (77, 85). It has been shown that SrtA will not accept linear stem peptides lacking glycine as a substrate (85) and that with the shortened interpeptides containing only one and three glycines in femAB and femB mutants, respectively, sorting of cell wall proteins is impaired (107). In sortase mutants (67, 68), virulence is reduced due to an impairment of the anchoring of cell surface proteins. In analogy, in femAB mutant strains, the truncated muropeptide side chains are expected to cause a reduction in virulence due to the concurrent inhibition of the sorting reaction.
A second sortase, SrtB, was found to specifically link IsdC (encoded by the iron-responsive surface determinant locus of S. aureus) to the cell wall via an NPQTN sequence (68).
It may be assumed that in other species as well a sortase catalyzes attachment of proteins to the cell wall via the branched peptides. In S. pneumoniae, neuraminidase contains a sorting signal, and several cell wall-localized proteins in enterococci also contain this signature sequence (73). A sortase has indeed been described recently in Listeria monocytogenes, whose cell wall peptides are directly cross-linked via meso-diaminopimelic acid (14), indicating that the substrate requirements for each sortase correspond to the species' peptidoglycan structure.
CONCLUSION
The FemABX protein family has emerged as a broadly distributed group of peptidoglycan stem peptide peptidyl transferases. The interpeptide serves as a means for cross-linking of the cell wall, mediated by the PBPs. Some PBPs appear to exhibit a marked substrate preference for the peptide side chain, as may be the case with PBP2x in S. pneumoniae. In S. aureus, the PBPs are unable to function at all if the interpeptide is lost. The reduction in length or the absence of the interpeptide impairs the structure of the cell wall and renders β-lactam-resistant strains that rely on low-affinity PBPs susceptible to the antibiotics. A subgroup of FemABX family members serve as immunity factors in producers of interpeptide-specific endopeptidases. In addition, the interpeptide plays an important role in cell separation and virulence.
A database search for FmhB homologs yields related sequences in a wide range of bacterial genomes. Species as diverse as Borrelia burgdorferi, Streptomyces coelicolor, and Clostridium perfringens contain a FemABX homolog (Table 1; Fig. 2). It is not surprising that all species containing a FemABX homolog also bear a peptide side chain in their peptidoglycan (Table 1), which makes it likely that all homologs encode functional FemABX family proteins. However, the function of these FemABX homologs needs to be confirmed experimentally. Interestingly, in bacteria such as Lactococcus lactis or E. faecium, which contain d-amino acids in their interpeptides, no FemABX homolog can be found. These amino acids are presumed to be attached by an entirely different but not yet identified mechanism (17, 88).
It appears that the reduction of β-lactam affinity has incurred structural restraints on the remodeled PBPs of S. aureus and S. pneumoniae. Their modified active site may exhibit a substrate preference for peptidoglycan precursors with a branched peptide chain. Unfortunately, as only the structures of S. pneumoniae PBP2x and E. faecium PBP5fm have been solved, this remains speculation. The precise requirements for branched peptide peptidoglycan precursors for the resistance level achieved in pneumococci, mediated by low-affinity PBPs with various levels of modification, will need to be carefully analyzed in an isogenic system. In addition, the importance of the branched peptide structure for β-lactam resistance in E. faecalis remains to be demonstrated.
The observation that S. aureus PBP2′ and E. faecium PBP5 are poor transpeptidases if the sensitive PBPs are masked by β-lactams (25, 92) indicates that these “rescue transpeptidases” (87) have reduced their affinities to β-lactams at the cost of efficiency. If the activities of these PBPs are further impaired by a suboptimal substrate, they are simply unable to perform their functions under antibiotic pressure, as is shown by the fem-type mutations in S. aureus and S. pneumoniae and the synergism between vancomycin and β-lactams in vancomycin-resistant enterococci.
The global spread of antimicrobial resistance has created a need for new agents for the treatment of bacterial infections. The knowledge that has been gained about the FemABX family of proteins suggests that they are a suitable target for the development of new antimicrobials. If an inhibitor of FemABX proteins was developed, it would be a potent and highly specific antimicrobial agent in those organisms in which the interpeptide structure is essential (60). Alternatively, in organisms in which the peptidoglycan interpeptide is dispensable, such as penicillin-resistant pneumococci, a FemABX inhibitor would certainly restore the activities of β-lactams. As a supporting strategy, the virulence of epidemic strains could be reduced (74) by preventing the attachment of cell wall-linked proteins via inhibition of the FemABX homologs and subsequent impairment of sorting of these virulence factors. In addition, peptidoglycan itself has been identified as a virulence factor eliciting host responses (113, 116). In analogy to mutation of femAB (64), it is conceivable that inhibition of FemABX factors might lead to a reduction in cell wall turnover or peptidoglycan “shedding” and thus reduce virulence. In the near future, as broad-spectrum antimicrobials become increasingly unreliable, the use of a FemABX inhibitor might be an option to be explored.
ADDENDUM IN PROOF
Recent publications of two important crystal structures provide a basis for further research: D. Lim and N. C. Strynadka, Nat. Struct. Biol. 9:870-876, 2002; T. Benson et al., Structure (Cambridge) 10:1107-1115, 2002.
Acknowledgments
Research in the laboratory of Brigitte Berger-Bächi is supported by Swiss National Science Foundation grant 32-63552.00.
We thank Regine Hakenbeck for helpful suggestions.
REFERENCES
- 1.Alborn, W. E. J., J. Hoskins, S. Unal, J. E. Flokowitsch, C. A. Hayes, J. E. Dotzlaf, W. K. Yeh, and P. L. Skatrud. 1996. Cloning and characterization of femA and femB from Staphylococcus epidermidis. Gene 180:177-181. [DOI] [PubMed] [Google Scholar]
- 2.al-Obeid, S., D. Billot-Klein, J. van Heijenoort, E. Collatz, and L. Gutmann. 1992. Replacement of the essential penicillin-binding protein 5 by high-molecular mass PBPs may explain vancomycin-beta-lactam synergy in low-level vancomycin-resistant Enterococcus faecium D366. FEMS Microbiol. Lett. 70:79-84. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Barber, M. 1961. Methicillin-resistant staphylococci. J. Clin. Pathol. 14:385-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barg, N., H. Chambers, and D. Kernodle. 1991. Borderline susceptibility to antistaphylococcal penicillins is not conferred exclusively by the overproduction of beta-lactamase. Antimicrob. Agents Chemother. 35:1975-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatson, S., G. Sloan, and R. Simmonds. 1998. Zoocin A immunity factor: a femA-like gene found in a group C streptococcus. FEMS Microbiol. Lett. 163:73-77. [DOI] [PubMed] [Google Scholar]
- 8.Berger-Bächi, B. 1989. Genetics of methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 23:671-673. [DOI] [PubMed] [Google Scholar]
- 9.Berger-Bächi, B. 1983. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J. Bacteriol. 154:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger-Bächi, B., L. Barberis-Maino, A. Strässle, and F. H. Kayser. 1989. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol. Gen. Genet. 219:263-269. [DOI] [PubMed] [Google Scholar]
- 11.Berger-Bächi, B., and S. Rohrer. 2002. Control of methicillin resistance in Staphylococcus aureus. Arch. Microbiol. 178:165-171. [DOI] [PubMed] [Google Scholar]
- 12.Berger-Bächi, B., A. Strässle, J. E. Gustafson, and F. H. Kayser. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beukes, M., and J. W. Hastings. 2001. Self-protection against cell wall hydrolysis in Streptococcus milleri NMSCC 061 and analysis of the millericin B operon. Appl. Environ. Microbiol. 67:3888-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. G. Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 15.Billot-Klein, D., D. Shlaes, D. Bryant, D. Bell, J. van Heijenoort, and L. Gutmann. 1996. Peptidoglycan structure of Enterococcus faecium expressing vancomycin resistance of the VanB type. Biochem. J. 313:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouhss, A., N. Josseaume, D. Allanic, M. Crouvoisier, L. Gutmann, J.-L. Mainardi, D. Mengin-Lecreulx, J. van Heijenoort, and M. Arthur. 2001. Identification of the UDP-MurNAc-pentapeptide:l-alanine ligase for synthesis of branched peptidoglycan precursors in Enterococcus faecalis. J. Bacteriol. 183:5122-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugg, T. 1999. Bacterial peptidoglycan biosynthesis and its inhibition, p. 241-294. In M. Pinto (ed.), Comprehensive natural products chemistry, vol. 3. Elsevier, Oxford, United Kingdom.
- 18.Bumsted, R. M., J. L. Dahl, D. Soll, and J. L. Strominger. 1968. Biosynthesis of the peptidoglycan of bacterial cell walls. X. Further study of the glycyl transfer ribonucleic acids active in peptidoglycan synthesis in Staphylococcus aureus. J. Biol. Chem. 243:779-782. [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 20.DeHart, H. P., H. E. Heath, L. S. Heath, P. A. LeBlanc, and G. L. Sloan. 1995. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 61:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jonge, B. L., S. Handwerger, and D. Gage. 1996. Altered peptidoglycan composition in vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 40:863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jonge, B. L. M., Y. S. Chang, N. Xu, and D. Gage. 1996. Effect of exogenous glycine on peptidoglycan composition and resistance in a methicillin-resistant Staphylococcus aureus strain. Antimicrob. Agents Chemother. 40:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jonge, B. L. M., D. Gage, and N. Xu. 2002. The carboxyl terminus of peptidoglycan stem peptides is a determinant for methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3151-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jonge, B. L. M., T. Sidow, Y. S. Chang, H. Labischinski, B. Berger-Bächi, D. A. Gage, and A. Tomasz. 1993. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J. Bacteriol. 175:2779-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jonge, B. L. M., and A. Tomasz. 1993. Abnormal peptidoglycan produced in a methicillin-resistant strain of Staphylococcus aureus grown in the presence of methicillin: functional role for penicillin-binding protein 2A in cell wall synthesis. Antimicrob. Agents Chemother. 37:342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lencastre, H., and A. Tomasz. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dessen, A., N. Mouz, E. Gordon, J. Hopkins, and O. Dideberg. 2001. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate. J. Biol. Chem. 276:45106-45112. [DOI] [PubMed] [Google Scholar]
- 28.Ehlert, K., W. Schröder, and H. Labischinski. 1997. Specificities of FemA and FemB for different glycine residues: FemB cannot substitute for FemA in staphylococcal peptidoglycan pentaglycine side chain formation. J. Bacteriol. 179:7573-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehlert, K., M. Tschierske, C. Mori, W. Schröder, and B. Berger-Bächi. 2000. Site-specific serine incorporation by Lif and Epr into positions 3 and 5 of the staphylococcal peptidoglycan interpeptide bridge. J. Bacteriol. 182:2635-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filipe, S. R., E. Severina, and A. Tomasz. 2000. Distribution of the mosaic structured murM genes among natural populations of Streptococcus pneumoniae. J. Bacteriol. 182:6798-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filipe, S. R., E. Severina, and A. Tomasz. 2001. Functional analysis of Streptococcus pneumoniae MurM reveals the region responsible for its specificity in the synthesis of branched cell wall peptides. J. Biol. Chem. 276:39618.. [DOI] [PubMed] [Google Scholar]
- 32.Filipe, S. R., E. Severina, and A. Tomasz. 2001. The role of murMN operon in penicillin resistance and antibiotic tolerance of Streptococcus pneumoniae. Microb. Drug Resist. 7:303-316. [DOI] [PubMed] [Google Scholar]
- 33.Filipe, S. R., and A. Tomasz. 2000. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc. Natl. Acad. Sci. USA 97:4891-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontana, R., M. Aldegheri, M. Ligozzi, H. Lopez, A. Sucari, and G. Satta. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 38:1980-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontana, R., P. Canepari, M. M. Lleo, and G. Satta. 1990. Mechanisms of resistance of enterococci to beta-lactam antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 9:103-105. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Bustos, J., and A. Tomasz. 1990. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc. Natl. Acad. Sci. USA 87:5415-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghuysen, J.-M. 1997. Penicillin-binding proteins. Wall peptidoglycan assembly and resistance to penicillin: facts, doubts and hopes. Int. J. Antimicrob. Agents 8:45-60. [DOI] [PubMed] [Google Scholar]
- 38.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon, E., N. Mouz, E. Duée, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 41.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutmann, L., S. al-Obeid, D. Billot-Klein, M. L. Guerrier, and E. Collatz. 1994. Synergy and resistance to synergy between beta-lactam antibiotics and glycopeptides against glycopeptide-resistant strains of Enterococcus faecium. Antimicrob. Agents Chemother. 38:824-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackbarth, C. J., and H. F. Chambers. 1989. Methicillin-resistant staphylococci: genetics and mechanisms of resistance. Antimicrob. Agents Chemother. 33:991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakenbeck, R., and J. Coyette. 1998. Resistant penicillin-binding proteins. Cell. Mol. Life Sci. 54:332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hakenbeck, R., T. Grebe, D. Zahner, and J. B. Stock. 1999. Beta-lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol. Microbiol. 33:673-678. [DOI] [PubMed] [Google Scholar]
- 47.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hegde, S. S., and T. E. Shrader. 2001. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J. Biol. Chem. 276:6998-7003. [DOI] [PubMed] [Google Scholar]
- 49.Henze, U., T. Sidow, J. Wecke, H. Labischinski, and B. Berger-Bächi. 1993. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 175:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henze, U. U., M. Roos, and B. Berger-Bächi. 1996. Effects of penicillin-binding protein 4 overproduction in Staphylococcus aureus. Microb. Drug Resist. Mech. Epidemiol. Dis. 2:193-199. [DOI] [PubMed] [Google Scholar]
- 51.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 52.Holt, J. 1994. Bergey's manual of determinative bacteriology, 9th ed. The Williams & Wilkins Co., Baltimore, Md.
- 53.Höltje, J. V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology (United Kingdom) 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 54.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamin, M., R. Hakenbeck, and J. M. Frère. 1993. Penicillin binding protein 2x as a major contributor to intrinsic beta-lactam resistance of Streptococcus pneumoniae. FEBS Lett. 331:101-104. [DOI] [PubMed] [Google Scholar]
- 56.Kamiryo, T., and M. Matsuhashi. 1972. The biosynthesis of the cross-linking peptides in the cell wall peptidoglycan of Staphylococcus aureus. J. Biol. Chem. 247:6306-6311. [PubMed] [Google Scholar]
- 57.Kamiryo, T., and M. Matsuhashi. 1969. Sequential addition of glycine from glycyl-tRNA to the lipid-linked precursors of cell wall peptidoglycan in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 36:215-222. [DOI] [PubMed] [Google Scholar]
- 58.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kell, C. M., U. K. Sharma, C. G. Dowson, C. Town, T. S. Balganesh, and B. G. Spratt. 1993. Deletion analysis of the essentiality of penicillin-binding protein-1A, penicillin-binding protein-2B and penicillin-binding protein-2X of Streptococcus pneumoniae. FEMS Microbiol. Lett. 106:171-175. [DOI] [PubMed] [Google Scholar]
- 60.Kopp, U., M. Roos, J. Wecke, and H. Labischinski. 1996. Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb. Drug Resist. 2:29-41. [DOI] [PubMed] [Google Scholar]
- 61.Laible, G., and R. Hakenbeck. 1991. Five independent combinations of mutations can result in low-affinity penicillin-binding protein 2x of Streptococcus pneumoniae. J. Bacteriol. 173:6986-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling, B. D., and B. Berger-Bächi. 1998. Increased overall antibiotic susceptibility in Staphylococcus aureus femAB null mutants. Antimicrob. Agents Chemother. 42:936-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maidhof, H., B. Reinicke, P. Blümel, B. Berger-Bächi, and H. Labischinski. 1991. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 173:3507-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuhashi, M., C. Dietrich, and J. Strominger. 1965. Incorporation of glycine into the cell wall glycopeptide in Staphylococcus aureus: role of sRNA and lipid intermediates. Proc. Natl. Acad. Sci. USA 54:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuhashi, M., C. P. Dietrich, and J. L. Strominger. 1967. Biosynthesis of the peptidoglycan of bacterial cell walls. III. The role of soluble ribonucleic acid and of lipid intermediates in glycine incorporation in Staphylococcus aureus. J. Biol. Chem. 242:3191-3206. [PubMed] [Google Scholar]
- 67.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 68.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mouz, N., E. Gordon, A. M. Di Guilmi, I. Petit, Y. Petillot, Y. Dupont, R. Hakenbeck, T. Vernet, and O. Dideberg. 1998. Identification of a structural determinant for resistance to beta-lactam antibiotics in gram-positive bacteria. Proc. Natl. Acad. Sci. USA 95:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murray, T., D. L. Popham, P. Setlow, A. M. Smith, K. P. Klugman, R. Hakenbeck, and J. Coyette. 1996. Identification and characterization of pbpC, the gene encoding Bacillus subtilis penicillin-binding protein 3. J. Bacteriol. 178:6001-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakagawa, J., S. Tamaki, S. Tomioka, and M. Matsuhashi. 1984. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. Penicillin-binding protein 1Bs of Escherichia coli with activities of transglycosylase and transpeptidase. J. Biol. Chem. 259:13937-13946. [PubMed] [Google Scholar]
- 72.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 73.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Novick, R. P. 1996. Possible alternatives to standard antibiotics in the management of infections with antibiotic-resistant organisms, p. 175-191. In C. F. Amabile-Cuevas (ed.), Antibiotic resistance: from molecular basics to therapeutic options. R. G. Landes Company, Austin, Tex.
- 75.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 76.Patti, J. M., and M. Höök. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6:752-758. [DOI] [PubMed] [Google Scholar]
- 77.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241-16248. [DOI] [PubMed] [Google Scholar]
- 78.Petinaki, E., G. Dimitracopoulos, and I. Spiliopoulou. 2001. Decreased affinity of PBP3 to methicillin in a clinical isolate of Staphylococcus epidermidis with borderline resistance to methicillin and free of the mecA gene. Microb. Drug Resist. 7:297-300. [DOI] [PubMed] [Google Scholar]
- 79.Pinho, M. G., S. R. Filipe, H. de Lencastre, and A. Tomasz. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183:6525-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piras, G., D. Raze, A. el Kharroubi, D. Hastir, S. Engelbert, J. Coyette, and J. M. Ghuysen. 1993. Cloning and sequencing of the low-affinity penicillin-binding protein 3r-encoding gene of Enterococcus hirae S185: modular design and structural organization of the protein. J. Bacteriol. 175:2844-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pootoolal, J., M. G. Thomas, C. G. Marshall, J. M. Neu, B. K. Hubbard, C. T. Walsh, and G. D. Wright. 2002. Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 99:8962-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raze, D., O. Dardenne, S. Hallut, M. Martinezbueno, J. Coyette, and J. M. Ghuysen. 1998. The gene encoding the low-affinity penicillin-binding protein 3r in Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob. Agents Chemother. 42:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reichmann, P., A. Konig, J. Linares, F. Alcaide, F. C. Tenover, L. McDougal, S. Swidsinski, and R. Hakenbeck. 1997. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J. Infect. Dis. 176:1001-1012. [DOI] [PubMed] [Google Scholar]
- 84.Rohrer, S., K. Ehlert, M. Tschierske, H. Labischinski, and B. Berger-Bächi. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. USA 96:9351-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ruzin, A., A. Severin, F. Ritacco, K. Tabei, G. Singh, P. A. Bradford, M. M. Siegel, S. J. Projan, D. M. Shlaes, A. M. Perry, H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J. Bacteriol. 184:2141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ryffel, C., A. Strässle, F. H. Kayser, and B. Berger-Bächi. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:724-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sauvage, E., F. Kerff, E. Fonzé, R. Hermann, B. Schoot, J.-P. Marquette, Y. Taburet, D. Prevost, J. Dumas, G. Leonard, P. Stefanic, J. Coyette, and P. Charlier. 2002. The 2.4 Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 59:1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Severin, A., A. M. Figueiredo, and A. Tomasz. 1996. Separation of abnormal cell wall composition from penicillin resistance through genetic transformation of Streptococcus pneumoniae. J. Bacteriol. 178:1788-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Severin, A., and A. Tomasz. 1996. Naturally occurring peptidoglycan variants of Streptococcus pneumoniae. J. Bacteriol. 178:168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sifaoui, F., M. Arthur, L. Rice, and L. Gutmann. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Signoretto, C., M. Boaretti, and P. Canepari. 1998. Peptidoglycan synthesis by Enterococcus faecalis penicillin binding protein 5. Arch. Microbiol. 170:185-190. [DOI] [PubMed] [Google Scholar]
- 93.Signoretto, C., P. Canepari, A. M. Perry, H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2000. Paradoxical effect of inserting, in Enterococcus faecalis penicillin-binding protein 5, an amino acid box responsible for low affinity for penicillin in Enterococcus faecium. Arch. Microbiol. 173:213-219. [DOI] [PubMed] [Google Scholar]
- 94.Simmonds, R. S., W. J. Simpson, and J. R. Tagg. 1997. Cloning and sequence analysis of zooA, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene 189:255-261. [DOI] [PubMed] [Google Scholar]
- 95.Smith, A. M., and K. P. Klugman. 2001. Alterations in MurM, a cell wall muropeptide branching enzyme, increase high-level penicillin and cephalosporin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:2393-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith, A. M., K. P. Klugman, R. Hakenbeck, and J. Coyette. 2000. Non-penicillin-binding protein mediated high-level penicillin and cephalosporin resistance in a Hungarian clone of Streptococcus pneumoniae. Microb. Drug Resist. 6:105-110. [DOI] [PubMed] [Google Scholar]
- 97.Strominger, J., and J. Ghuysen. 1967. Mechanisms of enzymatic bacteriolysis. Cell walls of bacteria are solubilized by action of either specific carbohydrases or specific peptidases. Science 156:213-221. [DOI] [PubMed] [Google Scholar]
- 98.Sugai, M. 1997. Peptidoglycan hydrolases of the staphylococci. J. Infect. Chemother. 3:113-127. [Google Scholar]
- 99.Sugai, M., T. Fujiwara, H. Komatsuzawa, and H. Suginaka. 1998. Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224:67-75. [DOI] [PubMed] [Google Scholar]
- 100.Sugai, M., T. Fujiwara, K. Ohta, H. Komatsuzawa, M. Ohara, and H. Suginaka. 1997. epr, which encodes glycylglycine endopeptidase resistance, is homologous to femAB and affects serine content of peptidoglycan cross bridges in Staphylococcus capitis and Staphylococcus aureus. J. Bacteriol. 179:4311-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugai, M., S. Yamada, S. Nakashima, H. Komatsuzawa, A. Matsumoto, T. Oshida, and H. Suginaka. 1997. Localized perforation of the cell wall by a major autolysin: atl gene products and the onset of penicillin-induced lysis of Staphylococcus aureus. J. Bacteriol. 179:2958-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thumm, G., and F. Götz. 1997. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 23:1251-1265. [DOI] [PubMed] [Google Scholar]
- 104.Tipper, D., and J. Strominger. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc. Natl. Acad. Sci. USA 54:1133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tomasz, A. 1986. Penicillin-binding proteins and the antibacterial effectiveness of beta-lactam antibiotics. Rev. Infect. Dis. 8(Suppl. 3):S260-S278. [DOI] [PubMed] [Google Scholar]
- 106.Tomasz, A., H. B. Drugeon, H. M. de Lencastre, D. Jabes, and L. McDougall. 1989. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob. Agents Chemother. 33:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ton-That, H., H. Labischinski, B. Berger-Bächi, and O. Schneewind. 1998. Anchor structure of staphylococcal surface proteins. III. Role of the femA, femB, and femX factors in anchoring surface proteins to the bacterial cell wall. J. Biol. Chem. 273:29143-29149. [DOI] [PubMed] [Google Scholar]
- 108.Tschierske, M., K. Ehlert, A. M. Stranden, and B. B. Berger. 1997. Lif, the lysostaphin immunity factor, complements FemB in staphylococcal peptidoglycan interpeptide bridge formation. FEMS Microbiol. Lett. 153:261-264. [DOI] [PubMed] [Google Scholar]
- 109.Tschierske, M., C. Mori, S. Rohrer, K. Ehlert, K. J. Shaw, and B. Berger-Bächi. 1999. Identification of three additional femAB-like open reading frames in Staphylococcus aureus. FEMS Microbiol. Lett. 171:97-102. [DOI] [PubMed] [Google Scholar]
- 110.Umemoto, T., T. Ota, H. Sagawa, K. Kato, H. Takada, M. Tsujimoto, A. Kawasaki, T. Ogawa, K. Harada, and S. Kotani. 1981. Chemical and biological properties of a peptidoglycan isolated from Treponema pallidum kazan. Infect. Immun. 31:767-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Heijenoort, J. 1994. Biosynthesis of the bacterial peptidoglycan unit. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 112.Vannuffel, P., M. Hersterspreute, M. Bouyer, B. Vandercam, M. Philippe, and J.-L. Gala. 1999. Molecular characterization of femA from Staphylococcus hominis and Staphylococcus saprophyticus, and femA-based discrimination of staphylococcal species. Res. Microbiol. 150:129-141. [DOI] [PubMed] [Google Scholar]
- 113.Wang, J. E., P. F. Jorgensen, M. Almlof, C. Thiemermann, S. J. Foster, A. O. Aasen, and R. Solberg. 2000. Peptidoglycan and lipoteichoic acid from Staphylococcus aureus induce tumor necrosis factor alpha, interleukin 6 (IL-6), and IL-10 production in both T cells and monocytes in a human whole blood model. Infect. Immun. 68:3965-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weber, B., K. Ehlert, A. Diehl, P. Reichmann, H. Labischinski, and R. Hakenbeck. 2000. The fib locus in Streptococcus pneumoniae is required for peptidoglycan crosslinking and PBP-mediated β-lactam resistance. FEMS Microbiol. Lett. 188:81-85. [DOI] [PubMed] [Google Scholar]
- 115.Yanagihara, Y., K. Kamisango, S. Yasuda, S. Kobayashi, I. Mifuchi, I. Azuma, Y. Yamamura, and R. Johnson. 1984. Chemical compositions of cell walls and polysaccharide fractions of spirochetes. Microbiol. Immunol. 28:535-544. [DOI] [PubMed] [Google Scholar]
- 116.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163:1-5. [PubMed] [Google Scholar]
- 117.Zhao, G., W. K. Yeh, R. H. Carnahan, J. Flokowitsch, T. I. Meier, W. E. Alborn, Jr., G. W. Becker, S. R. Jaskunas, A. M. Smith, K. P. Klugman, R. Hakenbeck, and J. Coyette. 1997. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to beta-lactam antibiotics. J. Bacteriol. 179:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]