Abstract
CP-31398, a styrylquinazoline, emerged from a high throughput screen for therapeutic agents that restore a wild-type-associated epitope (monoclonal antibody 1620) on the DNA-binding domain of the p53 protein. We found that CP-31398 can not only restore p53 function in mutant p53-expressing cells but also significantly increase the protein level and promote the activity of wild-type p53 in multiple human cell lines, including ATM-null cells. Cells treated with CP-31398 undergo either cell cycle arrest or apoptosis. Further investigation showed that CP-31398 blocks the ubiquitination and degradation of p53 but not in human papillomavirus E6-expressing cells. Of note, CP-31398 does not block the physical association between p53 and MDM2 in vivo. Moreover, unlike the DNA-damaging agent adriamycin, which induces strong phosphorylation of p53 on serines 15 and 20, CP-31398 exposure leads to no measurable phosphorylation on these sites. We found that CP-31398 could also stabilize exogenous p53 in p53 mutant, wild-type, and p53-null human cells, even in MDM2-null p53−/− mouse embryonic fibroblasts. Our results suggest a model wherein CP-31398-mediated stabilization of p53 may result from reduced ubiquitination, leading to high levels of transcriptionally active p53. Further understanding of this mechanism may lead to novel strategies for p53 stabilization and tumor suppression in cancers, even those with absent ARF or high MDM2 expression.
The tumor suppressor protein p53 is a potent inducer of apoptosis and cell cycle arrest in response to various cellular stresses, such as DNA damage, hypoxia, or hyperproliferative signals (2, 12, 36, 41, 45). Mutations in the p53 genes are found in about half of all human tumors (22). A large portion of the remaining tumors, although expressing wild-type p53, is believed to be defective in the pathway of p53-mediated tumor suppression due to accelerated degradation or ineffective stabilization of the wild-type p53.
Restoration or enhancement of p53 function in tumor cells is a logical approach for cancer therapy because it targets the major difference between normal and cancer cells (44). The most straightforward strategy is to provide exogenous p53 to tumor cells. Several clinical trials that use various vectors to transduce tumor cells with the human p53 cDNA are ongoing but are limited by transfection efficiency and the rapid degradation of p53 protein. Peptides are also being developed with the ability to reactivate mutant p53 (15, 37), but little progress has been made in the clinic because of the cost and difficulty in large-scale synthesis. Most recently, small molecules, due to the advantages of possible large-scale chemical synthesis and easier delivery in vivo, are being pursued as a potentially feasible strategy for cancer therapy (8, 14, 21, 42, 48). Among the very limited compounds reported, CP-31398 was the first that emerged from a screen of a chemical library. This prototype compound can maintain p53 in a conformation that is associated with active p53. Furthermore, CP-31398 can rescue some mutant p53s to a wild-type conformation and therefore restore the p53 functions of cell cycle arrest or apoptosis (14). It was recently reported that CP-31398 not only can affect cells with mutant p53 but also can elevate the steady-state wild-type p53 to high levels equivalent to those observed following DNA damage (40, 42). These findings have recently been confirmed (31).
The regulation of p53 activity is mainly posttranslational. Stabilization is an essential step for p53 to function efficiently in response to cellular stresses or checkpoints. Under physiological conditions, p53 is expressed at low or undetectable levels with a half-life of approximately 10 to 20 min in most cells. This rapid degradation is at least in part mediated by the ubiquitination pathway following the interaction of MDM2 with the N terminus of p53 (2, 6, 13, 19, 25, 32). Interestingly, it has been recently demonstrated that MDM2 can catalyze p53 ubiquitination within a conformationally flexible region of the p53 DNA-binding domain, suggesting a link between ubiquitination and the conformational status of p53 (39). DNA damage, caused by UV light or ionizing irradiation, results in stabilization of endogenous p53 through a series of physiological responses, including ataxia telangiectasia mutated (ATM)/ATR activation, phosphorylation of p53 and blockage of the binding of MDM2 to the p53 N terminus (5, 7, 9, 27, 38). Activation of some oncogenes like c-myc, ras, or E2F1 also stabilizes p53 by the ARF-MDM2 pathway, in which case ARF binds to MDM2 and releases p53 from MDM2 association (23, 24). Another well-known category of p53 stabilizers is proteasome inhibitors that prevent the degradation of the ubiquitinated p53 (1, 3, 26), as well as other ubiquitinated proteins, by the 26S proteosomes.
In the present study, we investigated the mechanism by which conformational stabilization of p53 by CP-31398 increases the p53 steady-state levels. The mechanism of increasing the cellular p53 concentration is clearly different from that which follows exposure to DNA-damaging agents, proteasome inhibitors, or activation of oncogenes. Further understanding of this mechanism may lead to novel strategies for p53 stabilization and tumor suppression in cancers, even those with absent ARF or high MDM2 expression.
MATERIALS AND METHODS
Cell lines.
Tumor cell lines with wild-type p53 were used, including the H460 non-small-cell lung carcinoma cell line, the PA-1 ovarian cancer cell line, and the HCT116/p53+/+ and the HCT116/p53−/− colon adenocarcinoma cell lines. H460, PA-1, and U2OS cells were from the American Type Culture Collection (Rockville, Md.), and HCT116 cells were from Bert Vogelstein (Johns Hopkins Unversity). The GM01389D ATM lymphoblastoid and the GM02184D control lymphoblastoid cell lines were obtained from Coriell Cell Repositories (Camden, N.J.) and grown in RPMI 1640 with 10% fetal bovine serum, 100 μg of penicillin/ml, and 100 μg of streptomycin/ml. The WI-38 human fibroblast cell was from the American Type Culture Collection and maintained in Dulbecco's minimal essential medium with 10% fetal bovine serum and antibiotics, 100 μg of penicillin/ml and 100 μg of streptomycin/ml. The mouse embryonic fibroblast (MEF) and MEF p53−/− MDM2−/−, the double knockout of p53 and MDM2, were kindly provided by Stephen Jones, University of Massachusetts Medical School.
Antibodies.
The anti-p53 antibodies Ab-2 (Oncogene), DO-1 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), and FL293 (Santa Cruz Biotechnology, Inc.) were used for immunoblotting of p53, detection of ubiquitinated p53, and p53 immunoprecipitation, respectively. Anti-MDM2 (Ab-1) was from Calbiochem/Oncogene Science (Boston, Mass.) and used for MDM2 immunoblotting. Anti-phospho-p53 antibodies against phosphorylation of ser15, ser20, and ser392 were obtained from Cell Signaling Technology (Beverly, Mass.). A rabbit anti-phospho-p53 at ser46 was kindly provided by Yoichi Taya (33), National Cancer Center Research Institute of Japan. Anti-Ub (FL76) from Santa Cruz Biotechnolology, Inc., was used for the detection of ubiquitinated proteins.
Adenovirus construction.
Ad/GFP-p53 and Ad/His-p53 were constructed by previously described methods (20). For Ad/GFP-p53 construction, green fluorescent protein (GFP) was fused to the N terminus of p53 and the open reading frame of the fused protein was inserted into the pAdTrack-CMV vector, which was kindly provided by Bert Vogelstein (Johns Hopkins University). For Ad/His-p53 construction, the His-p53 was generated by inserting the p53 cDNA into the plasmid pcDNA4/HisMax vector (Invitrogen). The fragment of HindIII and EcoRV from the plasmid pcDNA4/HisMax/p53, containing the QBI SP163 translational enhancer, polyhistidine (His6), and in-frame p53, was inserted into the pAdTrack-CMV vector. The pAdTrack-CMV constructs were recombined with pAdEasy-1, provided by Bert Vogelstein, in Escherichia coli BJ5183 cells to get recombined pADs with the exogenous inserts. The recombined adenoviruses were replicated and amplified in 293 cells by transfection of the pAD plasmid DNAs. The amplified viruses were purified by cesium chloride gradient ultracentrifugation and stored at −20°C in a buffer containing 25% (vol/vol) glycerol, 10 mM Tris (pH 8.0), 100 mM NaCl, 0.1% bovine serum albumin, and 1 mM MgCl2. Ad/E6 was previously described (34).
Western and Northern blotting.
For Western blotting, cells were collected and protein concentrations were quantified by the Bio-Rad (Hercules, Calif.) protein assay prior to sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE). Proteins were transferred to a polyvinylidene difluoride membrane (Immunobilon-P; Millipore Corporation, Bedford, Mass.) with a semidry transfer apparatus from Bio-Rad Laboratories. The membranes with transferred proteins were blotted with 10% (wt/vol) nonfat dry milk and then subsequently subjected to the primary antibody and horseradish peroxidase-labeled secondary antibody. The signal was visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Little Chalfont, England) and exposed to an X-ray film.
For Northern blotting, total RNA was purified with the RNeasy mini kit (Qiagen, Valencia, Calif.). After an agarose-formaldehyde gel electrophoresis, the RNAs were transferred to a nitrocellulose membrane (Zeta-Probe GT genomic tested blotting membranes; Bio-Rad Laboratories), probed with 32P-labeled probes of human p53 and MDM2 cDNA, and exposed to an X-ray film.
In vivo ubiquitination assay.
Cells were grown in a six-well plate to 80% confluence. CP-31398 was added at a final concentration of 15 μg/ml and incubated at 37°C for 1 h. Then ALLN, a proteosomal inhibitor (Sigma, St. Louis, Mo.), was added at a final concentration of 50 μM and incubated for 4 h. The plate with cells was placed on ice, and boiled SDS sample buffer was added after washing the plate twice with ice-cold phosphate-buffered saline. Lysates were collected for SDS-PAGE, and ubiquitinated p53s were detected by immunoblotting with the DO-1 antibody (Santa Cruz Biotechnology, Inc.).
Active caspase 3 assay.
Cells in a six-well plate were trypsinized and collected in 15-ml centrifuge tubes. The collected cells were fixed and stained for active caspase 3 with a Cytofix/Cytoperm kit from BD Biosciences Pharmingen (San Diego, Calif.) according to the manufacturer's instructions. The stained cells were subjected to flow cytometry (Beckman Coulter Epics Elite) counting for the active caspase 3-positive cells.
RESULTS
CP-31398 stabilizes wild-type p53 protein independent of ATM and MDM2.
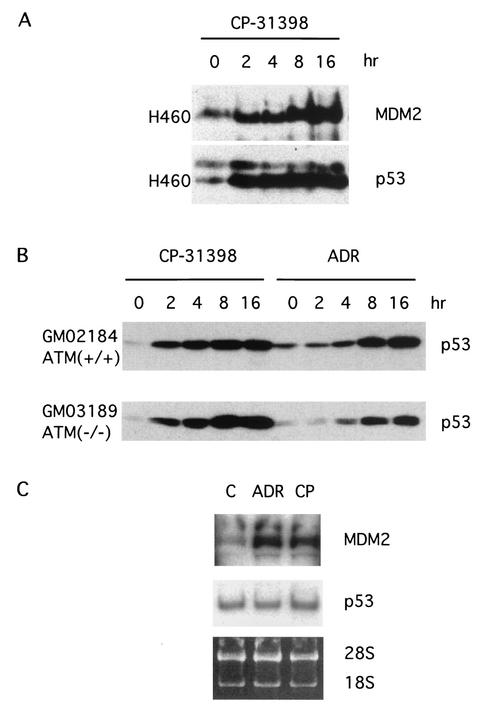
It was previously found that CP-31398 not only can rescue the DNA-binding activity of the mutant p53 in SW480 cells and induce cell cycle arrest and/or apoptosis in cells with mutant p53 but can also increase the steady-state levels of wild-type p53 and induce cells with wild-type p53 to undergo cell cycle arrest and/or apoptosis (42). To explore the mechanism of p53 accumulation, we first confirmed the effect of CP-31398 on wild-type p53 protein and excluded an effect of CP-31398 on p53 mRNA levels (Fig. 1). Less than 1 h after CP-31398 was added to the cell culture medium at a final concentration of 15 μg/ml, p53 protein levels were elevated in all the cell lines with wild-type p53 tested. These included H460 (Fig. 1A), HCT116, U2OS, PA-1, diploid fibroblast WI38 cells (data not shown), and lymphoblastoid cells (Fig. 1B). The accumulation of p53 in the CP-31398-treated cells appears to be due to posttranslational regulation because the half-life of p53 in these cells was significantly prolonged (42) and no significant change was found in the p53 mRNA levels (Fig. 1C). As p53 stabilization is essential and important in p53-mediated cell cycle arrest and apoptosis, we explored further the mechanism by which CP-31398 exposure results in accumulation of wild-type p53. We first speculated that CP-31398 might act as a DNA-damaging agent as has been recently proposed (35). As ATM represents an important link in the pathway of DNA-damage-induced p53-dependent checkpoint signaling, we treated the ATM−/− cell line GM03189D, a lymphoblastoid cell line established from a patient with ataxia telangiectasia, to test this possibility. As shown in Fig. 1B (lower panel), CP-31398 rapidly increased p53 to a high level in this ATM−/− cell line, while p53 elevation was delayed following treatment by the DNA-damaging agent adriamycin. Both CP-31398 and adriamycin induced high levels of p53 in an ATM+/+ cell line (Fig. 1B, upper panel).
FIG. 1.
CP-31398 stabilizes wild-type p53. (A and B) H460 cells, as well as ATM-positive 2184 and ATM-null 3189 lymphoblasts, were treated with CP-31398 at a final concentration of 15 μg/ml. Adriamycin (ADR) was used as a DNA damage control at a concentration of 0.2 μg/ml. Cells were collected at the indicated time points and subjected to SDS-PAGE. p53 and MDM2 were detected by anti-p53 (Ab-2; Calbiochem) and anti-MDM2 (Ab-1; Calbiochem) antibodies, respectively. Actin was probed as a loading control (data not shown). (C) Northern blot of the total RNA from CP-31398 (CP)-treated H460 cells with the probes of MDM2 and p53, respectively. Lane C, control.
Induction of p53 by CP-31398 does not involve phosphorylation of ser20 or ser15.
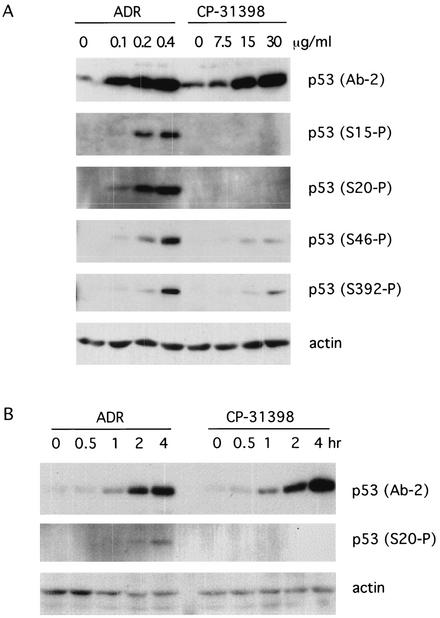
Because there are kinases and pathways other than ATM that also lead to p53 stabilization (e.g., Chk2 or ATR), we investigated the status of p53 phosphorylation in CP-31398-exposed cells. We chose several antibodies against phosphorylated p53, including ones that recognize phosphorylation at ser15, ser20, ser46, and ser392, respectively, to check whether p53 is phosphorylated at these sites. H460 cells were treated with CP-31398 or adriamycin (as a DNA damage control) for 4 h, and cell lysates were subjected to SDS-PAGE and probed with antiphosphorylated p53 antibodies. As shown in Fig. 2A, CP-31398 exposure did not lead to phosphorylation of p53 at ser15 and ser20, with minimal changes at ser46 and ser392, while adriamycin induced efficient dose-dependent phosphorylation of p53 at all of these sites. In order to rule out the possibility of transient phosphorylation of ser15 or 20, which might occur at an early stage, a time course of p53 phosphorylation was examined. Phosphorylation of ser20 appeared at 2 h after treatment with adriamycin (Fig. 2B), phosphorylation of ser15 appeared at 4 h after adriamycin treatment (data not shown), and no phosphorylation of either ser15 or 20 was observed after treatment with CP-31398 during the time course. The phosphorylation of p53 (5, 9), especially the phosphorylation of ser20, has been identified as critical for UV- and irradiation-induced p53 stabilization, as it prevents the binding of MDM2 to p53. However, phosphorylation of ser20 as well as ser15 does not appear to be involved in the accumulation of p53 following CP-31398 exposure. These results indicate that CP-31398 exposure leads to wild-type p53 protein accumulation by a mechanism different from that following DNA damage and suggests that CP-31398 may not be a DNA-damaging agent at concentrations required for p53 activity.
FIG. 2.
CP-31398 does not phosphorylate p53 at ser15 or ser20. (A) Adriamycin (ADR) or CP-31398 was added to H460 cell culture at various concentrations and incubated for 4 h. Cell lysates were subjected to SDS-PAGE and probed with anti-p53 (Ab-2) and anti-phospho-p53 antibodies against ser15, ser20, ser46, and ser392, respectively. (B) H460 cells were treated with ADR at a concentration of 0.2 μg/ml or CP-31398 at a concentration of 15 μg/ml. Cells were lysed at different time points after treatment and subjected to SDS-PAGE and immunoblotting with anti-phospho-ser20 of p53.
CP-31398 does not disrupt MDM2 binding to p53.
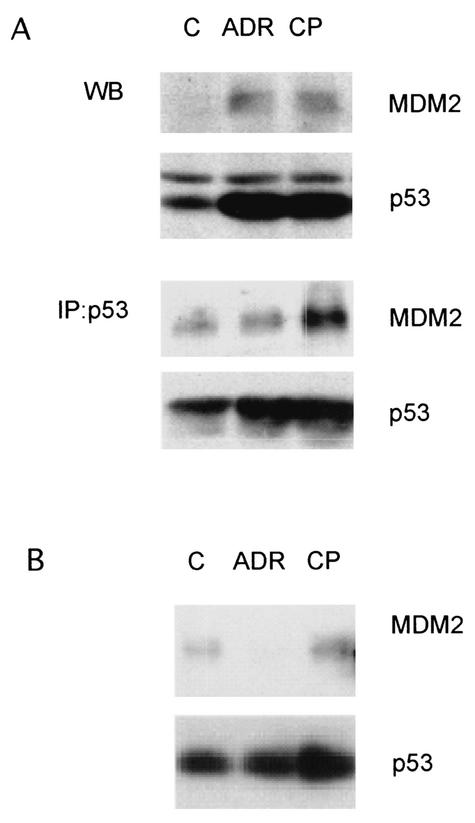
MDM2 is one of the critical posttranslational regulators of p53 (13, 19, 25). MDM2 binds to the N-terminal transcription activation domain of p53 and serves as an E3 ligase to target p53 for ubiquitination and proteasomal degradation. MDM2 is also a transcriptional target of p53, thereby acting in a negative feedback loop to regulate p53 function (4). In the CP-31398-treated cells, MDM2 levels became elevated in a manner that correlated with p53 accumulation (Fig. 1A and C). As CP-31398 exposure did not lead to phosphorylation of p53 at ser20, we speculated that it might not prevent the binding of MDM2 to the p53 N terminus. To test this hypothesis and to determine whether MDM2 still binds to p53 after CP-31398 treatment, we immunoprecipitated p53 with an antibody against full-length p53 to test whether MDM2 could be coimmunoprecipitated. As shown in Fig. 3A, more MDM2 was coimmunoprecipitated with p53 following CP-31398 treatment, whereas no more MDM2 was associated with p53 after the treatment with adriamycin. To confirm these observations, we infected H460 lung cancer cells with an adenovirus carrying a six-His-tagged p53 and treated the infected cells with CP-31398 or adriamycin. The six-His-tagged p53 was precipitated with Ni-nitrilotriacetic acid agarose and subjected to SDS-PAGE. As shown in Fig. 3B, more MDM2 was coprecipitated after the treatment with CP-31398 while less MDM2 associated with p53 after the treatment with adriamycin. These experiments revealed that CP-31398 does not disrupt the binding of MDM2 to p53. It remains unclear why the MDM2-bound p53 is transcriptionally active and capable of targeting gene activation.
FIG. 3.
CP-31398 does not prevent the binding of MDM2 to p53. (A) H460 cells were treated with adriamycin (ADR) or CP-31398 (CP) for 4 h. p53 was immunoprecipitated (IP) with anti-p53 (FL-393; Santa Cruz Biotech). The immunoprecipitated complexes were subjected to SDS-PAGE and probed with anti-p53 (Ab-2; Calbiochem) and anti-MDM2 (Ab-1; Calbiochem) antibodies. (B) H460 cells were infected with Ad/His-p53 and treated with adriamycin or CP-31398. p53 was precipitated with Ni-nitrilotriacetic acid resin prior to SDS-PAGE. MDM2 and p53 were probed with the same antibodies as in panel A. WB, Western blotting; C, control.
CP-31398 inhibits the ubiquitination of p53.
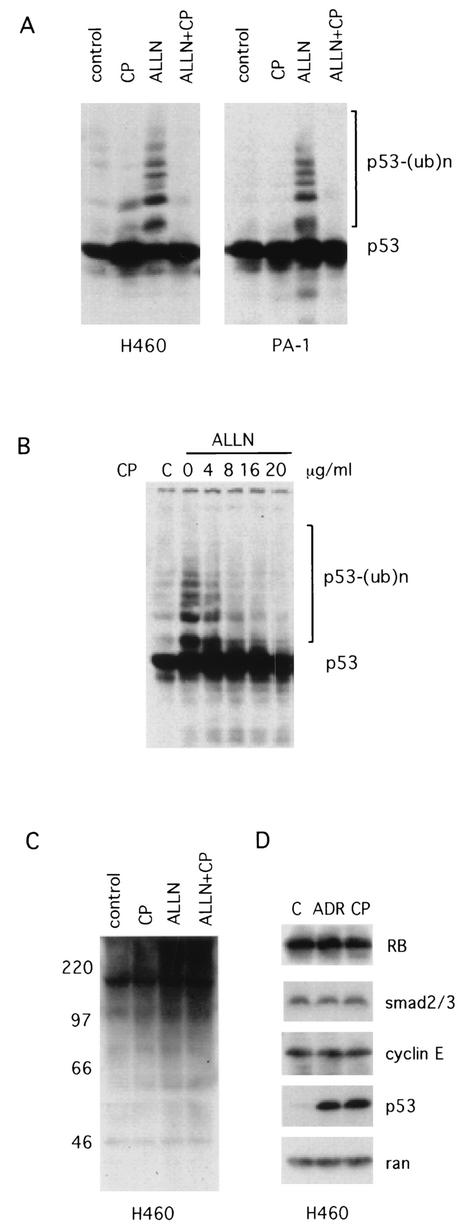
Because MDM2 levels rise after CP-31398 exposure and CP-31398 does not disrupt the association between MDM2 and p53, we hypothesized that CP-31398 may block ubiquitination or degradation of p53 at a step beyond MDM2-p53 interaction. We found no evidence that CP-31398 is a proteasome inhibitor because we did not observe any ubiquitinated p53 ladders in the CP-31398-treated cells with various p53 antibodies (Fig. 1A and B, 2, 3A, and 4 and data not shown). Since CP-31398 has been shown to influence the conformation of the DNA-binding domain and recent evidence suggested that ubiquitination of p53 occurs within a conformationally flexible region of the DNA-binding domain (39), we investigated whether CP-31398 could block ubiquitination even in the presence of MDM2. To analyze p53 ubiquitination in the CP-31398-treated cells, we exposed the H460 and HCT116 cells to CP-31398 (15 μg/ml) for 1 h and then ALLN, a proteasome inhibitor, was added at a final concentration of 50 μM and the cells were incubated for 4 h prior to analysis of the lysates. ALLN blocks the degradation of ubiquitinated proteins and consequently increases the level of ubiquitinated proteins that can be detected by immunoblotting. The CP-31398- and ALLN-treated cells were lysed and subjected to SDS-PAGE. p53 and its ubiquitinated forms were analyzed by immunoblotting with a monoclonal antibody against p53, DO-1. As shown in Fig. 4A, cells treated with ALLN alone showed a typical pattern of ubiquitinated p53 ladders above the native p53 band. However, the ubiquitinated p53 ladders were completely absent in cells treated with either CP-31398 alone or in combination with ALLN (Fig. 4A). We further showed that the inhibitory effect of CP-31398 on p53 ubiquitination was dose dependent (Fig. 4B). The inhibitory effect of CP-31398 appeared at a concentration of 4 μg/ml and reached its maximum effect at a concentration of 15 μg/ml, at which point the p53 protein accumulated to a high level.
FIG. 4.
CP-31398 inhibits the ubiquitination of p53. (A) H460 and PA-1 cells were treated with CP-31398 (CP) at a concentration of 15 μg/ml for 1 h, and then ALLN was added at a concentration of 50 μM. Four hours later, cells were lysed for Western blotting with DO-1 against p53. (B) Dose-dependent inhibition of p53 ubiquitination by CP-31398. (C) Same H460 cell lysates as in panel A, probed with an anti-ubiquitin antibody (Santa Cruz). (D) Immunoblots against pRB, smad2/3, and cyclin E in H460 cells after treatment with adriamycin (ADR) or CP-31398. Lanes C, control.
To test whether the inhibition of ubiquitination by CP-31398 is p53 specific or universal, we blotted total cell lysates with an anti-ubiquitin antibody. As shown in Fig. 4C, the lanes showed the expected smearing with some punctuated bands and there was no obvious change in the CP-31398-treated cells compared to untreated cells. To confirm further that CP-31398 is not a universal ubiquitination inhibitor, we checked the levels of several other rapidly degraded proteins, such as pRB, smad2/3, and cyclin E, after CP-31398 treatment. As shown in Fig. 4D, the levels of these randomly picked proteins were not changed by the treatment with CP-31398. These results indicate that CP-31398 specifically inhibits p53 ubiquitination, suggesting that the compound is affecting p53 rather than the ubiquitination machinery.
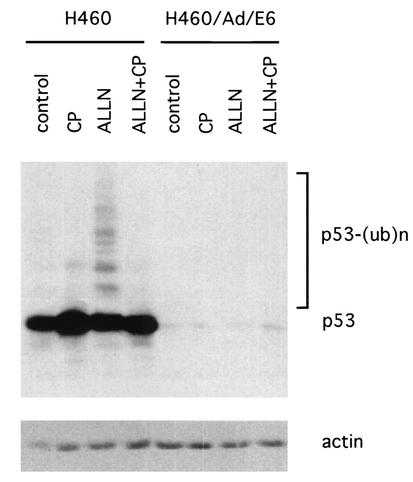
We also tested whether CP-31398 can block human papillomavirus (HPV) E6-induced p53 ubiquitination. The E6 proteins of HPV, both type 16 and type 18, are known to bind to p53 and degrade p53 by recruiting an E6-associated protein, which exists in most mammalian cells. E6-associated protein, with a HECT domain possessing ubiquitin ligase activity, can degrade p53 through the ubiquitin-proteasome pathway. H460 cells were infected with an adenovirus carrying E6 (34) for 12 h and then treated with CP-31398 together with ALLN. CP-31398 did not rescue E6-induced p53 degradation (Fig. 5). Of interest, the proteasome inhibitor ALLN also could not effectively block the observed E6-mediated p53 degradation. This phenomenon is consistent with the idea there may be a difference in the pathways between MDM2- and E6-induced p53 ubiquitination and degradation.
FIG. 5.
CP-31398 (CP) does not block p53 degradation by HPV E6. H460 and AdH460 Eb-infected cells were treated with CP-31398 at a concentration of 15 μg/ml for 1 h, and then ALLN was added at a concentration of 50 μM. Four hours later, cells were lysed for Western blotting with DO-1 against p53.
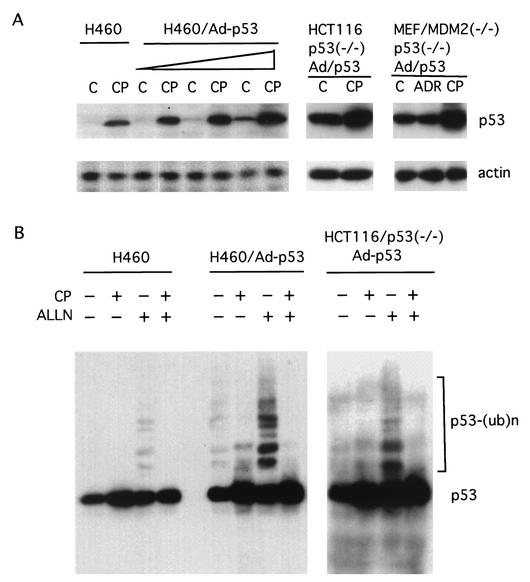
CP-31398 stabilizes exogenous p53, even in MDM2−/− cells.
In order to further examine the mechanism by which CP-31398 stabilizes p53, we tested whether CP-31398 could also stabilize exogenous p53. We infected H460 cells, HCT116/p53−/− cells, and p53−/− MDM2−/− MEFs with an adenovirus expressing human p53 for 6 h and then treated the cells with CP-31398 at a concentration of 15 μg/ml for 4 h. To our surprise, CP-31398 could not only stabilize exogenous p53 in MDM2-containing cells, such as H460 and HCT116 cells, but also stabilize p53 in MDM2 knockout cells (Fig. 6A). Moreover, CP-31398 stabilized exogenous p53 by inhibiting its ubiquitination (Fig. 6B). Taken together, these data suggest that MDM2 is unlikely to be the primary target involved in the stabilization of p53 following CP-31398 exposure. CP-31398-induced wild-type p53 accumulation appears to occur at a step subsequent to MDM2 binding and is associated with reduced accumulation of ubiquitinated p53.
FIG. 6.
CP-31398 stabilizes exogenous p53 by inhibiting its ubiquitination. (A) H460 cells were infected with Ad/p53 at multiplicities of infection (MOI) of 2.5, 5, and 10 for 12 h. CP-31398 (CP) was then added to the medium at a concentration of 15 μg/ml and incubated for 8 h. Cells were lysed for SDS-PAGE. p53 was probed with Ab-2. Also shown are p53−/− HCT116 cells and p53−/− MDM2−/− MEF cells infected with Ad/p53 (MOI-5) and treated with CP-31398. C, control. (B) H460 cells were infected with Ad/p53 at a MOI of 5 (second panel). Twelve hours after the infection, CP-31398 was added and incubated for 1 h and then ALLN was added at a concentration of 50 μg/ml. Four hours later, cells were lysed for Western blotting by DO-1 against p53. Also shown are results performed in HCT116/p53−/− cells (third panel) infected and treated as described for the H460 cells in the second panel.
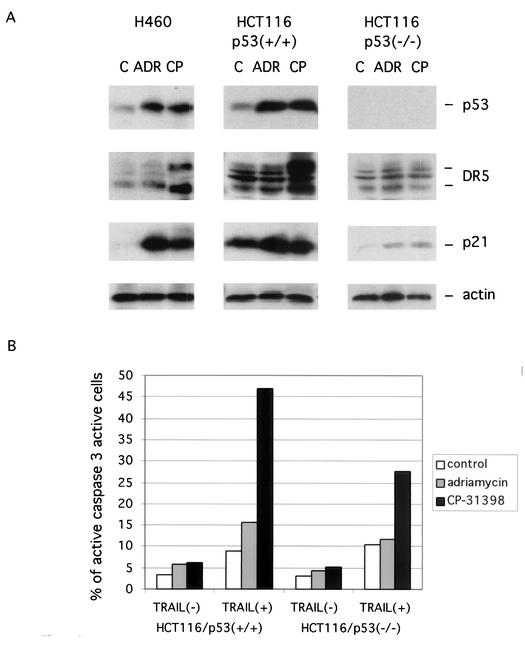
p53 in CP-31398-treated cells preferentially induces KILLER/DR5 versus p21 expression.
Since CP-31398-mediated accumulation of p53 is distinct from that observed following DNA damage (i.e., the mechanism does not involve phosphorylation of ser15 or ser20 and does not lead to disruption of the MDM2 binding to p53), we compared the function of p53 in cells treated with CP-31398 or with a DNA-damaging agent, adriamycin. The elevation of MDM2, both protein (Fig. 1A) and mRNA (Fig. 1C), levels in CP-31398- or adriamycin-treated cells indicates that both agents enhance p53 activity. We further examined other p53 targets, p21 and KILLER/DR5, two representative mediators of p53-induced cell cycle arrest and apoptosis, respectively. We treated H460 and HCT116 and HCT116/p53−/− cells with CP-31398 or adriamycin for 8 h and then collected the cell lysates for Western blotting. As shown in Fig. 7A, CP-31398 induced a high level of KILLER/DR5 while adriamycin did not induce much expression of DR5. However, although CP-31398 also induced the expression of p21 compared to the control cells, it was not as effective as adriamycin at p21 induction in either of the cell lines tested. CP-31398 induction of KILLER/DR5 and p21 was p53 dependent because there were only trace level changes in the expression of the two proteins in the p53−/− HCT116 cells. High expression of KILLER/DR5 induced by CP-31398 rendered the cells more sensitive (Fig. 7B) to the cytotoxic ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which binds to the KILLER/DR5 cell death receptor. These results suggest that conformational constraints and subsequent ubiquitination status of p53 can allow for preferential activation of various p53-responsive genes.
FIG. 7.
CP-31398 differentially induces p53 target gene expression and enhances sensitivity to TRAIL. (A) Cells were treated with CP-31398 (CP) or adriamycin (ADR) for 8 h and probed with antibodies as indicated. C, control. (B) HCT116/p53+/+ and HCT116/p53−/− cells were treated with CP-31398 or adriamycin for 4 h, then TRAIL was added at a final concentration of 20 ng/ml, and cells were incubated for another 4 h. Cells were collected for active caspase 3 staining, and positive cells were counted by fluorescence-activated cell sorter analysis.
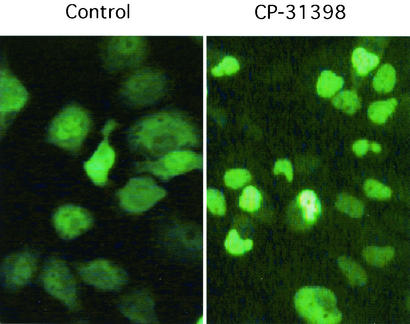
CP-31398 influences the accumulation and nuclear localization of exogenous wild-type p53.
As MDM2 and its binding to p53 were elevated after the CP-31398 treatment, we examined whether the subcellular localization of p53 may be influenced because it is reported that MDM2 promotes p53 export from the nucleus (16, 18). We constructed an adenovirus expressing GFP-fused p53 at the N terminus for tracking the subcellular localization of p53. As shown in Fig. 8, after 12 h of infection with Ad/GFP-p53, the p53 appeared mainly in the cytoplasm of H460 cells but after 4 h of subsequent treatment with CP-31398, p53 appeared primarily in the nucleus. In other cell lines tested, which included HCT116 and WI-38 cells, although p53 appeared mainly in the nucleus from the very beginning after Ad/GFP-p53 infection without obvious cytoplasm distribution, the signal, after the treatment of CP-31398, appeared much brighter in the nucleus (data not shown). The stabilization and increased nuclear localization of exogenous p53 in CP-31398-exposed cells may have applicability to the enhancement of the efficacy of exogenous p53 gene therapy.
FIG. 8.
CP-31398 promotes p53 nuclear accumulation. H460 cells were infected with Ad/GFP-p53. Twelve hours after the infection, CP-31398 was added at a concentration of 15 μg/ml and incubated for 6 h. Cells were photographed under a phase-contrast laser microscope.
DISCUSSION
CP-31398 was identified from a chemical library and represents a promising strategy for anticancer therapy. Whether the effect of CP-31398 on cell growth is dependent on p53 has been controversial. It was previously reported that CP-31398 could rescue some mutant p53 protein by restoring a wild-type conformation and could induce cell cycle arrest or apoptosis by activating p53 target genes (14, 42). Recently, it was reported that CP-31398 may not bind to p53 and may inhibit cell growth independently of p53 (35). A model was suggested in which CP-31398 may be incorporated into p53 during its biosynthesis and may not then dissociate from p53. In experiments investigating the mechanism of action of CP-31398, we were surprised to find that CP-31398 can increase wild-type p53 levels (42). We report here that CP-31398 inhibits the accumulation of ubiquitinated p53 and stabilizes p53 to a level equivalent to that of DNA damage but by a different mechanism, which may be related to the effects of the compound on p53 conformation.
The accumulation of p53 is an early and critical step for p53 activation. Under physiological conditions, p53 is under strict control with a very short half-life, in part due to the MDM2-p53 autoregulatory feedback loop (19, 25, 49). MDM2 binds to the N-terminal transactivation domain of p53 with its p53-binding domain, and the RING domain of MDM2 catalyses the ubiquitination of p53 (13). The ubiquitinated p53 is then degraded by the 26S proteosomes either in the cytoplasm or in the nucleus (16, 47). MDM2 is also a transcriptional target of p53 (4). Therefore, to activate p53, the MDM2-p53 feedback loop must be interrupted. In the case of DNA damage, p53 is highly phosphorylated by a number of protein kinases including ATM/ATR and Chk2 (11). Phosphorylation of ser20 has been reported to be crucial for the DNA damage-induced stabilization of p53 (9, 10, 43). Our experiments show that CP-31398 induced similar levels of p53 stabilization in both ATM+/+ and ATM−/− cells, whereas DNA damage-induced p53 stabilization was delayed in ATM−/− cells compared to the ATM+/+ control cells. Further experiments showed that the inhibition of p53 ubiquitination by CP-31398 does not involve the phosphorylation of ser20, as well as ser15, and does not prevent the binding of MDM2 to p53. Although the binding of MDM2 to p53 is essential for MDM2-mediated ubiquitination of p53, it is not necessarily predicted that the binding will always lead to ubiquitination. For example, MDM2 can bind to the p53 family member p73 but cannot ubiquitinate and degrade it (50). It is reported that replacement of a sequence of amino acids of p53 from 92 to 112 with the homologous p73 sequence would block the MDM2-induced p53 ubiquitination (17). Moreover, stabilization of exogenous p53 in MDM2 knockout cells suggests that MDM2 is unlikely to be the primary target involved in CP-31398-induced p53 accumulation. In addition, our recent preliminary experiments show that p63 and p73, two other p53 family members, could also be stabilized by CP-31398 (data not shown). These two p53 family members are not MDM2 targets for degradation, although MDM2 can bind to them (17, 29). Our results indicate that CP-31398 stabilizes p53 through a unique and novel pathway. Understanding this pathway for p53 stabilization may lead to new strategies for cell growth inhibition and tumor suppression.
Of interest, CP-31398 induces an extremely high level of KILLER/DR5, one of the receptors of TRAIL, which links p53 to the pathway of receptor-mediated apoptosis (46). The induction of DR5 in the CP-31398-treated cells is p53 dependent because CP-31398 does not significantly induce expression of DR5 in p53-null cells. As a consequence, CP-31398 enhances greatly the TRAIL-induced cell death. For comparison, the DNA-damaging agent adriamycin does not induce significant expression of DR5 by 8 h and, therefore, it does not significantly enhance the TRAIL-induced apoptosis following short exposure. Also, a p53/DR5-independent pathway may exist in CP-31398-induced apoptosis because CP-31398 appears to induce slightly more apoptosis than the control and adriamycin-treated p53-null cells, but this effect is much less than what is observed in the wild-type p53-containing cells.
In CP-31398-treated cells, p53 accumulates in the nucleus in all of the cell lines tested following exogenous adenovirus-mediated expression of a GFP-p53 fusion protein. The enhanced fluorescence signal of GFP-p53 in H460 cells clearly transferred from the cytoplasm to the nucleus after the treatment with CP-31398. Whether the stabilization of p53 could be due to changes in the subcellular localization of p53 is not certain. We favor a model wherein the localization of p53 in the nucleus is due to the conformational stabilization of p53 because CP-31398 blocks the ubiquitination of p53 and C-terminal ubiquitination of p53 is reported to contribute to its nuclear export (30). In our studies, increased nuclear localization and decreased ubiquitination of p53 occurred despite an increased MDM2 level and increased binding between p53 and MDM2.
Our studies provide evidence for a novel mechanism for wild-type p53 accumulation that occurs at a step subsequent to MDM2 binding that is associated with reduced accumulation of ubiquitinated p53. The CP-31398-stabilized p53, associated with MDM2, is localized in the nucleus and is transcriptionally active with an apparently altered target gene selectivity. The precise step at which p53 ubiquitination is blocked remains to be identified, i.e., CP-31398 may constrain p53 in a conformation that does not allow for access to the ubiquitination site within the DNA-binding domain, or CP-31398 may possibly stimulate the activity of the recently described p53-deubiquitinating enzyme HAUSP (28). The altered selectivity of p53 in target gene activation is also not yet clear and may involve altered conformation of p53, altered posttranslational modification, or association with histone acetylases. Our results support the idea that it may be possible to stabilize and alter p53 function by using drugs at a step beyond its interaction with MDM2. This possibility suggests a novel strategy for drug development in cancer therapy that may not be hindered by MDM2 amplification or ARF deletion.
REFERENCES
- 1.An, W. G., Y. Chuman, T. Fojo, and M. V. Blagosklonny. 1998. Inhibitors of transcription, proteasome inhibitors, and DNA-damaging drugs differentially affect feedback of p53 degradation. Exp. Cell Res. 244:54-60. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, D., and A. Liefshitz. 2001. Potential of the proteasomal inhibitor MG-132 as an anticancer agent, alone and in combination. Anticancer Res. 21:3941-3947. [PubMed] [Google Scholar]
- 4.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bean, L. J., and G. R. Stark. 2001. Phosphorylation of serines 15 and 37 is necessary for efficient accumulation of p53 following irradiation with UV. Oncogene 20:1076-1084. [DOI] [PubMed] [Google Scholar]
- 6.Blagosklonny, M. V. 1997. Loss of function and p53 protein stabilization. Oncogene 15:1889-1893. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, C., E. Tobiasch, M. Litfen, H. J. Rahmsdorf, and P. Herrlich. 1999. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene 18:1723-1732. [DOI] [PubMed] [Google Scholar]
- 8.Bykov, V. J., N. Issaeva, A. Shilov, M. Hultcrantz, E. Pugacheva, P. Chumakov, J. Bergman, K. G. Wiman, and G. Selivanova. 2002. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8:282-288. [DOI] [PubMed] [Google Scholar]
- 9.Chehab, N. H., A. Malikzay, E. S. Stavridi, and T. D. Halazonetis. 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA 96:13777-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumaz, N., D. M. Milne, L. J. Jardine, and D. W. Meek. 2001. Critical roles for the serine 20, but not the serine 15, phosphorylation site and for the polyproline domain in regulating p53 turnover. Biochem. J. 359:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durocher, D., and S. P. Jackson. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13:225-231. [DOI] [PubMed] [Google Scholar]
- 12.El-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 13.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 14.Foster, B. A., H. A. Coffey, M. J. Morin, and F. Rastinejad. 1999. Pharmacological rescue of mutant p53 conformation and function. Science 286:2507-2510. [DOI] [PubMed] [Google Scholar]
- 15.Friedler, A., L. O. Hansson, D. B. Veprintsev, S. M. Freund, T. M. Rippin, P. V. Nikolova, M. R. Proctor, S. Rudiger, and A. R. Fersht. 2002. A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proc. Natl. Acad. Sci. USA 99:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 17.Gu, J., D. Chen, J. Rosenblum, R. M. Rubin, and Z. M. Yuan. 2000. Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol. Cell. Biol. 20:1243-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, J., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol. Cell. Biol. 21:8533-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 20.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hietanen, S., S. Lain, E. Krausz, C. Blattner, and D. P. Lane. 2000. Activation of p53 in cervical carcinoma cells by small molecules. Proc. Natl. Acad. Sci. USA 97:8501-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollstein, M., K. Rice, M. S. Greenblatt, T. Soussi, R. Fuchs, T. Sorlie, E. Hovig, B. Smith-Sorensen, R. Montesano, and C. C. Harris. 1994. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 22:3551-3555. [PMC free article] [PubMed] [Google Scholar]
- 23.Honda, R., and H. Yasuda. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamijo, T., J. D. Weber, G. Zambetti, F. Zindy, M. F. Roussel, and C. J. Sherr. 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 95:8292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 26.Kubbutat, M. H., and K. H. Vousden. 1997. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol. Cell. Biol. 17:460-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latonen, L., Y. Taya, and M. Laiho. 2001. UV-radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene 20:6784-6793. [DOI] [PubMed] [Google Scholar]
- 28.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648-653. [DOI] [PubMed] [Google Scholar]
- 29.Little, N. A., and A. G. Jochemsen. 2001. Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene 20:4576-4580. [DOI] [PubMed] [Google Scholar]
- 30.Lohrum, M. A., D. B. Woods, R. L. Ludwig, E. Balint, and K. H. Vousden. 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luu, Y., J. Bush, K. J. Cheung, Jr., and G. Li. 2002. The p53 stabilizing compound CP-31398 induces apoptosis by activating the intrinsic Bax/mitochondrial/caspase-9 pathway. Exp. Cell Res. 276:214-222. [DOI] [PubMed] [Google Scholar]
- 32.Maki, C. G., J. M. Huibregtse, and P. M. Howley. 1996. In vivo ubiquitination and proteasome-mediated degradation of p53(1). Cancer Res. 56:2649-2654. [PubMed] [Google Scholar]
- 33.Okamura, S., H. Arakawa, T. Tanaka, H. Nakanishi, C. C. Ng, Y. Taya, M. Monden, and Y. Nakamura. 2001. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8:85-94. [DOI] [PubMed] [Google Scholar]
- 34.Prabhu, N. S., K. Somasundaram, K. Satyamoorthy, M. Herlyn, and W. S. El-Deiry. 1998. p73beta, unlike p53, suppresses growth and induces apoptosis of human papillomavirus E6-expressing cancer cells. Int. J. Oncol. 13:5-9. [DOI] [PubMed] [Google Scholar]
- 35.Rippin, T. M., V. J. Bykov, S. M. Freund, G. Selivanova, K. G. Wiman, and A. R. Fersht. 2002. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene 21:2119-2129. [DOI] [PubMed] [Google Scholar]
- 36.Sansom, O. J., and A. R. Clarke. 2000. P53 null mice: damaging the hypothesis? Mutat. Res. 452:149-162. [DOI] [PubMed] [Google Scholar]
- 37.Selivanova, G., L. Ryabchenko, E. Jansson, V. Iotsova, and K. G. Wiman. 1999. Reactivation of mutant p53 through interaction of a C-terminal peptide with the core domain. Mol. Cell. Biol. 19:3395-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu, H., L. R. Burch, A. J. Smith, D. Dornan, M. Wallace, K. L. Ball, and T. R. Hupp. 2002. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J. Biol. Chem. 277:28446-28458. [DOI] [PubMed] [Google Scholar]
- 40.Smith, M. L., and A. J. Fornace, Jr. 2002. Chemotherapeutic targeting of p53. Cancer Biol. Ther. 1:56-57. [DOI] [PubMed] [Google Scholar]
- 41.Soussi, T. 2000. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann. N. Y. Acad. Sci. 910:121-139. [DOI] [PubMed] [Google Scholar]
- 42.Takimoto, R., W. Wang, D. T. Dicker, F. Rastinejad, J. Lyssikatos, and W. S. El-Deiry. 2002. The mutant p53-conformation modifying drug, CP-31398, can induce apoptosis of human cancer cells and can stabilize wild-type p53 protein. Cancer Biol. Ther. 1:47-57. [DOI] [PubMed] [Google Scholar]
- 43.Unger, T., T. Juven-Gershon, E. Moallem, M. Berger, R. Vogt Sionov, G. Lozano, M. Oren, and Y. Haupt. 1999. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogelstein, B., and K. W. Kinzler. 2001. Achilles' heel of cancer? Nature 412:865-866. [DOI] [PubMed] [Google Scholar]
- 45.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 46.Wu, G. S., T. F. Burns, E. R. McDonald III, W. Jiang, R. Meng, I. D. Krantz, G. Kao, D. D. Gan, J. Y. Zhou, R. Muschel, S. R. Hamilton, N. B. Spinner, S. Markowitz, G. Wu, and W. S. El-Deiry. 1997. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 17:141-143. [DOI] [PubMed] [Google Scholar]
- 47.Xirodimas, D. P., C. W. Stephen, and D. P. Lane. 2001. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 48.Ye, R., A. Bodero, B. B. Zhou, K. K. Khanna, M. F. Lavin, and S. P. Lees-Miller. 2001. The plant isoflavenoid genistein activates p53 and Chk2 in an ATM-dependent manner. J. Biol. Chem. 276:4828-4833. [DOI] [PubMed] [Google Scholar]
- 49.Yu, Z. K., R. K. Geyer, and C. G. Maki. 2000. MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene 19:5892-5897. [DOI] [PubMed] [Google Scholar]
- 50.Zeng, X., L. Chen, C. A. Jost, R. Maya, D. Keller, X. Wang, W. G. Kaelin, Jr., M. Oren, J. Chen, and H. Lu. 1999. MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol. 19:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]