Abstract
Coactivator recruitment by activation function 2 (AF2) in the steroid receptor ligand binding domain takes place through binding of an LXXLL amphipathic α-helical motif at the AF2 hydrophobic surface. The androgen receptor (AR) and certain AR coregulators are distinguished by an FXXLF motif that interacts selectively with the AR AF2 site. Here we show that LXXLL and FXXLF motif interactions with steroid receptors are modulated by oppositely charged residues flanking the motifs and charge clusters bordering AF2 in the ligand binding domain. An increased number of charged residues flanking AF2 in the ligand binding domain complement the two previously characterized charge clamp residues in coactivator recruitment. The data suggest a model whereby coactivator recruitment to the receptor AF2 surface is initiated by complementary charge interactions that reflect a reversal of the acidic activation domain-coactivator interaction model.
Nuclear receptors are hormone-dependent transcription factors that bind steroids and other hormones, such as thyroid hormone, retinoic acid, and vitamin D, to regulate cell proliferation and differentiation, development, and homeostasis. Steroid receptors share a modular arrangement of structurally conserved domains, including the ligand binding domain (LBD) in the carboxyl-terminal region, a central DNA binding domain, and a variable NH2-terminal region (8). Most receptor LBDs are transcriptionally active when bound to agonist, an activity that results from activation function 2 (AF2). NH2-terminal regions of some receptors also contain the transactivation domain, activation function 1. While the structural basis for coactivator recruitment by activation function 1 is largely unknown, studies have indicated a general mechanism for AF2.
Nuclear receptors interact with p160 coactivators through the AF2 region of the LBD (16, 51). AF2 resides in a hydrophobic cleft created by helices 3, 4, 5, and 12. The hydrophobic residues that comprise AF2 were initially identified as a coactivator binding site by scanning surface mutagenesis of the thyroid hormone receptor (TRβ) (14) and confirmed in cocrystal structures (3, 11, 45, 50). Agonist binding induces a conformational shift in helix 12 (43) that completes the AF2 hydrophobic surface (11, 14), allowing p160 coactivator LXXLL motif binding (11, 23). Binding of certain ligands can alter the position of helix 12, thereby changing the specificity for LXXLL motif binding. For example, ERα binding of raloxifene, a tissue-selective synthetic estrogen antagonist (5), or 4-hydroxytamoxifen (50) causes helix 12 to bind the core static region of the AF2 hydrophobic groove, whereas in the presence of the agonist 17β-estradiol, helix 12 contributes to the recognition surface of AF2 for LXXLL motif binding. AF2 recruits the p160 coactivators steroid receptor coactivator 1 (SRC1), transcriptional intermediary factor 2 (also known as TIF2, SRC2, and GRIP1), and steroid receptor coactivator 3 (also known as SRC3, TRAM1, ACTR, AIB1, RAC3, and p/CIP) (16, 41), which are reported to have intrinsic histone acetyltransferase activity and associate with p300 and CBP, coactivators with more potent acetyltransferase activity (9, 46, 52). Accumulation of acetylase activity at enhancer-promoter regions of regulated genes results in histone modification and chromatin remodeling to facilitate transcription initiation (16, 41).
In contrast to other nuclear receptors, the AF2 region of the androgen receptor (AR) is a weak-interaction site for LXXLL motifs of p160 coactivators and preferentially binds an FXXLF motif that is present in the AR NH2-terminal region and in certain coregulators shown to associate with AR in the presence of androgen (19-20, 64). Like the LXXLL motif, interactions of the AR NH2-terminal FXXLF and the FXXLF motifs of AR-associated proteins involve binding of a conserved amphipathic α-helix with the complementary hydrophobic surface of AF2. Other protein-protein interaction motifs similar to the AR FXXLF motif have been described, such as FXXLW, required for p53-MDM2 complex formation (33), and FXXAL, required for the interaction of VP16 with TAFII31 (55). These signature motifs of gene-regulatory proteins provide specificity in protein-protein interactions. Indeed, the AR FXXLF motif binding is specific for the AF2 region of AR and mediates a strong androgen-dependent, NH2-terminal interaction with the carboxyl-terminal region (N/C interaction) (19) which is required for AR transactivation of androgen-regulated genes, such as prostate-specific antigen (21). While the FXXLF motif is predicted to form an α-helical structure that resembles the LXXLL motifs of p160 coactivators, it was not known what features of these α-helical regions initiate the interaction and establish the polarity of binding at the AF2 surface. In this report we show that binding of FXXLF and LXXLL to the AF2 of steroid receptors depends on clusters of charged residues flanking the AF2 interface and oppositely charged residues flanking the α-helical motifs. In addition, we show that interaction of the AR FXXLF motif with AF2 is attenuated by a conserved positively charged arginine residue flanking the FXXLF binding motif. The data suggest that multiple charge interactions initiate and modulate contact with the AF2 surface and serve to position the α-helical motifs for subsequent hydrophobic interactions at the core of AF2. We propose a charge polarity mechanism for AF2 recruitment of the LXXLL motifs of p160 coactivators and the FXXLF motifs in AR coregulators that represents a reversal of the acidic activation domain-coactivator recruitment model.
MATERIALS AND METHODS
Plasmids.
GAL4-DNA binding domain peptide fusions were constructed as described previously (20) by annealing complementary oligonucleotides and cloning into pGALO expressing the Saccharomyces cerevisiae GAL4 DNA binding domain amino acid residues 1 to 147. The GAL-AR624-919 and VP-AR1-660 mutants were created using a double PCR mutagenesis strategy where the VP fusion proteins contain VP16 activation domain residues 411 to 456. pCMX-hRXRα was provided by Ronald M. Evans, the Salk Institute for Biological Studies. pCMX-VP-F-hTRβ was provided by David D. Moore, Baylor College of Medicine. VP16-PR-A, VP16-ERα-LBD, pVP16-ERβ, pVP16-VDR, and the luciferase reporter 5XGAL4-Luc3 were provided by Donald P. McDonnell, Duke University.
Cell culture and two-hybrid assays.
Human hepatocellular carcinoma HepG2 cells were obtained from the American Type Culture Collection and maintained, transfected, and analyzed for luciferase activity as described previously (20). Ligand dissociation rate studies were performed with COS cells as described previously (19). The two-hybrid assay for interactions between GAL4 fusion peptides and full-length AR or VP16 fusion proteins was previously described (19, 34, 35).
AR LBD purification and isothermal titration calorimetry.
The AR LBD (amino acid residues 663 to 919) was expressed as a glutathione S-transferase (GST) fusion protein from pGEX-KG in Escherichia coli BL21(DE3) in the presence of 10 μM dihydrotestosterone (DHT). GST-AR-LBD was purified by affinity chromatography on glutathione-Sepharose and released from GST by digestion with thrombin as described previously (39) except that phosphate-buffered saline was used as the sonication and thrombin digestion buffer.
Isothermal titration calorimetry measurements were performed using an OMEGA titration microcalorimeter from Microcal (Northampton, Mass.) at the University of North Carolina Macromolecular Interactions Facility. Peptides were synthesized by the Peptide Synthesis Facility of the University of North Carolina and purified by high-performance liquid chromatography. The AR LBD and synthetic peptides were dissolved in phosphate-buffered saline, and measurements were performed at 26°C. Successive 5-μl aliquots of peptide (0.2 to 1 mM) were added to a solution containing a fixed amount of AR LBD (10 to 20 μM). Each titration experiment consisted of 30 to 50 injections of peptide into the calorimetric sample cell containing 1.34 ml of purified AR LBD protein solution. The thermal effect in calories of each addition was recorded, and the data were fit by least-squares regression using ORIGIN software (Microcal). The experimental approach provides a measure of binding in solution in terms of free energy (ΔG), enthalpy (ΔH), and entropy (ΔS) (4, 6, 63), where multiple sequential binding reactions are associated with a change in free energy (ΔG), the magnitude of which reflects binding affinity (6). Given the number of moles of AR LBD in solution and the moles of peptide added during the titration, stoichiometry and Kd can be obtained. The shape of the binding curve, i.e., the fraction of bound versus free peptide, is a measure of cooperativity. The standard free energy change for the AR LBD-peptide complex is related to the equilibrium association constant (Ka) (ΔG = −RT ln Ka). The free energy has enthalpic (ΔH) and entropic (ΔS) components (ΔG = ΔH − TΔS) of which the former is obtained with good accuracy from the magnitude of the thermal effect. The entropy was calculated using the last equation.
LBD structure models.
The space-filled models of the AR and ERα LBDs were determined based on previously published structural coordinates (48, 50) and were modeled using the RASMOL program (49).
RESULTS
Effect of AR FXXLF motif-flanking charged residues in binding the AR LBD.
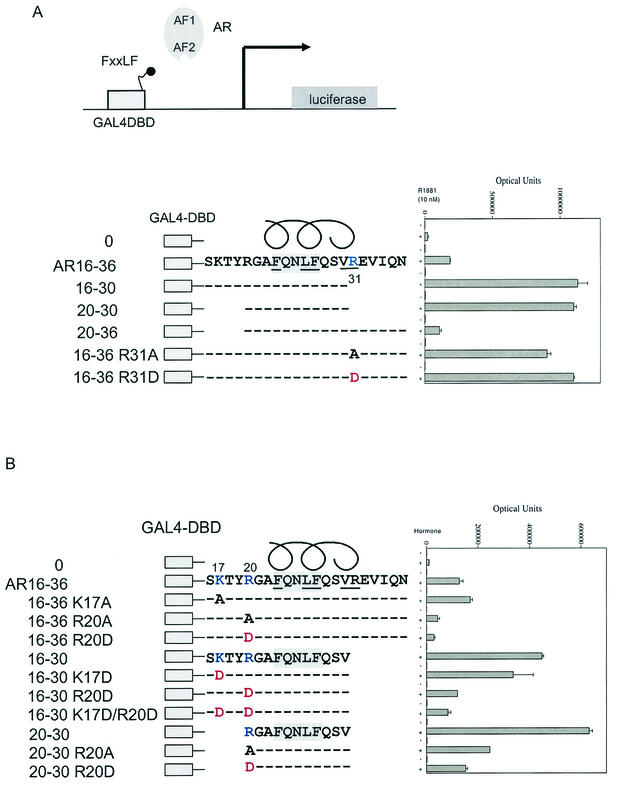
The FXXLF motif 23FQNLF27 in the AR NH2 terminus binds AF2 in the presence of androgen, resulting in the AR NH2-terminal and carboxyl-terminal N/C interaction (19). Based on the cocrystal structure of the ERα LBD-TIF2 LXXLL peptide (50) and the predicted α-helical structure of the FXXLF motif, residues F23, L26, and F27 on one face of the α-helix are expected to contact the nonpolar hydrophobic residues of AF2. The importance of these residues in AR-mediated transactivation was demonstrated previously (19). We tested several charged residues flanking the FXXLF motif that are predicted to contact the LBD surface near AF2 for their role in modulating the N/C interaction. Deletion of residues carboxyl to the AR NH2-terminal FXXLF core sequence in the GAL-AR FXXLF fusion peptides 16 to 30 and 20 to 30 increased binding to AR in a mammalian two-hybrid assay using a GAL-luciferase reporter (Fig. 1A). In contrast, no increase in activity was observed with deletion of sequence amino-terminal to R20 in the fusion peptide GAL-AR20-36. Changing R31 to alanine carboxyl-terminal to the core sequence increased FXXLF motif binding, which was increased further by replacing R31 with negatively charged aspartic acid. The attenuating effect of R31 on FXXLF motif binding to AF2 was supported by the ligand dissociation rate, a property that reflects FXXLF motif binding to AF2 in the N/C interaction in full-length AR (19). The dissociation half-time of [3H]R1881 from AR-R31D determined at 37°C (120 ± 3 min) was slower than that for wild-type AR (98 ± 3 min). The results indicate that the attenuating effect of R31 on binding of FXXLF to AF2 detected in the peptide interaction assay is also observed as part of the N/C interaction of full-length AR.
FIG. 1.
Effect of charged residues flanking the AR FXXLF in binding AR. (A) Mammalian two-hybrid assays were performed with HepG2 cells by coexpressing GAL-AR peptides containing the GAL4 DNA binding domain (GAL4-DBD) and the indicated AR NH2-terminal FXXLF motif amino acid wild-type or mutant sequence. GAL4-AR-FXXLF peptides (0.05 μg) were cotransfected using Effectine (Qiagen) with pCMVhAR (0.05 μg) coding for full-length human AR and the 5XGAL4Luc3 reporter vector (0.1 μg) in 12-well plates containing 0.2 × 106 HepG2 cells. Transfected cells were incubated for 24 h in the absence and presence of 10 nM R1881 (methyltrienolone), a synthetic androgen. (B) Two-hybrid peptide interaction assays were performed with HepG2 cells as described previously using GAL4-AR peptides with the FXXLF motif region with wild-type or mutant sequence. The GAL-FXXLF peptide vectors with the indicated amino acid residue number and mutations were cotransfected with pCMVhAR to test the role of positively charged residues NH2-terminal to the AR NH2-terminal FXXLF motif.
A similar mutagenesis strategy was used to establish the role of positively charged residues amino-terminal to the AR NH2-terminal FXXLF motif. K17 is predicted to lie outside the α-helical region, and a deletion including this residue (Fig. 1A, AR20-30) had little effect on the FXXLF interaction with AF2 in full-length AR. Similarly, changing K17 to alanine or aspartic acid had little effect on binding of AR16-36 or AR16-30 to AF2 (Fig. 1B). In contrast, changing R20 to alanine or aspartic acid in these peptides and in AR20-30 reduced binding by more than 50%. The data suggest that R20 and R31 contact the AR LBD surface to modulate FXXLF motif binding at AF2. R20 amino-terminal to the FXXLF core motif enhances binding of FXXLF to AF2, whereas R31 carboxyl to the core sequence weakens FXXLF motif binding to AF2. The flanking residues R20 and R31 are therefore important in FXXLF motif interaction with AF2 and have a role in mediating the N/C interaction.
Charged clusters flanking AR AF2 mediate interactions with the FXXLF and LXXLL motifs.
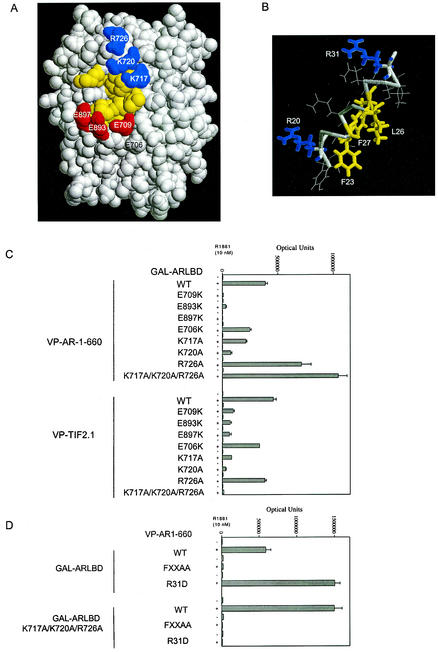
The AR LBD crystal structure (39, 48) and the ERα-LXXLL motif cocrystal structure (50) suggest that F23 of the FXXLF motif contacts the lower part of the AF2 hydrophobic surface (Fig. 2A), and L26 and F27 contact the upper part of the AF2 hydrophobic cleft. The predicted ideal α-helical structure of the FXXLF motif is shown in Fig. 2B. Data presented above support these models and place charged residues R20 and R31 flanking the FXXLF motif (Fig. 2B) on the surface facing the hydrophobic residues and thus are predicted to contact the flanking region of AF2 (Fig. 2A).
FIG.2.
Effect of charge clusters flanking AF2 in the AR LBD on AR-FXXLF and TIF2-LXXLL motif binding. (A) A space-filled model of the DHT-bound AR LBD is based on structural coordinates of rat AR646-901 (48), which has amino acid sequence identical to that of human AR664-919 (37). Positively charged residues K717, K720, and R726 flanking AF2 are shown in blue, negatively charged residues E709, E893, and E897 are shown in red, and AF2 hydrophobic residues are shown in yellow. DHT bound in the core structure would not be evident on the surface. (B) Ideal α-helical structure of the AR NH2-terminal FXXLF motif. The model helix was built using the Biopolymer module of the InsightII molecular modeling system from Accelrys Inc. (www.accelrys.com). (C) Two-hybrid assays were performed with HepG2 cells using GAL-AR-LBD (0.15 μg) coding for the GAL4 DNA binding domain fused to AR LBD containing residues 624 to 919 with wild-type (WT) or the indicated mutant sequences. GAL-AR-LBD was cotransfected with the 5XGAL4Luc3 reporter vector (0.1 μg) and VP-AR1-660 (0.15 μg) containing the VP16 transactivation domain and AR NH2-terminal residues 1 to 660 or VP-TIF2.1 containing the three-LXXLL motif region in TIF2. Assays were performed with HepG2 cells in the absence and presence of 10 nM R1881. (D) Two-hybrid assays were performed using VP-AR1-660 containing wild-type sequence or the 23FQNLF27-to-FQNAA (FXXAA) or R31D mutation. GAL-AR-LBD expressed residues 624 to 919 with wild-type sequence or with a triple mutation, K717A/K720A/R726A, as indicated. HepG2 cells were cotransfected with wild-type and mutant VPAR1-660 together with wild-type or mutant GAL-AR-LBD and the 5XGAL4Luc3 reporter vector. Cells were incubated for 24 h in the absence and presence of 10 nM 1881, and numbers of luciferase optical units were determined.
We determined the functional importance of residues flanking AF2 on the interaction with the FXXLF motif. The AF2 flanking region contains two oppositely charged amino acid clusters. Two of the conserved residues, AR-E897 and AR-K720, at opposite ends of AF2 in the AR LBD, are shared by members of the steroid receptor family and correspond to residues in estrogen receptor α (ERα) and peroxisome proliferator-activated receptor γ (PPARγ) that were proposed to function as a charge clamp, forming hydrogen bonds with the peptide backbone of the LXXLL peptide (45). E897 was critical for AR transcriptional activation; however, K720 was relatively unimportant (22). From the crystal structure of the AR LBD (39, 48), E897 is positioned near E893 and E709, forming a negative charge cluster on one side of AF2 (Fig. 2A). On the opposite side of AF2, residues K720, K717, and R726 form a positive-charge cluster (Fig. 2A). Using a mammalian two-hybrid peptide interaction assay, we tested the functional importance of these residues in the interaction with the AR NH2-terminal FXXLF motif and the LXXLL motif of the p160 coactivators.
The AR FXXLF motif sequence in VP-AR1-660, a fusion protein that contains the VP16 activation domain and AR NH2-terminal region, binds AF2 in GAL-AR-LBD in the presence of androgen (Fig. 2C). Mutagenesis of several residues flanking AF2 in GAL-AR-LBD showed that binding of the AR FXXLF motif in VP-AR1-660 and the LXXLL motif region in VP-TIF2.1 (TIF2 residues 624 to 1287) depended on each of the negatively charged residues E709, E893, and E897 flanking AF2. Mutation at E706 was less disruptive of binding. Mutation of positively charged residues K717 and K720 indicated a greater role in TIF2 LXXLL motif binding than in binding of the AR FXXLF motif. The R726A mutation increased the binding of VP-AR1-660 but had little effect on TIF2 LXXLL motif binding. A triple mutant of the positive-charge patch changing K717, K720, and R726 to alanine greatly increased FXXLF binding but eliminated LXXLL binding.
The attenuating effect of R31 in AR FXXLF motif binding shown above was also evident in the androgen-dependent N/C interaction assay using the VP-AR1-660 and GAL-AR-LBD (AR residues 624 to 919) fusion proteins. The R31D mutation in VP-AR1-660 or a triple mutation to neutralize the lysine and arginine residues in GAL-AR-LBD increased the AR N/C interaction (Fig. 2D). This contrasted with the loss of binding to an FXXAA mutant (Fig. 2D) that was shown previously to eliminate FXXLF binding to AF2 (19). Surprisingly, binding of the AR-R31D mutant was undetectable with the GAL-AR-LBD triple mutant where the positive charges were neutralized. This requirement for the positive-charge patch in AR-R31D binding suggests properties similar to VP-TIF2.1 LXXLL motif binding (see Fig. 2C).
The results indicate that negatively charged residues E709, E893, and E897 flanking AF2 promote FXXLF and LXXLL motif binding, whereas the positively charged cluster comprised of residues K717, K720, and R726 is required for TIF2 LXXLL motif binding but attenuates AR NH2-terminal FXXLF motif binding. Furthermore, in addition to the AR charge clamp residues E897 and K720 proposed previously for corresponding residues in ERα, additional charged residues are required for AR recruitment of 160 coactivators and for the FXXLF-mediated N/C interaction.
Role of polar charge distribution flanking the FXXLF and LXXLL motifs in AR coregulators.
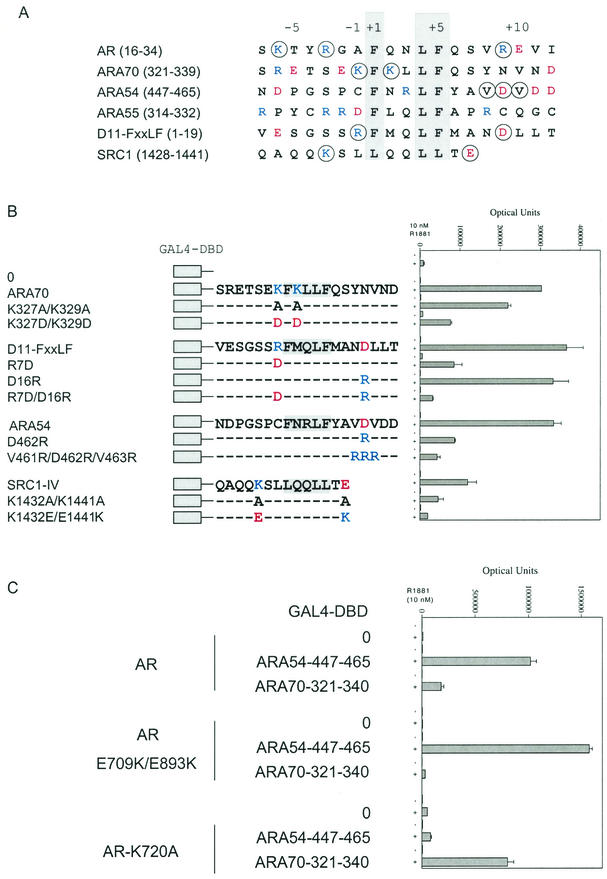
Sequence alignment of the FXXLF and LXXLL motif regions that mediate interaction of several coregulatory proteins with AR indicates that positively charged residues K and R predominate amino-terminal to the core sequence, and negatively charged residues E and D predominate carboxyl-terminal to the core sequence (Fig. 3A). The overall negative-to-positive charge ratios of the peptides shown in Fig. 3A are 0.55 left and 3.5 right of the core sequence. To assess the charge distribution requirements for FXXLF and LXXLL motif binding to AR, mutagenesis was performed at residues (Fig. 3A) flanking the FXXLF motifs in the AR coregulators ARA70, ARA54, and ARA55 (20, 25) as well as in a D11 FXXLF peptide (7, 20) and SRC1 carboxyl-terminal LXXLL-IV (44).
FIG.3.
Requirement for charged residues flanking the FXXLF and LXXLL motifs in AR coregulator interaction with AR. (A) FXXLF and LXXLL motifs and flanking sequence of regions that bind AR AF2. Shown are human AR amino acid residues 16 to 34 that contain the FXXLF motif sequence 23FQNLF27, which mediates the N/C interaction (19), and the FXXLF motif regions from AR coregulators ARA70 (residues 321 to 339), ARA54 (residues 447 to 465), and ARA55 (residues 314 to 332) (20). Also shown are the D11-FXXLF peptide (20) and SRC1 carboxyl-terminal LXXLL-IV motif (residues 1428 to 1441) (47). Consensus residues of the binding motifs are shaded, and the relative positions of amino acids are numbered from the start of the core sequence. Residues that were mutated are circled. Basic residues K and R are shown in blue, and acidic residues D and E are shown in red. (B) Two-hybrid peptide interactions with AR. The GAL-ARA70 peptide contained ARA70 residues 321 to 340 with wild-type or mutant sequence as indicated. GAL-D11-FXXLF contained sequence derived from the D11-LXXLL peptide (44) that was changed to an FXXLF motif (20) with wild-type or mutant sequence. Numbering of the D11 peptide was left to right from the beginning of the peptide. The GAL-ARA54 peptide contained residues 447 to 465 with wild-type or mutant sequence. GAL-SRC1-IV contained residues 1428 to 1441 including the fourth and carboxyl-terminal LXXLL motif with WT or mutant sequence. GAL peptide vectors were cotransfected with pCMVhAR coding for full-length human AR and the 5XGAL4Luc3 reporter vector in HepG2 cells as described. Cells were incubated in the absence and presence of 10 nM R1881 for 24 h, and numbers of luciferase optical units were determined. (C) Two-hybrid interaction assays were performed as described above using full-length wild-type AR and AR mutants E709K/E898K or K720A, cotransfected either with empty parent vector (GAL4-DBD-0) or with the GAL4-DBD-peptide fusion proteins. These were cotransfected with GAL-ARA54-447-465 or GAL-ARA70-321-340. Luciferase activity was determined in the absence and presence of 10 nM R1881 as shown.
In two-hybrid interaction assays using full-length AR, mutations to alanine at two positively charged residues, K327 and K329, reduced binding of the ARA70 FXXLF motif to AR compared to the wild-type control (Fig. 3B). A further reduction in luciferase activity was seen when these residues were changed to aspartic acid. A requirement for positively charged residues amino-terminal to the core motif was supported by the interaction of the D11-FXXLF R7D mutant peptide, by SRC1-IV mutations to alanine at K1432 and E1441, and by the activity of a second double mutant (K1432E/E1441K) in which the positions of the positively and negatively charged residues were exchanged. The contribution of negative charges carboxyl-terminal to the core sequence was less evident with the D16R mutation in D11-FXXLF. Binding of the D11-FXXLF peptide depended instead on R7, amino-terminal to the core sequence. The double mutant R7D and D16R in D11-FXXLF almost eliminated binding to AR. Binding of the most strongly AR-interacting peptide, ARA54 (20), was greatly reduced by changing D462 to arginine or changing V461, D462, and V463 to arginine on the carboxyl-terminal side of the core sequence. The results, in agreement with results of the AR FXXLF motif, indicate that flanking positively charged residues amino-terminal to the FXXLF core sequence or negatively charged residues carboxyl-terminal to the FXXLF core sequence promote motif binding to AF2.
The relative importance of complementary charge groups flanking the FXXLF peptides and AF2 was well supported by AR binding of the AR coregulator ARA54 and ARA70 FXXLF motifs. ARA54 lacks positively charged residues amino-terminal of the core sequence (Fig. 3A) and depended instead on negatively charged D462 carboxyl-terminal to the core sequence for FXXLF motif binding (Fig. 3B). As a result, the AF2 positively charged K720 residue was required for ARA54 FXXLF motif binding, but the negative cluster E709 and E893 was not, since AR-K720A but not AR-E709K/E893K eliminated ARA54 FXXLF motif binding (Fig. 3C). Conversely, ARA70 lacks flanking negatively charged residues carboxyl-terminal to the FXXLF motif (Fig. 3A). Binding of ARA70 to AF2 depended on residues K327 and K329 amino-terminal and within the FXXLF motif (Fig. 3B) as previously reported (64) and on AR AF2 residues E709 and E893, but not on K720 (Fig. 3C). The data demonstrate that negatively charged residues flanking AF2 mediate interactions with positively charged residues amino-terminal of the FXXLF core motif, and positively charged residues flanking AF2 mediate interactions with the negatively charged residues carboxyl to the FXXLF core motif.
Charge distribution requirements for binding of the LXXLL motif to steroid receptors.
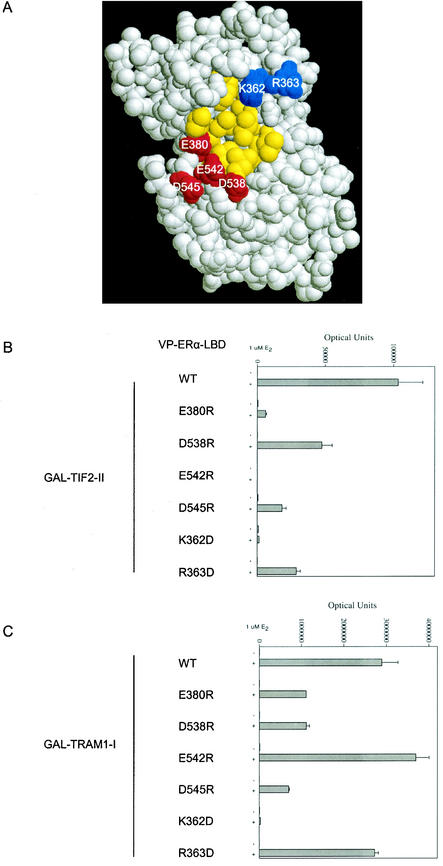
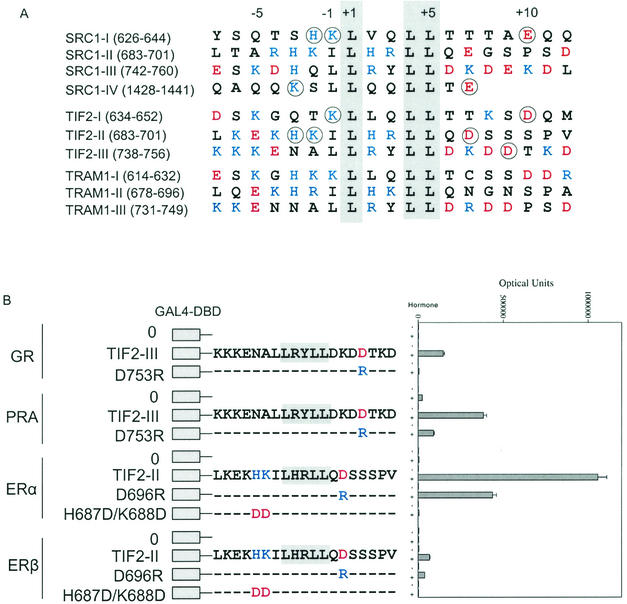
The LBD of the AR, glucocorticoid (GR), progesterone (PR), and mineralocorticoid receptors are closely related structurally with conservation of the positively and negatively charged clusters flanking AF2 (1, 58, 59, 61). ERα and ERβ are less related to this group but have a charge polarity flanking AF2 that resembles that for AR (Fig. 2A; also see Fig. 5A). ERβ differs from ERα in lacking D545, and R363 is replaced by lysine (32). We tested whether the charge requirements identified for binding of the FXXLF motif to AR AF2 apply to binding of the p160 coactivator LXXLL motif to other steroid receptors.
FIG.5.
Role of charged residues flanking AF2 in ERα LBD in TIF2-LXXLL-II binding. In panel A, the space-filled model of the ERα LBD was based on structural coordinates of ERα amino acids 297 to 554 in the presence of diethylstilbestrol (50). Positively charged residues K362 and R363 are shown in blue, negatively charged residues E380, D538, E542, and D545 are shown in red, and AF2 hydrophobic residues are shown in yellow. Two-hybrid peptide interaction assays were performed by cotransfecting the 5XGAL4Luc3 reporter vector, VP-ERα-LBD with wild-type (WT) or mutant sequence, with either GAL-TIF2-LXXLL-II (B) or GAL-TRAM1-LXXLL-I (C). Cells were incubated in the absence and presence of 1 μM 17β-estradiol (E2), and luciferase activity was determined as described in Materials and Methods.
The negative-to-positive charge ratios flanking the p160 coactivator LXXLL motifs shown in Fig. 4A are 0.31 left of the core motif and 2.9 right of the core sequence. These ratios are similar to those flanking the FXXLF motifs shown in Fig. 3A (0.55 and 3.5, respectively). The charge requirements for binding of the p160 coactivator LXXLL motif to other full-length steroid receptors were tested by mutagenesis of residues flanking the LXXLL motifs (circled in Fig. 4A). GR and PR preferentially bind the third TIF2 LXXLL motif (TIF2-LXXLL-III) relative to the other LXXLL motifs, whereas ERα and ERβ preferentially bind TIF2-LXXLL-II (data not shown) and were tested accordingly. Converting D753 to arginine in the TIF2-LXXLL-III peptide, a residue that corresponds to R31 flanking the AR FXXLF motif, abolished LXXLL motif binding to full-length GR and greatly reduced binding to PR (Fig. 4B). Converting D696 to arginine in TIF2-LXXLL-II reduced binding to ERα and ERβ. A double mutant in TIF2-LXXLL-II, converting H687 and K688 to aspartic acid, abolished binding to ERα and ERβ (Fig. 4B). The data provide strong evidence for a polar charge distribution for optimal LXXLL motif binding to AF2 in other steroid receptors. The data indicate a requirement for positively charged residues amino-terminal to, and negatively charged residues carboxyl-terminal to, the LXXLL core sequence and support the functional importance of complementary charge polarity in FXXLF and LXXLL motif-mediated coactivator recruitment.
FIG. 4.
Flanking charged residue requirements of p160 coactivator LXXLL motif binding to steroid receptors. (A) Amino acid sequence of the LXXLL motif regions in the human p160 coactivators SRC1 (47), TIF2 (29, 56), and TRAM1 (53). Basic residues K, R, and H are shown in blue, and acidic residues D and E are shown in red. The conserved LXXLL motif is shaded, and the residues that were mutated are circled. The LXXLL motifs are numbered relative to the start of the core motif. (B) Two-hybrid interaction assay of TIF2-LXXLL peptides II and III with full-length steroid receptors. GAL fusion peptide vectors coding for TIF2-LXXLL-II and TIF2-LXXLL-III were cotransfected with the 5XGAL4Luc3 reporter vector and expression vectors for GR (pCMVhGR), PR-A (VP-PR-A), ERα (VP-ERα-LBD coding for residues 312 to 595), and ERβ (VP-ERβ full-length residues 1 to 530). Transfected HepG2 cells were incubated in the absence or presence of 10 nM dexamethasone for GR, 10 nM R5020 for PR-A, and 1 μM 17β-estradiol for ERα and ERβ.
The ERα AF2 region is flanked by positively and negatively charged clusters similar to that observed for AR (Fig. 2A and 5A). ERα AF2-flanking residues E542 and K362 were shown previously to be required for SRC1 LXXLL motif binding (26, 50). Two-hybrid assays were performed using a GAL fusion peptide containing the second LXXLL motif of TIF2 (GAL-TIF2-LXXLL-II) or the first LXXLL motif of TRAM1 (GAL-TRAM-LXXLL-I) and VP-ERα-LBD with wild-type and mutant sequence. The TIF2 second LXXLL motif and the TRAM1 (ACTR) first LXXLL motif are the principal binding motifs that mediate recruitment by ERα (10). We show here for TIF2 LXXLL-II that in addition to the conserved charge clamp residues, ERα AF2-flanking residues E380, D538, D545, and R363 are important for LXXLL motif binding (Fig. 5B). For TRAM1 LXXLL-I, the conserved charge clamp residue E542 is not important, while E380, D538, and D545 are required for binding (Fig. 5C). The results support the steroid receptor family requirement for charged residues flanking AF2 in LXXLL motif binding. In addition, the previously reported charge clamp residue E542 in ERα is not required for all coactivator LXXLL motif binding.
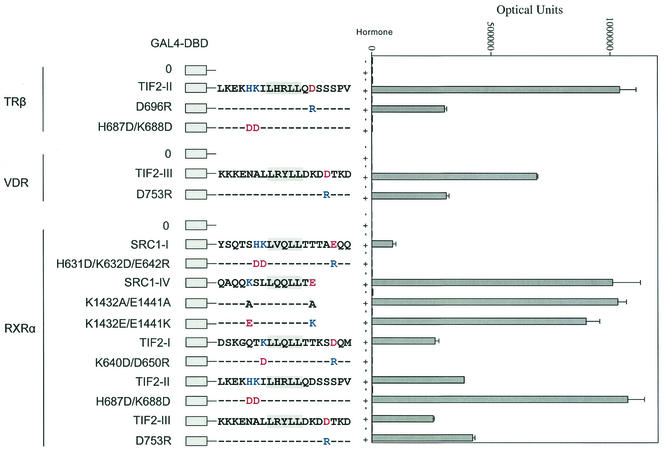
Charge distribution requirements for nonsteroid nuclear receptors.
We tested whether a similar polar charge distribution was required for binding of the LXXLL motif to nonsteroid nuclear receptors, where the AF2 sites have a less distinct pattern of charged residues relative to the steroid receptors (2, 13). We tested binding of the most strongly interacting LXXLL motifs of TIF2 and SRC1 as GAL-peptide fusions in binding assays with full-length TRβ, the vitamin D receptor (VDR), and RXRα (Fig. 6). Replacing aspartic acid with arginine carboxyl-terminal to TIF2-LXXLL-II (D696R) and TIF2-LXXLL-III (D753R) reduced binding to TRβ and VDR, respectively. In contrast, this mutation (D753R) slightly increased binding of TIF2-LXXLL-III to RXRα. Changing residues H687 and K688 to aspartic acid amino-terminal to TIF2-LXXLL-II abolished binding to TRβ but increased binding to RXRα. Neutralizing or reversing the charges flanking SRC1-LXXLL-IV (K1432 and E1441 to alanine or exchanging the charge positions) had little influence on RXRα binding (Fig. 6). A triple mutation, H631D/K632D/E642R, in SRC1 LXXLL-I or a double mutation, K640D/D650R, in TIF2 LXXLL-I abolished the interaction with RXRα. The results indicate that the requirements for complementary charge polarity that mediates AF2 interaction with the LXXLL motif apply to TRβ and VDR but apply less well to the nonsteroid nuclear receptor RXRα.
FIG. 6.
Role of charged residues flanking the LXXLL motifs in binding nonsteroid nuclear receptors. Two-hybrid interaction assays were performed using TIF2 and SRC1 LXXLL peptides with TRβ, VDR, and RXRα. GAL-TIF2-LXXLL-I, -II, and -III or GAL-SRC1-LXXLL-I and -IV expression vectors coding for wild-type or mutant sequence were cotransfected with pCMX-VP-F-hTRβ, pVP16-VDR, or VP16-hRXRα. Cells were incubated in the absence and presence of 1 μM triiodothyronine for TRβ, 100 nM 1,25-dihydroxyvitamin D for VDR, and 1 μM 9-cis-retinoic acid for RXRα for 24 h prior to determining luciferase activity. Basic residues K, R, and H are shown in blue, and acidic residues D and E are shown in red.
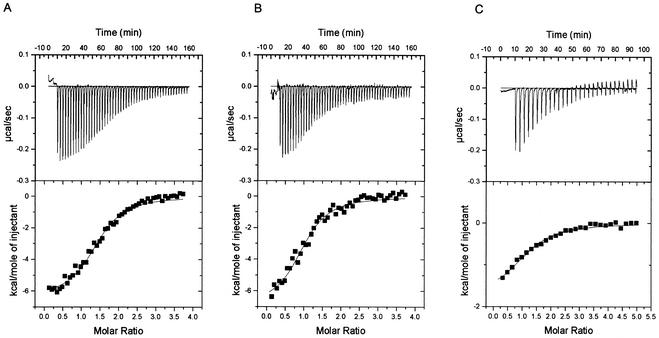
Binding affinity of the FXXLF and LXXLL motifs for the AR LBD.
We used isothermal titration calorimetry to measure the equilibrium binding affinities and thermodynamic properties of binding of the AR LBD to the FXXLF peptides derived from ARA54 and the AR NH2-terminal domain and to LXXLL-III from TIF2/GRIP1 (Fig. 7). Measurements were made by sequential injections of peptide into solutions containing the AR LBD. As the peptide-AR LBD titration progressed and the AR LBD became saturated, less of the added peptide combined with the AR LBD and the thermal effect was smaller. From the change in thermal effect during the course of titration, the equilibrium dissociation constant, Kd, was computed by standard methods (6). The data indicate the presence of a single binding site with no clear sign of cooperativity, with a Kd of 0.9 to 1.2 μM for the two FXXLF peptides and 6.0 μM for TIF2-LXXLL-III. Binding of the AR FXXLF NH2-terminal core peptide to the LBD had a greater enthalpic term (ΔH = −5.7 kcal/mol) (Table 1) than was observed with TIF2 LXXLL-III (ΔH = −1.8 kcal/mol). The entropy (ΔS) was −1.4 and 7.9 cal/mol for FXXLF peptide binding compared to 18.1 cal/mol for the TIF2 LXXLL-III peptide.
FIG. 7.
Isothermal titration calorimetry measurements of FXXLF and LXXLL peptide binding to the AR LBD. Binding parameters were determined as described in Materials and Methods by titrating ARA54 FXXLF peptide 447-NDPGSPCFNRLFYAVDVDD-465 (A), the AR NH2-terminal FXXLF peptide 20-RGAFQNLFQSV-30 (B), and the TIF2 LXXLL-III peptide 738-KKKENALLRYLLDKDDTK-755 (C) into solutions containing human AR LBD fragment (amino acid residues 663 to 919). Multiple sequential injections (50, 50, and 30, respectively) were performed for each of the peptide-AR LBD interactions. Thermograms are shown in the upper panels, and binding isotherms are shown below.
TABLE 1.
Thermodynamic properties of AR LBD binding of FXXLF and LXXLL peptides by isothermal titration calorimetry at 26°Ca
| Peptide | Kd (μM) | Ka (105 M−1) | ΔH (kcal/mol) | ΔS (cal/mol) | ΔG (kcal/mol) | N |
|---|---|---|---|---|---|---|
| ARA54-FXXLF | 0.9 ± 0.2 | 11.7 ± 1.5 | −8.7 ± 2.3 | −1.4 ± 7.3 | −8.3 ± 0.1 | 1.0 ± 0.3 |
| AR-NH2-FXXLF | 1.2 ± 0.2 | 8.4 ± 0.2 | −5.7 ± 1.2 | 7.9 ± 4.5 | −8.1 ± 0.1 | 1.1 ± 0.1 |
| TIF2-LXXLL-III | 6.0 ± 1.1 | 1.7 ± 0.3 | −1.8 ± 0 | 18.1 ± 0.5 | −7.2 ± 0.1 | 1.4 ± 0.2 |
Binding parameters were determined as described in Materials and Methods and the legend to Fig. 7 by titrating ARA54 FXXLF peptide residues 447 to 465, AR NH2-terminal FXXLF peptide residues 20 to 30, and TIF2 LXXLL-III peptide residues 738 to 755 with the AR LBD using isothermal titration calorimetry. N, number of peptide binding sites per LBD.
A possible interpretation of these changes is as follows. Consider the binding of the ARA54-FXXLF and TIF2-LXXLL-III peptides to the AR LBD (Table 1) that differed the most in ΔG. The ΔG value was about 1 kcal/mol, which was small but significant. ARA54-FXXLF (Kd = 0.9 μM) was more tightly bound than TIF2-LXXLL-III (Kd = 6 μM) to the AR LBD, in agreement with the two-hybrid results. A negative ΔH means that heat is produced as a result of the binding reaction. The difference in ΔH of these two peptides was much larger than the difference in ΔG and suggests fewer contacts, perhaps between polar groups for the TIF2-LXXLL-III peptide-AR LBD complex. The positive change in entropy, which produces a largely compensating term −TΔS in the free energy, indicates that the weaker TIF2-LXXLL-III-AR LBD complex is less ordered, which is not an unreasonable conclusion if fewer contacts are present. A more tightly bound FXXLF-AR LBD complex would be more rigid and therefore would have lower entropy, whereas a less tightly bound peptide, such as TIF2-LXXLL-III, would have fewer favorable contacts, be more disordered, and have greater entropy.
DISCUSSION
Charge clamp residues.
The opposite ends of the AF2 hydrophobic groove in the LBD of nuclear receptors contain two highly conserved charged residues. These so-called charge clamp residues are thought to assist in orienting LXXLL motif binding to AF2. K362 in ERα was initially shown to be highly conserved among nuclear receptors and required for transcriptional activity and coactivator recruitment (26). It was suggested that K362 is the only essential positively charged residue in the predominantly hydrophobic coactivator binding surface (26, 38). ERα also contains E542 separated from K362 by 15 Å, which is sufficient to accommodate the 11-Å axial length of the LXXLL binding region (50). Interaction of the LXXLL motif with the AF2 core is stabilized by these highly conserved charged residues.
Here we present evidence that additional, well-ordered charged residues are present flanking AF2 in the AR LBD that facilitate coactivator recruitment and binding specificity. The increased number of charged residues adjacent to AF2 in AR is conserved in GR and PR relative to other nuclear receptors (3, 61). The importance of these residues was recently demonstrated by the cocrystal structure of an F602S glucocorticoid LBD mutant bound to dexamethasone and the LXXLL-III peptide of TIF2 (3). The mutation was introduced to increase solubility of the GR LBD necessary for its purification while retaining wild-type transcriptional activity. In addition to the conserved GR residues E755 in helix 12 and K579 in helix 3, which correspond to the first reported charge clamp, the charged residues R585 and D590 in human GR are conserved in human AR (R726 and D731) and human PR (R740 and D745) and contribute to the specificity of LXXLL motif binding (3). In the cocrystal structure of the GR LBD and the LXXLL-III peptide, these residues form hydrogen bonds with R + 2 within the core motif and D + 6 in the p160 coactivator, where L + 1 is the first residue of the care motif. Based on GR crystal results, D731 in the AR LBD may form a second charge clamp with residue R + 2 of TIF2-LXXLL-III, K + 2 of ARA70, or R + 3 of ARA54. However, D731 will not form this interaction with the AR NH2-terminal FXXLF motif because of the absence of a positively charged residue at the +2 or +3 position.
We show that the initially reported charge clamp residue flanking AF2 in the ERα LBD that forms hydrogen bonds with the peptide backbone does not fully account for LXXLL motif binding. E542 in ERα is required for binding LXXLL-II of TIF2 (Fig. 5A) and TRAM1 (data not shown), but not for binding LXXLL-I of TRAM1. TIF2 LXXLL-II and TRAM1 LXXLL-I and II bind ERα to a similar extent based on two-hybrid interaction results (data not shown). K362 was also not required for RIP140 LXXLL motif binding (26).
Similarities exist in the first reported charge clamp residues adjacent to AF2 in other receptors. K362 and E542 in ERα correspond to K288 and E457 in TR-β1 (11), K292 and E462 in PPARα (62), and K301 and E471 in PPARγ (45), each facilitating tight hydrophobic packing of LXXLL helix residues in the LBD pocket. Crystallographic evidence indicates that the glutamic acid in helix 12 forms hydrogen bonds to backbone amides of leucine 1 of the LXXLL motif and the amino-terminal adjacent residue, whereas lysine in helix 3 forms hydrogen bonds to the backbone carbonyls of leucine 4 and 5 of the motif (45). E471 in one PPARγ monomer forms hydrogen bonds with K632 and L633 in SRC1 LXXLL-I (632KLVGLLTT639) and with K688, I689, and L690 in SRC1 LXXLL-II (688KILHRLL693) in the second monomer of the PPARγ dimer (45). Two LXXLL motifs in the same SRC1 molecule interact with both LBDs to stabilize the PPAR-RXR and RXR-RAR heterodimers and are dependent on the spacer distance between the motifs (15, 60). Shared binding of motifs by a receptor dimer has not been demonstrated with steroid receptors.
One function of the charged residues is in “helix capping” through hydrogen bonding to stabilize the hydrophobic interaction of the LXXLL motif with the AF2 core and to limit the length of the LXXLL motif amphipathic helix (11, 45, 50). In the absence of coactivator, the side chains of these charged residues are exposed to the solvent (5). However, in contrast to the steroid receptors, the crystal structures of PPARγ (45) and RXRα (13) show a less organized pattern of charged residues and lack the additional positive and negative charge clusters that characterize the AF2 regions of AR, GR, and PR. Whether the primary role of the charge clusters flanking AF2 is in stabilizing the LXXLL helix and/or facilitating initial LXXLL helix binding, these charged residues are clearly indispensable to coactivator recruitment.
Residues flanking the LXXLL/FXXLF motifs.
p160 coactivator LXXLL motif binding shows specificity for nuclear receptors (40) in LXXLL motif preference, the number and spacer distance between LXXLL motifs (11, 12, 40), and in the contribution of flanking sequence. Here we show that the charge requirements of sequence flanking the FXXLF and LXXLL motifs adhere to the following criteria for AF2 binding. Positively charged residues are favored at position −1, −2, or −3 relative to the FXXLF and LXXLL core sequence (where the first F and L are +1, respectively) and/or negatively charged residues at position +8 or +9. Charged residues amino- and carboxyl-terminal of the motif apparently function alone or together to facilitate coactivator binding. Neutral or positively charged residues occur at positions +2 and +3 within the core motifs, with neutral or negatively charged residues at +6 and +7. These criteria for FXXLF and LXXLL motif binding to AF2 are supported by the classification of high- and low-affinity LXXLL peptides (24) and for LXXLL peptides obtained by phage display (7) and apply to the nuclear receptor family, including TRβ and VDR but to a less extent for nuclear receptor RXRα.
In addition to the experimental evidence presented for AR, other evidence supports these protein interaction criteria. Lysine residues amino-terminal to LXXLL-I and -II correspond to R20 flanking the AR FXXLF motif but occupy slightly different positions. Early studies on ERα indicated that neutralization of a basic residue amino-terminal to SRC1 LXXLL-II did not diminish coactivator binding (38), and specificity depended only on motif sequence (24). However, three basic residues amino-terminal to the core LXXLL motif facilitated docking of the LXXLL motif (38). The PPARγ LBD-LXXLL peptide cocrystal structure indicates that these lysine residues are solvent exposed and do not directly contact PPARγ (45). Similarly, the carboxyl-terminal L636 and T639 of SRC1 LXXLL-I and L693 and L694 of LXXLL-II form hydrogen bonds with a highly conserved K301 in the PPARγ LBD, which corresponds to K720 in AR. Hydrogen bonds in the PPARγ-coactivator peptide complex form with backbone amides of the LXXLL motifs and backbone carbonyls in the LBD. Charged residues D590 and R585 in the GR LBD form hydrogen bonds with side chains of R + 2 and D + 6 in the LXXLL-III motif of TIF2. Mutation of GR D590 and R585 to alanine and the charged residues flanking LXXLL-III of TIF2 reduced the peptide interaction and transactivation by the GR LBD (3). The γ-carboxylate group of E542 in ERα forms a hydrogen bond(s) to amides in the LXXLL-II helix, and the ɛ-amino group of K362 forms a hydrogen bond(s) with carbonyls in the LXXLL helix (50).
Binding affinity of the core AR NH2-terminal FXXLF peptide to the AR LBD (Kd, 1.2 μM) measured by isothermal titration calorimetry is fivefold higher than the affinity for the LXXLL-III motif of TIF2 (6.0 μM). Calorimetry measurements support the two-hybrid interaction data and indicate preferential AR binding of the FXXLF motif over the preferred LXXLL-III motif of TIF2. The AR LBD binding affinities for the FXXLF motifs are in the same range as LXXLL motif binding by other receptor LBDs. The binding affinity of GRIP1 LXXLL-II to TRβ (Kd, 0.8 μM) was higher than the binding affinity for LXXLL-III (Kd, 3.2 μM) and LXXLL-I (30 μM) measured by GST fusion protein affinity in vitro binding (11). Binding of LXXLL-I and -II with TRβ showed that the flanking sequence of LXXLL-II contributes to higher-affinity binding (11). A 13-residue peptide containing LXXLL-II binds the agonist-bound ERα LBD with a 50% inhibitory concentration of <0.4 μM (50). Positively charged residues amino-terminal to the LXXLL motif interacted favorably with negatively charged residues carboxyl-terminal in the TRβ LBD (11). Binding of GRIP1 LXXLL-III to GR and AR was increased by GRIP1 residues 1011 to 1121, suggesting that additional contacts with the LBD facilitate coactivator binding to nuclear receptors (28) and that binding affinities determined using short peptides may underestimate the strength of coactivator interactions. Considering the local concentration of the AR NH2-terminal FXXLF motif in the AR dimer bound to androgen response element DNA and the higher affinity of the FXXLF motif, competitive inhibition of p160 coactivator binding might result from the androgen-induced N/C interaction. Indeed, experimental evidence supports this androgen-dependent competitive inhibition of p160 coactivator binding by the AR NH2-terminal FXXLF motif (18).
Electrostatic interactions in p160 coactivator recruitment.
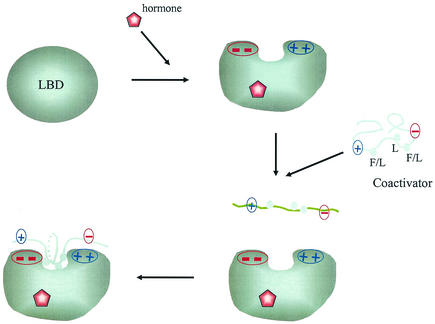
We propose that additional charged residues both in the steroid receptor LBD on opposite sides of the hydrophobic core of AF2 and flanking the FXXLF and LXXLL motif regions are involved in an electrostatic attractive mechanism to recruit the p160 coactivators (Fig. 8). Negatively charged residues carboxyl-terminal to the FXXLF and LXXLL motifs and positively charged residues amino-terminal to the core motifs interact with oppositely charged residues adjacent to AF2 so that the hydrophobic interaction between the AF2 core region and the conserved LXXLL motif is complemented by electrostatic interactions. In the case of AR and as recently shown for GR, charge clusters formed by additional residues near AF2 appear to contribute to the affinity and specificity of coactivator binding. This electrostatic attraction model provides a mechanistic explanation for different nuclear receptors showing preferences for LXXLL motif binding (11, 28, 40). Further, in AR the orientation of binding of the FXXLF motif to AF2 may be established in part by attractive forces that cause formation of an antiparallel dimer.
FIG. 8.
Schematic diagram of the charge polarity interaction model for steroid and nuclear hormone receptor recruitment of coactivators and for the AR N/C interaction. Hormone binding induces the formation of the AF2 hydrophobic cleft on the surface of the LBD that is flanked by clusters of oppositely charged residues. The charge patches on either side of AF2 interact with charged residues flanking the FXXLF or LXXLL motif sequences to mediate the androgen-dependent N/C interaction or steroid receptor coactivator recruitment. The less-ordered FXXLF or LXXLL motif is recruited to the AF2 interaction surface by complementary charges on either side of AF2, resulting in stabilization of the α-helix interface at the AF2 hydrophobic cleft.
The charge polarity model can account for several previous observations relating to coactivator recruitment. TIF2-LXXLL-II mutations changing H687 and K688 to aspartic acid disrupted binding of the LXXLL motif to ERα. The cocrystal structure of the TIF2-LXXLL-II peptide-ERα LBD showed that LXXLL motif residues H687 and K688 at positions −2 and −3 relative to the core sequence directly contact AF2 residues E542 and E380 in the ERα LBD (50). The charge polarity model for FXXLF and LXXLL motif recruitment by AF2 also explains the weak interaction of the AR coregulatory protein ARA55 (20). D320 in position −1 and R328 in position 8 of the FXXLF core peptide of ARA55 are predicted to cause charge repulsion at the AR AF2 surface. Binding of the ARA55 FXXLF peptide to AR AF2 was negligible in mammalian two-hybrid assays (20). Reversing the charge distribution in an ARA55 peptide double mutant increased binding to AF2 by threefold over that of the wild-type ARA55 control (data not shown).
The influence of charge interactions in recruiting the FXXLF or LXXLL motifs of coactivators and mediating the AR N/C interaction makes posttranslational modification of flanking charged residues a potential regulatory mechanism for hormone-induced gene transcription. The charge polarity model proposed here for coactivator recruitment is consistent with the effect of acetylation on ERα interaction with LXXLL-I of ACTR (10). Acetylation of K629 and K630 immediately preceding LXXLL-I of ACTR neutralizes the positive charges and terminates hormone-induced gene activation by causing dissociation of the receptor-bound p160 coactivator. In support of the model proposed here, it was speculated that the AF2-flanking charged residues E380 in helix 5 and D538 in helix 12 of ERα are involved in the electrostatic recruitment of oppositely charged residues flanking the LXXLL motif in ACTR (10). Similarly, phosphorylation of serine or threonine could alter an electrostatic interaction by introducing a negative charge. We found that mutations in ARA70 that converted two candidate phosphorylation sites, T324 and S325, to aspartic acid disrupted FXXLF motif binding to AR (data not shown). Phosphorylation of S884 carboxyl-terminal to the LXXLL motif of TRBP defined its selectivity for ERα and TRβ coactivator binding (31). Thus, the charge polarity model for coactivator recruitment and for the N/C interaction appears to establish an additional regulatory mechanism for steroid hormone action.
Charge-driven reactions were postulated previously in the heterodimerization and coactivator recruitment of the RXRα/PPARγ heterodimer (15). The asymmetric intradimer interaction that occurs within RXRα is facilitated by a salt bridge between E352 and R348, both in helix 7 of RXRα, and is strengthened by the presence of E431 in helix 10. An interdimer salt bridge occurs between K431 in helix 10 of RXRα with carboxyl-terminal Y477 in the AF2 helix 12 of PPARγ. In addition, K356 in RXRα interacts with a negatively charged region at E407 on the surface of PPARγ (15). Residues flanking the LXXLL motifs may also be involved in salt bridge formation and contribute to nuclear receptor specificity of coactivator recruitment, although these residues did not form structure in the PPARγ-coactivator peptide cocrystal (45). These multiple-charge interactions and salt bridges in the PPARγ/RXRα heterodimer between oppositely charged residues promote coactivator recruitment to AF2 and support the role of electrostatic interactions in LXXLL motif binding.
The AR-specific FXXLF motif.
The attenuating effect of R31 carboxyl-terminal to the AR NH2-terminal FXXLF motif in the AR N/C interaction appears to result from charge repulsion at the positively charged patch formed by K720, K717, and R726 flanking AF2. Comparison of the cocrystal structure of the TIF2-LXXLL-II peptide with the ERα LBD (50) and the predicted α-helical structure of the AR FXXLF motif suggests that AR R31 lies toward the AF2 surface in the androgen-dependent N/C interaction, where it can be influenced by the positively charged cluster in the LBD. TIF2-LXXLL-III has aspartic acid at position +9 rather than R31 in the AR FXXLF motif, which provides an explanation for disruption of TIF2 binding by the AR-K720A mutant (22). K362 in ERα corresponds to AR K720, which is also required to recruit p160 coactivators (22, 26). R31 is conserved among vertebrate AR and Xenopus in an otherwise poorly conserved AR NH2-terminal region, suggesting an important function (21). Charge repulsion between R31 flanking the AR NH2-terminal FXXLF motif and the positively charged AF2 patch constrains the androgen-induced N/C interaction. Because the AR N/C interaction competes with and represses p160 coactivator recruitment at AF2 (18), a less-than-optimal AR N/C interaction could allow coactivator access to AF2 under certain cell conditions. Recruitment of overexpressed p160 coactivators by AR AF2 may occur in recurrent prostate cancer (17).
It could be argued that the AR NH2-terminal FXXLF motif serves as an intra- or intermolecular mimic of the LXXLL motif. But unlike a similar role for the LXXML motif in helix 12 of ERα that binds to AF2 in the presence of antagonist, the FXXLF motif binds AF2 in the AR LBD specifically in the presence of the biologically active androgens, testosterone and dihydrotestosterone (19), and the synthetic anabolic steroids (30). Interaction of FXXLF with AF2 may contribute to the predominant activity of the NH2-terminal activation function 1 in AR. Considering the high affinity of FXXLF motif binding relative to the LXXLL motifs of p160 coactivators and its local concentration during AR dimerization and DNA binding, normal physiological levels of p160 coactivators may not activate AR through AF2.
In conclusion, the LBDs of nuclear receptors contain transcriptional activation domains that form similar highly structured arrangements of α-helices in response to hormone binding. As an activation domain, the LBD is large (∼250 amino acids) compared to smaller unstructured activation domains (36). The AF2 active surface of the nuclear receptor LBD is made up of hydrophobic and charged residues which are brought together by agonist binding (57). Transcriptional activation domains of mammalian regulatory proteins generally have little sequence homology and occur as random coils that acquire secondary structure after binding coactivators. Of the acidic, glutamine-rich and proline-rich classes of transcriptional activation domains (42), acidic activation domains enriched in aspartic and glutamic acid residues have been studied most extensively (54). Site-directed mutagenesis revealed that hydrophobic residues are critical for function and individual acidic residues are less important. Binding of coactivators induces acidic activation domains to form amphipathic α-helices (55). A two-step model proposed for acidic activation domains involves long-range interactions between negatively charged residues of an unstructured activation domain and positive charges of a coactivator to initiate recruitment. An amphipathic α-helix is then induced in the activation domain, and stable hydrophobic interactions occur with the coactivator (27). Based on studies in this report, a model for coactivator recruitment by steroid receptors is presented in which the recruitment of FXXLF and LXXLL coactivator motifs by the AF2 region of steroid receptors reflects a reversal of the acidic transcriptional activation domain-coactivator interaction model. Charge interactions between unstructured coactivator binding motifs and clusters of oppositely charged residues in the highly structured AF2 region initiate the orientation and recruitment of FXXLF and LXXLL motif binding by AF2. Subsequent hydrophobic residues of the FXXLF and LXXLL amphipathic helix establish nonpolar contacts with the hydrophobic cleft of AF2 to stabilize the receptor-coactivator complex. In both models, charge interactions initiate binding between structured and unstructured interacting partners.
Acknowledgments
This work was supported by Public Health Service Grant HD16910 from the National Institutes of Child Health and Development, by cooperative agreement U54-HD35041 as part of the Specialized Cooperative Centers Program in Reproductive Research of the National Institutes of Health, by the United States Army Medical Research and Material Command Grant DAMD17-00-1-0094, and by the International Training and Research in Population and Health Program supported by the Fogarty International Center and National Institutes of Child Health and Development, National Institutes of Health.
We thank Lori W. Lee, John T. Minges, Andrew T. Hnat, K. Michelle Cobb and De-Ying Zang for excellent technical assistance, Ashutosh Tripathy, Scientific Director of the University of North Carolina at Chapel Hill Macromolecular Interactions Facility, for assistance with isothermal titration calorimetry, Jan Hermans for interpretation of the thermodynamic binding results, Brenda Temple for modeling the FXXLF helix, and Frank S. French for reviewing the manuscript. Plasmids were generously provided by Donald P. McDonnell, David D. Moore, and Ronald M. Evans.
REFERENCES
- 1.Arriza, J. L., C. Weinberger, G. Cerelli, T. M. Glaser, B. L. Handelin, D. E. Housman, and R. M. Evans. 1987. Cloning of human mineralocorticoid receptor complementary DNA: structural and functional kinship with the glucocorticoid receptor. Science 237:268-275. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A. R., D. P. McDonnell, M. Hughes, T. M. Crisp, D. J. Mangelsdorf, M. R. Haussler, J. W. Pike, J. Shine, and B. W. O'Malley. 1988. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. USA 85:3294-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bledsoe, R. K., V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, and H. E. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93-105. [DOI] [PubMed] [Google Scholar]
- 4.Bradshaw, J. M., R. A. Grucza, J. E. Ladbury, and G. Waksman. 1998. Probing the “two-pronged plug two-holed socket” model for the mechanism of binding of the Src SH2 domain to phosphotyrosyl peptides: a thermodynamic study. Biochemistry 37:9083-9090. [DOI] [PubMed] [Google Scholar]
- 5.Brzozowski, A., A. Pike, Z. Dauter, R. Hubbard, T. Bonn, O. Engstrom, L. Ohman, G. Greene, J. Gustafsson, and M. Carlquist. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753-758. [DOI] [PubMed] [Google Scholar]
- 6.Bundle, D. R., and B. W. Sigurskjold. 1994. Determination of accurate thermodynamics of binding by titration microcalorimetry. Meth. Enzym. 247:288-305. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C., J. D. Norris, H. Gron, L. A. Paige, P. T. Hamilton, D. J. Kenan, D. Fowlkes, and D. P. McDonnell. 1999. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol. Cell Biol. 19:8226-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chawla, A., J. J. Repa, R. M. Evans, and D. J. Mangelsdorf. 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866-1870. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 11.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, X. F., C. M. Anderson, H. Ma, H. Hong, R. M. Uht, P. J. Kushner, and M. R. Stallcup. 1998. Nuclear receptor binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol. Endocrinol. 12:302-313. [DOI] [PubMed] [Google Scholar]
- 13.Egea, P. F., A. Mitschler, N. Rochel, M. Ruff, P. Chambon, and D. Moras. 2000. Crystal structure of the human RXR alpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 19:2592-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, W., R. C. Ribeiro, R. L. Wagner, H. Nguyen, J. W. Apriletti, R. J. Fletterick, J. D. Baxter, P. J. Kushner, and B. L. West. 1998. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 280:1747-1749. [DOI] [PubMed] [Google Scholar]
- 15.Gampe, R. T., V. G. Montana, M. H. Lambert, A. B. Miller, R. K. Bledsoe, M. V. Milburn, S. A. Kliewer, T. M. Willson, and H. E. Xu. 2000. Asymmetry in the PPARγ/RARα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 5:545-555. [DOI] [PubMed] [Google Scholar]
- 16.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 17.Gregory, C. W., B. He, R. T. Johnson, O. H. Ford, J. L. Mohler, F. S. French, and E. M. Wilson. 2001. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 61:4315-4319. [PubMed] [Google Scholar]
- 18.He, B., N. T. Bowen, J. T. Minges, and E. M. Wilson. 2001. Androgen-induced NH2- and COOH-terminal interaction inhibits p160 coactivator recruitment by activation function 2. J. Biol. Chem. 276:42293-42301. [DOI] [PubMed] [Google Scholar]
- 19.He, B., J. A. Kemppainen, and E. M. Wilson. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275:22986-22994. [DOI] [PubMed] [Google Scholar]
- 20.He, B., J. T. Minges, L. W. Lee, and E. M. Wilson. 2002. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem. 277:10226-10235. [DOI] [PubMed] [Google Scholar]
- 21.He, B., L. W. Lee, J. T. Minges, and E. M. Wilson. 2002. Dependence of selective gene activation on the androgen receptor NH2- and carboxyl-terminal interaction. J. Biol. Chem. 277:25631-25639. [DOI] [PubMed] [Google Scholar]
- 22.He, B., J. A. Kemppainen, J. J. Voegel, H. Gronemeyer, and E. M. Wilson. 1999. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 274:37219-37225. [DOI] [PubMed] [Google Scholar]
- 23.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 24.Heery, D. M., S. Hoare, S. Hussain, M. G. Parker, and H. Sheppard. 2001. Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors. J. Biol. Chem. 276:6695-6702. [DOI] [PubMed] [Google Scholar]
- 25.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocrine Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 26.Henttu, P. M., E. Kalkhoven, and M. G. Parker. 1997. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol. Cell. Biol. 17:1832-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann, S., K. D. Berndt, and A. P. Wright. 2001. How transcriptional activators bind target proteins. J. Biol. Chem. 276:40127-40132. [DOI] [PubMed] [Google Scholar]
- 28.Hong, H., B. D. Darimont, H. Ma, L. Yang, K. R. Yamamoto, and M. R. Stallcup. 1999. An additional region of coactivator GRIP1 required for interaction with the hormone binding domains of a subset of nuclear receptors. J. Biol. Chem. 274:3496-3502. [DOI] [PubMed] [Google Scholar]
- 29.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemppainen, J. A., E. Langley, C. I. Wong, K. Bobseine, W. R. Kelce, and E. M. Wilson. 1999. Distinguishing androgen receptor agonists and antagonists: distinct mechanisms of activation by medroxyprogesterone acetate and dihydrotestosterone. Mol. Endocrinol. 13:440-454. [DOI] [PubMed] [Google Scholar]
- 31.Ko, L., G. R., Cardona, T. Iwasaki, K. S. Bramlett, T. P. Burris, and W. W. Chin. 2002. Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol. Endocrinol. 16:128-140. [DOI] [PubMed] [Google Scholar]
- 32.Kuiper, G. G., E. Enmark, M. Pelto-Huikko, S. Nilsson, and J. A. Gustafsson. 1996. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. USA 93:5925-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 34.Langley, E., J. A. Kemppainen, and E. M. Wilson. 1998. Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J. Biol. Chem. 273:92-101. [DOI] [PubMed] [Google Scholar]
- 35.Langley, E., Z. X. Zhou, and E. M. Wilson. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270:29983-29990. [DOI] [PubMed] [Google Scholar]
- 36.Lu, X., A. Z. Ansari, and M. Ptashne. 2000. An artificial transcriptional activating region with unusual properties. Proc. Natl. Acad. Sci. USA 97:1988-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubahn, D. B., D. R. Joseph, M. Sar, J. A. Tan, H. N. Higgs, R. E. Larson, F. S. French, and E. M. Wilson. 1988. The human androgen receptor: complementary DNA cloning, sequence analysis and gene expression in prostate. Mol. Endocrinol. 2:1265-1275. [DOI] [PubMed] [Google Scholar]
- 38.Mak, H. Y., S. Hoare, P. M. A. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matias, P. M., P. Donner, R. Coelho, M. Thomaz, C. Peixoto, S. Mecedo, N. Otto, S. Joschko, P. Scholz, A. Wegg, S. Basler, M. Schafer, U. Egner, and M. A. Carrondo. 2000. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J. Biol. Chem. 275:26164-26171. [DOI] [PubMed] [Google Scholar]
- 40.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 43.Moras, D., and H. Gronemeyer. 1998. The nuclear receptor ligand binding domain: structure and function. Curr. Opin. Cell Biol. 10:384-391. [DOI] [PubMed] [Google Scholar]
- 44.Needham, M., S. Raines, J. McPheat, C. Stacey, J. Ellston, S. Hoare, and M. Parker. 2000. Differential interaction of steroid hormone receptors with LXXLL motifs in SRC-1a depends on residues flanking the motif. J. Steroid Biochem. Mol. Biol. 72:35-46. [DOI] [PubMed] [Google Scholar]
- 45.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, G. M. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 46.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 47.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 48.Sack, J. S., K. F. Kish, C. Wang, R. M. Attar, S. E. Kiefer, Y. An, G. Y. Wu, J. E. Scheffler, M. E. Salvati, S. R. Krystek, R. Weinmann, and H. M. Einspahr. 2001. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc. Natl. Acad. Sci. USA 98:4904-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 50.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937. [DOI] [PubMed] [Google Scholar]
- 51.Shibata, H., T. E. Spencer, S. A. Onate, G. Jenster, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Role of coactivators and corepressors in the mechanism of steroid/thyroid receptor action. Rec. Prog. Horm. Res. 52:141-165. [PubMed] [Google Scholar]
- 52.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 53.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 54.Tasset, D., L. Tora, C. Fromental, E. Scheer, and P. Chambon. 1990. Distinct classes of transcriptional activating domains function by different mechanism. Cell 62:1177-1187. [DOI] [PubMed] [Google Scholar]
- 55.Uesugi, M., O. Nyanguile, H. Lu, A. J. Levine, and G. L. Verdine. 1997. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277:1310-1313. [DOI] [PubMed] [Google Scholar]
- 56.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner, R. L., J. W. Apriletti, M. E. McGrath, B. L. West, J. D. Baxter, and R. J. Fletterick. 1995. A structural role for hormone in the thyroid hormone receptor. Nature 378:690-697. [DOI] [PubMed] [Google Scholar]
- 58.Webster, N. J., S. Green, D. Tasset, M. Ponglikitmongkol, and P. Chambon. 1989. The transcriptional activation function located in the hormone-binding domain of the human estrogen receptor is not encoded in a single exon. EMBO J. 8:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinberger, C., S. M. Hollenberg, E. S. Ong, J. M. Harmon, S. T. Brower, J. Cidlowski, E. B. Thompson, M. G. Rosenfeld, and R. M. Evans. 1985. Identification of human glucocorticoid receptor complementary DNA clones by epitope selection. Science 228:740-742. [DOI] [PubMed] [Google Scholar]
- 60.Westin, S., R. Kurokawa, R. T. Nolte, G. B. Wisely, E. M. McInerney, D. W. Rose, M. V. Milburn, M. G. Rosenfeld, and C. K. Glass. 1998. Interactions controlling the assembly of nuclear receptor heterodimers and coactivators. Nature 395:199-202. [DOI] [PubMed] [Google Scholar]
- 61.Williams, S. P., and P. B. Sigler. 1998. Atomic structure of progesterone complexed with its receptor. Nature 393:392-396. [DOI] [PubMed] [Google Scholar]
- 62.Xu, H. E., M. H. Lambert, V. G. Montana, K. D. Plunket, L. B. Moore, J. L. Collins, J. A. Oplinger, S. A. Kliewer, R. T. Gampe, D. D. McKee, J. T. Moore, and T. M. Willson. 2001. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA 98:13919-13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye, H., and H. Wu. 2000. Thermodynamic characterization of the interaction between TRAF2 and tumor necrosis factor receptor peptides by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA 97:8961-8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, Z. X., B. He, S. H. Hall, E. M. Wilson, and F. S. French. 2002. Domain interactions between coregulator ARA70 and the androgen receptor. Mol. Endocrinol. 16:287-300. [DOI] [PubMed] [Google Scholar]