Abstract
Transcriptional activation of the yeast HO gene involves the sequential action of DNA-binding and chromatin-modifying factors. Here we examine the role of the SAGA complex and the Nhp6 architectural transcription factor in HO regulation. Our data suggest that these factors regulate binding of the TATA-binding protein (TBP) to the promoter. A gcn5 mutation, eliminating the histone acetyltransferase present in SAGA, reduces the transcription of HO, but expression is restored in a gcn5 spt3 double mutant. We conclude that the major role of Gcn5 in HO activation is to overcome repression by Spt3. Spt3 is also part of SAGA, and thus two proteins in the same regulatory complex can have opposing roles in transcriptional regulation. Chromatin immunoprecipitation experiments show that TBP binding to HO is very weak in wild-type cells but markedly increased in an spt3 mutant, indicating that Spt3 reduces HO expression by inhibiting TBP binding. In contrast, it has been shown previously that Spt3 stimulates TBP binding to the GAL1 promoter as well as GAL1 expression, and thus, Spt3 regulates these promoters differently. We also find genetic interactions between TBP and either Gcn5 or the high-mobility-group protein Nhp6, including multicopy suppression and synthetic lethality. These results suggest that, while Spt3 acts to inhibit TBP interaction with the HO promoter, Gcn5 and Nhp6 act to promote TBP binding. The result of these interactions is to limit TBP binding and HO expression to a short period within the cell cycle. Furthermore, the synthetic lethality resulting from combining a gcn5 mutation with specific TBP point mutations can be suppressed by the overexpression of transcription factor IIA (TFIIA), suggesting that histone acetylation by Gcn5 can stimulate transcription by promoting the formation of a TBP/TFIIA complex.
Binding of the TATA-binding protein (TBP) to promoters is an essential event in transcriptional activation by RNA polymerase II (22, 37). In vitro studies have shown that binding by TBP is followed by that of transcription factor IIA (TFIIA) and TFIIB and that this TBP/TFIIA/TFIIB/DNA complex can then recruit other factors, resulting in the formation of a preinitiation complex. Thus, regulation of DNA binding by TBP could be a critical mechanism for regulating gene expression (41).
The SAGA complex has at least 14 subunits and regulates transcriptional activity by modulating chromatin structure (52, 59). Genetic analysis suggests that SAGA is encoded by three groups of genes. Deletion of the SPT7 or SPT20 gene causes severe growth defects. Other SAGA genes in this group (TRA1 and TBP-associated factors [TAFs]) are essential for viability, but these genes encode proteins that are also present in other transcriptional regulatory complexes. It is believed that Spt7 and Spt20 are part of the core of SAGA, because spt7 and spt20 mutations affect the structural integrity of the complex. In contrast, the Gcn5 and Spt3 modules may function on the periphery of SAGA, as mutations in these genes result in an intact SAGA complex. These mutants have modest but distinct phenotypes, suggesting different functions (42, 53). GCN5 encodes a histone acetyltransferase (8), and it is required for chromatin acetylation at promoters in vivo (28).
Spt3 has been shown to physically interact with TBP, and genetic experiments show allele-specific interactions between SPT3 and TBP (19). Spt3 is required for expression of the GAL1 gene, and chromatin immunoprecipitation experiments show that TBP binding to the GAL1 promoter requires SPT3 (17). Experiments with specific alleles of Spt3 and TBP show that a specific interaction between these proteins is required for GAL1 activation (30). While Spt3 stimulates TBP binding to the GAL1 promoter, other experiments suggest that Spt3 can act oppositely, inhibiting TBP binding to the HIS3 and TRP3 promoters (3).
High-mobility-group (HMG) proteins are small, abundant chromatin proteins that bend DNA sharply and modulate gene expression (10). The yeast Nhp6 HMG-like factor is encoded by two redundant genes, NHP6A and NHP6B. HO expression is reduced in an nhp6a nhp6b mutant, and genetic analysis suggests that Nhp6 and Gcn5 function in the same pathway of HO activation (60). Several experiments by Paull et al. (38) suggest that Nhp6 stimulates transcription by promoting the formation of preinitiation complexes. In vivo studies with chimeric promoter constructs suggest that Nhp6 acts at core promoters, and in vitro binding experiments show that Nhp6 stimulates the formation of a TBP/TFIIA/DNA complex that has an increased affinity for TFIIB. Since formation of a TBP/TFIIA/TFIIB/DNA complex is required for transcriptional initiation, Nhp6 may stimulate transcription by promoting formation of this complex.
The transcriptional activation of the yeast HO gene is preceded by the sequential binding of factors (6, 11, 12). First, the Swi5 DNA-binding factor binds far upstream and facilitates binding of the Swi/Snf and Mediator complexes. These factors promote the binding of the SAGA complex containing the Gcn5 histone acetyltransferase, resulting in changes in histone acetylation at the HO promoter (26). Finally, the SBF DNA-binding factor, composed of the Swi4 and Swi6 factors, binds to the promoter and it is believed that SBF ultimately activates HO transcription.
In this report, we provide evidence that Gcn5 and Nhp6 promote expression of the yeast HO gene via TBP. We also show that Spt3 acts to inhibit HO expression by blocking TBP binding to the HO promoter. Interactions among these factors are important for regulating other yeast genes, as indicated by the observation of multiple genetic interactions between TBP and both Gcn5 and Nhp6 and the suppression of mutant growth defects by either an spt3 mutation or by TFIIA overexpression.
MATERIALS AND METHODS
All strains listed in Table 1 are isogenic in the W303 background (55), except for FY61 and FY1006, which are S288c strains kindly provided by Fred Winston (20). Standard genetic methods were used for strain construction (43, 46). W303 strains with disruptions in ahc1, gcn5, nhp6a, nhp6b, sin4, spt20, spt3, spt8, and swi6 have been described previously (16, 18, 58, 60), and these were crossed to other W303 strains to produce the strains used in this study.
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| DY150 | MATaade2 can1 his3 leu2 trp1 ura3 |
| DY151 | MATα ade2 can1 his3 leu2 trp1 ura3 |
| DY1696 | MATasin4::URA3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2381 | MATα nhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2382 | MATanhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY3398 | MATaade2 can1 his3 leu2 trp1 |
| DY5199 | MATα gcn5::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5454 | MATα ho(URS2Δ) ade2 can1 his3 leu2 trp1 ura3 |
| DY5457 | MATα gcn5::TRP1 ho(URS2Δ) ade2 can1 his3 leu2 trp1 ura3 |
| DY5925 | MATagcn5::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY5926 | MATα gcn5::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY5929 | MATasin4::URA3 gcn5::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY6155 | MATanhp6a::KanMX nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY6178 | MATaspt8::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY6219 | MATaspt3::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY6277 | MATagcn5::HIS3 spt3::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY6404 | MATaahc1::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY6422 | MATagcn5::HIS3 spt8::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY6441 | MATanhp6a::KanMX nhp6b::ADE2 ade2 can1 his3 leu2 trp1 ura3 |
| DY6603 | MATagcn5::HIS3 URA3::GCN5(E173Q) ade2 can1 his3 leu2 trp1 |
| DY6707 | MATaspt20::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY6728 | MATanhp6a::URA3 nhp6b::HIS3 spt8::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY6758 | MATaswi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY6760 | MATα swi6::TRP1 spt3::ADE2 ade2 can1 his3 leu2 trp1 ura3 |
| DY6771 | MATα nhp6a::URA3 nhp6b::HIS3 gcn5::TRP1 spt8::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY6776 | MATanhp6a::KanMX nhp6b::ADE2 gcn5::HIS3 spt3::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY6778 | MATanhp6a::KanMX nhp6b::ADE2 spt3::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY6857 | MATα gcn5::HIS3 nhp6a::KanMX nhp6b::ADE2 ade2 can1 his3 leu2 trp1 ura3 |
| DY7202 | MATα ho(URS2Δ) spt3::ADE2 gcn5::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY7206 | MATα ho(URS2Δ) spt3::ADE2 ade2 can1 his3 leu2 trp1 ura3 |
| DY7242 | MATaspt15::LEU2 + SPT15(YCp-URA3) ade2 ade3 can1 his3 leu2 trp1 ura3 |
| DY7244 | MATα nhp6a::KanMX nhp6b::HIS3 spt15::LEU2 + SPT15(YCp-URA3) ade2 ade3 can1 his3 leu2 lys2 trp1 ura3 |
| DY7247 | MATaGALp::CDC20::ADE2 SPT15-HA::URA3 spt3::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY7250 | MATα GALp::CDC20::ADE2 SPT15-HA::URA3 ade2 can1 his3 leu2 trp1 ura3 |
| DY7515 | MATα gcn5::HIS3 spt15::LEU2 + SPT15(YCp-URA3) ade2 can1 his3 leu2 trp1 ura3 |
| DY7593 | MATaspt15::LEU2 + spt15-21(YCp-URA3) ade2 can1 his3 leu2 trp1 ura3 |
| DY7592 | MATα spt3::ADE2 URA3::spt3-401 ade2 can1 his3 leu2 trp1 |
| DY7593 | MATaspt3::ADE2 URA3::spt3-401 spt15::LEU2 + spt15-21(YCp-URA3) ade2 can1 his3 leu2 trp1 |
| DY7596 | MATα gcn5::HIS3 spt3::ADE2 URA3::spt3-401 ade2 can1 his3 leu2 trp1 |
| DY7597 | MATα gcn5::HIS3 spt15::LEU2 + spt15-21(YCp-URA3) ade2 can1 his3 leu2 trp1 ura3 |
| DY7598 | MATagcn5::HIS3 spt3::ADE2 URA3::spt3-401 spt15::LEU2 + spt15-21(YCp-URA3) ade2 can1 his3 leu2 trp1 |
| DY7604 | MATα gcn5::HIS3 spt3::ADE2 spt15::LEU2 + spt15-21(YCp-URA3) ade2 can1 his3 leu2 trp1 ura3 |
| DY7723 | MATanhp6a::KanMX nhp6b::HIS3 spt3::ADE2 spt15::LEU2 + SPT15(YCp-URA3) ade2 can1 his3 leu2 trp1 ura3 |
| DY8158 | MATagcn5::HIS3 spt15::KanMX + SPT15(YCp-URA3) ade2 can1 his3 leu2 trp1 ura3 |
| FY61 | MATahis4-917δ leu2 ura3 |
| FY1006 | MATaspt7::LEU2 his4-917δ lys2-173R2 leu2 |
The integrated GALp::CDC20 allele marked with ADE2 has been described previously (6), as has the SPT15-hemagglutinin (HA)-epitope-tagged allele marked with URA3 (29), provided by Kevin Struhl. A “marker swap” strategy (14), using plasmids M3926 (leu2::KanMX), M3927 (ura3::KanMX), M2371 (his3::ADE2), and M3938 (trp1::ADE2), was used to change disruption markers. By using this approach, spt15::LEU2 was converted to spt15::KanMX, nhp6a::URA3 was converted to nhp6a::KanMX, nhp6b::HIS3 was converted to nhp6b::ADE2, and spt3::TRP1 was converted to spt3::ADE2.
A strain with the ho(URS2Δ) promoter deletion (removing nucleotides [nt] −929 to −129 from ATG) was made in two steps by first inserting URA3 into the promoter and then replacing URA3 with the ho(URS2Δ) construct by selection on 5-fluoroorotic acid (5-FOA). Plasmid pDE124-1, provided by Fred Winston, was cleaved with EcoRV to integrate the spt3-401 allele at the URA3 locus. Plasmid gcn5(E173Q)-pRS306, provided by Shelley Berger, was cleaved with NsiI to integrate the gcn5(E173Q) allele at the URA3 locus. The SPT15 gene was disrupted with plasmid pKA23, provided by Karen Arndt. All gene disruptions and promoter replacements were confirmed by Southern blot analysis.
Plasmids are described in Table 2. Cells were grown in yeast extract-peptone-dextrose (YEPD) medium (46) at 30°C, except where the use of higher temperatures is noted or where synthetic complete medium with 2% glucose supplemented with adenine, uracil, and amino acids, as appropriate, but lacking essential components was used to select for plasmids. 5-FOA medium was prepared as described previously (7).
TABLE 2.
Plasmid list
| Plasmid | Description | Source or reference |
|---|---|---|
| YEp351 | YEp-LEU2 vector | 23 |
| pRS314 | YCp-TRP1 vector | 48 |
| YEplac195 | YEp-LEU2 vector | 21 |
| gcn5(E173Q) | gcn5(E173Q) in pRS306 | 56 |
| pSH223 | TBP (wild type) in YEp-LEU2 plasmid | Steve Hahn |
| pDE28-6 | TBP (wild type) in YCp-URA3 plasmid | 19 |
| pTM8 | TBP (wild type) in YCp-TRP1 plasmid | 25 |
| pDE58-1 | TBP(G174E) (spt15-21) in YCp-TRP1 plasmid | 19 |
| M4492 | TBP (F237D) in YCp-TRP1 plasmid | 49 |
| M4493 | TBP (K138T,Y139A) in YCp-TRP1 plasmid | 50 |
| M4495 | TBP (E188A) in YCp-TRP1 plasmid | 31 |
| pSH346 | TFIIA in YEp351 | Steve Hahn |
| pKA23 | spt15::LEU2 disruptor | 2 |
| M2371 | his3::ADE2 marker swap converter | Manuscript in preparation |
| M3926 | leu2::KanMX marker swap converter | Manuscript in preparation |
| M3927 | ura3::KanMX marker swap converter | Manuscript in preparation |
| M3938 | trp1::ADE2 marker swap converter | Manuscript in preparation |
RNA levels were determined by S1 nuclease protection assays with the HO and CMD1 probes as described previously (5). Strains with the GAL1:CDC20 allele were synchronized by removing galactose to arrest cells in mitosis, and then galactose was added to release from the arrest, as described previously (6). Chromatin immunoprecipitations were performed as described previously (6) by using 12CA5 monoclonal antibody to the HA epitope.
RESULTS
Gcn5 is required for HO activation to counteract Spt3 repression.
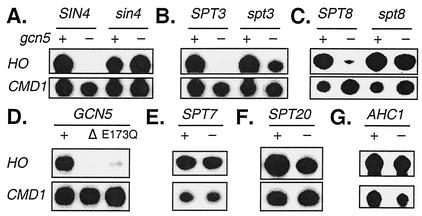
The sequential recruitment of multiple transcription factors is required for activation of the yeast HO gene (12). SAGA is one of these factors, and a mutation in the GCN5 histone acetyltransferase subunit reduces HO expression (39). This defect in HO activation can be suppressed by mutations in SIN3, RPD3, or SIN4 (60). Sin3 and Rpd3 are part of a histone deacetylase complex, while Sin4 is present in two distinct transcriptional regulatory complexes, SAGA itself and the Mediator (32; P. Grant and J. Workman, personal communication). To address the mechanism by which a sin4 mutation suppresses the Gcn5 requirement for HO expression, we asked whether mutations in other Mediator or SAGA components had similar effects. Mutations in Mediator genes GAL11, MED2, and HRS1, which are in the Sin4 subcomplex of the Mediator, result in reduced HO expression, which is opposite to the effect of a sin4 mutation (6). In contrast, mutations in the genes encoding SAGA components Spt3 and Spt8 did not reduce HO expression (Fig. 1A). Importantly, these spt mutations suppressed the gcn5 defect and HO expression in the gcn5 spt3 and gcn5 spt8 strains resembled that of gcn5 sin4 mutants (Fig. 1A through C). This suggests that Sin4 affects HO regulation through its role as a member of SAGA.
FIG. 1.
spt mutations allow HO expression in the absence of Gcn5. S1 nuclease protection assays were performed with probes specific for HO and CMD1 (internal control). RNAs were prepared from the following strains: (A) DY150, DY5925, DY1696, and DY5929; (B) DY150, DY5925, DY6219, and DY6277; (C) DY150, DY5925, DY6178, and DY6422; (D) DY151, DY5926, and DY6603; (E) FY61 and FY1006; (F) DY150 and DY6707; (G) DY150 and DY6404.
These experiments have been done using strains with a gcn5 gene disruption. It is possible that the histone acetyltransferase activity of Gcn5 is not required for HO activation but that the mere presence of the Gcn5 polypeptide as a component of SAGA would be sufficient for HO expression. To address this question, we constructed an isogenic strain with a Gcn5(E173Q) mutation that eliminates the histone acetyltransferase catalytic activity (56). The experiment depicted in Fig. 1D shows that the Gcn5 catalytic mutant does not express HO and thus the histone acetyltransferase activity is required for HO expression.
The fact that spt3 and spt8 mutations allow HO to be expressed in the absence of the Gcn5 histone acetyltransferase suggests that Gcn5's major role is to overcome the repression caused by Spt3 and Spt8. If this were true, then spt7 or spt20 mutations, which disrupt the structural integrity of SAGA (53), should not affect HO expression, since it would result in the loss of both a repressor and the factor responsible for overcoming the repression. In fact, HO expression was not affected by spt7 or spt20 mutations (Fig. 1E and F) and thus, eliminating SAGA entirely does not affect HO expression. In addition to being a component of SAGA, Gcn5 is also present in a second complex, ADA, and an ahc1 mutation disrupts the integrity of the ADA complex (18). HO expression was unaffected in an ahc1 mutant (Fig. 1G), indicating that the ADA complex is not required for HO expression. These experiments support a model in which Gcn5 functions within SAGA to overcome the repression meditated by SAGA components such as Sin4, Spt3, and Spt8.
spt3 effect is independent of SBF.
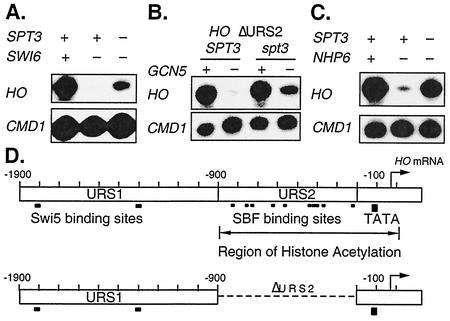
The SBF DNA-binding factor, composed of the Swi4 and Swi6 subunits, is the last factor recruited to the HO promoter, but SBF does not bind to HO in a gcn5 mutant (12). HO is expressed in a swi6 sin4 double mutant (60), and thus, sin4 suppresses the swi6 defect. The experiment depicted in Fig. 2A shows that an spt3 mutation also partially suppressed swi6. These results indicate that a major function of the SBF activator is to overcome the repression caused by Sin4 and Spt3 in SAGA.
FIG. 2.
spt3 effect is independent of SBF. S1 nuclease protection assays were performed with probes specific for HO and CMD1 (internal control). RNAs were prepared from the following strains: (A) DY150, DY6758, and DY6760; (B) DY5454, DY5457, DY7206, and DY7202; (C) DY150, DY2382, and DY6778. (D) The upper map shows the positions of the Swi5-binding sites in URS1, the SBF binding sites in URS2, and the TATA element of the HO promoter, as well as the region that is subject to GCN5-dependent histone acetylation (26). The lower map shows the ΔURS2 version of the HO promoter.
The HO promoter has been divided into three regions: the URS1 region (nt −1900 to −1000), where the Swi5 factor binds; the URS2 region (nt −900 to −200), where the SBF factor binds; and the TATA region (36). Histones in the URS2 and TATA regions of the HO promoter, but not in URS1, are acetylated in a Gcn5-dependent manner (26). To address which promoter regions confer Gcn5 dependence on the HO promoter, we deleted the URS2 region of the promoter and found that the HO ΔURS2 promoter required Gcn5 for activation (Fig. 2B). Importantly, an spt3 mutation could suppress the requirement for Gcn5 for activation of the HO ΔURS2 promoter to a similar extent as the native promoter (compare Fig. 2B and 1B). The HO ΔURS2 promoter contains the URS1 and TATA regions, but of these, only the TATA region is acetylated by Gcn5 (26). Additionally, while an spt20 mutation did not affect HO (Fig. 1E), expression of a HO-lacZ reporter is reduced in an spt20 (ada5) mutant (34) although this reporter contains the CYC1 TATA region instead of the HO TATA region. These results suggest that it is the HO TATA region that is the target of regulation by Gcn5 and Spt3.
spt3 mutation allows HO expression in an nhp6 mutant.
Nhp6 is a small HMG-like protein encoded by two redundant genes, NHP6A and NHP6B. Nhp6 is required for HO expression, as HO is not expressed in an nhp6a nhp6b mutant (60). Because a sin4 mutation allows HO to be expressed in the absence of either Gcn5 or Nhp6, we tested whether an spt3 mutation also suppresses nhp6. HO was expressed in both the nhp6a nhp6b spt3 (Fig. 2C) and nhp6a nhp6b spt8 (data not shown) strains, and thus, spt3 or spt8 can suppress the requirement for either Gcn5 or Nhp6 in HO activation, similar to sin4. Furthermore, overexpression of Nhp6B partially suppresses the gcn5 defect in HO expression (60), extending the genetic interactions between GCN5 and NHP6.
Suppression of growth phenotypes by spt3 and spt8.
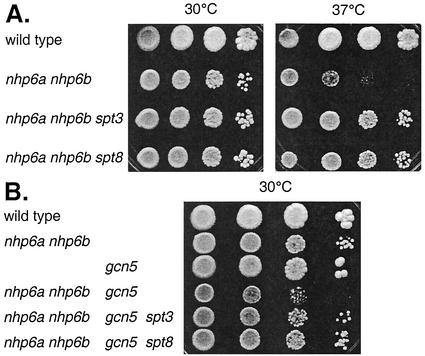
Mutations in SPT3 and SPT8 allow HO to be expressed in the absence of either Gcn5 or Nhp6, but is this suppression a general phenomenon? A nhp6a nhp6b double-mutant strain is defective for growth at 37°C (13), but this temperature sensitivity was suppressed by spt3 or spt8 mutations (Fig. 3A). Furthermore, combining a gcn5 mutation with nhp6a nhp6b resulted in a severe growth defect in the gcn5 nhp6a nhp6b strain (Fig. 3B) (60), suggesting that Gcn5 and Nhp6 have functional targets in common. However, the gcn5 nhp6a nhp6b growth defect could be suppressed by spt3 and spt8 mutations (Fig. 3B) and also by a sin4 mutation (60). These results again suggest that Spt3 and Spt8 act in opposition to Gcn5 and Nhp6 at certain critical promoters.
FIG. 3.
Suppression of growth phenotypes by spt3. (A) The following strains were plated on YEPD medium and grown for 5 days at 37°C: DY2381, DY6778, and DY6728. (B) The following strains were plated on YEPD medium and grown for 4 days at 30°C: DY6857, DY6776, and DY6771.
TBP as the critical target of regulation.
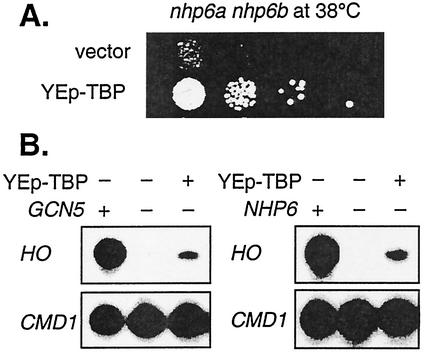
Several lines of evidence support a model in which TBP is the critical target of this regulation by Spt3. Spt3 interacts with TBP in both biochemical and genetic experiments (17, 19), and it has been suggested that Spt3 inhibits TBP binding at some promoters (3). We found that overexpression of TBP from a multicopy plasmid allowed an nhp6a nhp6b mutant to grow at 38°C (Fig. 4A). This restoration of growth in the absence of Nhp6 by high-copy-number TBP was consistent with the hypothesis that Nhp6 normally promotes TBP binding. TBP overexpression also partially restored HO expression in both gcn5 and nhp6a nhp6b mutants (Fig. 4B), and similarly, Nhp6B overexpression increases HO expression in a gcn5 mutant (60).
FIG. 4.
Suppression by overexpression of TBP. (A) Strain DY6155 (nhp6a nhp6b) was transformed with plasmids YEp351 (YEp-LEU2 vector) or pSH223 (YEp-LEU2 with TBP), and dilutions were plated on YEPD and grown for 2 days at 38°C. (B) RNA was prepared from cells grown without uracil to select for plasmids and used for S1 nuclease protection assays. RNAs were prepared from the following strains: DY3398 (wild type) with YEplac195 vector, DY5199 (gcn5) with YEplac195 vector, DY5199 (gcn5) with pJG18-2 (YEp-URA3 with TBP), DY3398 (wild type) with YEplac195 vector, DY6441 (nhp6a nhp6b) with YEplac195 vector, and DY6441 (nhp6a nhp6b) with pJG18-2 (YEp-URA3 with TBP).
Recent work has shown that the temperature sensitivity of nhp6a nhp6b mutants is due in part to defects in the transcription of RNA polymerase III genes (27, 33, 35). The suppression of nhp6a nhhp6b mutants by YEp-TBP is consistent with the fact that TBP is also part of the RNA polymerase III factor TFIIIB (24). The fact that TBP overexpression partially suppresses the nhp6 defect in HO expression (Fig. 4B) suggests that the genetic interaction between Nhp6 and TBP also affects RNA polymerase II transcription.
To analyze Nhp6's role in regulating TBP binding to the TATA element by the chromatin immunoprecipitation method, we attempted to construct an nhp6a nhp6b mutant bearing an HA-epitope-tagged version of TBP. However, we found that this nhp6a nhp6b HA-TBP strain was inviable, suggesting that the HA epitope tag at the N terminus diminishes TBP function in a way not tolerated in the absence of Nhp6. We also tagged TBP at the C terminus, and this TBP-HA allele was also synthetically lethal with nhp6. To further analyze these allele-specific effects, we determined whether other previously characterized viable TBP mutations were synthetically lethal with nhp6. We constructed a strain with the SPT15 (encoding TBP), NHP6A, and NHP6B genes deleted; TBP is essential for viability, and this strain is kept alive with wild-type TBP on a YCp-URA3 plasmid. Since strains with a wild-type URA3 gene cannot grow on media containing 5-FOA (7) and the plasmid supplies the essential TBP protein, this strain cannot grow on 5-FOA (Fig. 5A, line 1). Introducing a TRP1 plasmid with the wild-type TBP gene allows for loss of the URA3 plasmid and growth on 5-FOA (Fig. 5A, line 2). The TBP(K138T, Y139A) and TBP(F237D) mutations eliminate interaction with TFIIA in vitro (49, 50), and plasmids with these TBP mutations did not permit growth of DY7244 on 5-FOA (Fig. 5A, lines 3 and 4). Importantly, these TBP mutations did support viability in an NHP6+ strain, as evidenced by growth on 5-FOA plates (Fig. 5B). Thus TBP(K138T, Y139A) and TBP(F237D) cannot support growth in a cell lacking Nhp6, consistent with the suggestion that Nhp6 stimulates TBP-TFIIA interaction (38). In contrast, the TBP(E188A) mutation, which affects interaction with TFIIB (31), was viable in the absence of Nhp6 (Fig. 5A). Finally, the TBP(G174E) mutation, which affects interaction with Spt3 (19), was also lethal without Nhp6 (Fig. 5A).
FIG. 5.
Genetic interactions with TBP. (A) Strain DY7244 (nhp6a nhp6b spt15 with wild-type TBP on a YCp-URA3 plasmid) was transformed with the indicated YCp-TRP1 plasmid, and dilutions were plated on either synthetic complete or 5-FOA plates and grown for 3 days at 30°C. (B) Strain DY7242 (spt15 with wild-type TBP on a YCp-URA3 plasmid) was transformed with the indicated YCp-TRP1 plasmid, and dilutions were plated on either synthetic complete or 5-FOA plates and grown for 3 days at 30°C. (C) Strain DY7723 (nhp6a nhp6b spt15 spt3 with wild-type TBP on a YCp-URA3 plasmid) was transformed with the indicated YCp-TRP1 plasmid, and dilutions were grown at 30°C on either YEPD plates for 3 days or 5-FOA plates for 4 days. (D) Strain DY7515 (gcn5 spt15 with wild-type TBP on a YCp-URA3 plasmid) was transformed with the indicated YCp-TRP1 plasmid, and dilutions were grown at 30°C on either YEPD plates for 3 days or 5-FOA plates for 4 days.
We have found that an spt3 mutation can suppress growth defects in both the nhp6a nhp6b and gcn5 nhp6a nhp6b strains (Fig. 3). We were therefore interested in determining whether an spt3 mutation could suppress the synthetic lethality observed between nhp6 and mutant TBP alleles. We constructed an spt15 nhp6a nhp6b spt3 strain with wild-type TBP on a YCp-URA3 plasmid. This strain was transformed with TRP1 plasmids carrying either wild-type TBP, TBP(K138T, Y139A), or no insert, and these transformed strains were then plated on 5-FOA medium to determine whether the cells were viable after loss of the YCp-URA3 plasmid with wild-type TBP. The TBP(K138T, Y139A) mutation was viable and reasonably healthy in the nhp6a nhp6b spt3 strain (Fig. 5C), while this same mutation was lethal in the nhp6a nhp6b SPT3 strain (Fig. 5A). This was not true of all TBP alleles, as spt3 suppressed the nhp6a nhp6b TBP(G174E) lethality only weakly and spt3 did not suppress the nhp6a nhp6b TBP(F237D) lethality at all (data not shown). This demonstrates that the lethality of the TBP(K138T, Y139A) mutation in the absence of Nhp6 could be suppressed by an spt3-null mutation and suggests that Nhp6 and Spt3 act in opposition on TBP.
We have seen that the absence of either Gcn5 or Nhp6 reduces HO expression, and these defects can be suppressed by sin4, spt3, or spt8. Additionally, there are synergistic effects of combining gcn5 with nhp6a nhp6b mutations. With this similarity in mind, we wanted to determine whether these TBP point mutations were lethal in the absence of the Gcn5 histone acetyltransferase. We constructed a gcn5 spt15 yeast strain, kept alive with wild-type TBP on a YCp-URA3 plasmid. This strain was transformed with the TRP1 plasmids containing the TBP alleles, and growth on 5-FOA in the absence of wild-type TBP on YCp-URA3 was assessed. The results showed that TBP(K138T, Y139A) is lethal in the gcn5 mutant (Fig. 5D), while the TBP(F237D) mutation showed no growth defect in absence of Gcn5 (data not shown). The TBP(G174E) allele showed significant genetic interactions with gcn5 and spt3, as described below.
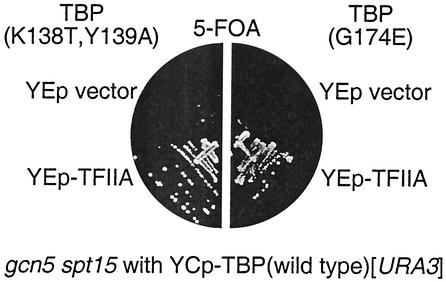
As the TBP(K138T, Y139A) mutation eliminates interaction with TFIIA in vitro (50), it seemed possible that overexpression of TFIIA might suppress the synthetic lethality observed with nhp6 or gcn5 mutations. A multicopy plasmid with TOA1 and TOA2, the two genes encoding the TFIIA subunits, was transformed into the nhp6 spt15 and gcn5 spt15 strains that had two TBP plasmids, a YCp-URA3 plasmid with wild-type TBP and a YCp-TRP1 plasmid with TBP(K138T, Y139A). The nhp6 strain with TBP(K138T, Y139A) and the YEp-TFIIA plasmid was unable to grow on 5-FOA medium (data not shown), and thus, overexpression of TFIIA cannot suppress this synthetic lethality. However, this YEp-TFIIA plasmid did allow the gcn5 spt15 strain with TBP(K138T, Y139A) to grow on 5-FOA (Fig. 6), and thus, TFIIA overexpression can suppress the lethality caused by the TBP(K138T, Y139A) mutation in the gcn5 mutant. TFIIA is required for this effect, as the YEp vector, without any gene insert, does not allow this strain to grow on 5-FOA. A similar experiment was done with the TBP(G174E) allele, and the results showed that TFIIA overexpression suppresses the synthetic growth defect in the gcn5 TBP(G174E) double mutant (Fig. 6).
FIG. 6.
TFIIA overexpression suppresses the gcn5 TBP(K138T, Y139A) synthetic lethality. Strain DY8158 (gcn5 spt15 with wild-type TBP on a YCp-URA3 plasmid) was transformed with either M4493 [TBP(K138T, Y139A) in YCp-TRP1] or pDE58-1 [TBP(G174E) in YCp-TRP1] and either YEp351 or pSH346 (LEU2), selecting for the TRP1 and LEU2 plasmids. Transformants were grown on 5-FOA plates for either 7 days at 25°C (left half) or 3 days at 30°C (right half).
These experiments show important genetic interactions between TBP and both Nhp6 and Gcn5. The TBP(K138T, Y139A) point mutation, which is viable in a NHP6A NHP6B GCN5 strain, is lethal in the absence of either Nhp6 or Gcn5. Additionally, TBP has been shown to interact with Spt3 (19), and an spt3 mutation suppresses the synthetic lethality of the TBP(K138T, Y139A) nhp6a nhp6b genotype, suggesting that Spt3 and Nhp6 function in opposing directions. Finally, overexpression of TFIIA overcomes the lethality caused by combining the gcn5 mutation with either TBP(K138T, Y139A) or TBP(G174E), suggesting that Gcn5 functions in vivo to promote the formation of a TBP/TFIIA complex.
spt3 mutations affect TBP binding to HO.
TBP and Spt3 physically interact, and genetic interactions between TBP and SPT3 mutations are highly allele specific, suggesting that this interaction is functionally important (19, 30). Spt3 promotes expression of the GAL1 gene (17), and thus, Spt3 functions as an activator at GAL1, although it inhibits HO expression. It should be noted that the experiments from the Winston lab (17, 30) used a strain background different from those used here. They examined TBP binding to the GAL1 promoter in strains with specific TBP and SPT3 mutations (30). TBP(G174E), expressed from the spt15-21 allele, appears to be defective for interaction with Spt3, as spt15-21 phenotypes can be suppressed by spt3-401 [Spt3(E240K)]. TBP binds to the GAL1 promoter under inducing conditions, and this binding requires Spt3 (17). The TBP(G174E) mutant does not bind to GAL1, but this defect is suppressed by Spt3(E240K), suggesting that TBP(G174E) and Spt3(E240K) interact like the two wild-type proteins (30).
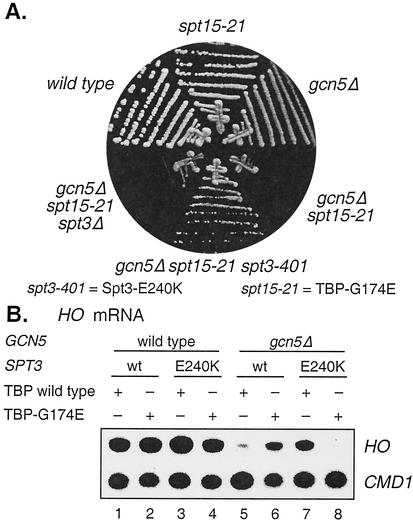
We investigated whether these specific alleles that affect the interaction between TBP and Spt3 would affect the regulation of HO expression by constructing isogenic strains differing at the GCN5, SPT3, and SPT15 (TBP) loci. Strains with a gcn5 or a spt15-21 [TBP(G174E)] single mutation grew well, but the gcn5 spt15-21 double mutant was extremely sick (Fig. 7A). This suggests that TBP(G174E), which shows no defect in binding DNA in vitro (19), has severe defects when chromatin is underacetylated due to the gcn5 mutation. We found that the spt3-401 mutation, but not a spt3-null mutation, suppresses this defect. One explanation for these results is that Spt3 stimulates TBP binding to certain promoters such as GAL1 (17), but wild-type Spt3 cannot interact with TBP(G174E) and thus cannot stimulate TBP binding. This stimulation of TBP binding by Spt3 becomes critical in a gcn5 mutant, and the ability of Spt3(E240K) to interact with TBP(G174E) restores healthy growth.
FIG. 7.
spt3-401 suppresses spt15-21 for growth and HO expression. (A) Isogenic strains were constructed, differing at the GCN5, SPT15 and SPT3 loci. The gcn5 spt15-21 double mutant had a severe growth defect, but this was suppressed specifically by the spt3-401 allele. The following strains were grown on YEPD medium for 2 days at 30°C: DY151, DY7593, DY5926, DY7597, DY7598, and DY7604. (B) RNA was prepared from the following strains and used for S1 nuclease protection assays: DY151, DY7593, DY7592, DY7594, DY5926, DY7597, DY7596, and DY7598.
HO expression was reduced in a gcn5 mutant (Fig. 7B, lane 5), but HO expression was restored largely in the gcn5 TBP(G174E) double mutant (lane 6). This result is consistent with the suggestion that TBP(G174E) does not interact with Spt3, and thus, TBP(G174E) is insensitive to repression by Spt3. Combining the TBP(G174E) and Spt3(E240K) mutations in the gcn5 background, where Spt3(E240K) can interact with and inhibit TBP(G174E), resulted in the loss of HO expression (lane 8). This allele-specific interaction provides strong support for the hypothesis that Spt3 inhibits TBP binding to HO. These results are similar to those of experiments with these alleles showing that Spt3 inhibits the basal expression of TRP3 and HIS3 (3).
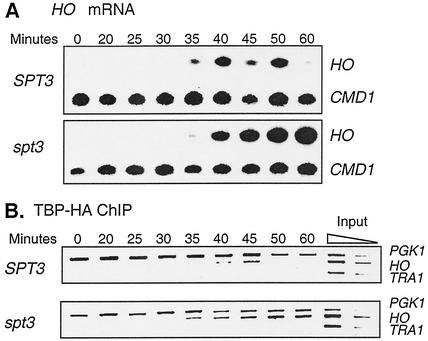
Although Spt3 promotes the binding of TBP to the GAL1 promoter (17), our data suggest that Spt3 inhibits TBP binding to HO TATA. Cosma et al. (12) reported that they were unable to detect TBP binding to HO by chromatin immunoprecipitation, a result that would be seen if TBP binds only very briefly to this cell-cycle-regulated promoter. We used chromatin immunoprecipitation assays to examine the binding of HA-epitope-tagged TBP to the HO promoter in SPT3 and spt3 strains that had been synchronized in the cell cycle by a CDC20 arrest-and-release protocol (6). At various time intervals following release from the cell cycle arrest, samples were taken for HO RNA measurement and for chromatin immunoprecipitation. RNA measurements showed a large increase in HO mRNA levels in the spt3 mutant compared to that in the wild type, particularly at the later time points (Fig. 8A). For the chromatin immunoprecipitation experiment, sheared chromatin was prepared, TBP-HA was immunoprecipitated, and the DNA present in the immunoprecipitated material was analyzed by PCR (Fig. 8B). HA-TBP binding to the PGK1 promoter and the TRA1 open reading frame were assessed as positive and negative controls, respectively. While only minimal binding of TBP-HA to HO was seen in the wild-type strain, consistent with a previous report (12), strong TBP-HA binding to HO was seen in the spt3 mutant. This experiment demonstrates that Spt3 inhibits the binding of TBP to the HO promoter.
FIG. 8.
Spt3 inhibits TBP binding to HO. SPT3 (DY7247) and spt3 (DY7250) strains with a TBP-HA tag were synchronized, and at timed intervals, samples were taken for RNA analysis and chromatin immunoprecipitation. (A) S1 nuclease protection assays showed that HO RNA levels were higher in the spt3 mutant. (B) Chromatin immunoprecipitation (ChIP) analysis showed increased TBP-HA binding to the HO TATA in spt3 mutants. PGK1 TATA and the TRA1 open reading frame served as positive and negative controls for the chromatin immunoprecipitation.
DISCUSSION
The Gcn5 histone acetyltransferase and the Nhp6 HMG protein are required for expression of the yeast HO gene, but a mutation in the SPT3 gene allows HO to be expressed in the absence of either Gcn5 or Nhp6. Spt3 interacts with the TBP, and our data suggest that Gcn5 and Nhp6 stimulate HO transcription by promoting TBP binding.
HO activation involves the sequential binding of transcription factors to the promoter (12). We suggest that TBP may be the last factor to bind to HO and that TBP binding may trigger HO transcription. In vitro binding experiments suggest that TBP is the first factor to bind DNA in the formation of a preinitiation complex (9). However, this is not necessarily the case at specific promoters in vivo, as TBP is the last factor to bind to the beta interferon promoter, after Mediator and RNA polymerase II (1). Mediator binds to the HO promoter very early, and binding of RNA polymerase to HO as assayed by chromatin immunoprecipitation is much more robust than that for TBP (6, 11). Further work is needed to determine the order of RNA polymerase II and TBP binding to the HO promoter. As cells approach the commitment point in the cell cycle (START), the Swi6 component of the SBF factor is activated, directly or indirectly, by the Cdc28 cyclin-dependent kinase (57). SBF already bound to the TATA proximal region of the HO promoter is now activated, and we suggest that it promotes binding of TBP, as Swi6 has been shown to interact with TBP-containing complexes (44).
In vitro studies show that some factors remain bound at the promoter after transcriptional initiation, including Mediator and TBP, and that these factors promote subsequent rounds of reinitiation (61). Mediator binds stably to HO (6, 11), and thus, continued TBP binding after initiation could promote rapid transcriptional reinitiation. However, TBP binds to HO only very transiently, with increased binding seen in an spt3 mutant (Fig. 8B). Thus Spt3 may function at HO to actively displace TBP from the promoter following transcriptional initiation. HO encodes an endonuclease, and it may be advantageous to limit expression of the endonuclease gene product by actively blocking the reinitiation pathway.
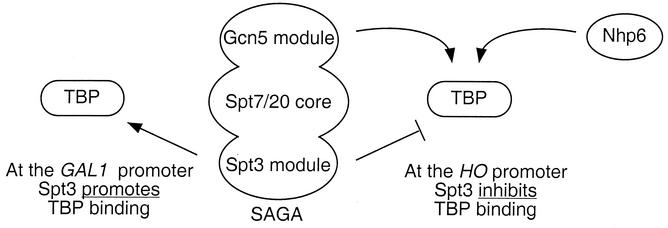
Our genetic data suggest that TBP binding can be stimulated by the Nhp6 HMG protein and the Gcn5 histone acetyltransferase (Fig. 9). These results include the suppression of defects caused by nhp6 and gcn5 mutations by overexpression of TBP (Fig. 4) and synthetic lethality caused by combining TBP mutations with either nhp6 or gcn5 (Fig. 5). How do these factors regulate TBP binding? Zhao and Herr (62) have recently shown that TBP binding to DNA is a two-step process, starting with an unstable complex containing unbent DNA that slowly isomerizes into a stable complex with bent DNA. HMG proteins bend DNA, and by doing so, an HMG protein could promote formation of the stable TBP/bent DNA complex. In vitro experiments show that mammalian HMG proteins stimulate TBP binding to DNA (15, 47, 54). Moreover, TFIIB stimulates the formation of a stable TBP/DNA complex (62), suggesting that association of TFIIB with the complex promotes the formation of the stable bent DNA form. Paull et al. (38) have shown that TFIIB has a higher affinity for a TBP/TFIIA/Nhp6 complex than for the complex lacking Nhp6. Taken together, these results suggest that Nhp6, by bending DNA, promotes the formation of the multiprotein complex of TBP, TFIIA, and TFIIB with DNA that is critical for transcriptional initiation (37).
FIG. 9.
Model for the differential regulation of TBP binding by Spt3. The Gcn5 module also contained Ada2 and Ada3. In addition to Spt7 and Spt20, the SAGA core also contained Ada1, TAF90, TAF61, TAF60, TAF25, TAF17, and Tra1. The Spt3 module also contained Spt8. At GAL1 and other promoters, Spt3 promoted TBP binding and Gcn5 was not required (4, 17). At HO, Gcn5 and Nhp6 stimulated TBP binding while Spt3 inhibited binding.
Our data suggest that Gcn5 promotes DNA binding by TBP. In support of this idea, it has been shown that histone acetylation stimulates TBP binding in vivo (45). The TBP(K138T, Y139A) and TBP(G174E) mutations, which are healthy on their own, showed marked growth defects in a gcn5 mutant (Fig. 5D and 7A). These mutant forms of TBP show no defect in DNA binding in vitro (19, 50), but these defects become apparent when chromatin is underacetylated in the gcn5 mutant. Importantly, this synthetic growth defect can be suppressed by overexpression of TFIIA (Fig. 6), suggesting that TFIIA overexpression suppresses this defect by promoting formation of the TBP/TFIIA/TFIIB/DNA complex. The TBP(G174E) mutation affects interaction with Spt3, and the TBP(G174E) gcn5 defect can also be suppressed by the compensatory Spt3(E240K) mutation, which restores interaction with TBP(G174E) (Fig. 7A). Spt3 stimulates TBP binding to the GAL1 promoter (17), and the defect caused by TBP(G174E) can be suppressed by Spt3(E240K) (30). In summary, we suggest that some TBP mutants have difficulty binding DNA in a gcn5 mutant when the template is underacetylated, but this can be suppressed by increased TFIIA levels or by restoration of the TBP-Spt3 interaction, each of which may promote formation of the TBP/TFIIA/TFIIB/DNA complex at certain promoters.
Certain TBP mutations show a strong synthetic growth defect in the absence of either Gcn5 or Nhp6 (Fig. 5). We suggest that TBP binding at some critical promoters can be stimulated by either Gcn5 or Nhp6 (Fig. 9), and thus, the defect in TBP binding in the gcn5 nhp6a nhp6b mutant results in a severe growth defect. This growth defect can be suppressed by mutations in sin4 (60), spt3, or spt8 (Fig. 2B), consistent with our hypothesis that TBP binding at some promoters can be inhibited by Spt3 and/or Spt8 in SAGA. Importantly, SAGA inhibits TBP binding to the HIS3 promoter in vitro, but this inhibition is not seen with SAGA lacking Spt3 or Spt8 (3). It is noteworthy that the Gcn5 and Spt3 proteins are both part of the same complex, SAGA, but they have opposing roles in the regulation of HO expression.
Promoter structure apparently plays a role in how Spt3 regulates transcription, because, in contrast to our results at HO, Spt3 stimulates TBP binding to the GAL1 promoter (17). These results suggest that Spt3 functions differently at distinct promoters (Fig. 9). HO is expressed briefly in the cell cycle, and Spt3 may function to limit HO expression. GAL1, when induced by a nonpreferred carbon source, is expressed at very high levels, and Spt3 may function to stabilize TBP binding and thereby promote reinitiation. A variant form of SAGA, called either SALSA or SLIK (40, 51), which lacks Spt8 and has a truncated Spt7 subunit, has been recently identified. It appears that SALSA/SLIK promotes transcriptional activation, and it may be involved in the activation of GAL1, because SPT8 is not required for GAL1 expression (4). However, there are genes that require both Spt3 and Spt8 for TBP to bind, suggesting that it is SAGA that functions at these promoters (4). Interestingly, some of these genes are still expressed in a gcn5 mutant. Further work is needed to decipher how promoter structure determines the requirements for Spt3 and Gcn5 for regulation at any given promoter.
Acknowledgments
We acknowledge the generosity of these colleagues in providing strains or plasmids: Karen Arndt, Shelley Berger, Tim Formosa, Steve Hahn, Rohinton Kamakaka, Tetsuro Kokubo, Laurie Stargell, Kevin Struhl, Jesper Svestrup, Fred Winston, and Jerry Workman. We also thank Brad Cairns, Tim Formosa, Warren Voth, Dennis Winge, and Fred Winston for helpful discussions and Patrick Grant and Jerry Workman for communicating unpublished results.
This work was supported by a grant from the National Institutes of Health awarded to D.J.S.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Arndt, K. M., S. L. Ricupero, D. M. Eisenmann, and F. Winston. 1992. Biochemical and genetic characterization of a yeast TFIID mutant that alters transcription in vivo and DNA binding in vitro. Mol. Cell. Biol. 12:2372-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., and M. R. Green. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhoite, L. T., and D. J. Stillman. 1998. Residues in the Swi5 zinc finger protein that mediate cooperative DNA-binding with the Pho2 homeodomain protein. Mol. Cell. Biol. 18:6436-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 8.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 9.Buratowski, S., S. Hahn, L. Guarente, and P. A. Sharp. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549-561. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 12.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 13.Costigan, C., D. Kolodrubetz, and M. Snyder. 1994. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 14:2391-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross, F. R. 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647-653. [DOI] [PubMed] [Google Scholar]
- 15.Das, D., and W. M. Scovell. 2001. The binding interaction of HMG-1 with the TATA-binding protein/TATA complex. J. Biol. Chem. 276:32597-32605. [DOI] [PubMed] [Google Scholar]
- 16.Donze, D., C. R. Adams, J. Rine, and R. T. Kamakaka. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates III, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 20.Gansheroff, L. J., C. Dollard, P. Tan, and F. Winston. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 22.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 24.Kassavetis, G. A., C. A. Joazeiro, M. Pisano, E. P. Geiduschek, T. Colbert, S. Hahn, and J. A. Blanco. 1992. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell 71:1055-1064. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, A., T. Miyake, Y. Ohyama, M. Kawaichi, and T. Kokubo. 2001. Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae. J. Biol. Chem. 276:395-405. [DOI] [PubMed] [Google Scholar]
- 26.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruppa, M., R. D. Moir, D. Kolodrubetz, and I. M. Willis. 2001. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell 7:309-318. [DOI] [PubMed] [Google Scholar]
- 28.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 30.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, M., and K. Struhl. 1997. A severely defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol. Cell. Biol. 17:1336-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane, D. J. Stillman, and R. D. Kornberg. 1995. Yeast global transcriptional repressors Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92:10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez, S., M. Livingstone-Zatchej, S. Jourdain, F. Thoma, A. Sentenac, and M.-C. Marsolier. 2001. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol. 21:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus, G. A., J. Horiuchi, N. Silverman, and L. Guarente. 1996. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol. Cell. Biol. 16:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, M. P., V. L. Gerlach, and D. A. Brow. 2001. A novel upstream RNA polymerase III promoter element becomes essential when the chromatin structure of the yeast U6 RNA gene is altered. Mol. Cell. Biol. 21:6429-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasmyth, K. 1985. At least 1400 base pairs of 5′-flanking DNA is required for the correct expression of the HO gene in yeast. Cell 42:213-223. [DOI] [PubMed] [Google Scholar]
- 37.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 38.Paull, T. T., M. Carey, and R. C. Johnson. 1996. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 10:2769-2781. [DOI] [PubMed] [Google Scholar]
- 39.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22:8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-302. [DOI] [PubMed] [Google Scholar]
- 44.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sewack, G. F., T. W. Ellis, and U. Hansen. 2001. Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol. 21:1404-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:1-21. [DOI] [PubMed] [Google Scholar]
- 47.Shykind, B. M., J. Kim, and P. A. Sharp. 1995. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 9:1354-1365. [DOI] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stargell, L. A., and K. Struhl. 1996. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol. Cell. Biol 16:4456-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stargell, L. A., and K. Struhl. 1995. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science 269:75-78. [DOI] [PubMed] [Google Scholar]
- 51.Sterner, D. E., R. Belotserkovskaya, and S. L. Berger. 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 99:11622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA- binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sutrias-Grau, M., M. E. Bianchi, and J. Bernues. 1999. High mobility group protein 1 interacts specifically with the core domain of human TATA box-binding protein and interferes with transcription factor IIB within the pre-initiation complex. J. Biol. Chem. 274:1628-1634. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 56.Trievel, R. C., J. R. Rojas, D. E. Sterner, R. N. Venkataramani, L. Wang, J. Zhou, C. D. Allis, S. L. Berger, and R. Marmorstein. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 96:8931-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wijnen, H., A. Landman, and B. Futcher. 2002. The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22:4402-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittschieben, B. O., J. Fellows, W. Du, D. J. Stillman, and J. Q. Svejstrup. 2000. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 19:3060-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 60.Yu, Y., P. Eriksson, and D. J. Stillman. 2000. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 20:2350-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, X., and W. Herr. 2002. A regulated two-step mechanism of TBP binding to DNA: a solvent-exposed surface of TBP inhibits TATA box recognition. Cell 108:615-627. [DOI] [PubMed] [Google Scholar]