Abstract
Individual members of the E2F/DP protein family control cell cycle progression by acting predominantly as an activator or repressor of transcription. In Drosophila melanogaster the E2f1, E2f2, Dp, and Rbf1 genes all contribute to replication control in ovarian follicle cells, which become 16C polyploid and subsequently undergo chorion gene amplification late in oogenesis. Mutation of E2f2, Dp, or Rbf1 causes ectopic DNA replication throughout the follicle cell genome during gene amplification cycles. Here we show by both reverse transcription-PCR and DNA microarray analysis that the transcripts of prereplication complex (pre-RC) genes are elevated compared to the wild type in E2f2, Dp, and Rbf1 mutant follicle cells. For some genes the magnitude of this transcriptional derepression is greater in Rbf1 than in E2f2 mutants. These differences correlate with differences in the magnitude of the replication defects in follicle cells, which attain an inappropriate 32C DNA content in both Rbf1 and Dp mutants but not in E2f2 mutants. The ectopic genomic replication of E2f2 mutant follicle cells can be suppressed by reducing the Orc2, Orc5, or Mcm2 gene dose by half, indicating that small changes in pre-RC gene expression can affect DNA synthesis in these cells. We conclude that RBF1 forms complexes with both E2F1/DP and E2F2/DP that cooperate to repress the expression of pre-RC genes, which helps confine DNA synthesis to sites of gene amplification. In contrast, E2F1 and E2F2 repressors function redundantly for some genes in the embryo. Thus, the relative functional contributions of E2F1 and E2F2 to gene expression and cell cycle control depends on the developmental context.
During animal development, individual cells choose different fates based on temporal and spatial molecular cues. The integration of these cues is essential for cells to behave in a manner commensurate with their fate. An excellent example of such behavior is the execution of a specialized cell cycle program during Drosophila melanogaster oogenesis. In order to achieve the biosynthetic capacity to rapidly synthesize a mature egg, cells in the ovary progress through a complex cell cycle program that includes cell division, polyploidization, and gene amplification (26, 30, 31, 39). A Drosophila ovary contains 14 to 16 ovarioles, each of which consists of a series of connected egg chambers at successive stages of oogenesis (37). Each egg chamber contains a cyst of 16 germ cells, one of which becomes the oocyte, while the rest differentiate to form polyploid nurse cells that support oocyte growth. The germ cell cyst is encapsulated within a single epithelial layer of somatic follicle cells. Follicle cells arise from two stem cells at the anterior end of each ovariole and proliferate through six rounds of mitotic cell division to generate this epithelium. These follicle cells subsequently begin endocycles through which they attain a polyploid 16C DNA content by stage 10 of oogenesis (5, 26). At this stage, the follicle cells cease endocycling and begin synchronous amplification of several distinct genomic loci, two of which contain a cluster of chorion genes that encode eggshell proteins. Gene amplification occurs through repeated use of replication origins within these loci, and an ca. 80-fold amplification has been measured for the third chromosome chorion locus (6, 29, 37). Disruption of the follicle cell cycle program, especially chorion gene amplification, prevents proper formation of the eggshell and causes sterility (23, 24).
Transitions between different stages of the follicle cell cycle program are under developmental control. For instance, loss of Notch signaling in follicle cells prevents the normal transition from the mitotic cell cycle to the endocycle (10, 27). Ecdysone signaling also contributes to the normal patterns of endocycle S phase and gene amplification in follicle cells (43). Insight into follicle cell cycle control has also been gained through a genetic analysis of the cell cycle machinery. Hypomorphic, female sterile alleles of genes such as Orc2, Mcm6, and chiffon (DBF4), which encode proteins required for the initiation of DNA replication, reduce the level of chorion gene amplification and cause production of a thin eggshell (23, 24, 36). Regulators of the initiation of DNA synthesis, such as cyclin E/cdk2 and E2F transcription factors, also play an important role in controlling follicle cell cycles (5, 7, 32). In the present study we examine the contribution that E2F family members make to the transition from endocycles to chorion gene amplification.
Functional E2F is a heterodimer consisting of a molecule of E2F bound to a molecule of DP. E2F affects cell cycle progression by acting as both a positive and negative regulator of the G1-to-S transition via the transcriptional control of genes encoding replication factors, including prereplication complex (pre-RC) components such as ORC and MCM (8, 15, 42). In the negative role, E2F is bound to a member of the pRb family of tumor suppressors and functions in a transcriptional repressor complex (2, 15, 18, 20). These complexes are the predominant form of E2F in early G1 and quiescent cells (17). In the positive role, pRb becomes phosphorylated by G1 cyclin/cdk's in response to proliferative signals and releases E2F/DP to act as a transcriptional activator (15).
The relative importance of the activator versus repressor roles for E2F in cell cycle control in vivo is unresolved (8). The components of E2F complexes are encoded by gene families (e.g., six E2Fs, two DPs, and three pRB family proteins are found in mammals), leading to the hypothesis that there is functional specialization within the E2F family. In this model, certain E2Fs act primarily as positive regulators of the cell cycle, while others act primarily as inhibitors (9, 17, 21, 22, 34, 45). Studies in Drosophila have provided some evidence for this model. The Drosophila genome contains two E2Fs (E2f1 and E2f2), a single Dp, and two pRb (Rbf1 and Rbf2) genes (15). E2f1, Dp, and Rbf1 are each essential, whereas E2f2 is not essential for viability but is required for normal female fertility (7, 11, 13, 14, 16, 33). Phenotypic analysis of these mutants suggests the possibility that E2F2/DP participates predominantly in repression, whereas E2F1/DP may be the predominant activator. Mutation of E2f2 extends the stage of development at which E2f1-null mutants die (i.e., from larvae to pupae), indicating that these two E2Fs functionally antagonize one another in vivo (16).
E2F and RBF also function in the control of the follicle cell cycle program. Viable, hypomorphic alleles of E2f1 and Dp decrease chorion gene amplification, producing eggs with a thin shell (32). Conversely, Rbf1 mutant follicle cells overamplify chorion genes (3). Interestingly, these phenotypes may be independent of transcription because E2F1 is found at chorion loci by chromatin immunoprecipitation experiments and E2F1, DP, and RBF1 all coimmunoprecipitate with ORC2 from ovary extracts (3). However, these data do not preclude a role for transcriptional regulation by E2F complexes in other aspects of follicle cell cycle control. One such role may be to prevent the inappropriate firing of nonchorion genomic origins of replication during gene amplification. In a subset of follicle cells in Dp, Rbf1, or E2f2 mutant egg chambers, DNA synthesis is not restricted to sites of gene amplification but rather occurs throughout the nucleus (3, 7, 32). Here we present molecular and genetic evidence suggesting that E2F1/DP and E2F2/DP cooperate in transcriptional repressor complexes with RBF1 to reduce the expression of pre-RC genes in follicle cells, which helps restrict DNA replication to sites of gene amplification. In addition, our data suggest that during embryogenesis these repressors function redundantly rather than cooperatively. Therefore, activator and repressor roles are not always strictly segregated between different E2F molecules in Drosophila, and the extent to which their similar repressor functions are both required depends on the developmental context.
MATERIALS AND METHODS
Fly stocks and genetics.
The null deletion alleles E2f2329 and E2f2G5.1, and the nearly null E2f21-188 allele, were described previously (7, 16). Rbf1120 is a hypomorphic allele that results from a P-element insertion in the 5′ untraslated region of the Rbf gene, whereas Rbf114 is a null deletion allele (3, 11). Dpa1 is an ethyl methanesulfonate-induced hypomorphic Arg-to-Cys missense mutation in the conserved DEF box (33). Df(2L)vg56 is a large deletion that uncovers the Dp genomic region (13). The null alleles Orc2γ3, Orc5l(2)34BG1 and ChifRA5 were kindly provided by Mike Botchan and John Tower, respectively (23, 24). The Mcm2rl74 allele was kindly provided by Jessica Treisman (41). For the genetic interactions with E2f2, Orc5l(2)34BG1, and ChifRA5were recombined onto the E2f2329 chromosome, and the recombinants were crossed to E2f21-188/CyO. In these crosses, all E2f2 mutant females are heterozygous for Orc5 or Chif. For Orc2 and Mcm2, males of genotype E2f21-188/+;Orc2γ3/+ and E2f21-188/+;Mcm2rl74/+ were mated with E2f2329/CyO females, and ovaries were dissected from all homozygous E2f2 mutant females (identified by the lack of CyO and the presence of w+ in the E2f21-188 allele). In these crosses half of the E2f2 mutants are predicted to be heterozygous for Orc2 or Mcm2, and therefore the percent suppression was calculated by doubling the number of egg chambers that exhibit a suppressed phenotype. To express green fluorescent protein (GFP) specifically in the follicle cells of E2f2 and Rbf1 mutant ovaries using the c323 GAL4 driver (28), c323/Y; E2f2329/CyO males were mated to E2f21-188/CyO;UAS-GFP/+ females and c323-Rbf14/Dp(1;Y)y2sc males were mated to Rbf120/FM7;UAS-GFP/+females, respectively. Embryos from Rbf14 germ line clones were generated as described previously (11). E2f2-null embryos were generated by crossing E2f2G5.1/E2f2329 P[E2f2−, Mpp6+] females to sibling males of the same genotype (both of these E2f2 alleles inactivate the neighboring, essential Mpp6 gene, thus necessitating inclusion of the P element carrying wild-type Mpp6 [7]). Even though E2f2 homozygous mutant females have reduced fertility (7, 16), they produce some embryos that exhibit no overt morphological defects at the stages needed to analyze E2F-dependent gene expression. For this allele combination, ∼60% of the eggs hatch. A total of 418 progeny from the cross between E2f21-188/ +;E2f1i2/+ males and E2f2329/CyO;E2f191/TM6 females were scored for the presence of the E2f21-188/E2f2329;E2f1i2/E2f191 double mutant population. Chi-square analysis was used to determine the level of significance of the lack of the E2F double mutant population. E2F1336-805 was engineered by PCR of E2F1 cDNA with the primers 5′-TACCTGCTCGAGGAATTCATGTCGTTGCGGCTGGAGCAACAGGAG-3′ and 5′-TCGTGTGGAGGTGGCCGTACGGAC-3′ and cloned into pUAST.
BrdU labeling and in situ hybridization and immunofluorescence.
Ovaries were dissected and pulse-labeled for 1 h in Schneiders medium with bromodeoxyuridine (BrdU) as described previously (26). Anti-BrdU antibodies (Becton Dickinson) were detected in situ by using cyanine 3- or rhodamine-conjugated goat anti-mouse secondary antibodies (Jackson Laboratories). DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole) at a final concentration of 1 μg/ml for 1 min. The extent of suppression of the E2f2 mutant follicle cell BrdU phenotypes by Orc2, Mcm2, and E2f1 was determined by counting the numbers of follicle cells undergoing genomic replication within a fixed area on 5 to 10 images of stage 13 egg chambers.
Embryos were collected for 4 h at room temperature, aged until the desired stage at either 25 or 18°C, fixed in 37% formaldehyde for 5 min, and subjected to in situ hybridization with antisense probes against Mcm3, Pcna, and RnrS transcripts (13).
Dissected ovaries were fixed in 6% formaldehyde for 15 min for immunostaining. Polyclonal rabbit anti-E2F1 (1:400) and mouse monoclonal anti-RBF1 (1:4) antibodies were kindly provided by Nick Dyson (16, 40) and were detected with goat anti-rabbit rhodamine (1:1,000; Molecular Probes) and goat anti-mouse cyanine 3 (1:300; Jackson Laboratories), respectively.
Isolation and analysis of follicle cells.
Follicle cells were isolated by trypsinization and filtration of ovaries dissected from ∼100 females/per experiment (4, 7). The filtration process assures that germ line cysts, which consist of nurse cells and the oocyte interconnected by ring canals, are eliminated because they cannot pass through the mesh. Microscopic examination of Hoechst 33342-stained, dissociated follicle cell preparations indicated that >95% of the cells were single cells, and therefore were not germ cells. The majority of the remaining cells were in small aggregates of incompletely dissociated polyploid follicle cells. The rest of the cells occur in even number aggregates that represent either diploid (i.e., 2C and 4C) germ cell cystoblasts or follicle cells, which cannot be distinguished microscopically. Thus, the germ cell contribution to the follicle cell preparations is exceedingly small. Wild-type and Rbf1, E2f2, and Dp mutant females were aged 2, 2, 5, and 7 to 8 days after eclosion, respectively, before dissection in order to obtain egg chambers from all stages of oogenesis. E2f2 and Dp mutants required longer maturation times because oogenesis is delayed. Follicle cell ploidy was determined by fluorescence-activated cell sorting (FACS) analysis of Hoechst 33342-stained cell preparations by using a MoFlo high-speed molecular flow cytometer with excitation at 364 nm for Hoechst stain and at 488 nm for GFP. To quantify mRNA levels, total follicle cell RNA prepared with Trizol reagent (Gibco-BRL) was used in 30-μl reverse transcription (RT) reactions by using the TaqMan PCR core reagent kit (Applied Biosystems/Roche). Rp49 was amplified simultaneously with Orc2, Orc5, or Mcm2 in a single reaction. RT-PCR consisted of an RT reaction at 48°C for 30 min, followed by 5 min at 95°C incubation, and then 40 cycles of PCR performed at 95°C for 15 s and 60°C for 1 min. The following primers and Taqman probes were used: Rp49 forward primer, 5′-TGCTAAGCTGTCGCACAAATG-3′; Rp49 reverse primer, 5′-CGATGTTGGGCATCAGATAC-3′; Rp49 probe, TET 5′-CGCAAGCCCAAGGGTATCGACAAC-3′ TAMRA; Orc5 forward primer, 5′-GCGGATGCAGACGGTCA-3′; Orc5 reverse primer, 5′-AAATGGCCAGCAAACGATCG-3′; Orc5 probe, FAM 5′-CGCCAGAGCCAAGACCACGGA-3′ TAMRA; Orc2 forward primer, 5′-CTGGCCTCCATTGATCACAT-3′; Orc2 reverse primer, 5′-ATGGCAGCATTGTTGTGCAG-3′; Orc2 probe, FAM 5′-CCCACCACGAGAAGTTGAAGCTGCA-3′ TAMRA; Mcm2 forward primer, 5′-CCAAGCTAACGAACATCGAC-3′; Mcm2 reverse primer, 5′-CCGTGGCAAAAGACTCCTGT-3′; and Mcm2 probe, FAM 5′-TCGCCAAGATGTCACGCCCAGCTG-3′ TAMRA.
Microarray analysis.
Two RNA samples prepared from follicle cells isolated as described above were obtained from control (one E2f2329/CyO and one yw67), E2f2 mutant (both E2f2329/E2f2329), and Rbf mutant (both Rbf14/Rbf120) females. RNA quality was assessed by nondenaturing agarose gel electrophoresis of rRNA bands. cDNA was synthesized by using a T7-(dT)24 primer and 7 μg of total RNA (Life Technologies). Biotinylated cRNA was generated from the cDNA by using the BioArray High Yield RNA transcript kit and subsequently fragmented in 40 mM Tris-acetate (pH 8.1), 100 mM potassium acetate, and 30 mM magnesium acetate at 94°C for 35 min. Then, 15 μg of fragmented cRNA was hybridized to a Drosophila genome array (part 900335; Affymetrix) in 0.05 μg of fragmented cRNA/μl; 50 pM control oligonucleotide B2; BioB, BioC, BioD, and cre hybridization controls at 1.5, 5, 25, and 100 pM, respectively; 0.1 mg of herring sperm DNA/ml; 0.5 mg of acetylated bovine serum albumin/ml; 100 mM morpholineethanesulfonic acid; 1 M NaCl; 20 mM EDTA; and 0.01% Tween 20. Arrays were hybridized for 16 h in a GeneChip Fluidics Station 400 and were washed and scanned with the Hewlett-Packard GeneArray scanner. During the washing, the cRNA probe was labeled with R-phycoerythrin streptavidin. Affymetrix GeneChip microarray suite 5.0 software was used for washing, scanning, and basic analysis. Sample quality was confirmed by examination of the 3′ to 5′ intensity ratios of certain genes.
Statistical analysis.
The significant differences between the averages of pairwise comparisons of the wild type and the E2f2 and Rbf1 mutants for pre-RC genes and replication genes in microarray experiments were determined with the sign rank test (44). The sign rank test is nonparametric and uses both ranks and signs of differences. To assess the significance in comparisons between genotypes in RT-PCR experiments, an unpaired, two-tailed Student t test was applied by using Microsoft Excel.
RESULTS
Mutation of Dp and Rbf1 causes similar ectopic follicle cell replication phenotypes that differ from the one caused by mutation of E2f2.
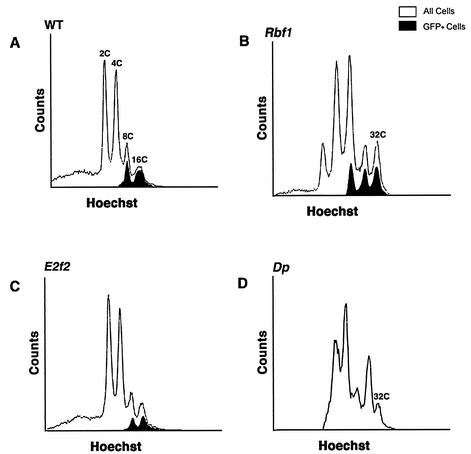
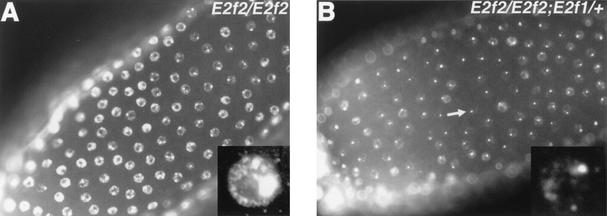
In stage 10b and older E2f2, Dp, and Rbf1 mutant egg chambers, BrdU incorporation is detected at sites of chorion gene amplification and also ectopically throughout the follicle cell nuclei. We previously postulated that this ectopic genomic replication is caused by the loss of E2F2/DP/RBF1 complexes (7). This was based both on the similarity in the BrdU incorporation phenotype among these mutant follicle cells (3, 7, 32) and on the observation that E2F2, DP, and RBF1 proteins coimmunoprecipitate (3, 16, 35, 40). The isolation of relatively pure populations of individual follicle cells from trypsinized ovaries (4) allows the determination of DNA content by flow cytometry and provides a way to evaluate the extent of DNA replication in addition to BrdU labeling. FACS analysis of wild-type follicle cells by this technique results in four distinct populations ranging from 2C to 16C (Fig. 1A). The 8C and 16C cell populations result from endocycles. In some experiments UAS-GFP was expressed in these cells by using a GAL4 driver (c323) that is activated specifically in follicle cells beginning at stage 9 (28), allowing us to detect all stage 9 and older follicle cells (Fig. 1A) (7). When this technique was applied to Rbf1 mutants, a 32C population of GFP+ follicle cells was observed (Fig. 1B). This indicated that the ectopic replication seen in stage 10b and older Rbf1 mutant egg chambers by BrdU labeling results in a complete round of S phase, as previously reported (3).
FIG. 1.
Increased ploidy in Rbf1 and Dp mutant follicle cells. Each panel contains a FACS profile of follicle cells isolated from different genotypes. Open traces indicate total cell population. Filled traces indicate the GFP+ subset of the total population that was marked by using the c323 GAL4 driver. (A) Wild-type follicle cells. Four distinct populations that represent 2C, 4C, 8C, and 16C DNA content were evaluated. In this experiment all follicle cells are genetically c323-GAL4/+ and UAS-GFP/+. Because of asynchrony in endocycles, some cells attain 8C and 16C before GFP is activated in stage 9. (B) Rbf120/Rbf14 mutant cells. (C) E2f2329/E2f2329 mutant cells. (D) Df(2R)vg56/Dpa1 mutant cells. In panels B to C only half of the follicle cell population is genetically UAS-GFP/+. Note that the 32C population is present in both Rbf1 and Dp mutants but not in the E2f2 mutants. The 32C population in Dp mutant cells was positively identified in five independent experiments.
If E2F2/DP/RBF1 complexes were the only E2F complexes that repressed genomic DNA synthesis in follicle cells, then the replication phenotype caused by mutation of each individual subunit should be indistinguishable by using different assays. However, E2f2 mutants do not contain a 32C population of follicle cells (Fig. 1B), even though the ectopic BrdU labeling phenotype is similar to Rbf1 (see Fig. 5B) (7). What could account for this phenotypic difference? One possibility is that the extent of ectopic DNA synthesis in E2f2 mutants (as detected by BrdU incorporation) is not as great as in Rbf1 mutants and does not result in a complete, additional round of S phase. This could occur if RBF1 is a component of two functionally nonredundant repressor complexes: one containing E2F1/DP and the other containing E2F2/DP. Accordingly, knocking out both complexes by mutation of Rbf1 would cause a more extensive genomic replication compared to knocking out a single RBF1 complex by mutation of E2f2. To test this hypothesis, we characterized follicle cells isolated from females that were hemizygous for a viable, female sterile allele of Dp (32). Since DP binds both E2F1 and E2F2 (16, 35), the Dp mutant phenotype should be more like Rbf1 than like E2f2. Indeed, a 32C population of Dp mutant follicle cells was detected by flow cytometry (Fig. 1D), although the peak was not as prominent as with Rbf1 mutants. This finding is consistent with the observation that the Dp ectopic genomic replication phenotype is not fully penetrant (32). These data support a model in which both E2F1/DP and E2F2/DP form repressor complexes with RBF1 that cooperate in suppressing the activity of nonchorion genomic replication origins after stage 10.
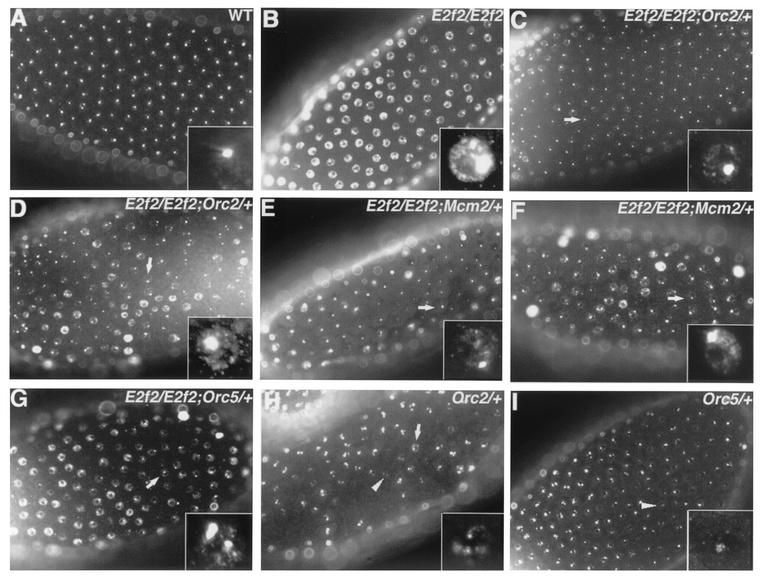
FIG. 5.
Suppression of E2f2 mutant follicle cell cycle phenotype by heterozygosity for Orc2, Orc5, or Mcm2. Each panel contains a stage 13 egg chamber labeled with BrdU. (A) Wild type. (B) E2f2329/E2f21-188 mutant. (C and D) E2f2329/E2f21-188;Orc2γ3/+ mutant. (E and F) E2f2329/E2f21-188;Mcm2rL74/+ mutant. (G) Orc5l(2)34BG1-E2f2329/E2f21-188 mutant. The arrows in panels C to G indicate follicle cells that are either not undergoing genomic replication or exhibit a reduced intensity of BrdU incorporation compared to E2f2 mutants. (H) Orc2γ3/+ mutant. (I) Orc5l(2)34BG1/+ mutant. Slight defects in chorion gene amplification are observed in Orc2γ3/+ and Orc5l(2)34BG1/+ heterozygous egg chambers. The arrow indicates a follicle cell undergoing aberrant genomic replication, and the arrowheads indicate reduced chorion gene amplification.
We attempted to corroborate our hypothesis that E2F1 and E2F2 perform overlapping repressor roles in follicle cells by making E2f1-E2f2 double mutants. However, the correct double-mutant genotype was never recovered (P < 0.001), even though the individual E2f2329/E2f21-188 null and E2F1i2/E2F191 hypomorphic genotypes used are each viable on their own. E2f191 is a null allele (14), and E2f1i2 synthesizes a truncated protein that lacks the RBF1 interaction domain at the COOH terminus (3). Thus, this E2f1-E2f2 allele combination is predicted to disrupt both RBF1 complexes and consequently phenocopies the lethality caused by Rbf1-null alleles (11). Therefore, the ability of RBF1 to form repressor complexes with both E2Fs may be necessary for Drosophila development.
E2F2 acts late in follicle cell development to suppress genomic replication.
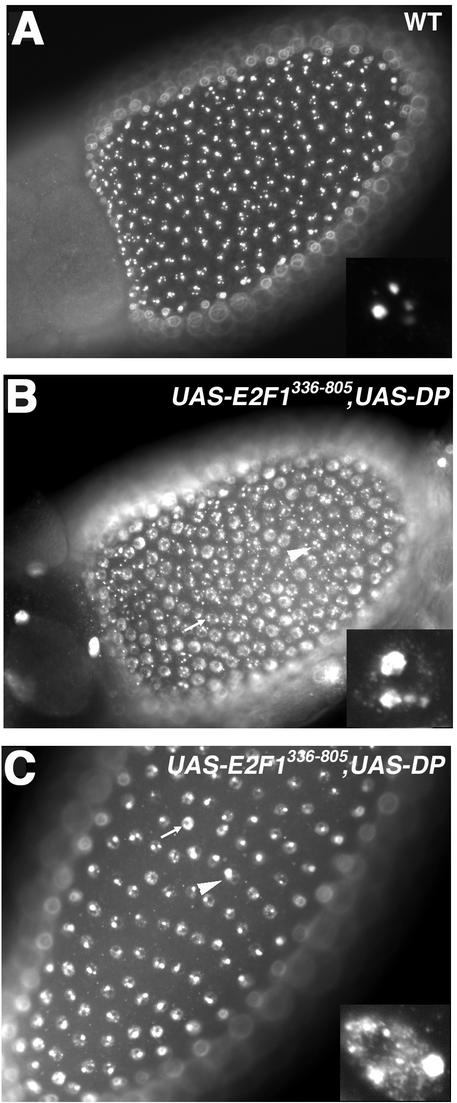
To investigate when during follicle cell development E2F2 functions, UAS-E2F2 was expressed in E2f2 mutant follicle cells by using the c323-GAL4 driver. In these females, there was no evidence of ectopic genomic replication in stage 10 and older egg chambers, as assayed by BrdU incorporation, indicating virtually 100% rescue of the mutant phenotype (not shown). This result suggests that E2F2 acts to inhibit genomic replication when follicle cells make the transition from endocycling to chorion gene amplification cycles. If the inhibition of genomic replication requires E2F2 to act as part of an RBF1 repressor complex, and/or other RBF1 containing complexes, then disruption of these complexes starting at stage 9 should cause an ectopic replication phenotype. Disruption of RBF/E2F complexes was achieved by overexpressing DP with a mutant E2F1 protein (E2F1336-805) lacking the DNA-binding domain but retaining the RBF interaction domain. Coexpression was used because E2F/DP heterodimers bind RB proteins more efficiently than E2F or DP monomers (12, 19). Coexpression of E2F1336−805 and DP activates E2F target gene expression in embryos, probably by titrating RBF, whereas expresion of either one alone does not (D. Lanzotti and R. Duronio, unpublished results). Expression of E2F1336-805 and DP with c323-GAL4 in follicle cells resulted in ectopic genomic DNA replication similar to that seen in E2f2 and Rbf1 mutants (Fig. 2). We interpret this as an indication that disruption of RBF complexes beginning at stage 9 is sufficient to trigger ectopic genomic replication.
FIG. 2.
E2F repressor complexes act late in follicle cell development. Each panel contains a BrdU-labeled egg chamber. (A) Stage 10 wild type; (B and C) stage 10 and stage 13 c323-GAL4/+;UAS-E2F1336-805 and UAS-DP/+, respectively. See Fig. 5A for a wild-type stage 13. The arrowheads and arrows indicate overamplification and aberrant genomic replication in follicle cells, respectively.
Genomic replication silencing in follicle cells relies on E2F-RBF transcriptional repression.
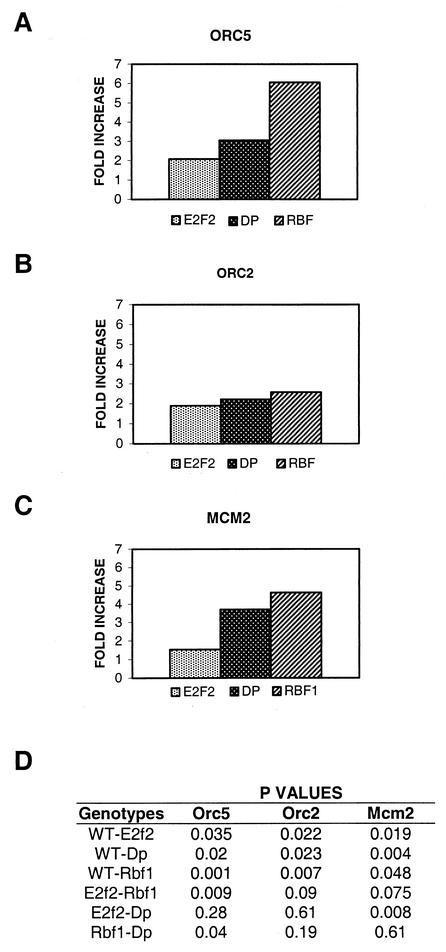
The mechanism by which E2F transcription factors regulate the cell cycle is thought to occur through changes in the expression of target genes (8, 15, 42). To determine whether this plays any role in the control of DNA synthesis in follicle cells, we analyzed the level of expression of various replication factor and pre-RC genes by using total cellular RNA isolated from follicle cells prepared from wild-type and E2f2, Rbf1, and Dp mutant ovaries (see Materials and Methods). Specific mRNAs were detected by using real-time fluorescence RT-PCR, which allows accurate quantification of differences among independent PCR products by measuring the fluorescence emitted from each DNA molecule as it is synthesized. Mutation of E2f2 resulted in a twofold increase in the expression of the Orc5 transcript relative to wild type (Fig. 3A), a finding that is in agreement with our previous results (7). In Dp and Rbf1 mutant follicle cells the amounts of Orc5 message were approximately three- and sixfold greater, respectively, than in the wild type (Fig. 3A). The level of Orc2 message increased approximately 2-fold in E2f2 mutants and 2.5-fold in both Dp and Rbf1 mutants (Fig. 3B). Similarly, Mcm2 transcript levels increased 1.5-fold in E2f2, 3.5-fold in Dp, and 4.5-fold in Rbf1 mutant follicle cells relative to the wild type (Fig. 3C). To evaluate these differences, P values were generated by using a Student t test (Fig. 3D). In each case the measurements made in the mutant genotypes were significantly different than in the wild type (P < 0.05). These data support the hypothesis that an increase in the steady-state level of pre-RC components contributes to the appearance of ectopic genomic DNA synthesis in the mutants. Moreover, the level of these transcripts were higher in Rbf1 mutants compared to E2f2 (P < 0.1), suggesting that both E2F1/DP/RBF1 and E2F2/DP/RBF1 repressor complexes are involved in the proper downregulation of these genes.
FIG. 3.
Orc2, Mcm2, and Orc5 transcripts are elevated in E2f2, Dp, and Rbf1 mutant follicle cells. Relative levels of Orc5 (A), Orc2 (B), and Mcm2 (C) transcripts were determined in wild-type and in E2f2329/E2f2329, Df(2R)vg56/Dpa1, and Rbf120/Rbf14 mutant total follicle cell RNA preparations by the TaqMan quantitative RT-PCR method. The PCR product was quantified by determining the fluorescence intensity at one cycle in the linear phase of the amplification. The fluorescence values of Orc5, Orc2, and Mcm2 were compared between different genotypes by normalizing them to the value obtained for Rp49. All experiments were performed in triplicate, except for Orc5 in wild-type, E2f2, and Rbf1 genotypes, which were performed four times. The data are presented as an average of the fold increases relative to the wild type for each of these experimental trials. (D) An unpaired, two-tailed Student t test was used to determine level of significance of changes in expression between different genotypes. P values for each pairwise combination of experiments is shown.
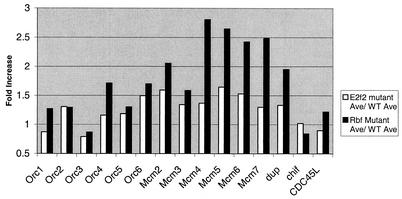
To determine whether an upregulation of expression applies to many components of the pre-RC, expression profiles were obtained from the wild-type and from the E2f2 and Rbf1 mutant follicle cells by using Affymetrix gene chips. The data are presented in Fig. 4 as a ratio of mutant/wild-type gene expression obtained from an average of two independent experiments for each genotype (see Materials and Methods). In general, pre-RC gene expression was elevated in both E2f2 and Rbf1 mutants compared to the wild type (P < 0.0003 and 0.000004, respectively). In addition, the levels of these transcripts in Rbf1 mutants were elevated compared to the E2f2 mutants (P < 0.0003). On average, Mcm and Orc gene expression was increased by 1.5- and 1.2-fold in E2f2 mutants and by 2.4- and 1.4-fold, respectively, in the Rbf1 mutants (Fig. 4 and Table 1). These data correlate with the RT-PCR data, although the magnitude of the increases as measured by the microarray experiments is less than that measured by real-time RT-PCR. The basis for this difference is not clear, although it could represent differences in sensitivity between the two techniques. Nevertheless, these data suggest that multiple genes encoding components of the pre-RC are subject to E2F2/RBF1 transcriptional repression in follicle cells, that these genes are regulated to different degrees, and that mutation of E2f2 and Rbf1 alter gene expression differently.
FIG. 4.
Prereplication component genes are elevated in E2f2 and Rbf1 mutant follicle cells. Each bar indicates the ratio of mutant to wild-type hybridization signal obtained from an average of two independent experiments for each follicle cell genotype.
TABLE 1.
Replication genes are derepressed in E2f2 and Rbf1 mutantsa
| Complex or factor | Ratio of mutant/wild-type hybridization signal
|
|
|---|---|---|
| E2f2 mutant | Rbf1 mutant | |
| Replication initiation complex | ||
| Orc1 | 0.9 | 1.3 |
| Orc2 | 1.3 | 1.3 |
| Orc3 (lat) | 0.8 | 0.9 |
| Orc4 | 1.2 | 1.7 |
| Orc5 | 1.2 | 1.3 |
| Orc6∗ | 1.5 | 1.7 |
| Mcm2∗ | 1.6 | 2.1 |
| Mcm3 | 1.3 | 1.6 |
| Mcm4 (dpa) | 1.4 | 2.8 |
| Mcm5∗ | 1.6 | 2.6 |
| Mcm6∗ | 1.5 | 2.4 |
| Mcm7 | 1.3 | 2.5 |
| dup (CBT1) | 1.3 | 2.0 |
| chif (DBF4) | 1.0 | 0.8 |
| CDC45L | 0.9 | 1.2 |
| Replication factors | ||
| awd (NDP kinase) | 0.9 | 0.9 |
| Dhfr | 1.0 | 1.2 |
| deoxynucleoside kinase (dnk) | 1.1 | 2.9 |
| nmdyn-D6 (NDP kinase) | 0.7 | 0.6 |
| nmdyn-D7 (NDP kinase) | 1.5 | 0.8 |
| RnrL | 1.3 | 2.4 |
| RnrS | 1.2 | 2.5 |
| Ts | 1.3 | 1.5 |
| PCNA (mus209) | 1.1 | 2.2 |
| CG10262 (PCNA2) | 0.5 | 0.6 |
| CG8142∗ (RfC) | 1.6 | 1.6 |
| Gnf1 (RfC) | 1.5 | 1.3 |
| RfC3 | 0.7 | 1.0 |
| RfC38 | 1.4 | 1.5 |
| RfC40 | 1.0 | 1.0 |
| DNApol-α180 | 1.2 | 1.4 |
| DNApol-α50∗ | 2.0 | 2.8 |
| DNApol-α73∗ | 1.7 | 1.9 |
| DNApol-δ | 1.3 | 0.8 |
| CG10489 (DNApol-ɛ) | 0.6 | 0.7 |
| DNApol-ɛ∗ | 1.7 | 1.7 |
| DNApol-γ35 | 0.8 | 0.9 |
| DNAprimase | 1.3 | 4.0 |
| RpA-30 | 1.0 | 1.7 |
| RpA-70 | 1.1 | 2.0 |
| Chromatin assembly | ||
| Acf1∗ | 1.6 | 1.5 |
| Caf1-105 | 1.2 | 1.4 |
| Caf1-180∗ | 1.6 | 2.8 |
| Caf1 | 1.2 | 1.4 |
| Iswi | 1.5 | 1.1 |
| Chromatid cohesion | ||
| Cap (SMC3) | 0.8 | 0.9 |
| mei-S332∗ | 1.8 | 2.3 |
| SA | 1.1 | 1.3 |
| SMC1 | 0.9 | 1.2 |
The data are presented as a ratio of mutant to wild-type hybridization signals obtained from an average of two independent DNA microarray experiments for each follicle cell genotype. Genes that exhibited a ≥1.5-fold increase in both E2f2 and Rbf1 mutant follicle cells are indicated by an asterisk.
To investigate whether there was an impact on the expression of other replication factors, 49 genes encoding various functions required for chromosome duplication during S phase were identified from the annotated Drosophila genome database, and their levels of expression were assessed by microarray analysis (Table 1). As with the pre-RC subset of this group, gene expression was in general elevated in both E2f2 and Rbf1 mutants compared to the wild type (P < 0.004 and < 0.0009, respectively), and gene expression in the Rbf1 mutants was elevated compared to E2f2 mutants (P < 0.002). Of the 49 replication genes listed in Table 1, 14 genes were upregulated by ≥1.5-fold in E2f2 mutants and 25 genes were upregulated by ≥1.5-fold in Rbf1 mutants. Of these, 11 were upregulated by ≥1.5-fold in both mutants (asterisks, Table 1). The fold increase in these 11 common genes ranged from 1.5 to 2.0 in E2f2 mutants and from 1.5 to 2.8 in Rbf1 mutants. These data indicate that replication factor gene transcription in follicle cells is regulated by E2F2 and RBF1 and are consistent with a model whereby the increased amounts of gene expression in Rbf1 versus E2f2 mutants contributes to the greater severity of the Rbf1 overreplication phenotype.
Reduction of the pre-RC gene dose suppresses ectopic genomic replication in E2f2 mutants.
The changes in replication factor gene expression detected in the E2f2 and Rbf1 mutant follicle cells were modest. To test whether small increases in pre-RC gene expression could affect the regulation of follicle cell DNA synthesis, we sought to determine whether a 50% reduction in gene dose of certain replication factors could suppress the ectopic genomic replication phenotype in E2f2 mutants. To test this, BrdU incorporation was analyzed in E2f2 mutant follicle cells that were also heterozygous for null alleles of Orc2, Orc5, Chiffon (Dbf4), or Mcm2. Stage 13 E2f2 mutant egg chambers were scored because at this stage nearly all follicle cells within a given egg chamber exhibit ectopic genomic replication, and this phenotype is fully penetrant (Fig. 5A and B). Orc2 or Mcm2 heterozygosity in an E2f2 mutant background resulted in suppression of the phenotype in ∼70% of the egg chambers (Fig. 5C to F). Suppression is evidenced by an approximate 65% reduction in the number of follicle cells within an egg chamber that display ectopic genomic replication (Fig. 5C and F) and by an apparent reduction in the intensity of the ectopic BrdU incorporation compared to E2f2 mutant follicle cells (Fig. 5D and F). Reduction of the Orc5 gene dose suppressed the phenotype in 36% of the stage 13 E2f2 mutant egg chambers (Fig. 5G). We were not able to detect any obvious suppression by using mutations in Chiffon (data not shown). These experiments suggest that inhibition of genomic origins of replication during gene amplification cycles is sensitive to twofold changes in the levels of ORC2, MCM2, and ORC5.
In contrast to the results obtained with E2f2, the ectopic genomic replication phenotype in Rbf mutant follicle cells was not suppressed by Orc2 heterozygosity (data not shown). This suggests that reducing the ORC2 levels by half cannot overcome the extent of transcriptional derepression in the Rbf1 mutant and is consistent with the greater severity of the Rbf1 overreplication phenotype relative to E2f2.
Taken together, these results indicate that relatively small changes in the amount of pre-RC components can affect replication control in follicle cells. This suggests that simple heterozygosity of some pre-RC components may affect DNA replication in follicle cells. To test this possibility, we examined BrdU incorporation patterns in stage 13 Orc5/+ and Orc2/+ egg chambers. In both genotypes, the intensity of BrdU-labeled foci was slightly reduced in certain follicle cells (Fig. 5H and I). Orc2/+ replication patterns were more abnormal than those in Orc5/+ egg chambers, displaying a more obvious cytological reduction in the replication of chorion gene clusters, as well as slight genomic replication in some follicle cells. These data further support the hypothesis that follicle cell DNA synthesis during amplification cycles is sensitive to fairly small changes in the cellular levels of pre-RC components.
E2F1 activator function stimulates the genomic replication phenotype observed in E2f2 mutant follicle cells.
In addition to functioning in conjunction with RBF1 in transcriptional repression, E2F1 also acts to stimulate replication factor gene expression when not bound to RBF1. Therefore, two factors could influence the extent of ectopic genomic replication in E2f2 mutant follicle cells. The first is loss of E2F2/DP/RBF1 repressor complexes, and the second is activation mediated by E2F1/DP complexes. If the E2f2 ectopic replication phenotype requires input from E2F1-mediated activation, then reducing the level of E2F1 function may suppress the phenotype. BrdU labeling revealed that 55% of stage 13 E2f2 mutant egg chambers that were also heterozygous for a E2f1 null allele contained ∼50% reduction in the number of follicle cells exhibiting ectopic genomic replication compared to E2f2 mutants (Fig. 6). This suggests that a shift in the endogenous complexes toward formation of E2F1/DP activators might be contributing to the E2f2 mutant phenotype by stimulating the transcription of replication genes in the absence of functional E2F2/DP/RBF1 repressor complexes. An increase in the level of E2F1 protein in E2f2 mutant follicle cells does not appear to contribute to this shift, since the level of E2F1 protein detected by immunostaining of whole egg chambers appeared indistinguishable between wild-type and E2f2 mutant follicle cells (data not shown).
FIG. 6.
Suppression of E2f2 mutant follicle cell cycle phenotype by mutations in E2f1. Both panels contain a stage 13 egg chamber pulse-labeled with BrdU. (A) E2f2329/E2f21-188 mutant; (B) E2f2329/E2f21-188; E2f91/+ mutant. The arrow indicates a follicle cell with a reduced level of BrdU incorporation compared to E2f2 mutants.
Developmental differences in the action of E2F1 and E2F2 repressors.
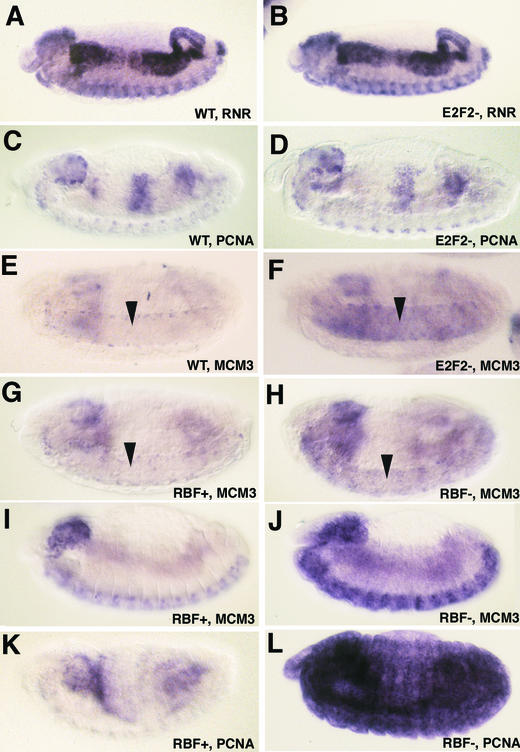
To determine whether E2F1 and E2F2 repressor activities cooperate at other times or in other tissues during development, the expression of E2F target genes was analyzed during embryogenesis. RnrS and Pcna are both expressed in late embryos coincidentally with replicating cells in a pattern that results from activation and repression mediated by E2F1 and RBF1 (Fig. 7A and C). In E2f1 and Dp mutant embryos, the expression of these two genes is lost (13, 14, 33) and, in embryos lacking Rbf1, both are hyperactivated (11) (shown for Pcna in Fig. 7L). Consequently, whereas E2F1 provides activator function for these two genes in the embryo, the repression of these genes is not affected by mutation of E2f1. To determine the contribution of E2F2, we generated embryos lacking both maternal and zygotic E2f2 (see Materials and Methods). Both the RnrS and Pcna expression pattern appeared similar between E2f2 mutant and wild-type embryos (Fig. 7B and D). The same result was obtained with Mcm2 (not shown). These data suggest that in the embryo E2F1 can alone provide both activation and repression activities for these genes in the absence of E2F2.
FIG. 7.
E2F1 and E2F2 repressors primarily act redundantly during embryogenesis. Each panel contains a germ band retracted embryo subject to in situ hybridization with the indicated probe. Anterior is at left, and dorsal at the top. (A, C, and E) Wild-type embryos. Panels A and C contain embryos at different stages in order to show two different representative patterns of the replication-asssociated gene expression program. (B, D, and F) Embryos lacking both maternal and zygotic E2f2. Note that the pattern of expression is identical to that of the wild type for RnrS and Pcna but not for Mcm3, which is derepressed in the CNS (arrows). The embryos in panels E and F are rotated slightly to the ventral surface. (G, I, and K) Phenotypically normal sibling control embryos of those shown in panels H, J, and L, respectively. (H, J, and L) Embryos lacking maternal and zygotic Rbf1 function. The focal plane in panels I to L is at the epidermis, rather than the interior tissues as in panels A to H, in order to show the ectopic expression in this tissue.
We previously observed that Mcm3 expression is derepressed in Dp mutant embryos in quiescent cells of the central nervous system (CNS) but not in quiescent epidermal cells (13). Both the loss of E2f2 (Fig. 7E and F) and the loss of Rbf1 (Fig. 7G and H) function cause a Mcm3 derepression phenotype in the CNS similar to Dp mutants, suggesting that an E2F2/DP/RBF1 complex participates in Mcm3 repression here. Because E2f1 is required for embryonic Mcm3 expression in the CNS (13), this result suggests that for some genes in the embryo E2F1 and E2F2 do not act redundantly, as is the case in follicle cells. Interestingly, the loss of Rbf1 causes detectable ectopic Mcm3 expression in the epidermis (Fig. 7I and J), although it is not as obvious as with Pcna (Fig. 7K and L). Since neither mutation of E2f1 or E2f2 causes ectopic epidermal expression of these genes, these data suggest that E2F1 and E2F2 act redundantly as repressors in this tissue. Therefore, the extent to which E2F1 and E2F2 act redundantly or cooperatively as repressors is tissue specific.
DISCUSSION
The relative contribution of the E2F activator versus repressor functions to cell cycle control in specific tissues during development is not completely understood. Previous data have suggested that Drosophila E2F1 primarily functions as an activator and that E2F2 functions as a repressor of transcription and consequently cell cycle progression (7, 16). Here we provide evidence that E2F1 and E2F2 are both required in ovarian follicle cells to repress transcription and inhibit DNA synthesis and therefore can also function in a similar way. Moreover, our analysis indicates that the requirements for E2F1 and E2F2 in embryos are not the same in every tissue.
During oogenesis follicle cells execute a complex cell cycle program that includes canonical mitotic cycles, endocycles, and finally chorion gene amplification (38). Chorion gene amplification requires two types of regulation: (i) the repeated firing of replication origins within chorion gene clusters and (ii) prevention of replication initiation at other genomic origins. Genes known to affect the first process encode components of the replication initiation complex (ORC proteins, Chiffon, and MCMs) (23, 24, 36), as well as E2F1, DP, and RBF1, which are likely bound to chromatin near chorion origins and regulate gene amplification independently of transcription (3, 33). The second process is also under E2F regulation, and mutations in E2f2, Dp, or Rbf1 result in ectopic genomic replication in follicle cells at a stage when only chorion origins should be active (3, 7, 33). However, genomic replication is more extensive in Rbf1 and Dp mutant follicle cells, which attain a 32C DNA content, than in E2f2 mutants, which do not. One possible explanation for this difference is that the complete inactivation of genomic replication is achieved by the cooperative activity of E2F2/DP/RBF1 and E2F1/DP/RBF1 repressor complexes. E2f2 loss of function disrupts only one of these complexes, whereas the loss of Rbf1 or Dp disrupts both. Therefore, the presence of E2F1 containing repressor complexes constrains the extent of overreplication in the E2f2 mutant cells. Since in mammalian cells E2Fs can regulate pRB levels (8, 15), another possibility is that loss of E2f2 causes an increase in the level of RBF1 protein, thereby driving the formation of more E2F1/RBF1/DP repressor complexes and making the E2f2 mutant phenotype less severe than Rbf or Dp. However, we consider this unlikely since there was no detectable difference in RBF1 immunostaining of E2F2 mutant follicle cells compared to wild type (not shown). The same result was obtained by immunoblotting of protein extracts from wild-type or E2f2 mutant larvae (40).
A large body of evidence indicates that the mechanism by which E2F complexes control DNA replication is via transcription (8, 15). Here and in our previous report (7) we demonstrate that E2f2 mutant follicle cells contain a two- to threefold increase in transcripts encoding pre-RC proteins such as Orc2, Orc5, and Mcm2 relative to wild type. This small increase appears to be significant, as a reduction of one functional copy of the Orc2, Orc5, or Mcm2 gene substantially suppressed the incidence of ectopic genomic replication in E2f2 mutant follicle cells. This suggests that increases in the levels of pre-RC components in the absence of E2F/RB repressors may increase the number of active origins, thereby causing ectopic DNA synthesis. In vitro studies have shown that activation of mammalian replication origins is sensitive to the ratio of initiation factors to DNA substrate, which in turn could regulate the number of origins activated (25). Previous cytological observations made by immunodetection of pre-RC subunits also support this model. In wild-type ovaries, ORC subunits are detected throughout the follicle cell nucleus during endocycles but are subsequently localized to chorion loci during gene amplification (33). In contrast, ORC proteins are not localized to sites of gene amplification in E2f2, Dp, and Rbf1 mutant follicle cells but are found throughout the nucleus (3, 7, 32). This suggests that pre-RC protein levels are inappropriately elevated. Similarly, a twofold increase in ORC1 protein levels is sufficient to cause mislocalization throughout the nucleus during gene amplification stages and also results in genomic replication (1). Taken together, these data suggest that transcriptional inhibition of pre-RC genes by both E2F/DP/RBF1 and E2F2/DP/RBF1 complexes contributes to the proper restriction of DNA synthesis to sites of gene amplification. The E2f2 replication phenotype is completely rescued by initiating expression of wild-type E2F2 at stage 9, suggesting that E2F2 can exert its effect just prior to the onset of chorion gene amplification. Consequently, the downregulation of transcription must result in a decrease of pre-RC proteins below a threshold necessary to support genomic replication between stages 9 and 10 (∼6 h).
Contrasting and overlapping roles for E2F family members during Drosophila development.
Recent experiments have indicated that E2F1 and E2F2 functionally oppose one another rather than act similarly (16). First, overexpression of E2F2 suppresses the rough eye phenotype caused by overexpression of E2F1. Second, E2f1 and E2f2 double mutants progress to a later stage of development (pupae) than do E2f1 single mutants (larvae), indicating that E2F2 contributes to the earlier lethality of E2f1 mutants. How can these results be reconciled with our observation in follicle cells? One possibility is that E2F2 acts as a dedicated repressor and is never an activator, whereas E2F1 can function as both an activator and a repressor. In this scenario, E2F2 can both antagonize E2F1 activator function and act redundantly with E2F1 for repression. The relative contribution of these interactions to gene expression and cell cycle control will depend on the genes being analyzed and the developmental context.
This model is supported by our analysis of different tissues. A dual role for E2F1 is evident in the follicle cells, where the reduction of the E2f1 gene dose in an E2f2 mutant background partially suppressed the ectopic replication phenotype, representing a positive contribution to replication by E2F1/DP. In the embryo, the replication-associated pattern of RnrS and Pcna expression requires E2f1, Dp, and Rbf1 but not E2f2. Because a second Drosophila pRB homolog called RBF2 only binds to E2F2 (40), the simplest interpretation is that the activation and repression of transcription responsible for this pattern is predominantly generated by E2F1 complexes. Moreover, the fact that RBF1 loss causes derepression of RnrS, Pcna, and Mcm3 in the epidermis but mutation of either E2f1 or E2f2 alone does not suggests that E2F1 and E2F2 can act redundantly to repress these genes in the embryo. This is consistent with biochemical data indicating that RBF1 coimmunoprecipitates with both E2F1 and E2F2 (16, 40). A similar redundancy in the inhibitory activities of E2F-1 and E2F-2 in T cells (46) and of E2F-4 and E2F-5 in embryonic fibroblasts (17) also occurs in the mouse.
Interestingly, redundancy between E2F1 and E2F2 repression of Mcm3 does not occur in all embryonic tissues. Mcm3 transcription is derepressed in the CNS of E2f2, Dp, and Rbf1 mutant embryos but not in E2f1 mutant embryos (13). These gene-specific requirements for the functions of various E2F complexes may reflect differences in how the cell cycles are controlled during development. That is, although the follicle cells rely on inhibiting pre-RC gene expression for replication control, this mode of regulation is not necessary for embryonic cell cycle control, including in the CNS. Thus, the relative contributions of individual E2F repressor and activator functions to gene expression and cell cycle regulation depend on the developmental context.
Acknowledgments
We thank Mike Botchan, Brian Calvi, Nick Dyson, John Tower, Jessica Treisman, and Terry Orr-Weaver for reagents; Larry Arnold for assistance with FACS analysis; Mike Vernon of the UNC Affymetrix Core Facility for processing the microarrays; Harry Hurd for contributions to statistical analysis of microarray data; Hyung-Suk Kim for helping with Taqman analysis; and Melissa Adams, Mitch McVey, and Bill Marzluff for comments on the manuscript.
S.C.S.K. is a Seeding Postdoctoral Innovators in Research and Education fellow supported by GM000678 funded through the Minority Opportunities in Research Division of NIH-GM. This work was supported by NIH training grant T32-CA71341-06 to W.O.W. and NIH grant GM57859 to R.J.D.
REFERENCES
- 1.Asano, M., and R. P. Wharton. 1999. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 18:2435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beijersbergen, R. L., R. M. Kerkhoven, L. Zhu, L. Carlee, P. M. Voorhoeve, and R. Bernards. 1994. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 8:2680-2690. [DOI] [PubMed] [Google Scholar]
- 3.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, Z., L. Subrahmanyan, M. Tworoger, L. LaTray, C. R. Liu, M. J. Li, G. van den Engh, and H. Ruohola-Baker. 1999. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc. Natl. Acad. Sci. USA 96:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvi, B. R., M. A. Lilly, and A. C. Spradling. 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12:734-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvi, B. R., and A. C. Spradling. 1999. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18:407-417. [DOI] [PubMed] [Google Scholar]
- 7.Cayirlioglu, P., P. C. Bonnette, M. R. Dickson, and R. J. Duronio. 2001. Drosophila E2F2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development 128:5085-5098. [DOI] [PubMed] [Google Scholar]
- 8.DeGregori, J. 2002. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim. Biophys. Acta 1602:131-150. [DOI] [PubMed] [Google Scholar]
- 9.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, W. M., C. Althauser, and H. Ruohola-Baker. 2001. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128:4737-4746. [DOI] [PubMed] [Google Scholar]
- 11.Du, W., and N. Dyson. 1999. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 18:916-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, W., M. Vidal, J. E. Xie, and N. Dyson. 1996. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10:1206-1218. [DOI] [PubMed] [Google Scholar]
- 13.Duronio, R. J., P. C. Bonnette, and P. H. O'Farrell. 1998. Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the G1-S transition. Mol. Cell. Biol. 18:141-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duronio, R. J., P. H. O'Farrell, J. E. Xie, A. Brook, and N. Dyson. 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 9:1445-1455. [DOI] [PubMed] [Google Scholar]
- 15.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 16.Frolov, M. V., D. S. Huen, O. Stevaux, D. Dimova, K. Balczarek-Strang, M. Elsdon, and N. J. Dyson. 2001. Functional antagonism between E2F family members. Genes Dev. 15:2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 18.Ginsberg, D., G. Vairo, T. Chittenden, Z. X. Xiao, G. Xu, K. L. Wydner, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 8:2665-2679. [DOI] [PubMed] [Google Scholar]
- 19.Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7:1850-1861. [DOI] [PubMed] [Google Scholar]
- 20.Hijmans, E. M., P. M. Voorhoeve, R. L. Beijersbergen, L. J. van 't Veer, and R. Bernards. 1995. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol. Cell. Biol. 15:3082-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690-703. [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, D. G., J. K. Schwarz, W. D. Cress, and J. R. Nevins. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365:349-352. [DOI] [PubMed] [Google Scholar]
- 23.Landis, G., R. Kelley, A. C. Spradling, and J. Tower. 1997. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc. Natl. Acad. Sci. USA 94:3888-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landis, G., and J. Tower. 1999. The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126:4281-4293. [DOI] [PubMed] [Google Scholar]
- 25.Li, C. J., J. A. Bogan, D. A. Natale, and M. L. DePamphilis. 2000. Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci. 113(Pt 5):887-898. [DOI] [PubMed] [Google Scholar]
- 26.Lilly, M. A., and A. C. Spradling. 1996. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10:2514-2526. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Schier, H., and D. St Johnston. 2001. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 15:1393-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manseau, L., A. Baradaran, D. Brower, A. Budhu, F. Elefant, H. Phan, A. V. Philp, M. Yang, D. Glover, K. Kaiser, K. Palter, and S. Selleck. 1997. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn 209:310-322. [DOI] [PubMed] [Google Scholar]
- 29.Orr-Weaver, T. L. 1991. Drosophila chorion genes: cracking the eggshell's secrets. Bioessays 13:97-105. [DOI] [PubMed] [Google Scholar]
- 30.Orr-Weaver, T. L., C. G. Johnston, and A. C. Spradling. 1989. The role of ACE3 in Drosophila chorion gene amplification. EMBO J. 8:4153-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr-Weaver, T. L., and A. C. Spradling. 1986. Drosophila chorion gene amplification requires an upstream region regulating s18 transcription. Mol. Cell. Biol. 6:4624-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Royzman, I., R. J. Austin, G. Bosco, S. P. Bell, and T. L. Orr-Weaver. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Royzman, I., A. J. Whittaker, and T. L. Orr-Weaver. 1997. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 11:1999-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sardet, C., M. Vidal, D. Cobrinik, Y. Geng, C. Onufryk, A. Chen, and R. A. Weinberg. 1995. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc. Natl. Acad. Sci. USA 92:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawado, T., M. Yamaguchi, Y. Nishimoto, K. Ohno, K. Sakaguchi, and A. Matsukage. 1998. dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 251:409-415. [DOI] [PubMed] [Google Scholar]
- 36.Schwed, G., N. May, Y. Pechersky, and B. R. Calvi. 2002. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spradling, A. C. 1993. Developmental genetics of oogenesis, p. 1-70. In M. Bate, and A. Martinez-Arias (ed.), The Development of Drosophila melanogaster, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Spradling, A. C. 1999. ORC binding, gene amplification, and the nature of metazoan replication origins. Genes Dev. 13:2619-2623. [DOI] [PubMed] [Google Scholar]
- 39.Spradling, A. C., D. V. de Cicco, B. T. Wakimoto, J. F. Levine, L. J. Kalfayan, and L. Cooley. 1987. Amplification of the X-linked Drosophila chorion gene cluster requires a region upstream from the s38 chorion gene. EMBO J. 6:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevaux, O., D. Dimova, M. V. Frolov, B. Taylor-Harding, E. Morris, and N. Dyson. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21:4927-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treisman, J. E., P. J. Follette, P. H. O'Farrell, and G. M. Rubin. 1995. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev. 9:1709-1715. [DOI] [PubMed] [Google Scholar]
- 42.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 43.Tzolovsky, G., W. M. Deng, T. Schlitt, and M. Bownes. 1999. The function of the broad-complex during Drosophila melanogaster oogenesis. Genetics 153:1371-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilcoxon, F. 1945. Individual comparisons by ranking methods. Biometrics Bull. 1:80-83. [PubMed] [Google Scholar]
- 45.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, J. W., S. J. Field, L. Gore, M. Thompson, H. Yang, Y. Fujiwara, R. D. Cardiff, M. Greenberg, S. H. Orkin, and J. DeGregori. 2001. E2F1 and E2F2 determine thresholds for antigen-induced T-cell proliferation and suppress tumorigenesis. Mol. Cell. Biol. 21:8547-8564. [DOI] [PMC free article] [PubMed] [Google Scholar]