Abstract
As viruses are reliant upon their host cell to serve as proper environments for their replication, many have evolved mechanisms to alter intracellular conditions to suit their own needs. For example, human cytomegalovirus induces quiescent cells to enter the cell cycle and then arrests them in late G1, before they enter the S phase, a cell cycle compartment that is presumably favorable for viral replication. Here we show that the protein product of the human cytomegalovirus UL82 gene, pp71, can accelerate the movement of cells through the G1 phase of the cell cycle. This activity would help infected cells reach the late G1 arrest point sooner and thus may stimulate the infectious cycle. pp71 also induces DNA synthesis in quiescent cells, but a pp71 mutant protein that is unable to induce quiescent cells to enter the cell cycle still retains the ability to accelerate the G1 phase. Thus, the mechanism through which pp71 accelerates G1 cell cycle progression appears to be distinct from the one that it employs to induce quiescent cells to exit G0 and subsequently enter the S phase.
Human cytomegalovirus (HCMV) is a betaherpesvirus that can cause serious disease in individuals with immature or compromised immune systems (reviewed in reference 44). Our lab has been exploring the complicated interplay between HCMV and its host cell during the lytic replication cycle in vitro. We have been most interested in virus-induced alterations in cellular gene expression (8, 79, 80) and cell cycle progression (35, 36).
Cell cycle transit (55, 64) in mammalian cells is controlled by a family of cyclin-dependent kinases (cdk's) whose activity is dependent upon the correct subset of phosphorylations and their physical association with a cyclin protein. The synthesis and degradation of cyclin proteins are tightly regulated, and this, as well as the action of the cdk inhibitors, contributes to the control of cell cycle progression. During the early G1 phase, as well as in the G0 compartment, cdk's are held inactive, either because their obligate cyclin partner has not yet been synthesized or because of the action of the cdk inhibitors. Because of this, the retinoblastoma tumor suppressor protein (Rb) is hypophosphorylated and is found in a complex with the E2F family of transcription factors (reviewed in reference 74). This complex represses transcription from E2F-responsive promoters, and since many genes required for S-phase progression respond to E2F, cellular DNA replication is prevented. As cells progress through G1, cdk4/cyclin D complexes become activated and phosphorylate Rb, liberating E2F, which can now activate transcription from promoters with E2F binding sites, such as the cyclin E promoter (49). Synthesis of this cyclin and its subsequent pairing with cdk2 produce an active kinase, which is thought to be important for induction of late-G1/early-S-phase events. As cells enter S phase, cyclin E-associated kinase activity decreases and cyclin A-associated kinase activity increases, leading to cellular DNA replication and further cell cycle progression.
HCMV infection of human fibroblasts induces profound changes in cell cycle regulation (reviewed in reference 29). Infection of asynchronous proliferating cells results in a cell cycle arrest in G1 (5, 13, 35). A cell cycle arrest in G2 has been observed in some experiments (25, 35) but not in others (62). HCMV infection of cells brought to quiescence either by contact inhibition or by serum starvation results in an elevation of the level of the protein and associated kinase activity of cyclin E, but not cyclin A (5), and does not stimulate host genomic DNA synthesis (5, 35). Thus, infection of quiescent cells stimulates their reentry into the cell cycle and progression through G1 phase, with an eventual cell cycle arrest at the G1/S border. This is consistent with earlier studies showing that HCMV infection induces the genes for dihydrofolate reductase (73) and thymidine kinase (15), which participate in nucleotide biosynthesis, and c-myc (4), which induces the expression of genes involved in polyamine and pyrimidine synthesis (3, 40). The G1/S compartment presumably offers a favorable environment for viral replication, since the precursors for DNA replication are available but not being consumed in the synthesis of the host cell's genomic DNA. The delay in the viral life cycle observed in cells infected during the S phase (62) supports this hypothesis.
Multiple HCMV proteins have been shown to modulate cell cycle progression. The IE2 protein, as well as IE1 (in the absence of a functional p53 pathway), can drive quiescent cells into the cell cycle (9, 76). Interestingly, IE2-stimulated cell cycle progression results in an arrest at the G1/S border (45, 75, 76). The mechanisms through which these alterations occur are unknown. However, the ability of IE2 to activate transcription from E2F-responsive promoters (66) may be responsible for the cell cycle stimulation afforded by this protein. Both IE1 and IE2 perform multiple functions, including the regulation of transcription (reviewed in reference 44) and inhibition of apoptosis (78), and they are mutagenic when expressed in cells (63). IE1 is a kinase that binds to (53) and phosphorylates (52) members of the Rb family. Two additional viral products can modulate cell cycle progression. The product of the UL69 gene arrests cells with a 2n DNA content (G1 phase or the G1/S border) through an unknown mechanism (36). We recently have shown that the UL82 gene product of HCMV, pp71, which is a component of infectious virions, stimulates quiescent cells to enter the cell cycle by degrading the hypophosphorylated members of the Rb family of tumor suppressors in a proteasome-dependent manner (26a).
In this report, we demonstrate that pp71 modulates cell cycle progression in cultures of actively growing asynchronous and synchronized cells by accelerating progression through the G1 phase of the cell cycle. This activity may be important to ensure that infected cells reach the late G1 arrest point in a timely fashion. The mechanism by which pp71 accelerates G1 progression appears to be distinct from its ability to stimulate the movement of quiescent cells out of the G0 compartment.
MATERIALS AND METHODS
Cell line propagation and synchronization.
U-2 OS, 293, Rat-1, and REF-52 rat fibroblasts and 12.1 mouse fibroblasts were maintained in Dulbecco's modification of Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; HyClone). U-2 OS cells were transfected as described previously (27), and 12.1 cells were transfected by electroporation. For serum starvation, cells were washed once with and then cultured in DMEM plus 0.1% FBS for 48 h (REF-52) or 72 h (Rat-1). Starved cells were stimulated with serum by changing the culture medium to DMEM plus 10% FBS. REF-52 cells synchronized in mitosis were generated by culturing asynchronous cells in medium containing 40 ng of nocodazole/ml for 5 h and collecting the loosely attached and floating cells, which were subsequently washed once in phosphate-buffered saline (PBS) and once in medium before plating.
Plasmids and recombinant adenoviruses.
pCDNA3 was from Invitrogen. pCMV71, pCMV71-R, pCGNpp65 (2) pCGN, pCGN71, pCMVE1A, pCMVE2F-1 (26a), and pBB14 (7, 28) have been described elsewhere. Recombinant adenoviruses were generated by the AdEasy system (21) and have been described elsewhere (26a). Adenovirus stocks were prepared and infectious titers were determined by optical absorbance as described previously (42).
Generation of pp71-expressing cell lines.
REF-52 cells were cotransfected with 5 μg of SacI-digested pCGN71 and 1 μg of SacI-digested pCDNA3 by electroporation. Forty-eight hours later, selection medium (DMEM-10% FBS-500 μg of G418/ml) was applied. Approximately 2 weeks later, resistant colonies were cloned and expanded.
Flow cytometry.
Cells were harvested, fixed in ethanol, and stained with propidium iodide (PI) as described previously (27, 28). To determine bromodeoxyuridine (BrDU) incorporation, ethanol-fixed cells were washed once with PBS-S (PBS plus 0.5% bovine serum albumin and 0.5% goat serum), and their DNA was denatured with 2 N HCl for 30 min at room temperature. After washing with PBS-S, residual acidity was neutralized with 0.1 M sodium borate (pH 8.5) and the cells were washed again with PBS-S prior to incubation for 30 min at room temperature with a 1.5-μg/ml solution of fluorescein isothiocyanate (FITC)-coupled anti-BrDU antibody. The cells were then washed twice with PBS-S and stained at 4°C for 30 min with 5 μg of PI/ml in PBS containing 100 μg of RNase A/ml. The low level of PI employed in these experiments is necessary to minimize quenching of the FITC signal (67; A. J. Beavis and R. F. Kalejta, unpublished observations). Flow cytometric data acquisition and analysis were performed as described previously (27, 28). For simultaneous BrDU and PI analysis, the BrDU signal was collected as an FL1 parameter, the PI was collected as an FL3 parameter, and spectral overlap was eliminated by electronic compensation.
Assays for gene expression and BrDU incorporation.
Transactivation of the HCMV major immediate-early promoter was examined by transient transfection of 12.1 cells and detection of luciferase activity by employing a Monolight 2010 luminometer. The number of light units is presented relative to the optical density of the sample at 260 nm to control for sample recovery. The reporter plasmid pGL3-HCMV-MIEP has been described previously (59). BrDU incorporation, detection by fluorescence microscopy, and the counting of nuclei have been described elsewhere (26a).
RESULTS
HCMV pp71 modulates cell cycle progression in asynchronous cell populations.
We developed a novel transient-transfection assay with the ability to identify proteins that affect cell cycle progression (27, 28). This assay utilizes a membrane-localized green fluorescent protein (GFP) fusion protein marker to identify the subpopulation of transfected cells and PI staining to measure DNA content (cell cycle position) by flow cytometry. A marker is required since only a minority of the cells actually receive and express the transfected DNA, and it is only in those cells that a biological effect is expected. We employ a membrane-localized enhanced GFP as the marker because GFP is a stable protein and easily detected by flow cytometry, but it must be anchored to an intracellular structure to prevent it from leaching out of the cell during the permeabilization of cells with ethanol in preparation for staining with PI (27, 28). One of our approaches to identifying HCMV genes that could alter cell cycle progression was to perform this assay with candidate viral genes that we thought were likely to be cell cycle regulators.
One such gene was UL82, which codes for the pp71 protein. This HCMV protein shares some characteristics with the herpes simplex virus type 1 VP16 protein (reviewed in reference 16). Both are incorporated into virions, transactivate viral immediate-early gene synthesis, and increase the infectivity of transfected viral genomic DNA. Since VP16 binds to the cell cycle modulator HCF (18), we reasoned that pp71 might also interact with regulators of the cell cycle, perhaps resulting in both alterations in cell cycle progression and enhanced infectivity.
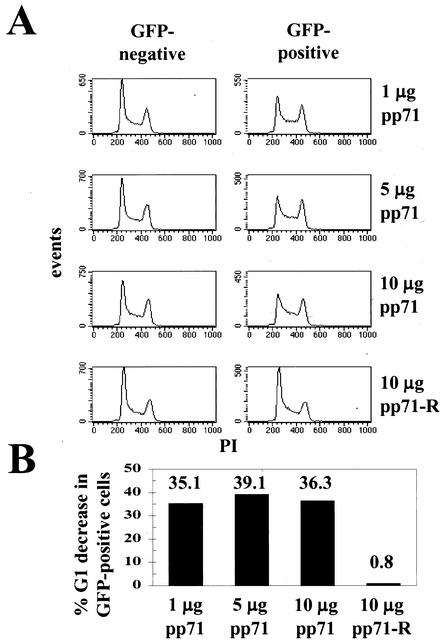
To test if pp71 could alter cell cycle progression, we employed in our transient-transfection assay increasing amounts of pCMV71, a plasmid that encodes the UL82 gene of the Towne strain of HCMV. Actively growing cells cotransfected with GFP and the pp71 expression vector showed altered cell cycle profiles in the GFP-positive cells (those that received the transfected DNA) (Fig. 1A). This cell population contained fewer cells in G1 and more in S phase than did the cells in the same transfection reaction that did not receive plasmid DNA (GFP negative). The effect was observed with each amount of pp71 plasmid examined. Transfection of a control plasmid with the pp71 coding region in reverse orientation (pCMV71-R) so as not to make a functional protein had no effect on cell cycle progression (Fig. 1A). To allow for a quantitative comparison between individual experiments, we calculated the decrease in the percentage of GFP-positive cells with a G1 DNA content compared to the G1 cells in the GFP-negative population of the same transfection (Fig. 1B). This demonstrated that transfection of asynchronous cells with pp71, but not with equal amounts of a vector unable to make a functional protein, could significantly decrease the percentage of cells in G1 (Fig. 1B).
FIG. 1.
pp71 alters cell cycle progression. (A) Asynchronous U-2 OS cells were transfected with 1 μg of the GFP marker plasmid (pBB14) and the indicated amount of the HCMV UL82 (pp71) expression plasmid pCMV71. Cells were harvested for analysis 48 h later. A plasmid with the gene for pp71 cloned in reverse orientation (pCMV71-R) was employed as a control. The DNA histograms of the untransfected (GFP-minus; average mean fluorescence intensity = 4.04 ± 0.24 arbitrary units) and transfected (GFP-plus; average mean fluorescence intensity = 178 ± 4.72 arbitrary units) cells are shown. The y axes of all the DNA histograms presented in this work are scaled according to the number of events displayed. (B) The DNA content histograms created in CellQuest and shown in panel A were quantitated with the MacDNA analysis software. The percent G1 decrease in GFP-positive cells was calculated from these values.
Either of two models can explain the decrease in G1 and the increase in S-phase cells observed when pp71 was expressed (Fig. 1). pp71 could inhibit progression through the S phase by arresting or dramatically slowing cell cycle progression. This first model predicts that, if cells were analyzed at different times after transfection, the DNA histograms would remain essentially unchanged and the number of cells in S phase would either remain constant or increase. In the second model, pp71 causes an acceleration through the G1 phase of the cell cycle. This would cause cells to reach the S phase prematurely, before they are fully prepared to replicate their DNA, and thus slow the progression of early-S-phase events, causing cells to accumulate. If pp71 were to function in this manner, cells sampled at different times after transfection should enter, traverse, and eventually leave the S phase.
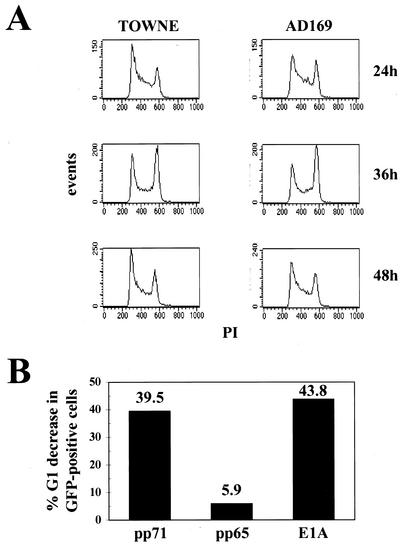
To distinguish between the two models, actively growing cells were transfected with pCMV71 or pCGN71, which expresses pp71 encoded by the Towne and AD169 strains of HCMV, respectively, and cultures were harvested for DNA content analysis at the indicated times (Fig. 2A). We observed a bolus of cells in the transfected cell population that moved through the cell cycle, entering the S phase at about 24 h; passing through S and into G2 by 36 h; and completing mitosis, traversing G1, and entering S phase again by 48 h after transfection. Because these cells continued to cycle, we conclude that the pp71-induced decrease in G1 and increase in S do not result from an arrest of the cells in the early S phase but rather from a stimulation of cell cycle progression, most probably in the G1 phase. Not surprisingly, both the Towne and AD169 pp71 proteins share this activity.
FIG. 2.
pp71 stimulates cell cycle progression. (A) Asynchronous U-2 OS cells were transfected with 1 μg of pBB14 and 10 μg of an expression plasmid for either the Towne (pCMV71) or AD169 (pCGN71) UL82 gene and harvested for analysis at the indicated times. DNA histograms of the GFP-positive (average mean fluorescence intensity = 111.77 ± 14.85) cells are shown. The average mean fluorescence intensity of the GFP-minus population was 2.03 ± 0.11 (data not shown). (B) Asynchronous U-2 OS cells were transfected with 1 μg of pBB14 and 10 μg of expression plasmids for the indicated gene, harvested 48 h after transfection, and analyzed by flow cytometry. The percent G1 decrease in GFP-positive cells was calculated from DNA content histograms (data not shown) created in CellQuest and quantitated in MacDNA.
Since we suspected that pp71 stimulated cell cycle progression through the G1 phase, we compared its effects on cells to those of the adenovirus E1A protein, a viral oncogene known to modulate G1 cell cycle progression (reviewed in reference 16). We also employed another HCMV tegument protein, pp65, as an additional negative control in our transient-transfection assay. The product of the UL83 gene, pp65, is modestly related in sequence to pp71 (48, 61) but does not act as a transcription factor, nor does it increase the infectivity of transfected viral genomic DNA (2). We found that E1A and pp71 were able to decrease the number of cells in the G1 phase by similar percentages, while transfection with pp65 had only a minor effect (Fig. 2B). The effect of pp71 in this experiment was quantitatively similar to the effect observed in Fig. 1.
pp71 accelerates progression through the G1 phase of the cell cycle.
The transfection experiments indicate that pp71 accelerates cell cycle progression through the G1 phase. Ectopic expression of cyclins A, D, and E, but not cyclin B, has also been shown elsewhere to accelerate cells through the G1 phase (1, 23, 26, 30, 46, 50, 51, 54, 56-58, 60, 77). In these experiments, cell lines were produced that express the cyclin to be tested. This allowed for a comparison not only of the percentages of cells in the G1 phase in asynchronous populations but also of the lengths of the G1 interval in synchronized cultures. Therefore, to confirm our initial result, we generated cell lines that constitutively express pp71.
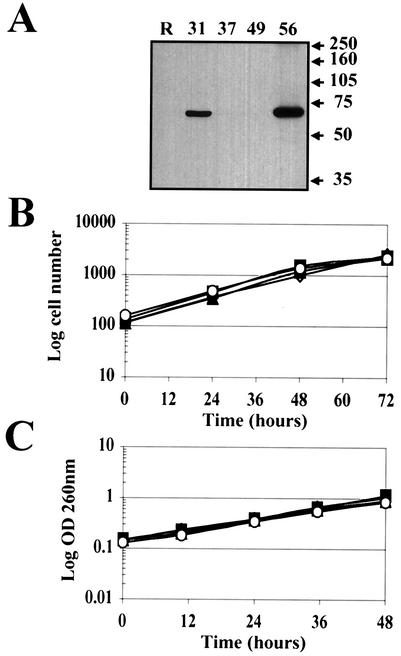
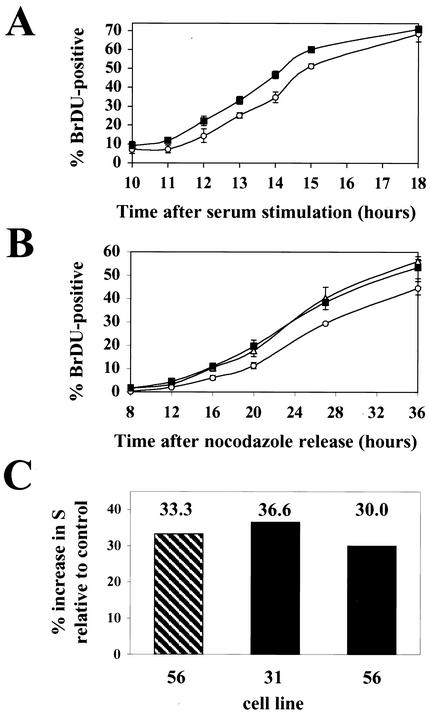
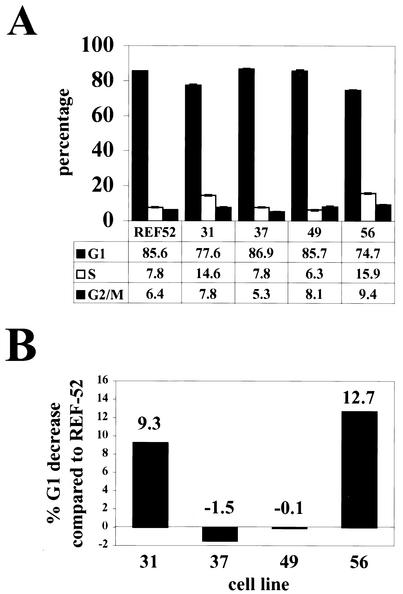
REF-52 rat fibroblasts were cotransfected with pCGN71, the AD169 pp71 expression vector, and pCDNA3, which expresses a neomycin resistance gene. G418-resistant colonies were cloned, and cell extracts were screened by Western blotting for pp71 expression employing an antihemagglutinin (anti-HA) antibody (Fig. 3A). We utilized two clones from the screen that express pp71 (clones 31 and 56) and two clones that do not (clones 37 and 49). Immunofluorescence microscopy employing both the HA antibody and an antiserum specific for pp71 (47) confirmed that pp71 was expressed in more than 99% of the cells of clones 31 and 56 and was localized to the nucleus as in transiently transfected and infected cells (data not shown). Furthermore, the pp71-expressing cells (lines 31 and 56), the parental cell line (REF-52), and the neomycin-resistant clones isolated in the screen which do not express pp71 (lines 37 and 49) all had similar doubling times when grown in standard medium with serum (Fig. 3B and C), demonstrating that pp71 does not alter the length of the complete cell cycle.
FIG. 3.
Cells that constitutively express pp71 divide at the same rate as do control cells. (A) REF-52 cells were cotransfected with pCDNA3 and pCGN71 as described in Materials and Methods, and colonies resistant to G418 were isolated and cloned. Whole-cell lysates prepared from the indicated clones were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted to a membrane, and probed with an anti-HA antibody. The parental REF-52 cells (R) were also analyzed. Molecular mass markers are shown in kilodaltons. (B) The indicated cell lines plated at a low density and fed fresh medium every 24 h were harvested at the indicated times, and the number of cells was determined by counting an aliquot of the cells on a hemocytometer. The pp71-expressing cell lines are 31 (open diamonds) and 56 (open circles), and the control cell lines are 37 (filled squares) and 49 (filled triangles). (C) Cells cultured as described above were harvested at the indicated times, and the optical density (OD) at 260 nm of an aliquot was determined. The optical density at 260 nm is indicative of the number of cells in a culture. Symbols are as in panel B.
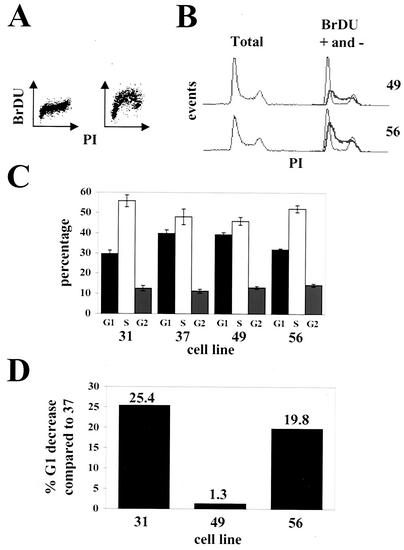
We analyzed by flow cytometry the DNA content of asynchronous cultures and found that the two pp71-expressing cell lines showed fewer cells in the G1 phase of the cell cycle than did the cell lines that do not express the protein. To accurately determine cell cycle distributions in these experiments, asynchronous cultures were pulsed with BrDU prior to sampling. The cells were stained with an FITC-coupled antibody specific for BrDU and with PI for DNA and subsequently analyzed by flow cytometry (Fig. 4A). Cell lines constitutively expressing pp71 contained a smaller percentage of cells in G1 than did those that do not express the protein. This is evidenced in the PI-stained histograms alone (Fig. 4B, left panel) in which pp71-expressing cells show a smaller G1 peak and more cells in early S, as well as when the BrDU-positive (Fig. 4B, right panel, thick gray lines) and BrDU-negative (Fig. 4B, right panel, black lines) subpopulations of cells are displayed together. This analysis indicates that the ratio of early-S-phase cells to G1-phase cells is substantially greater in cells expressing pp71 than in those that do not. The percentage of cells in each cell cycle phase was determined from dual-color dot plots (as displayed in Fig. 4A) of PI and BrDU incorporation (Fig. 4C), showing that pp71-expressing cells spend less time in G1 (and more in S) than do cells that do not express the protein.
FIG. 4.
Asynchronous cultures of cells that constitutively express pp71 have altered cell cycle profiles. (A) Asynchronous cell cultures pulsed with BrDU prior to harvest were stained as described in Materials and Methods. Representative dual-color dot plots of control cells not pulsed with BrDU (left) and pulsed cells (right) are shown. (B) DNA histograms for the indicated cell lines are displayed. On the left are total DNA histograms obtained from the PI-stained cells (BrDU-positive and -negative cells combined). On the right, the PI profiles of the BrDU-positive (gray lines) and BrDU-negative (black lines) are separated. Gating was based on dual-color dot plots as in panel A. (C) The percentage of cells in each phase of the cell cycle was calculated from dual-color dot plots. The analysis was performed in triplicate, and standard deviations are shown. (D) The percent decreases in the percentages of G1 cells for the indicated cell lines compared to that for cell line 37 are shown.
In these experiments, pp71 advanced cells through G1 and into S phase about 23% faster than normal cells (Fig. 4D). The observed G1 acceleration in cells constitutively expressing pp71 is slightly less than in the transiently transfected cells, most likely due to a lower level of pp71 expression. However, it is similar in magnitude to the same effect seen in cells either constitutively or inducibly expressing ectopic cyclin proteins. For example, cyclin D has been found elsewhere to accelerate G1 with different efficiencies, 10 to 15% (56), 25% (26), 28% (30), or 22 to 31% (23). Cyclin A accelerated G1 by 12 to 15% (58), and cyclin E accelerated G1 by 25% (56) or 33% (50).
We also demonstrated the ability of pp71 to accelerate cell cycle progression through G1 in synchronized cultures (Fig. 5). Cells were synchronized in G0 by serum starvation for 48 h and released into the cell cycle by being fed with medium containing 10% FBS. At the indicated times, cultures were pulsed with BrDU for 15 min prior to harvesting. The cells were stained with an FITC-coupled anti-BrDU antibody and PI and analyzed by flow cytometry, and the number of BrDU-positive cells at each time point was determined. The pp71-expressing cells entered the S phase approximately 1 h before the nonexpressing cells did (Fig. 5A). The pp71-expressing cells also reached S phase prior to nonexpressing cells when released from a mitotic block imposed by nocodazole. Cells expressing pp71 were found to enter S phase approximately 4 h before cells that did not express the protein (Fig. 5B), again indicating that, in pp71-expressing cells, the G1 interval was shortened. It should be noted that, in this experiment, the length of time required for the cells to reach S is somewhat longer than expected, probably because the cells recover slowly from the mitotic block. A similar effect was observed when the acceleration of the G1 phase by the cyclin proteins was analyzed in rat fibroblasts (50, 54). The percent increase in S phase by pp71 in synchronized cultures (Fig. 5C) is similar in magnitude to the percent G1 decrease observed in asynchronous cultures (Fig. 4) and the transient-transfection assay (Fig. 1B and 2B), as well as with the effects of the cyclin proteins (23, 26, 30, 50, 56, 58).
FIG. 5.
pp71 accelerates cells through the G1 phase of the cell cycle. (A) REF-52 cells (open circles) and the pp71-expressing cell line 56 (filled squares) were synchronized in G0 by serum starvation for 48 h and then stimulated with serum in the presence of BrDU. At the indicated times cells were harvested and the percentage of BrDU-positive cells was determined by flow cytometry. (B) Cells synchronized in mitosis with nocodazole as described in Materials and Methods were prepared from the pp71-expressing cell lines 31 (open triangles) and 56 (filled squares) and the control cell line 37 (open circles). They were washed and replated onto coverslips in the presence of BrDU. At the indicated times, coverslips were harvested, fixed, and subsequently stained for BrDU incorporation. BrDU-positive cells were counted by fluorescence microscopic examination of at least 500 nuclei per time point. (C) Shown are the percent increases in cells in S phase for the indicated pp71-expressing cell lines from the G0 (hatched bar) and mitotic (solid bars) synchronization experiments. The percent increase in cells in S phase was calculated by dividing the difference of the percentage of BrDU-positive cells from the pp71-expressing cell lines and the control cells by the percentage of BrDU-positive cells in the control cell line at the time point where those cultures were 30% BrDU positive.
Cells overexpressing ectopic cyclin proteins were also impaired in their ability to achieve quiescence upon serum withdrawal (50, 56). To determine if the same was true of the pp71-expressing cells, we cultured them in medium with 0.1% FBS for 48 h before harvesting the cells and analyzing their DNA content by flow cytometry. As was the case with the cyclin proteins, cells expressing pp71 were impaired in their ability to achieve quiescence compared to cells that did not express the viral protein and showed a higher percentage of cells in the S phase (Fig. 6A). The magnitude of the decrease in G1 (Fig. 6B) was reproducible and somewhat less than the 16% effect observed with the cyclin E protein (50).
FIG. 6.
Cells expressing pp71 fail to achieve quiescence with the same kinetics as those of wild-type cells. (A) The indicated cell lines were plated and serum deprived as described in Materials and Methods. The DNA content in cells 48 h after starvation was determined by flow cytometry. The percentage of cells in each cell cycle phase is presented with error bars denoting the standard deviations. (B) The percent decrease in G1 was calculated relative to that for the parental REF-52 cells for each cell line from the data in panel A.
These experiments demonstrate that pp71 can accelerate the passage of growing cells through the G1 phase of the cell cycle, as well as decrease their ability to enter a quiescent state upon serum withdrawal.
The ability of pp71 to accelerate G1 progression is substantially distinct from its ability to induce G0 cells to enter the cell cycle.
We have recently identified a second cell cycle effect of pp71 expression: pp71 stimulates quiescent cells to enter the cell cycle and progress into the S phase (26a). An LxCxD sequence is present in pp71 that is similar to the LxCxE sequence (11) in simian virus 40 T antigen, adenovirus E1A, and papillomavirus E7. This motif mediates interactions of the tumor virus proteins with the retinoblastoma family of tumor suppressors, Rb, p107, and p130 (reviewed in reference 12). The hypophosphorylated forms of these proteins bind to and block the activity of the E2F family of transcription factors, which stimulate the expression of many genes required for entry into the S phase (reviewed in reference 14). Thus, Rb-E2F complexes can arrest the cell cycle in G1. Normally, the cdk-dependent phosphorylation of the Rb family members disrupts their complexes with E2F, resulting in cell cycle stimulation. This association is also disrupted upon the binding of T antigen, E1A, or E7 to the Rb family member. We have shown elsewhere that pp71 interacts with the hypophosphorylated forms of all three members of the Rb family, it induces their degradation in a proteasome-dependent manner, and a mutation (C219G) in the LxCxD motif eliminates the ability of pp71 to degrade the Rb family members and to drive quiescent cells into the cell cycle (26a).
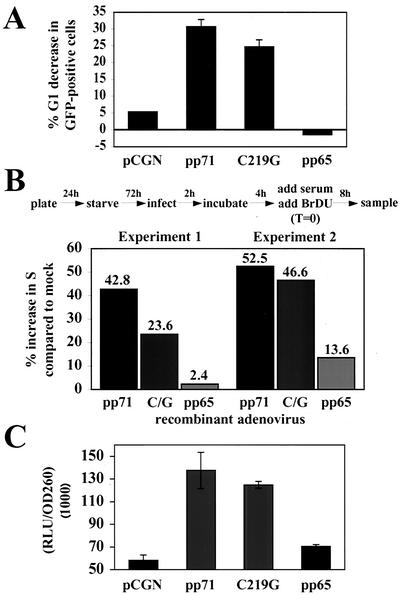
The ability of pp71 to direct the degradation of the Rb family proteins raised the possibility that disruption of the Rb-E2F pathway was the mechanism by which pp71 accelerates cells through the G1 phase of the cell cycle. To test this hypothesis, we asked if the pp71 C219G mutant, which fails to degrade the Rb family members and does not drive quiescent cells into the cell cycle, was still able to perform this function. We first employed our transient-transfection assay and found that the C219G pp71 mutant retained the ability to accelerate the G1 phase, although with a slightly decreased magnitude compared to that for the wild-type protein (Fig. 7A). As controls, the empty vector (pCGN) and the pp65 protein were tested, and they had no effect on G1 progression. We confirmed this result in synchronized cultures. Quiescent cells were infected with recombinant adenoviruses expressing wild-type pp71, the pp71 C219G mutant, or pp65, and 6 h later cells were stimulated with 10% FBS to reenter the cell cycle. Entry into the S phase was monitored by BrDU incorporation. Both pp71 and the C219G mutant were able to accelerate progression through the G1 phase of the cell cycle, but pp65 was not (Fig. 7B). The wild-type protein had a somewhat stronger effect on the cell cycle than did the C219G mutant, but this is likely due to some stimulation of the quiescent cells by wild-type pp71 during the 6-h incubation to allow for gene expression from the infecting recombinant adenovirus genomes.
FIG. 7.
The ability to disrupt the Rb pathway is not required for the pp71-induced acceleration of the G1 phase and activation of the viral major immediate-early promoter. (A) Transient-transfection assays of U-2 OS cells were performed as described above. The percent decreases in G1 of the GFP-positive cells after transfection with the indicated plasmids are shown, with error bars indicating the standard deviations. (B) Rat-1 cells grown on coverslips were serum starved, infected with recombinant adenoviruses expressing the indicated proteins, and labeled with BrDU according to the protocol shown above the graph and described in the text. At 8 h, coverslips were removed and treated as for Fig. 5B. The percent increase in S phase compared to that for mock-infected cells was calculated as for Fig. 5C. Results of two individual experiments are shown. C/G, C219G. (C) 12.1 mouse fibroblasts were transfected with a luciferase reporter driven by the HCMV major immediate-early promoter and the indicated effector plasmid. The ratios of the luciferase activities to the optical densities at 260 nm of aliquots of the sample are shown, with error bars indicating standard deviations. RLU, relative light units.
We cannot rule out the possibility that the ability of pp71 to attack the Rb-E2F pathway contributes to the acceleration in G1 that we observe. However, the continued activity of the C219G mutant argues that a different function of pp71 must be responsible for the majority of the effect. pp71 may direct the degradation of proteins other than the Rb family in an LxCxD-independent manner. Alternatively, pp71 might stimulate G1 progression through its activity as a transcription factor. The wild-type protein stimulates the activity of the viral major immediate-early promoter as well as other viral and cellular promoters (33), and here we show that the C219G mutant retains the ability to activate the HCMV major immediate-early promoter (Fig. 7C). Thus, at least for the major immediate-early promoter, regulation of the Rb pathway is not required for pp71 to act as a transcription factor. Therefore, it is possible that pp71 influences expression of one or more cellular proteins that regulate the rate of progression through the G1 phase.
DISCUSSION
Multiple functions have been attributed to pp71. First, it transactivates gene expression from both viral and cellular promoters (33). Second, it increases the infectivity of transfected viral genomic DNA (2). Third, it directs degradation of the Rb family of proteins, inducing cells to move from G0 to the S phase of the cell cycle (26a). Fourth, we show here that it accelerates progression of cells through the G1 compartment (Fig. 1, 2, 4, 5, and 7). Although it could play a minor role, degradation of Rb family members is not the major cause of its effect on the rate of G1 progression (Fig. 7). The overexpression of cyclins also accelerates progression through the G1 compartment (23, 26, 30, 50, 56, 58), and the magnitude of their effects on the rate of progression is similar to that observed here for pp71.
The cyclins and pp71 are not the only proteins that accelerate cells through the G1 phase. The signaling molecules ras, raf, src (34), and rad (70); cdk6 (19); and the acute myeloid leukemia-1 (AML-1) protein (68) also share this function. In addition, human T-cell leukemia virus type 1 Tax (31) and human papillomavirus E7 (38) also accelerate G1 phase. The mechanisms through which these proteins advance G1 are not completely understood. G1 acceleration by the cyclins and ras was correlated with premature Rb phosphorylation (23, 34, 57), and kinase activity was required for cdk6 to accelerate G1 (19). Most interestingly, AML-1 proteins mutated in the transactivation or DNA binding domains that have lost the ability to regulate transcription also fail to accelerate G1 (68). As the ability to modulate gene expression is required for AML-1 to advance G1, the same might be true for pp71, explaining why the C219G mutant, which transactivates the major immediate-early promoter (Fig. 7C) but does not regulate the Rb-E2F pathway, still accelerates G1 (Fig. 7).
Whereas expression of pp71 accelerated the G1 phase, it did not affect the overall cell cycle time (Fig. 3B and C) because cells were slowed in progressing through the S phase (Fig. 1A and 4D). Similar results were obtained for other proteins that shorten G1 (1, 23, 26, 30, 50, 56, 68), but proteins that accelerated passage through this compartment also shortened the cell cycle time (31, 34, 54, 60). Whether these differences are results of the expression level of the proteins, of cell type differences, or of unique mechanisms of G1 acceleration has not been determined.
Expression of pp71 has at least two effects on the cell cycle: the stimulation of quiescent cells to reenter the cell cycle and the acceleration of cycling cells through the G1 phase. pp71 attacks the Rb pathway, degrading the hypophosphorylated forms of Rb, p107, and p130 in a proteasome-dependent manner (26a). This degradation stimulates quiescent cells to reenter the cell cycle and proceed to the S phase, and it is possible that it contributes to the ability of pp71 to accelerate progression through the G1 phase of the cell cycle (Fig. 7). Interestingly, however, the major component of the G1 acceleration afforded by pp71 does not involve its ability to attack the Rb pathway, because a pp71 mutant that fails to degrade the Rb family members retains the ability to accelerate movement through the G1 phase (Fig. 7). The G1 acceleration mediated by pp71 could result from an ability to direct the degradation of another as yet unidentified cell cycle regulatory protein, from its ability to regulate transcription, or from a currently unknown function of pp71.
As noted above, HCMV encodes four proteins, IE1, IE2, pUL69, and pp71, known to alter cell cycle progression (reviewed in references 10 and 29). Modulation of the cell cycle likely facilitates the lytic cycle of the virus by forcing the cell into a state favorable for viral replication. The genes encoding three of the proteins, pUL69, pp71, and IE1, have been deleted from the virus, and each has been shown elsewhere to be required for efficient lytic replication at low multiplicities of infection (6, 20, 43). Although a virus lacking IE2 has not been generated, an F plasmid carrying the viral genome with a deletion of IE2 does not produce infectious virus upon transfection into permissive cells (37). Moreover, a virus with a temperature-sensitive allele of IE2 that is impaired in its ability to activate transcription does not grow at the restrictive temperature and fails to accumulate early mRNAs (22). Thus, each of the gene products known to modulate the cell cycle is required for efficient viral replication in vitro. However, because these proteins each have multiple activities, experiments with null mutants cannot reliably examine the specific affects of the cell cycle regulatory activities of these proteins. More sophisticated mutants will be needed, and even then, functional redundancy among viral gene products might confound the interpretation of genetic experiments.
There is precedent for a role of cell cycle regulatory proteins in herpesvirus latency. The cyclin D homologue of murine gammaherpesvirus 68 can induce cell cycle progression and is oncogenic (71) but is not required for lytic replication in either cycling or quiescent cells. Interestingly, viral cyclin mutants were able to establish latency at a similar rate as wild-type virus but were unable to reactivate from a latent infection (24, 72). Thus, it was proposed that the viral cyclin is required for altering the intracellular environment to aid in reactivation from latency but is not required during the lytic replication cycle.
Perhaps pp71 functions during reactivation of latent infections and its ability to degrade the Rb family and stimulate the cell cycle is required for this effect. HCMV DNA can be detected in monocytes from infected individuals, but these cells are not permissive for viral replication because of a block to viral immediate-early gene synthesis (39, 69). Upon differentiation to macrophages, immediate-early viral genes are synthesized and a lytic replication cycle ensues (41, 65, 69). The Rb proteins are known to affect the differentiation status of cells (reviewed in reference 32), and since pp71 targets the Rb family and induces viral immediate-early gene synthesis, it is a strong candidate for a regulator of HCMV latency. Our lab has recently developed an in vitro latency assay (17) that should be useful, in conjunction with viral mutants with defined mutant alleles of the viral cell cycle regulators such as pp71, in determining the roles of these proteins during latency.
Acknowledgments
We thank Jill Bechtel for help in screening the pp71-expressing cell lines and Andy Beavis for help with the flow cytometry.
This work was supported by a grant from the NIH (CA82396).
REFERENCES
- 1.Ando, K., F. Ajchenbaum-Cymbalista, and J. D. Griffin. Regulation of G1/S transition by cyclins D2 and D3 in hematopoietic cells. Proc. Natl. Acad. Sci. USA 90:9571-9575. [DOI] [PMC free article] [PubMed]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldogh, I., S. AbuBakar, and T. Albrecht. 1990. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science 247:561-564. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., and T. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brideau, A. D., B. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo, J. P., and T. F. Kowalik. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19-34. [DOI] [PubMed] [Google Scholar]
- 11.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Classon, M., and N. Dyson. 2001. p107 and p130: versatile proteins with interesting pockets. Exp. Cell Res. 264:135-147. [DOI] [PubMed] [Google Scholar]
- 13.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, N. 1998. The regulation of E2F proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 15.Estes, J. E., and E.-S. Huang. 1977. Stimulation of cellular thymidine kinases by human cytomegalovirus. J. Virol. 24:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 17.Goodrum, F., C. Jordan, K. High, and T. Shenk. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc. Natl. Acad. Sci. USA 99:16255-16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto, H., S. Motomura, A. C. Wilson, R. N. Freiman, Y. Nakabeppu, K. Fukushima, M. Fujishima, W. Herr, and T. Nishimoto. 1997. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 11:726-737. [DOI] [PubMed] [Google Scholar]
- 19.Grossel, M. J., G. L. Baker, and P. W. Hinds. 1999. cdk6 can shorten G1 phase dependent upon the N-terminal INK4 interaction domain. J. Biol. Chem. 274:29960-29967. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, T.-C., S. Zhou, L. T. DaCosta, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heider, J. A., W. A. Bresnahan, and T. Shenk. 2002. Construction of a rationally designed human cytomegalovirus variant encoding a temperature-sensitive immediate-early 2 protein. Proc. Natl. Acad. Sci. USA 99:3141-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzinger, T., and S. I. Reed. 1998. Cyclin D3 is rate-limiting for the G1/S phase transition in fibroblasts. J. Biol. Chem. 273:14958-14961. [DOI] [PubMed] [Google Scholar]
- 24.Hoge, A. T., S. B. Hendrickson, and W. H. Burns. 2000. Murine gammaherpesvirus 68 cyclin D homologue is required for efficient reactivation from latency. J. Virol. 74:7016-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jault, F. M., J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, W., S. M. Kahn, P. Zhou, Y. J. Zhang, A. M. Cacace, A. S. Infante, S. Doi, R. M. Santella, and I. B. Weinstein. 1993. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene 8:3447-3457. [PubMed] [Google Scholar]
- 26a.Kalejta, R. F., J. T. Bechtel, and T. Shenk. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 27.Kalejta, R. F., T. Shenk, and A. J. Beavis. 1997. Use of a membrane-localized green fluorescent protein allows simultaneous identification of transfected cells and cell cycle analysis by flow cytometry. Cytometry 29:286-291. [DOI] [PubMed] [Google Scholar]
- 28.Kalejta, R. F., A. D. Brideau, B. W. Banfield, and A. J. Beavis. 1999. An integral membrane green fluorescent protein marker, Us9-GFP, is quantitatively retained in cells during propidium iodide-based cell cycle analysis by flow cytometry. Exp. Cell Res. 248:322-328. [DOI] [PubMed] [Google Scholar]
- 29.Kalejta, R. F., and T. Shenk. 2002. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 7:d295-d306. [DOI] [PubMed] [Google Scholar]
- 30.Kato, J., and C. J. Sherr. 1993. Inhibition of granulocyte differentiation by G1 cyclins D2 and D3, but not D1. Proc. Natl. Acad. Sci. USA 90:11513-11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemoine, F. J., and S. J. Marriott. 2001. Accelerated G1 phase progression induced by the human T cell leukemia virus type 1 (HTLV-1) Tax oncoprotein. J. Biol. Chem. 276:31851-31857. [DOI] [PubMed] [Google Scholar]
- 32.Lipinski, M. M., and T. Jacks. 1999. The retinoblastoma gene family in differentiation and development. Oncogene 55:7873-7882. [DOI] [PubMed] [Google Scholar]
- 33.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, J. J., J. R. Chao, M. C. Jiang, S. Y. Ng, J. J. Y. Yen, and H. F. Yang-Yen. 1995. Ras transformation results in elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol. Cell. Biol. 15:3654-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in the G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, L. G., G. W. Demers, and D. A. Galloway. 1998. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J. Virol. 72:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendelson, M., S. Monard, P. Sissons, and J. Sinclair. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 77:3099-3102. [DOI] [PubMed] [Google Scholar]
- 40.Miltenberger, R. J., K. A. Sukow, and P. J. Farnham. 1995. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol. Cell. Biol. 15:2527-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minton, E. J., C. Tysoe, J. H. Sinclair, and J. G. P. Sissons. 1994. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J. Virol. 68:4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1 (491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 45.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musgrove, E. A., C. S. L. Lee, M. F. Buckley, and R. L. Sutherland. 1994. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc. Natl. Acad. Sci. USA 91:8022-8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325-338. [DOI] [PubMed] [Google Scholar]
- 48.Nowak, B., A. Gmeiner, P. Sarnow, A. J. Levine, and B. Fleckenstein. 1984. Physical mapping of human cytomegalovirus genes: identification of DNA sequences coding for a virion phosphoprotein of 71 kDa and a viral 65-kDa polypeptide. Virology 134:91-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146-12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohtsubo, M., and J. M. Roberts. 1993. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science 259:1908-1912. [DOI] [PubMed] [Google Scholar]
- 51.Ohtsubo, M., A. M. Theodoras, J. Schumacher, J. M. Roberts, and M. Pagano. 1995. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15:2612-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pajovic, S., E. L. Wong, A. R. Black, and J. C. Azizkhan. 1997. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol. Cell. Biol. 17:6459-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E.-S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quelle, D. E., R. A. Ashmun, S. A. Shurtleff, J. Kato, D. Bar-Sagi, M. F. Roussel, and C. J. Sherr. 1993. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7:1559-1571. [DOI] [PubMed] [Google Scholar]
- 55.Reed, S. I., E. Bailly, V. Dulic, L. Hengst, D. Resnitzky, and J. Slingerland. 1994. G1 control in mammalian cells. J. Cell Sci. Suppl. 18:69-73. [DOI] [PubMed] [Google Scholar]
- 56.Resnitzky, D., M. Gossen, H. Bujard, and S. I. Reed. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Resnitzky, D., and S. I. Reed. 1995. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol. 15:3463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Resnitzky, D., L. Hengst, and S. I. Reed. 1995. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol. Cell. Biol. 15:4347-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenberg, A. R., F. Zindy, F. L. Deist, H. Mouly, P. Metezeau, C. Brechot, and E. Lamas. 1995. Overexpression of human cyclin A advances entry into S phase. Oncogene 10:1501-1509. [PubMed] [Google Scholar]
- 61.Ruger, B., S. Klages, B. Walla, J. Albrecht, B. Fleckenstein, P. Tomlinson, and B. Barrell. 1987. Primary structure and transcription of the genes coding for the two virion phosphoproteins pp65 and pp71 of human cytomegalovirus. J. Virol. 61:446-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen, Y., H. Zhu, and T. Shenk. 1997. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc. Natl. Acad. Sci. USA 94:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherr, C. J., and J. M. Roberts. 1995. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149-1163. [DOI] [PubMed] [Google Scholar]
- 65.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogenic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 66.Song, Y. J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 99:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stokke, T., K. Solberg, P. DeAngelis, and H. B. Steen. 1998. Propidium iodide quenches the fluorescence of TdT-incorporated FITC-labeled dUTP in apoptotic cells. Cytometry 33:428-434. [PubMed] [Google Scholar]
- 68.Strom, D. K., J. Nip, J. J. Westendorf, B. Lingg, B. Lutterbach, J. R. Downing, N. Lenny, and S. W. Hiebert. 2000. Expression of the AML-1 oncogene shortens the G1 phase of the cell cycle. J. Biol. Chem. 275:3438-3445. [DOI] [PubMed] [Google Scholar]
- 69.Taylor-Wiedeman, J., P. Sissons, and J. Sinclair. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tseng, Y. H., D. Vicent, J. Zhu, Y. Niu, A. Adeyinka, J. S. Moyers, P. H. Watson, and C. R. Kahn. 2001. Regulation of growth and tumorigenicity of breast cancer cells by the low molecular weight GTPase Rad and Nm23. Cancer Res. 61:2071-2079. [PubMed] [Google Scholar]
- 71.Van Dyk, L. F., J. L. Hess, J. D. Katz, M. Jacoby, S. H. Speck, and H. W. Virgin IV. 1999. The murine gammaherpesvirus 68 v-cyclin gene is an oncogene that promotes cell cycle progression in primary lymphocytes. J. Virol. 73:5110-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Dyk, L. F., H. W. Virgin IV, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wade, M., T. F. Kowalik, M. Mudryj, E.-S. Huang, and J. C. Azizkhan. 1992. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol. Cell. Biol. 12:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 75.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiebusch, L., and C. Hagemeier. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin-dependent kinase activation. EMBO J. 20:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wimmel, A., F. C. Lucibello, A. Sewing, S. Adolph, and R. Muller. 1994. Inducible acceleration of G1 progression through tetracycline-regulated expression of human cyclin E. Oncogene 9:995-997. [PubMed] [Google Scholar]
- 78.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to asses the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]