Abstract
The Cajal (coiled) body (CB) is a structure enriched in proteins involved in mRNA, rRNA, and snRNA metabolism. CBs have been shown to interact with specific histone and snRNA gene loci. To examine the potential role of CBs in U2 snRNA metabolism, we used a variety of genomic and oligonucleotide probes to visualize in situ newly synthesized U2 snRNA relative to U2 loci and CBs. Results demonstrate that long spacer sequences between U2 coding repeats are transcribed, supporting other recent evidence that U2 transcription proceeds past the 3′ box. The presence of bright foci of this U2 locus RNA differed between alleles within the same nucleus; however, this did not correlate with the loci's association with a CB. Experiments with specific oligonucleotide probes revealed signal for preU2 RNA within CBs. PreU2 was also detected in the locus-associated RNA foci, whereas sequences 3′ of preU2 were found only in these foci, not in CBs. This suggests that a longer primary transcript is processed before entry into CBs. Although this work shows that direct contact of a U2 locus with a CB is not simply correlated with RNA at that locus, it provides the first evidence of new preU2 transcripts within CBs. We also show that, in contrast to CBs, SMN gems do not associate with U2 gene loci and do not contain preU2. Because other evidence indicates that preU2 is processed in the cytoplasm before assembly into snRNPs, results point to an involvement of CBs in modification or transport of preU2 RNA.
INTRODUCTION
First described in 1903 (Ramon y Cajal, 1903), the subnuclear structure known as the coiled or “Cajal” bodies (CBs) are roughly spherical structures found in many cell types, numbering one to five per nucleus (Gall et al., 1995; Matera, 1998, 1999; reviewed by Schul et al., 1998a). Numerous snRNAs and proteins of both nuclear and nucleolar origin are enriched in the CB. These include factors involved in transcription and/or processing of mRNAs, snRNAs, histone mRNA, and rRNA (Jordan et al., 1997; Schul et al., 1998b; Abbott et al., 1999; Gall et al., 1999).
CBs have been reported to interact with other subnuclear bodies. The association between CBs and the nucleolus are well established (Raska et al., 1990; Malatesta et al., 1994; Bohmann et al., 1995), and transient interactions between CBs and cleavage bodies (Schul et al., 1996) and PML domains (Grande et al., 1996; Ishov and Maul, 1996) have also been reported. The SMN (survival motor neuron) protein, the gene for which is deleted in spinal muscular atrophy, is consistently enriched in foci termed “SMN gems.” Gems are often very closely associated or even coincident with CBs (Liu and Dreyfuss, 1996); in many cultured cells studied, CBs and gems appear to be coincident (Carvalho et al., 1999). SMN has been implicated in snRNP biogenesis, supporting the idea that gems (and potentially CBs) may be involved in snRNP production (Pellizzoni et al., 1998). An extension of this hypothesis is that CBs are sites of preassembly of pol I, II, and III transcription/RNA processing complexes, or “transcriptosomes” (Gall et al., 1999).
Apart from their potential role in snRNP biogenesis or recycling, observations showing a relationship between CBs and specific gene loci suggest that CBs could have a more direct relationship to the expression and nuclear metabolism of specific types of RNAs. Several years ago, it was demonstrated that sphere organelles of amphibian germinal vesicles associate with histone gene loci (Gall et al., 1981; Callan et al., 1991). More recently, we and others have shown that mammalian CBs preferentially associate with several different snRNA, snoRNA, and histone gene loci (Frey and Matera, 1995; Smith et al., 1995; Gao et al., 1997; Jacobs et al., 1999). In the HeLa cell line we studied, ∼45% of U2 gene loci are positioned at the immediate periphery of the CB, with a subset of CBs associated with multiple U2 loci and/or with U2 and U1 loci simultaneously (Smith et al., 1995). These findings raise the question of whether the interaction of CBs with specific gene loci, and potentially other nuclear bodies, is related to the metabolism of the corresponding RNAs.
Here we investigate the important question of whether the variable, apparently transient association of U2 genes with CBs reflects differences in RNA detected at different loci. We report that a large RNA transcript is derived from the tandemly repeated U2 genes, including a region well beyond the 199-base pair (bp) coding region of U2 snRNA. We also report the first visualization of preU2 RNA distribution within a nuclear structure with the use of a series of oligonucleotide probes. We find surprising evidence that preU2 RNA is concentrated within CBs, implicating CBs directly in some aspect of U2 RNA metabolism, which we suggest could involve modification of newly synthesized U2 snRNA. Finally, because of the close association of SMN gems to CBs, we determined whether these structures are also preferentially associated with the U2 gene and with preU2 RNA. Our results show that the gems, which are physically distinct from CBs in the HeLa cells studied here, show no specific interaction with the U2 gene or the precursor U2 RNA.
MATERIALS AND METHODS
Antibodies and DNA Probes
To detect the tandemly repeated U2 locus and RNA, we used a 6.1-kilobase (kb) U2 locus probe, pTP18, cloned in the laboratory of Alan Weiner (Van Arsdell and Weiner, 1984). A plasmid (pMRG3U2) containing only the U2 coding sequence (Jacobson et al., 1993) was obtained from Thoru Pederson (University of Massachusetts, Worcester). 5′ biotinylated oligonucleotide probes were purchased from Keystone Laboratories (Foster City, CA).
A rabbit polyclonal antibody against the CB factor p80 coilin (R288) (Raska et al., 1991), kindly provided by Edward Chan (Scripps Research Institute), was used to visualize CBs. 2B1, a mAb to SMN (Liu and Dreyfuss, 1996), was a gift from Gideon Dreyfuss (University of Pennsylvania).
Cell Culture and Fixation
Experiments were done with the use of S3 HeLa cells or human diploid fibroblasts WI38. Cells were grown on coverslips in DMEM plus 10% FCS with 10 mg/ml pen/strep. Before fixation, cells were washed first at room temperature and then on ice in HBSS (Life Technologies/BRL, Grand Island, NY) followed by successive washes on ice in cytoskeletal buffer (Fey et al., 1986) before a 0.5- to 5.0-min extraction in ice-cold cytoskeletal buffer containing 0.5% Triton X-100 plus 2 mM vanadyl adenosine (BRL). Cells were then immediately fixed in 4% paraformaldehyde in 1× PBS (pH 7.4) for 10 min and stored at 4°C in 70% ethanol.
Fluorescence In Situ Hybridization and Immunofluorescence Staining
A detailed hybridization protocol can be found elsewhere (Johnson et al., 1991). To target DNA alone, RNA was removed before hybridization by denaturation in 0.07 N NaOH. For RNA hybridizations, cells were simply dehydrated through ethanol. All hybridizations were for 3 h to overnight at 37°C. Washes were done in 50% formamide/2× SSC, 2× SSC, and 1× SSC. To colocalize the U2 gene and its primary transcripts, a sequential hybridization protocol was performed as described previously (Xing et al., 1995). Oligonucleotide hybridizations were done with 5 ng of biotinylated oligonucleotide probe in 25% formamide. Washes were done in 15% formamide/2× SSC, 2× SSC, and 1× SSC.
Antibody staining was carried out with antibody diluted 1:500 in 4× SSC, 1% BSA for 1 h. Three washes (4× SSC, 4× SSC/0.1% Triton X-100, 4× SSC) of 15 min each were then done. The antibody was detected with the use of a fluorescein-, rhodamine-, or AMCA-tagged secondary antibody (Jackson Laboratories, Bar Harbor, ME) diluted 1:500 in 4× SSC, 1% BSA. Ordinarily, in experiments with both hybridization and immunofluorescence, the coilin staining was done first and fixed again because hybridization resulted in a reduction of coilin staining.
Microscopy and Image Analysis
Images were captured with the use of a Photometrics (Tucson, AZ) P-250 cooled charge-coupled device camera and either the WHIP (G.W. Hannaway & Associates, Boulder, CO) or MetaMorph (Universal Imaging, West Chester, PA) image-processing package. The microscope was a Zeiss (Thornwood, NY) Axioplan with a 100× plan-apo 1.4 numerical aperture objective, a triple band-pass filter set (63000, Chroma, Brattleboro, VT), and a Z-axis motorized stage (LEP, Hawthorne, NY). The analysis of gene and/or RNA position relative to CBs was done by first viewing the target signal under a single filter set to verify the efficiency of hybridization and then switching to a separate filter set to verify coilin staining before viewing simultaneously. For each experiment, at least two investigators scored the slide separately.
For three-dimensional analysis in Figure 4, a focal series of 20 images at intervals of 0.15 μm were captured. The image stack was then processed with the Scanalytics CELLview 2.0 nearest neighbor deblurring algorithm to remove out-of-focus light. The area of interest (∼3 μm × 3 μm × 2 μm) in the stack was rendered with Brick of Bytes version 1.2 (Graphics and Visualization Laboratory, Army High-Performance Computing Research Center, University of Minnesota).
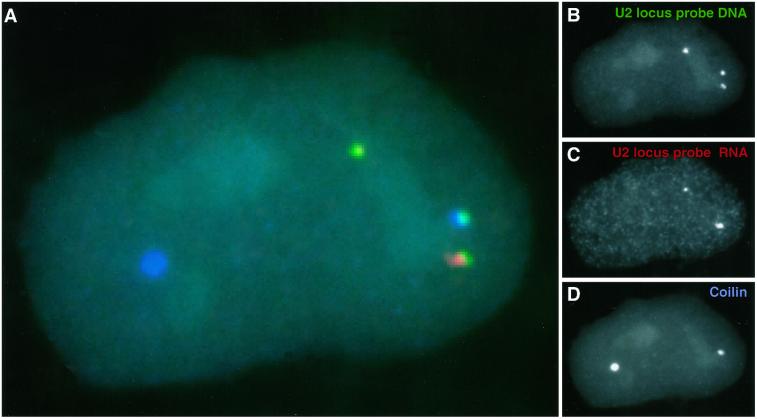
Figure 4.
Three-dimensional visualization of CB, U2 gene locus, and RNA. CB (blue) associated with two U2 loci (green) and RNA from the U2 locus (red). The close association of the CB and the U2 gene locus is evident, whereas the RNA foci do not appear to be as closely associated with the CB.
RESULTS
Detection of RNA Transcripts from the U2 Gene Locus
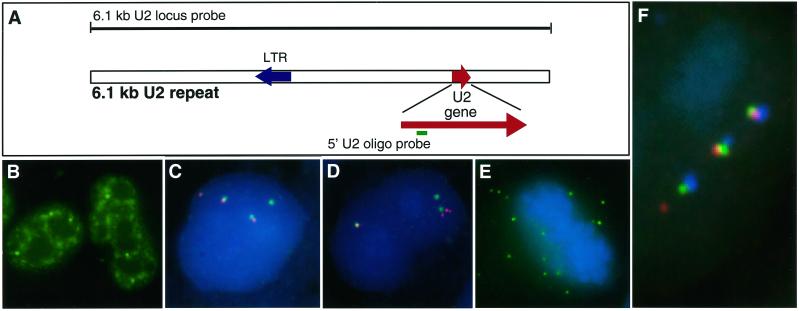
A U2 oligonucleotide probe (Figure 1A) detects a broad nucleoplasmic distribution of mature U2 RNA (Figure 1B). This pattern is consistent with the concentration of U2 snRNPs in CBs and in a “speckled” pattern corresponding to splicing factor–rich domains, as described previously (Carmo-Fonseca et al., 1992; Huang and Spector, 1992). Given the abundance of mature U2 RNA, the challenge was to discriminate new versus mature U2 transcripts. We initially investigated whether newly synthesized transcripts from the U2 locus could be detected in association with the gene with a sequential RNA/DNA hybridization procedure in which the same probe sequence is used to detect RNA and DNA in two distinct colors (Xing et al., 1995). For this purpose, we used as a probe the 6.1-kb locus sequence for the U2 tandem repeat, which includes the U2 gene as well as intergenic sequences (Figure 1A). Results revealed bright focal RNA signals often tightly associated with U2 DNA signal but not completely overlapping it (Figure 1, C and D). The presence or amount of RNA signal was variable, with some U2 loci often associated with a large, bright RNA focus (easily seen through the microscope) but other U2 loci associated with little or no detectable RNA. This variability between RNA signals did not appear to be technical, because within a single nucleus the RNA signals varied markedly between alleles, whereas the corresponding DNA signals were far more uniform (Figure 1, C and D). Although variation in RNA signal intensity was apparent in most cells, ∼30% of HeLa nuclei had at least one U2 allele with which no RNA focus was visible above background.
Figure 1.
Detection of RNA transcripts from the U2 locus. (A) Diagram of the 6.1-kb U2 repeat and positions of the U2 locus probe and 5′ U2 oligonucleotide probe. See Figure 5 for the sequence of the 5′ U2 oligonucleotide. (B) Distribution of U2 snRNA in HeLa nucleus detected with the U2-specific oligonucleotide. (C and D) HeLa cell nucleus hybridized for DNA (green) and RNA (red) with the 6.1-kb U2 gene locus probe shows gene foci without significant RNA signals associated or not associated with a DNA signal. There are three U2 loci in HeLa cells. (E) U2 locus RNA hybridization (green) in a metaphase cell. Numerous discrete foci are evident. (F) HeLa cell nucleus hybridized with the U2 locus probe for U2 DNA (green), RNA (red), and stained with an anti-coilin antibody to visualize CBs (blue). This nucleus shows a CB with each of the three gene locus foci as well as a separate RNA signal, possibly separated from the nearest gene locus.
Remarkably, in ∼30% of cells, discrete RNA signals were observed spatially separate from the DNA loci, sometimes by a considerable distance (Figure 1, C, D, and F). The brightest RNA foci were generally juxtaposed to the gene; however, even when separate from the gene, the RNA accumulations maintained a tight round focus, suggesting some structural constraint to their free diffusion. In a large fraction of cells, there were more U2 RNA foci than U2 DNA signals, further indicating that these foci are not always at sites of transcription. Rather, the results indicate that these “packets” of RNA from the U2 locus have detached and moved from their site of transcription, a phenomenon that has not been seen for any of numerous premRNAs yet studied. These round RNA accumulations are distinct from the more diffuse or track-like distributions reported for (pre)-mRNAs such as collagen and actin (Xing et al., 1995). Additionally, these RNA foci are clearly visible in the cytoplasm of many dividing cells (Figure 1E), further supporting the concept of packets of RNA that persist independent of the gene.
A Large Transcript from the U2 Locus Extends beyond the 3′ Box of “PreU2 RNA”
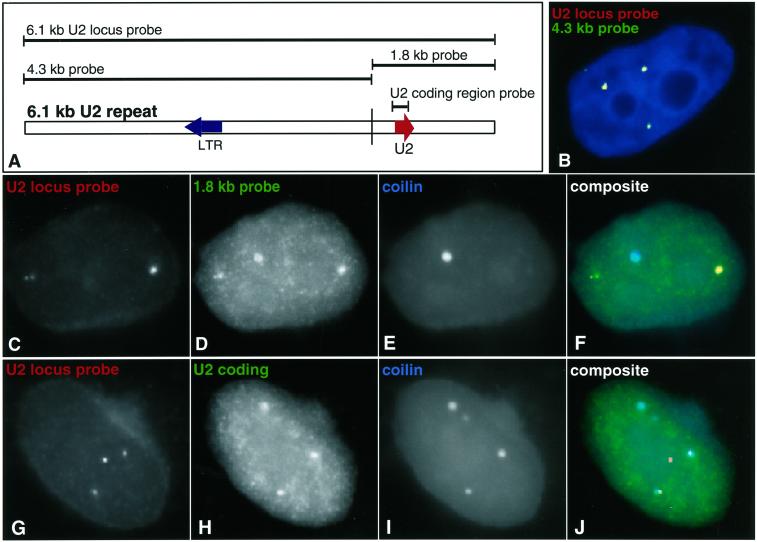
The U2 locus probe detected bright foci of RNA with the gene, yet not the mature U2 transcripts within the CB (Figure 1F). Although this probe contains the mature U2 sequences, we reasoned that the probe complexity must be too high to efficiently detect the mature U2 sequence, which constitutes only 3% of the U2 locus probe sequence. The influence of probe complexity on the sensitivity of detection has been demonstrated directly (Johnson et al., 1993). Our results further suggested that foci of “U2 locus RNA” likely contain a larger transcript, more readily detected by the U2 locus probe. This hypothesis was borne out by a series of analyses with the use of the probes shown in Figure 2A. The 6.1-kb probe was cut, creating a 1.8-kb fragment containing sequences encoding U2 snRNA and a 4.3-kb sequence containing no known U2 RNA coding sequence (Figure 2A). These probes detected distinct RNA distributions. The 4.3-kb probe detected the U2 locus RNA foci seen with the 6.1-kb probe (Figure 2B), thus proving that sequences >1 kb beyond the U2 coding region are transcribed. The 1.8-kb probe (containing U2 RNA as 14% of the sequence) detected both U2 locus RNA foci and faint signal in CBs (Figure 2, C–F). An additional probe containing only the 188-bp mature U2 snRNA sequence detected only snRNA within the CB and very weakly in speckles, but not the U2 locus RNA foci (Figure 2, G–J). Although the small U2 coding sequence probe was seen only in the CB, this is likely because it represents such a small portion of the larger transcript within the U2 locus RNA foci, as further suggested below.
Figure 2.
The U2 locus transcribes more than preU2 RNA. (A) Map of the U2 repeat indicating different probes. (B) A 4.3-kb fragment from the U2 gene locus (red) that does not contain any U2 coding sequences overlaps the focus of RNA detected with the full-length U2 locus probe. Overlap appears yellow. This indicates that DNA outside of the U2 coding sequence is transcribed. (C–F) A 1.8-kb probe detects both U2 RNA in CBs and the RNA focus seen with the full-length probe. (C) U2 RNA signals seen with a full-length 6.1-kb probe. (D) U2 RNA detection with a 1.8-kb probe containing the U2 gene. (E) CB detection. (F) Composite of all three colors. (G–J) U2 coding sequence detects only U2 in CBs with faint nucleoplasmic speckle staining. (G) U2 RNA signals seen with a full-length 6.1-kb probe. (H) U2 RNA detection with a U2 coding sequence probe. (I) CB detection. (J) Composite of all three colors.
Do Differences in U2 Locus RNA Foci Correlate with CB Association?
Using a combination of immunofluorescence for coilin and RNA FISH with the U2 locus probe, we determined the level of association between CBs and the newly synthesized transcripts from the U2 locus. Interestingly, the frequency with which the U2 locus RNA foci associated with CBs was ∼40%, very similar to the frequency with which we found the U2 DNA loci associated with this structure in the same HeLa cells. Also similar to the gene (Smith et al., 1995), the RNA foci localized precisely at the periphery of the CB, with little or no overlap (Figure 1F).
The similar percentages of DNA and RNA signals that associate with CBs might appear to suggest a correlation in which gene loci that associate with CBs have U2 locus RNA foci. To address this directly required simultaneous detection of U2 locus DNA, RNA, and CBs in three distinct colors, as illustrated in Figures 1F and 3. Initial analysis suggested that there was not a consistent correlation between the amount of RNA present and whether the DNA was associated with a CB. In quantifying this relationship in a large cell sample, we scored cells in which there were very clear differences in RNA at different loci, particularly those in which one locus appeared negative for RNA foci. Scoring these triple-label experiments showed that the presence or amount of locus RNA signal did not correlate with the association of that DNA locus with a visible CB (Table 1). In fact, within individual nuclei, examples were seen in which one U2 locus had no detectable RNA and yet was associated with a CB, whereas another locus with a large RNA focus was not (Figure 3, A–D). These results show that the presence (or absence) of a detectable RNA accumulation at the U2 locus does not correlate with or depend on its CB association. Because we cannot strictly infer the transcriptional status of the U2 locus by the absence of these RNA foci, we do not conclude that gene loci without detectable RNA are transcriptionally silent. Nonetheless, we clearly see U2 locus RNA emanating from DNA loci that are not associated with CBs.
Figure 3.
CB association with the U2 gene is not dependent on the presence of U2 locus RNA. (A) Three-color image shows one CB (D, blue) associating with a U2 gene locus (B, green) in the absence of any detectable RNA (C, red), whereas another gene has a significant RNA focus but no CB.
Table 1.
CB association with U2 gene loci relative to RNA accumulation
| Gene only with CB (no RNA detected) | RNA with CB (RNA signal separate from gene) | Gene plus RNA with CB | |
|---|---|---|---|
| Percent of U2 signals scored | 43.3 (52/120)a | 5.0 (2/40) | 51.0 (125/245) |
Three-color detection of U2 gene loci (DNA), RNA (using the 6.1-kb U2 locus probe), and CBs (Fig. 3) was used to determine if CB association was related to the presence of detectable RNA accumulations. The difference in CB association seen in genes with and without detectable RNA accumulations (51 vs. 43%) was not deemed to be significant. RNA signals separated from the genes did not appear to associate with CBs at significant levels.
Number associated/number scored.
Aspects of these results indicate that CBs interact most closely with the DNA locus itself, rather than with the RNA foci. Specifically, U2 DNA signals frequently associate with CB in the absence of detectable U2 RNA foci, but the reverse is not true, i.e., packets of U2 locus RNA rarely associate with CBs in the absence of the gene (Table 1). Additionally, when an RNA focus is oriented to one side of a gene, a CB, if present, typically contacts the gene, but not necessarily the RNA focus (Figures 3 and 4). A representative example of the three-dimensional analysis of U2 gene locus, RNA, and CB is demonstrated by three-dimensional rendering of a three-color DNA/RNA/coilin experiment (Figure 4). The intimate relationship of the two U2 gene loci with the outer edge of the CB is apparent, whereas the RNA foci are not so closely associated (and neither appears to enter the interior of the CB). These results are most consistent with the CB interacting directly with the gene, irrespective of whether a detectable RNA accumulation is present.
PreU2 RNA, but Not the Longer U2 Locus Transcript, Is Found within the CB
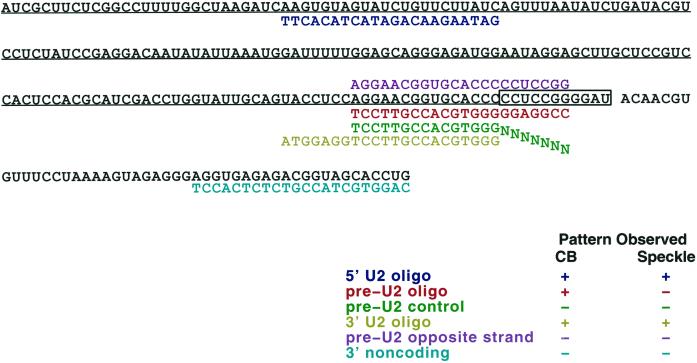
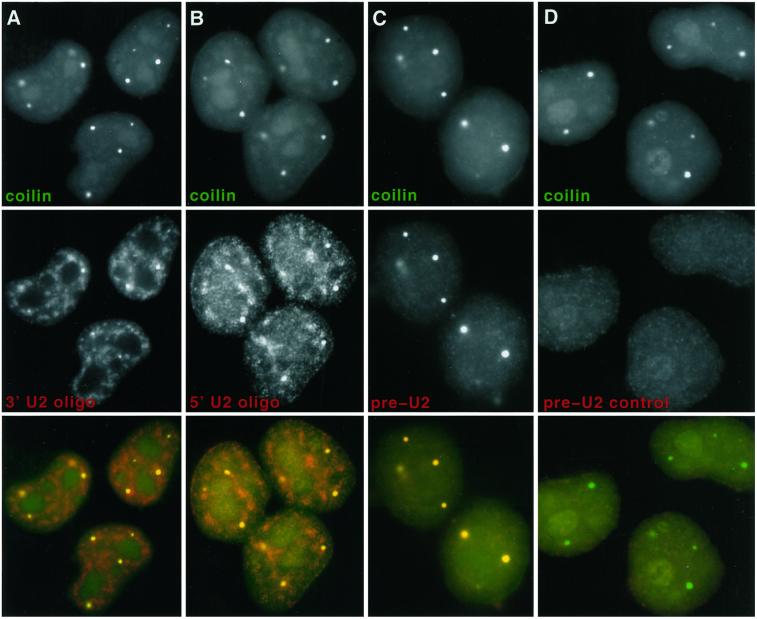
A second strategy was then used to visualize the distribution of immature U2 RNA transcripts within the nucleus, based on specific detection of an 11-nucleotide sequence at the 3′ end of preU2, which is cleaved in the cytoplasm to produce the mature U2 transcript (Wieben et al., 1985). To distinguish between preU2 and mature U2 RNA, we generated oligonucleotides designed to detect this 3′ tail (boxed sequence in Figure 5). A set of oligonucleotide probes was used, each 22 nucleotides long: two that would detect both mature U2 and preU2, one specific to the preU2, and a control oligonucleotide that illustrates the specificity of the preU2 oligonucleotide (Figure 5). The preU2 control oligonucleotide is identical to the preU2 oligonucleotide for the first 15 nucleotides but differs in 7 nucleotides specific to the preU2 tail, providing a negative control to ensure that the 15 nucleotides complementary to the mature U2 are insufficient f eU2 oligonucleotide hybridization produces a signal that overlaps CBs, as seen with anti-coilin antibody. In contrast, the preU2 control produces little or no nuclear signal (Figure 6D). The 5′ and 3′ mature U2 oligonucleotides detect RNA in CBs but also U2 in a more diffuse speckled pattern, corresponding to splicing factor–rich domains known to contain snRNPs (Huang and Spector, 1992) (Figure 6, A and B). Although 15 nucleotides of the 3′ U2 oligonucleotide overlap the preU2 sequence, the two probes produce a different pattern by in situ hybridization. The fact that the preU2 probe did not detect the more diffuse speckled signal seen with both 5′ and 3′ mature U2 oligonucleotides provides further evidence of its specificity for preU2 versus mature U2 RNA. Additionally, the lack of CB staining with the preU2 control shows that the preU2 colocalization with CBs is not an artifact of “bleed-through” of the coilin detection. This experiment was repeated several times with similar results.
Figure 5.
Map of U2 RNA indicating positions of probe oligonucleotides (see Figures 6–8). The underlined sequence is the U2 coding region. The boxed sequence at the 3′ end of U2 represents preU2 tail that is removed in the cytoplasm. The box below the sequence map indicates the name of each oligonucleotide (color coded) and whether or not it detects RNA in CBs and/or splicing factor–rich speckles.
Figure 6.
CBs contain preU2 RNA. (Column A) 5′ U2 oligonucleotide (red) recognizes U2 in CBs (green) and more diffuse U2 in speckles (SC-35 domains). (Column B) 3′ U2 oligonucleotide (red) also recognizes U2 in CBs (green) and more diffuse U2 in speckles. (Column C) The preU2 oligonucleotide (red) detects preU2 in CBs (green) only. (Column D) The preU2 control oligonucleotide (red) does not produce any signal above background.
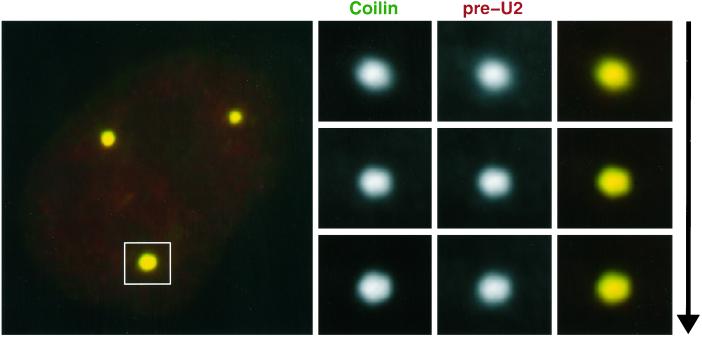
As shown in Figure 7, three-dimensional analysis with the use of optical sectioning confirms what was apparent from two-dimensional visualization: that the preU2 RNA signal is clearly within the CB, not just around it or over it. Interestingly, the preU2 RNA is detected in all CBs, rather than only in CBs associated with the U2 gene loci (see DISCUSSION). The preU2 signal was reasonably intense and easily visible without image processing. To estimate the relative amounts of preU2 and mature U2 in CBs, the average fluorescence signal with each probe was measured under identical conditions. Although this procedure provides only a rough estimate, we measured the preU2 signal within CBs at ∼10–20% of the U2 signal. Thus, it appears that both mature and preU2 RNAs concentrate in CBs, whereas only the mature U2 is found in the rest of the speckled pattern.
Figure 7.
PreU2 is found throughout the CB. To show conclusively that preU2 is inside the CB and not next to or around it, we performed a three-dimensional analysis of overlap between the preU2 signal and coilin staining. Shown are three optical sections through a large CB that are ∼0.2 μm apart. These sections indicate that the preU2 and coilin signals are completely coincident within the CB.
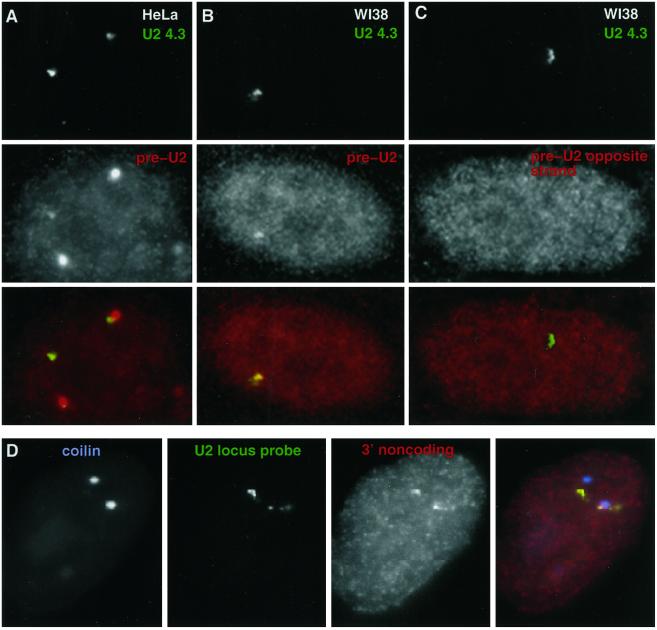
Having demonstrated a sensitive oligonucleotide assay for preU2 RNA, we then used this to determine directly that foci of U2 locus RNA (seen with the large U2 locus probe) contain preU2 RNA. Because it has been reported that there is a putative LTR promoter in each U2 locus tandem repeat that points in the direction opposite the U2 gene (Figure 1A) (Pavelitz et al., 1995), we used strand-specific oligonucleotide probes to investigate whether one or both DNA strands produced RNA. These experiments were also done in WI38 cells, which rarely contain CBs, to avoid confusion regarding whether preU2 signal was in the locus RNA foci or the CB. Using the preU2 oligonucleotide probe or its opposite-strand counterpart (Figure 5), we found that the bright U2 locus RNA foci contain preU2 but not opposite-strand RNA (Figure 8, A–C). The preU2 oligonucleotide signals were faint but usually visible through the microscope and clearly apparent by digital imaging, as seen in Figure 8, A and B. Almost all of the very bright U2 locus RNA signals (generally associated with the gene) contained preU2 signal. However, the opposite-strand probe did not produce a signal above background (Figure 8C), indicating that the U2 locus RNA foci contain preU2 transcribed from the U2 promoter and not from the putative LTR.
Figure 8.
The U2 locus RNA accumulation contains preU2. Cohybridizations with the U2 4.3-kb probe (green) and preU2 (red) or preU2 opposite-strand (red) oligonucleotide probes in HeLa cells (column A) and WI38 fibroblasts (columns B and C). In the HeLa cells, preU2 signal (red) detects CBs and fainter signals (arrows) overlapping the RNA foci detected with the U2 4.3-kb probe (green). WI38 fibroblasts do not have CBs but show a similar overlap of preU2 signal with U2 4.3-kb signal. The preU2 opposite-strand probe does not produce a signal above background. (Row D) 3′ noncoding oligonucleotide probe (red) that hybridizes to a region 27 nucleotides downstream of the end of the preU2 sequence produces a signal that overlaps the RNA focus seen with the 4.3-kb U2 probe (green) but does not overlap CBs (blue). See Figure 5 for sequences and positions of oligonucleotide probes.
Hybridization with an oligonucleotide probe to a sequence ∼30 nucleotides downstream of the U2 coding region (3′ noncoding oligonucleotide; Figure 5) produced a signal that overlaps the U2 locus RNA signal but did not overlap CBs, as does preU2 (Figure 8D). This is further evidence that most of the primary U2 transcripts extend past the 3′ box, as was reported recently by Cuello et al. (1999). However, our results also make the important point that the longer 3′ sequences are not present on the preU2 RNA within CBs. As discussed below, this suggests that somewhere between the gene locus and the CB, sequences that extend beyond the 3′ box are removed from a larger precursor transcript.
Do SMN Gems Associate with U2 snRNA Genes or PreU2 RNA?
Finally, we determined whether nuclear gems mirrored the relationships with U2 seen for CBs. Although in many cell types gems and CBs appear to be coincident (Carvalho et al., 1999), in the HeLa cells studied here they were clearly distinct and separate entities in most cells. Hence, it was compelling to address whether the gems showed similar relationships to U2 genes and RNAs. In our HeLa cells, ∼68% of nuclei had detectable gems, indicated by the accumulation of SMN protein. These nuclei averaged 3.3 CBs and 2.1 SMN gems. Interestingly, gems not associated with CBs showed no preferential relationship with the U2 loci (Table 2). This point further corroborates the specificity and potential functional significance of the U2 loci's very frequent association with CBs. We also examined whether the U2 loci associated with gems in WI38 fibroblasts in which CBs are absent, but we found no specific interaction with U2 loci (Table 3). Although these diploid fibroblasts rarely contain visible CBs, as in HeLa cells ∼66% of nuclei contained at least 1 gem (averaging ∼2.1/nucleus). These results further suggest that CBs and gems are different entities, although they appear to be coincident in many cell types. Finally, we found no evidence of preU2 RNA within the nuclear gems (our unpublished results), suggesting a potential functional difference between CBs and gems in the metabolism of U2 snRNA.
Table 2.
Gems associate with U2 gene loci via CBs
| Percent with CB only | Percent with U2 only | Percent CB/SMN/U2 | |
|---|---|---|---|
| SMN | 34.5 (20/58)a | 6.9 (4/58) | 36.2 (21/58) |
| CB | – | 53.8 (63/117) | – |
The percentage of SMN gems associated with only CBs or with CBs plus U2 gene loci is far greater than the percentage of gems found only with U2 loci, indicating that CB-independent gems do not associate with U2 loci.
Number associated/number scored.
Table 3.
Association of SMN gems with the U2 gene locus is cell dependent
| Percent gems associated with U2 locus | Percent gems close to U2 locus | |
|---|---|---|
| WI38 | 0 (0/42)a | 4.8 (2/42) |
| HeLa | 14.8 (12/81) | 18.5 (15/81) |
SMN gems do not associate with U2 gene loci in WI38 fibroblasts in which CBs are rare, whereas they are often associated with or very close to (less than ∼0.5 μm) CBs in HeLa cells. Approximately 54% of CBs are associated with U2 loci in these HeLa cells (Table 2).
Number associated/number scored.
DISCUSSION
This work provides a number of insights into the interaction of CBs with the U2 gene locus and its RNA products, including several unexpected findings, the most surprising of which is the detection of substantial preU2 signal within CBs. This may initially appear inconsistent with earlier results showing trimethyl-guanosine cap structures, indicative of mature snRNAs, within CBs (Raska et al., 1991; Carmo-Fonseca et al., 1992). However, these results are not at all inconsistent given that we show both mature and immature U2 RNA within CBs. Similarly, our results suggest an explanation for how the presence of preU2 RNA in CBs can be reconciled with their apparent paucity of uridine incorporation (Raska, 1995; Jordan et al., 1997; Schul et al., 1998c), long thought to indicate that they contain no newly synthesized RNA. Our results agree with uridine-labeling studies that most of the U2 in the CB is mature and that transcription per se does not occur within the CB. Moreover, our results also indicate that an unexpectedly large U2 transcript is made outside the CB and that only a much smaller preU2 RNA transcript enters the CB. Hence, only a very small amount of the RNA mass would be found inside the CB, which may be difficult to detect by uridine labeling. Certainly, some caution is warranted in interpreting the results of oligonucleotide hybridizations; however, both the absence of staining with the negative control probe and the clearly distinct distribution of the preU2 RNA and mature U2 RNA oligonucleotides strongly indicate specificity for preU2 RNA. Although we would not conclude that the results presented here provide unequivocal proof that preU2 RNA is within CBs, they clearly provide very strong and unexpected evidence for this important point. This evidence leads us to consider new ways of thinking about potential functions of the CB.
Are CBs Involved in PreU2 RNA Modifications and/or Transport?
Earlier studies indicate that the newly synthesized U2 transcripts must go to the cytoplasm for 5′ cap trimethylation, 3′ end cleavage, and assembly into snRNPs (Wieben et al., 1985; Mattaj, 1988). If preU2 transcripts cannot be assembled or used in the CB, then their presence in the CB is more likely related to some aspect of preU2 RNA metabolism or transport. Although modification of the 5′ cap and cleavage of the 11- to 12-bp 3′ tail occur in the cytoplasm, recent evidence indicates that preU2 RNA must undergo other modifications that occur in the nucleus, including methylation and pseudouridylation (Yu et al., 1998). It is not known whether these modifications occur before or after the RNA goes to the cytoplasm, but factors involved in these modifications have been found in the nucleolar cell fraction, which includes both CBs and nucleoli. Hence, preU2 RNA may either undergo modification in the CB or the CB may transport the preU2 RNA to the nucleolus (for modification) or the nuclear envelope (for cytoplasmic processing).
It is important to note that no similar concentration of preU2 is seen in SMN gems, providing a clue to a functional distinction between these two entities. Not only do the gems (when distinct from the CB) not label for preU2 RNA, they also show no association with U2 gene loci. Given the similar size and number of CBs and gems, this not only provides a clue to a functional distinction between these bodies but also corroborates the specificity of the CB's interaction with U2 genes.
The U2 Gene Locus Produces a Large Transcript
The U2 gene locus transcript was previously thought to be a small 200-nucleotide preU2 RNA (Wieben et al., 1985; Dahlberg and Lund, 1988). A recent report shows that the human U2 gene produces transcripts most of which are at least 250 nucleotides longer than preU2, with some (∼18%) more than 600 nucleotides longer (Cuello et al., 1999). The U2 locus transcripts seen here support and extend this finding and suggest that some of the transcripts could be even larger, given that we detect RNA with a probe that begins ∼1.5 kb downstream from the U2 gene. These results also raise the possibility that the many tandem 6.1-kb U2 repeats could be transcribed as a single large transcript that is then rapidly processed. Although there is no previous evidence for this type of transcript from the U2 locus, the histone gene cluster in newt oocytes is transcribed as a multigene transcript (Bromley and Gall, 1987), as are snoRNA gene clusters in maize (Leader et al., 1997; Shaw et al., 1998). Alternatively, the U2 locus RNA could contain transcripts other than those initiated at the U2 promoter. The U2 repeat unit does contain a putative viral LTR sequence that could possibly support transcription (Pavelitz et al., 1995). However, our strand-specific probes (Figure 7) do not find evidence for this possibility, and to our knowledge there has been no evidence presented for any other transcribed sequences in this repeat.
Another novel observation of the U2 DNA and RNA hybridizations is the separation of some RNA foci from the gene locus (Figure 1, C, D, and F). It is possible that the RNA packets that have detached from the gene could be nonfunctional transcripts in the process of being degraded; however, they may also be sites where a larger U2 RNA precursor is undergoing some step in nuclear processing. Thus, our results indicate that there is a large transcript produced by the U2 locus and that this focus of RNA contains, at least in part, preU2 (Figure 8). Although the exact nature of the RNA in this focus is unclear at this time, we believe that it still provides a marker indicative of the transcriptional activity of the U2 locus.
Foci of U2 Locus RNA Show No Simple Correlation with CBs
Given that in our HeLa cells only ∼45% of U2 gene loci associate with a CB at any given time, it was important to determine whether differences would be observed in RNA at these loci. Interestingly, we do find marked differences in the amount of RNA associated with different U2 loci within the same cell. Although some variability in RNA amount is observed between alleles for protein-coding genes, for the U2 locus these allelic differences are much more frequent and pronounced. Although we cannot rule out the possibility that our approach may miss low levels of the nascent preU2 transcripts on the gene, the presence or absence of RNA foci at different alleles did not simply correlate with the gene's association with a CB (Figure 3 and Table 1).
Our results indicate that U2 gene expression is independent the gene's immediate association with the CB. However, this association could still be involved in U2 RNA regulation within a population. Production of the snRNAs is reportedly regulated by dosage compensation, which keeps their relative levels constant (Mangin et al., 1985). It remains possible that the CB/gene interaction is involved in regulating the production (up or down) of basal cellular metabolic components, analogous to a “thermostat” that indicates and integrates basal metabolic needs within the cell. CB associations could regulate a U2 locus but, because our observations are just a snapshot in time, the presence or absence of RNA foci would not necessarily reflect the dynamic transcriptional status of the U2 genes.
A recent report (Frey et al., 1999) concluded that there is a positive correlation between CB association and U2 gene expression, suggesting that the association is not just fortuitous but functionally significant. This was based on an analysis of the frequency (within a cell population) of CB–U2 gene associations and the general cellular level of snRNA detected by primer extension. Although the results from that study are consistent with RNA at a given locus mediating interactions with the CB, this is not shown directly. Our direct visualization of the genes, RNA, and CBs suggests that neither the presence nor absence of RNA is correlated with the gene's interaction with the CBs. The fact that we see no correlation between U2 RNA foci at the locus and its association with the CB does not fit easily with the hypothesis that sustained association with the CB is required to maintain gene activity or gene repression. However, our data do not rule out some involvement in regulation, but they indicate that, if such regulation exists, it would involve a mechanism in which transient interaction with the CB results in a sustained impact on gene expression.
Our findings leave open the possibility that the CB interaction may be with the gene rather than its RNA, because genes without detectable RNA foci frequently associate but RNA foci without genes only rarely contact CBs. Our three-dimensional analysis of gene, RNA, and CB also suggests a closer spatial association with the gene locus than with the RNA foci (Figure 4).
Because preU2 RNA is in all CBs but only a subset is associated with gene loci, these puzzling results preclude a simple model in which only the CBs associated with U2 genes contain preU2. We can envision three possibilities that could account for this finding. The first would involve the formation of CBs at U2 gene loci. The idea that U2 loci are involved in the nucleation or biogenesis of CBs was previously suggested based on the appearance of very small CBs at some U2 loci (Smith et al., 1995). Second, CBs could rapidly move through the nucleus. The CB has often been described as a kinetic structure, and most evidence would suggest movement between different associations in the nucleus. Our observations are only a snapshot in time, so it could conceivably be difficult to catch a CB without preU2 in it, especially if the larger CBs coalesce from smaller ones moving together from different locations. A recent study that used a GFP–U2B fusion protein in plant cells reports CB movement and coalescence (Boudonck et al., 1999). This finding is in agreement with recent preliminary results from our laboratory with the use of GFP–coilin in HeLa cells (R. Tam, unpublished observation). Third, the preU2 could move to the CB by diffusion or a directed process. If so, then the CB associations with the U2 gene locus would not necessarily be related to any potential role in preU2 metabolism, but it could still reflect a distinct function related to gene regulation.
The CB is a dynamic nuclear structure that may not have a singular function but rather multiple interrelated functions. The potential involvement of CBs in the assembly of snRNPs and RNA metabolic complexes could be unrelated to the presence of preU2 RNA in CBs or their specific association with U2 genes. However, we favor the view that the Cajal (coiled) body is a structure that reflects and may serve to integrate and balance multiple functions related to the biogenesis of RNA metabolic components.
ACKNOWLEDGMENTS
We thank Meg Byron, Carol Johnson, and John McNeil for their excellent technical assistance. We also thank Dr. Lindsay Shopland for her critical review of the manuscript and Dr. Philip Moen for his help with oligonucleotide hybridization. This study was made possible by grants from the National Institutes of Health (GM49254) and the Muscular Dystrophy Association to J.B.L. and a development grant from the Muscular Dystrophy Association to K.P.S. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the Muscular Dystrophy Association.
REFERENCES
- Abbott J, Marzluff WF, Gall JG. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Lamond A. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley SE, Gall JG. Transcription of the histone loci on lampbrush chromosomes of the newt Notophthalmus viridescens. Chromosoma. 1987;95:396–402. doi: 10.1007/BF00333990. [DOI] [PubMed] [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello P, Boyd DC, Dye MJ, Proudfoot NJ, Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 1999;18:2867–2877. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg JE, Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel ML, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Heidelberg, Germany: Springer-Verlag; 1988. pp. 38–70. [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The non-chromatin substructures of the nucleus: the ribonucleoprotein RNP-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells [published erratum appears in Proc. Natl. Acad. Sci. USA (1995) 92, 8532] Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort HT, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector D. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell. 1999;10:1653–1663. doi: 10.1091/mbc.10.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Rhoadhouse M, Pederson T. U2 small nuclear RNA 3′ end formation is directed by a critical internal structure distinct from the processing site. Mol Cell Biol. 1993;13:1119–1129. doi: 10.1128/mcb.13.2.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CV, Cool DE, Glaccum MB, Green N, Fischer EH, Bruskin A, Hill DE, Lawrence JB. Isolation and mapping of human T-cell protein tyrosine phosphatase sequences: localization of genes and pseudogenes discriminated using fluorescence hybridization with genomic versus cDNA probes. Genomics. 1993;16:619–629. doi: 10.1006/geno.1993.1239. [DOI] [PubMed] [Google Scholar]
- Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: implication for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- Jordan P, Cunha C, Carmo-Fonseca M. The cdk7-cyclin H-MAT1 complex associated with TFIIH is localized in coiled bodies. Mol Biol Cell. 1997;8:1207–1217. doi: 10.1091/mbc.8.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader DJ, Clark GP, Watters J, Beven AF, Shaw PJ, Brown JW. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Amalric F, Luhrmann R, Vogel P, Fakan S. Is the coiled body involved in the nucleolar functions? Exp Cell Res. 1994;211:415–419. doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- Mangin M, Ares M, Jr, Weiner AM. U1 small nuclear RNA genes are subject to dosage compensation in mouse cells. Science. 1985;229:272–275. doi: 10.1126/science.2409601. [DOI] [PubMed] [Google Scholar]
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. UsnRNP assembly and transport. In: Birnstiel ML, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Heidelberg, Germany: Springer-Verlag; 1988. pp. 100–114. [Google Scholar]
- Pavelitz T, Rusche L, Matera AG, Scharf JM, Weiner AM. Concerted evolution of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. EMBO J. 1995;14:169–177. doi: 10.1002/j.1460-2075.1995.tb06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal SR. Un sencillo metodo de coloracion seletiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados y invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J Cell Biochem. 1995;59:11–26. doi: 10.1002/jcb.240590103. [DOI] [PubMed] [Google Scholar]
- Raska I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Raska I, Ochs RL, Andrade LEC, Chan EKL, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Schul W, de Jong L, van Driel R. Nuclear neighbors: the spatial and functional organization of genes and nuclear domains. J Cell Biochem. 1998a;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Schul W, Groenhout B, Koberna K, Takagaki Y, Jenny A, Manders E, Raska I, van Driel R, de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell. 1998b;9:1025–1036. doi: 10.1091/mbc.9.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp Cell Res. 1998c;238:1–12. doi: 10.1006/excr.1997.3808. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Beven AF, Leader DJ, Brown JW. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Van Arsdell SW, Weiner AM. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol. 1984;4:492–499. doi: 10.1128/mcb.4.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieben ED, Nenninger JM, Pederson T. Ribonucleoprotein organization of eukaryotic RNA. XXXII. U2 small nuclear RNA precursors and their accurate 3′ processing in vitro as ribonucleoprotein particles. J Mol Biol. 1985;183:69–78. doi: 10.1016/0022-2836(85)90281-5. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil JA, Lawrence JB. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]