Abstract
The replication of positive-strand RNA viruses involves not only viral proteins but also multiple cellular proteins and intracellular membranes. In both plant cells and the yeast Saccharomyces cerevisiae, brome mosaic virus (BMV), a member of the alphavirus-like superfamily, replicates its RNA in endoplasmic reticulum (ER)-associated complexes containing viral 1a and 2a proteins. Prior to negative-strand RNA synthesis, 1a localizes to ER membranes and recruits both positive-strand BMV RNA templates and the polymerase-like 2a protein to ER membranes. Here, we show that BMV RNA replication in S. cerevisiae is markedly inhibited by a mutation in the host YDJ1 gene, which encodes a chaperone Ydj1p related to Escherichia coli DnaJ. In the ydj1 mutant, negative-strand RNA accumulation was inhibited even though 1a protein associated with membranes and the positive-strand RNA3 replication template and 2a protein were recruited to membranes as in wild-type cells. In addition, we found that in ydj1 mutant cells but not wild-type cells, a fraction of 2a protein accumulated in a membrane-free but insoluble, rapidly sedimenting form. These and other results show that Ydj1p is involved in forming BMV replication complexes active in negative-strand RNA synthesis and suggest that a chaperone system involving Ydj1p participates in 2a protein folding or assembly into the active replication complex.
Genetic and biochemical results suggest that the replication of RNA viruses involves numerous host factors (reviewed in references 5 and 36). Identifying such host factors and elucidating their function are therefore necessary to understand the molecular mechanisms of RNA virus replication. To facilitate such host factor analysis, it is advantageous to use a genetically tractable host with strong cell biology and biochemistry. One such host is the yeast Saccharomyces cerevisiae. Studies of intrinsic double-stranded RNA replicons in S. cerevisiae have provided significant information not only on the replication mechanisms of these replicons but also on a wide variety of cellular processes (reviewed in reference 51). Among higher eukaryotic positive-strand RNA viruses, brome mosaic virus (BMV), which belongs to the alphavirus-like superfamily, and flock house virus have been shown to replicate their RNA, synthesize mRNA, and assemble virions in S. cerevisiae (29, 41).
The genome of BMV consists of three 5′-capped messenger- sense RNAs. RNA1 and RNA2 encode replication proteins 1a and 2a, respectively. 1a contains domains implicated in RNA helicase and RNA capping functions, and 2a contains an RNA-dependent RNA polymerase (RdRp) domain. These three domains are shared with the members of alphavirus-like superfamily (reviewed in reference 1). 1a interacts with 2a in vivo and in vitro (11, 18, 31, 32, 40) and is targeted to the endoplasmic reticulum (ER) membranes in plant cells and in yeast (43, 44). RNA3 encodes two proteins, the 3a protein, which is necessary for cell-to-cell movement of the virus in plants, and the coat protein. The coat protein is translated from subgenomic RNA4 that is produced by internal initiation of RNA synthesis from full-length negative-strand RNA3 (1).
Prior to negative-strand RNA synthesis of BMV, 1a forms spherules budding into the ER lumen, and BMV RNA templates are sequestered in the spherules in a state isolated from cytoplasmic machineries for translation and mRNA degradation (12, 30, 47). Template selection by 1a requires a box B containing sequence present in the 5′ noncoding regions (NCRs) of RNA1 and RNA2 or in the intercistronic region of RNA3 (12, 50) and host protein Lsm1p in trans (17). 1a also recruits the 2a polymerase into the spherules (11, 44, 47). Initiation of negative-strand RNA synthesis requires these steps and a specific membrane lipid composition (38). In the spherules, negative-strand RNAs are synthesized, retained, and used as templates for the synthesis of positive-strand RNAs that are to be exported to the cytoplasm (47).
Previously, to identify host factors required for BMV RNA replication, Ishikawa et al. isolated yeast mutants in which BMV RNA replication was inhibited (26). Here we report the results of analysis of one of those yeast mutants, mab3. We find that the mutant had a defect in YDJ1, a gene encoding a cytoplasmic Escherichia coli DnaJ homologue (3, 8, 10, 14). The encoded protein Ydj1p is known to assist the function of molecular chaperones Hsp70 (13, 14, 15, 39) and Hsp90 (34) and is involved in protein folding (16, 39), protein translocation across membranes (3, 7), assembly of macromolecular complexes (23), and protein degradation (37). In this study, we investigated how the mab3/ydj1 mutation affected BMV RNA replication.
MATERIALS AND METHODS
Yeast strains and cell growth.
Yeast strain YMI04 was described previously (26). ydj1i yeast was constructed from YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) by two-step gene replacement (4) with the 3.2-kbp SphI-BamHI genomic DNA fragment encompassing the mutation site (initiation codon of Ydj1p) obtained from the mab3 mutant by gap repair (4). Yeast cultures were grown at 30°C in a defined synthetic medium containing 2% glucose or 2% galactose as indicated. Relevant amino acids were omitted to maintain DNA plasmids (4). To insure galactose induction, yeast cells grown in glucose medium were transferred to synthetic galactose liquid medium for 2 days and were then transferred once more to fresh galactose liquid medium and grown to mid-logarithmic phase (optical density at 600 nm = 0.4 to 0.7) unless otherwise stated. Cells were harvested by centrifugation, and the resulting cell pellets were stored at −80°C until RNA or protein extraction.
Plasmids.
The 1a and 2a proteins were expressed from pB1CT19 and pB2CT15, respectively (29). BMV RNA3, B3GUS, and B3URA3 were expressed from pB3RQ39 (42), pB3MI22 (26), and pB3MI8 (27), respectively. B3CAT RNA and luciferase mRNA were synthesized by in vitro transcription from pB3CA101 (29) and pGEM-luc (Promega), respectively. p1148 was constructed by inserting the 3.2-kb XhoI-ApaI fragment encompassing the wild-type (wt) YDJ1 gene between the SalI-XbaI sites of a centromeric plasmid vector, YCplac22 (20). pYDJ1 was constructed by subcloning the 3.2-kb SphI-BamHI fragment of p1148 between the SphI-BamHI sites of YCplac33 (20).
RNA and protein analysis.
Extraction and Northern hybridization analysis of total yeast RNA were performed as described previously (12, 29). 32P-labeled RNA probes to detect positive- and negative-strand BMV RNA3 and -4 were generated from pBCPSN1 (28). The hybridization signals were detected and quantified using a bioimaging analyzer (model no. BAS1000; Fuji Photo Film, Tokyo, Japan). Protein extraction, electrophoresis, Western blot analysis, and subcellular fractionation of yeast cell lysates were performed as described previously (11). Polyclonal anti-1a and monoclonal anti-2a antibodies (43) and monoclonal anti-phosphoglycerate kinase antibody (Molecular Probes) were used at 10,000-, 3,000-, and 500-fold dilutions, respectively.
Measurement of the activity of reporter gene products.
A β-glucuronidase (GUS) enzymatic assay was performed as described previously (26). Protein concentrations of lysates were determined with a protein assay kit (Bio-Rad). Synthesis of capped mRNAs, RNA transfection into yeast, and enzymatic assays of chloramphenicol acetyltransferase (CAT) and luciferase were performed as described previously (29, 38).
BMV RdRp assay.
BMV RdRp extract was prepared from a protease-deficient BJ5465 yeast (obtained from the Yeast Genetic Stock Center, University of California at Berkeley) expressing BMV 1a and 2a and replicating B3URA3 as described previously (42), except that intact yeast cells were disrupted by using glass beads as described by Iizuka et al. (25). To reduce the concentration of Ydj1p, BMV RdRp extract was loaded onto a DEAE Bio-Gel A (Bio-Rad) column equilibrated by buffer D (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 25 mM dithiothreitol, 1 mM EGTA, 1 mM benzamidine-HCl, 15% [vol/vol] glycerol, 25 μg of aprotinin/ml, 5 μg of leupeptin/ml, 5 μg of pepstatin/ml) containing 2% (vol/vol) Triton X-100 (buffer DT). The column was washed with buffer DT containing 150 mM KCl, and BMV RdRp was eluted with buffer DT containing 250 mM KCl. Polyclonal antibodies against Ydj1p were prepared by injecting E. coli-expressed, hexahistidine-tagged Ydj1p into rabbits. To deplete Ydj1p from the DEAE-purified RdRp extract, the RdRp extract was mixed with affinity-purified anti-Ydj1p antibody bound to protein A-Sepharose 4 Fast Flow (Amersham Pharmacia Biotech) and incubated at 4°C for 1 h. After the removal of the Sepharose beads by centrifugation, the supernatant was used for RdRp assays and Western blotting to confirm the depletion of Ydj1p. RdRp assays were performed using exogenously added BMV virion RNA as a template as described by Quadt et al. (42). Using the pET system, Ydj1p was expressed in E. coli and purified as described by Cyr et al. (15). The concentration of the purified Ydj1p was determined by comparing the band intensity on a Coomassie brilliant blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide electrophoresis gel with that of known amounts of marker protein. The purified Ydj1p (1.4 nM) activated the ATPase activity of Ssa1p (Hsp70) (0.2 nM) approximately threefold, confirming Ydj1p functionality.
RESULTS
To identify host factors involved in BMV RNA replication and transcription, we previously constructed and used yeast strain YMI04, which expresses 1a and 2a from a constitutive ADH1 promoter and replication-competent BMV RNA3 derivatives B3URA3 and B3GUS (in which the coat protein open reading frame was replaced by that for yeast uracil biosynthetic genes URA3 and E. coli GUS, respectively) from a galactose-inducible GAL1 promoter (26). Upon galactose induction, positive-strand RNA3 derivatives are synthesized by host RNA polymerase II and serve as templates for 1a- and 2a-dependent replication and transcription of subgenomic mRNAs (Fig. 1), resulting in Ura3p and GUS expression. Ura3p and GUS expression in this system is thus completely dependent on BMV RNA replication (to make negative-strand RNA) and subgenomic mRNA synthesis (27). From UV-mutagenized YMI04, mutants with simultaneous reduction of Ura3p and GUS expression in galactose medium were isolated (26). One such mutant, mab3, was recessive and unable to grow at 37°C and showed reduced accumulation of positive- and negative-strand BMV RNA3 replication products but increased accumulation of 2a (26).
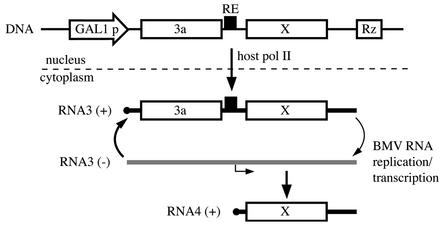
FIG. 1.
Schematic diagram of DNA-based expression of BMV RNA3 replicons and subsequent BMV-directed, RNA-dependent RNA replication and subgenomic mRNA synthesis. The cDNA-based RNA3 launching cassette includes BMV NCRs, the 3a gene, an intergenic replication enhancer (RE), a 5′-end-flanking GAL1 promoter, and 3′-end-flanking ribozymes (Rz) from hepatitis delta virus or hammerhead ribozyme. X represents the BMV coat protein gene or its replacements, the coding sequences for GUS or Ura3p. Upon galactose induction, cellular RNA polymerase II-dependent transcription synthesizes positive-strand RNA3 transcripts that serve as the templates for 1a- and 2a-dependent RNA3 replication and synthesis of subgenomic mRNA (RNA4) required for expression of the coat gene or its replacements.
MAB3 encodes Ydj1p.
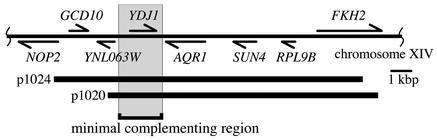
To identify the MAB3 gene, the mab3 mutant was transformed with a yeast genomic library (45) and two transformants were isolated that grow at 37°C. These transformants harbored plasmid p1020 or p1024 containing overlapping yeast genomic DNA fragments derived from chromosome XIV (Fig. 2). Deletion mapping showed that an AflII-XhoI DNA segment of 2.1 kb containing a single complete open reading frame of YDJ1 complemented the mab3 growth defect at 37°C and BMV-replication-dependent GUS expression from in vivo-transcribed B3GUS (Fig. 2 and data not shown). The same 2.1-kb genomic DNA fragment containing YDJ1 also restored the reduced accumulation of positive- and negative-strand RNA replication products to wt levels in the original mab3 mutant (data not shown).
FIG. 2.
Cloning of the MAB3/YDJ1 gene. A region of S. cerevisiae chromosome XIV corresponding to base coordinates 511000 to 495000 is shown. Arrows indicate the positions of genes and the directions of transcription. The regions carried by yeast genomic clones p1024 and p1020, which complemented the temperature-sensitive growth and low-level BMV-directed GUS gene expression in the mab3 mutant, are shown by thick lines. The square bracket shows the minimal complementing region corresponding to the 2.1-kb AflII-XhoI fragment containing the complete YDJ1 open reading frame.
To determine whether YDJ1 was the originally mutated gene or an extragenic suppressor acting on another mutant gene, the relevant genomic DNA region was cloned from the mab3 mutant by gap repair (4). Sequencing of this DNA revealed a single base substitution at the predicted Ydj1p start codon (ATG to ATA) in the mab3 mutant. While AUA can serve as an initiation codon at a low efficiency (21), Ydj1p was not detected in the mutant (Fig. 3E) and the growth phenotype of the mab3 yeast resembled that of a ydj1 deletion mutant (8). Therefore, this mutation inhibits Ydj1p production almost completely.
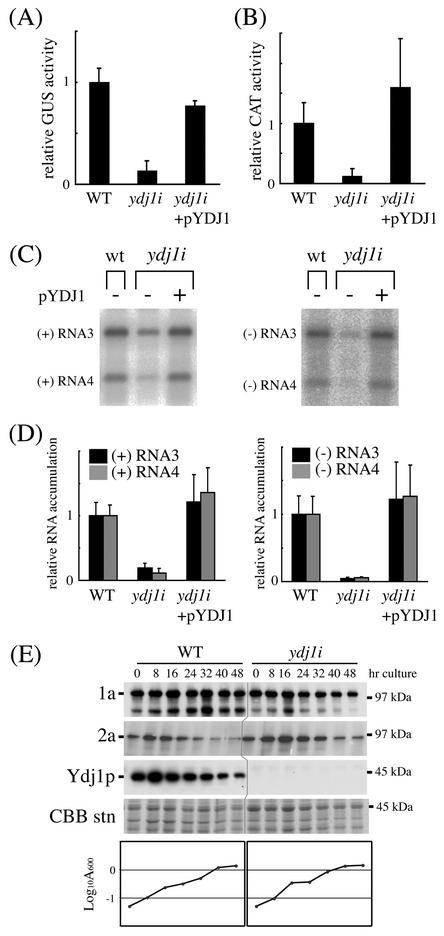
FIG. 3.
The phenotype of the ydj1i yeast. The 1a and 2a proteins were expressed from pB1CT19 and pB2CT15, respectively, in wt (YPH500) and ydj1i yeast with (+) and without (−) pYDJ1. (A) BMV-directed GUS expression from a DNA platform. B3GUS was expressed in vivo from a GAL1 promoter. Cell cultures were grown in a defined galactose liquid medium, and GUS activity was determined as described in Materials and Methods. The histogram shows averages and standard deviations of relative GUS activity (the average for wt = 1) from three or four transformants. (B) BMV-directed CAT expression from directly transfected B3CAT RNA. Cells were cotransfected with in vitro-synthesized B3CAT and luciferase RNA, and CAT and luciferase activity was determined as described in Materials and Methods. The histogram shows averages and standard deviations of relative ratios of CAT activity to luciferase activity (the average for wt = 1) from four transformants. (C and D) Accumulation of BMV RNA3-related RNAs. BMV RNA3 was expressed in vivo from a GAL1 promoter.Cell cultures were grown in a defined galactose liquid medium, and RNA was analyzed as described in Materials and Methods. Northern blots were probed with 32P-labeled RNA hybridizing with positive-strand (left panels) and negative-strand (right panels) BMV RNA3 and -4 and autoradiographed. A representative result of Northern hybridization is shown in panel C. Histograms in panel D show averages and standard deviations of relative RNA accumulation (the average for wt = 1) from five or three transformants. The accumulation of each RNA species was quantified and normalized to that of actin mRNA. (E) Accumulation of the 1a and 2a proteins. Two-day cultures of yeast in a defined galactose liquid medium were inoculated in a fresh galactose medium to give an initial A600 = 0.05 and grown at 30°C as described in Materials and Methods. At each time point, total protein was extracted and subjected to Western analyses using antibodies against 1a, 2a, or Ydj1p. Parts of Coomassie brilliant blue-stained gels (from 0.1% SDS-9% polyacrylamide gel electrophoresis ) are shown to indicate the amount of loaded protein. Positions of molecular mass markers are shown at the right. Growth curves are also shown. Similar results were obtained in two other independent experiments.
To further test the possibility that the mab3 phenotype was derived from the ydj1 mutation alone, this mutation was introduced into a wt yeast (YPH500) by two-step gene replacement (4). The resulting ydj1 isogenic strain, referred to as ydj1i, reproduced the following phenotypes of the original mab3 mutant. First, cells were unable to grow at 37°C. Second, BMV-directed GUS expression from B3GUS RNA introduced by in vivo transcription by host RNA polymerase II was reduced (original mab3 expression = 18% of that of wt; ydj1i expression = 13% of wt) (Fig. 3A). Third, BMV-directed CAT expression from B3CAT RNA introduced by direct transfection of in vitro-synthesized capped RNA was reduced (original mab3 expression = 28% of that of wt; ydj1i expression = 12% of wt) (Fig. 3B), indicating that this defect is independent of DNA-directed transcription. Fourth, the accumulation of BMV RNA3 replication products (positive- and negative-strand RNA3 and -4) was reduced (Fig. 3C and D). All of these phenotypes were simultaneously suppressed by introduction of a plasmid carrying the wt YDJ1 gene (Fig. 3A to D). These results indicate that the ydj1 mutation alone caused most of the phenotypes of the mab3 mutant strain. Accordingly, we used the ydj1i strain for further analysis to eliminate any effects (caused by potential mutations outside the YDJ1 gene) which might have been generated in the original mab3 mutant strain upon mutagenesis. In contrast to the original mab3 mutant, in which 2a protein strongly overaccumulated, 2a accumulation in ydj1i yeast was only slightly higher than that in wt yeast (Fig. 3E), suggesting that this phenotype was not caused by the ydj1 mutation alone.
ydj1 mutation inhibits negative-strand RNA3 accumulation.
In experiments using the wt RNA3 as a model replicon, the cyclical nature of RNA replication (Fig. 1) means that specific inhibition of either positive- or negative-strand RNA3 results in reduced accumulation of both strands. Moreover, wt RNA3 expression of BMV coat protein, which stabilizes positive-strand RNA3 and -4 by encapsidation (35, 50), further complicates interpretation of RNA analysis data.
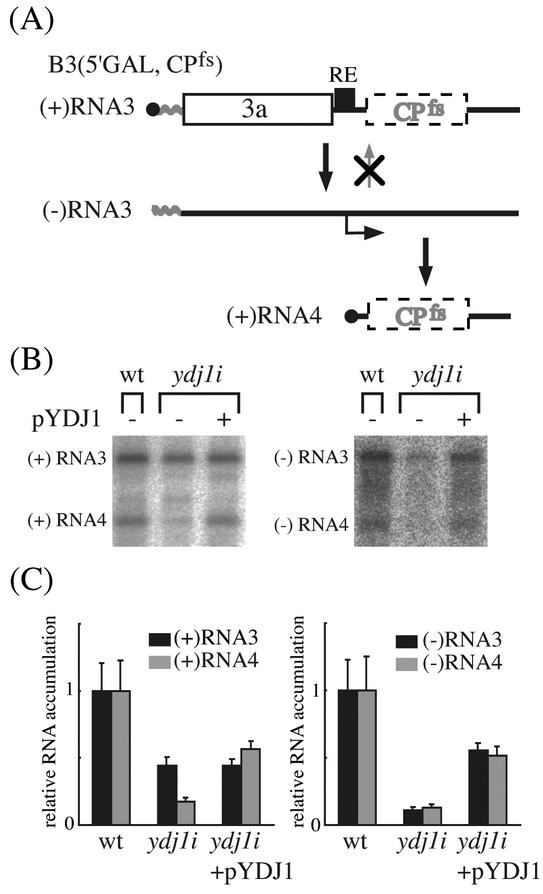
Thus, to examine negative-strand RNA synthesis directly, we utilized a model RNA B3(5′GAL, CPfs) in which the wt BMV 5′ NCR was replaced with the 5′ NCR of the yeast GAL1 mRNA and coat protein expression was eliminated by a 4-base frame-shifting insertion immediately after the initiating AUG codon (Fig. 4A) (38). The resulting B3(5′GAL, CPfs) transcript lacks a cis-acting signal for positive-strand synthesis and so was supplied solely by DNA-dependent transcription. As shown in Fig. 4B and C, positive-strand B3(5′GAL, CPfs) RNA accumulated in ydj1i yeast to 44% of the wt level. In contrast, negative-strand RNA3 and RNA4 accumulation was 11% and 13% of the wt level, respectively (Fig. 4C). Positive-strand RNA4, which is synthesized using negative-strand RNA3 as a template (Fig. 1), was also reduced (18% of wt) (Fig. 4B and C). The reduced accumulation of negative-strand RNA3 and positive-strand RNA4 was recovered, if not completely (to 55% of wt level for negative-strand RNA3 and to 56% of wt level for positive-strand RNA4), by introducing the wt YDJ1 gene on a plasmid. These results suggested that BMV RNA replication is inhibited at or before negative-strand RNA synthesis in ydj1i yeast.
FIG. 4.
Inhibition of accumulation of negative-strand B3(5′GAL, CPfs) in ydj1i yeast. (A) Schematic representation of BMV-directed RNA synthesis pathway for a BMV RNA3 derivative, B3(5′GAL, CPfs). B3(5′GAL, CPfs) was constructed by replacing the viral 5′ NCR of B3CPfs (38) with the 5′ NCR of yeast GAL1 mRNA and was deficient in the initiation of positive-strand RNA synthesis. (B and C) Accumulation of positive- and negative-strand B3(5′GAL, CPfs). B3(5′GAL, CPfs) was expressed in vivo from a GAL1 promoter. Cell cultures were grown in a defined galactose liquid medium and RNA was extracted from the cells and analyzed by Northern hybridization as described in Materials and Methods. Northern blots were probed with 32P-labeled RNA hybridizing with positive-strand (left panels) and negative-strand (right panels) BMV RNA3 and -4 and autoradiographed. A representative result of Northern hybridization is shown in panel B. Histograms in panel C show averages and standard deviations of relative RNA accumulation (the average for wt = 1) from three transformants. The accumulation of each RNA species was quantified and normalized to that of actin mRNA.
1a-dependent recruitment of RNA3 and 2a to membranes in ydj1i yeast.
1a recruits BMV positive-strand RNA replication templates to membranes and stabilizes them, and this process appears to reflect recruitment of the template RNA away from translation and into RNA replication (12, 50). 1a also recruits the otherwise soluble, polymerase-like 2a protein to membranes (11). These processes, which are thought to take place prior to negative-strand synthesis, are crucial for BMV RNA replication (47). So we next examined whether the ydj1 mutation affects 1a-dependent stabilization of positive-strand RNA3 and membrane association of RNA3 and 2a.
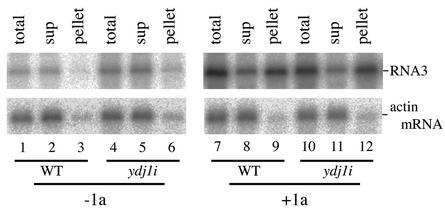
To examine 1a-dependent RNA3 stabilization and recruitment to membranes, wt and ydj1i yeast strains expressing RNA3 from the GAL1 promoter with or without 1a were established. Northern blot analysis showed that 1a coexpression increased RNA3 accumulation approximately 10-fold in wt yeast and approximately 7-fold in ydj1i yeast (Fig. 5 [compare lane 7 to lane 1 for wt and lane 10 to lane 4 for ydj1i] and data not shown). To examine 1a-dependent RNA3 recruitment to membranes, the same yeast strains were treated with lyticase and the resulting spheroplasts were lysed osmotically. The lysate was centrifuged at 10,000 × g to give a membrane-rich pellet and a cytoplasmic supernatant, from which RNA was extracted. Northern analysis demonstrated that, like actin mRNA, the most RNA3 was detected in the supernatant fraction in wt and ydj1i yeast in the absence of 1a (Fig. 5, lanes 1 to 3 for wt and lanes 4 to 6 for ydj1i). In contrast, in the presence of 1a, a majority of RNA3 fractionated to the pellet both in wt and ydj1i yeast, while actin mRNA remained predominantly in the supernatant (Fig. 5, lanes 7 to 9 for wt and lanes 10 to 12 for ydj1i). These results suggest that 1a-dependent RNA3 stabilization and recruitment to membranes was not significantly affected by the ydj1 mutation.
FIG. 5.
Effects of ydj1 mutation on 1a-dependent recruitment of RNA3 to membranes. Yeasts expressing BMV RNA3 with (+1a) or without (−1a) 1a protein were grown in defined galactose medium, spheroplasted, and lysed osmotically. The lysate was then centrifuged at 10,000 × g to yield pellet and supernatant (sup) fractions as described by Chen and Ahlquist (11). RNA was purified by phenol-chloroform extraction and ethanol precipitation, and an equal percentage of each fraction was analyzed by Northern blot hybridization to detect positive-strand RNA3 or cellular actin mRNA. In the lanes labeled “total,” RNA from the unfractionated lysate of the same volume as the supernatant fraction was analyzed.
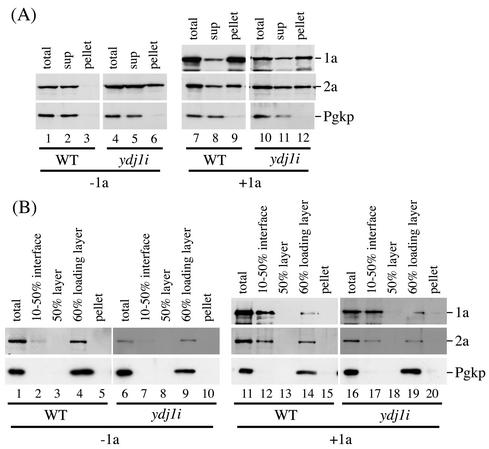
To examine the 1a-dependent recruitment of 2a to membranes, cell lysates from wt and ydj1i yeast expressing 2a either alone or with 1a were prepared and fractionated as described above. A soluble cytoplasmic protein, phosphoglycerate kinase, was followed as a fractionation control (Fig. 6A). In wt cells, 2a fractionated mainly to the supernatant when expressed alone (Fig. 6A, lanes 1 to 3) but fractionated to both the membrane-containing pellet and the supernatant when expressed with 1a (Fig. 6A, lanes 7 to 9). In contrast, in ydj1i yeast, a significant amount of 2a pelleted even in the absence of 1a (Fig. 6A, lanes 4 to 6). Since Ydj1p is involved in protein folding (39) and proteasome-dependent degradation of denatured proteins (37), the 2a protein detected in the pellet fraction from ydj1i yeast expressing 2a alone might represent denatured, insoluble 2a. When 1a was coexpressed, the fraction of 2a protein in the pellet was higher than in ydj1i yeast expressing 2a alone (Fig. 6A, lanes 10 to 12). Therefore, it appeared that a fraction of 2a likely was recruited to membranes by 1a.
FIG. 6.
Effect of ydj1 mutation on 1a-dependent recruitment of 2a protein to membranes. Yeast cells expressing 2a in the presence (+1a) or absence (−1a) of 1a were grown, spheroplasted, and lysed osmotically as described by Chen and Ahlquist (11). (A) The lysate was then centrifuged as indicated in the legend to Fig. 5 to yield pellet and supernatant (sup). An equal percentage of each fraction was separated by 0.1% SDS-9% PAGE, and Western analysis was performed with antibodies against 1a or 2a proteins or phosphoglycerate kinase (Pgkp), a cytosolic protein marker. (B) The lysate was subjected to a flotation analysis in a discontinuous sucrose gradient as described by Ahola and Ahlquist (2), with slight modifications. Briefly, 1 ml of the lysate was mixed with 5 ml of 67% (wt/wt) sucrose in HN buffer containing protease inhibitors (2) in an SW40Ti ultracentrifuge tube (Beckman) and overlaid by 5 ml of 50% (wt/wt) sucrose and 1 ml of 10% (wt/wt) sucrose in NH buffer containing protease inhibitors (2). After centrifugation at an average speed of 150,000 × g for ca. 17 h, a membrane fraction (top 2.5 ml; 10 to 50% interface layer), an intermediate fraction (next 2.5 ml; 50% layer), and the sample loading layer fraction (bottom 7 ml; 60% loading layer) were recovered. The same percentage of each fraction was analyzed by SDS-PAGE and Western blotting with the antibodies against 1a or 2a proteins or phosphoglycerate kinase.
To further test whether any 2a protein associated with membranes in a 1a-dependent manner in ydj1i yeast, we isolated crude membranes from lysates of yeast spheroplasts by flotation in discontinuous sucrose gradients and examined the amount of 2a protein in the membrane fractions. As shown in Fig. 6B for both wt (lanes 11 to 15) and ydj1i (lanes 16 to 20) yeast, similar amounts of 2a protein were present in the membrane fraction (10 to 50% sucrose interface layer; lane 12 for wt and lane 17 for ydj1i) and the 60% sucrose loading layer (lane 14 for wt and lane 19 for ydj1i) in the presence of 1a, whereas much smaller amounts of 2a were present in the membrane fraction in the absence of 1a than in the presence of 1a (for wt, see lanes 1 to 5 and compare lane 2 with the other lanes; for ydj1i, see lanes 6 to 10 and compare lane 7 with the other lanes). Thus, 1a induced similar fractions of 2a to associate with membranes in ydj1i and wt yeast. The results also indicate that the 10,000 × g-precipitable 2a protein found in ydj1i yeast expressing 2a alone is largely non-membrane-bound.
Effect of Ydj1p addition and depletion on BMV RdRp activity in vitro.
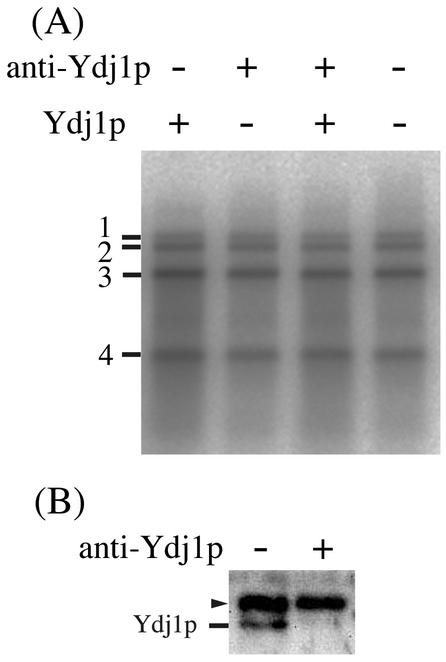
Detergent-solubilized membrane fractions prepared from yeast coexpressing 1a and 2a and harboring replicating RNA3 contain a BMV-specific RdRp activity that synthesizes negative-strand RNA copies of exogenously added BMV RNA (Fig. 7A) (42). We examined whether addition or depletion of Ydj1p affected the in vitro activity of RdRp prepared from wt yeast expressing 1a, 2a, and RNA3. Approximately 10 ng of Ydj1p was contained in 50 μl of typical RdRp reaction mixture, as estimated by comparing band intensities of Western blots with those of known amounts of purified Ydj1p. To this 50-μl reaction mixture, we either added 500 ng of E. coli-expressed and purified Ydj1p or depleted Ydj1p by treating the RdRp with anti-Ydj1p antibody bound to protein A-Sepharose (Fig. 7B). However, the pattern of negative-strand RNA synthesis from exogenously added BMV virion RNA template was not changed by either treatment (Fig. 7A). The specific activity of detergent-solubilized RdRp from ydj1i yeast expressing 1a, 2a, and RNA3 was lower than that from wt yeast and was not increased by the addition of E. coli-expressed and purified Ydj1p (data not shown).
FIG. 7.
Effects of addition and depletion of Ydj1p on in vitro BMV RdRp activity. BMV RdRp extract was prepared from protease-deficient BJ5465 yeast expressing 1a and 2a proteins and a replicating BMV RNA3 derivative, B3URA3 RNA, and purified with DEAE-Bio gel A column chromatography. (A) Purified BMV RdRp (50 μl) was treated with protein A-Sepharose-conjugated anti-Ydj1p antibody (+) or protein A-Sepharose without antibodies (−), followed by addition of 500 ng of E. coli-expressed, purified Ydj1p or phosphate-buffered saline. The RdRp reaction procedure was performed with exogenously added BMV virion RNA as a template in the presence of [α-32P]UTP as described by Quadt et al. (42). The product of the RdRp reaction was treated with S1 nuclease and analyzed by electrophoresis on a 1% agarose gel and by autoradiography. Positions of double-stranded BMV RNAs 1 to 4 are indicated in the figure. (B) Confirmation of depletion of Ydj1p. Purified BMV RdRp, treated with protein A-Sepharose-conjugated anti-Ydj1p antibody (+) or just protein A-Sepharose (−), was subjected to SDS-PAGE followed by Western analysis with anti-Ydj1p antibodies. The position of the Ydj1p band is indicated. Arrowhead shows a background signal presumably derived from leaked protein A.
A possible reason for the failure of added Ydj1p to affect RdRp activity was that E. coli-expressed Ydj1p lacks the farnesylation found with Ydj1p in yeast (8). However, this posttranslational modification is not essential for BMV RNA replication, since we found that the nonfarnesylatable Ydj1p mutant S406p (9) complemented the mab3 mutation as well as wt Ydj1p (Y. Tomita, T. Mizuno, S. Naito, and M. Ishikawa, unpublished results). Thus, while Ydj1p contributes significantly to normal BMV RNA replication in vivo (Fig. 3), these results suggested that Ydj1p is not needed in vitro to maintain the activity of preformed RdRp (see Discussion).
DISCUSSION
In this report, we have shown that a mutation in the yeast YDJ1 gene changing the initiation codon ATG to ATA inhibits BMV RNA replication. Since the mutation is recessive and dramatically suppresses Ydj1p accumulation (Fig. 3E), the phenotype is likely caused by the loss of normal Ydj1p functions. Ydj1p is one of the two major homologues of E. coli DnaJ in the cytoplasm of S. cerevisiae (10, 14). Functioning with Hsp90 (34) or the Ssa family of Hsp70 (13, 15, 39), Ydj1p assists protein folding (16, 39), protein translocation across membranes (3, 7), assembly of macromolecular complexes (23), and protein degradation (37). While the possibility that the ydj1 mutation indirectly affects BMV RNA replication (because of the pleiotropic nature of this mutation) cannot be excluded, our data support the possibility that Ydj1p directly affects BMV RNA replication through interaction with the 2a protein, as discussed below.
Control of the state of the 2a protein by Ydj1p.
In ydj1i yeast, the 2a protein took a 10,000 × g-precipitable form that was not found in wt cells (Fig. 6A; compare lanes 3 and 6). This 10,000 × g-precipitable 2a is consistent with an aggregated form of 2a protein. Independent evidence for a potential for 2a self-aggregation was found in another recent study (11). DnaJ family members bind hydrophobic surfaces of substrate proteins and target them to DnaK (Hsp70) (46), preventing irreversible aggregation and facilitating correct folding. In the folding of 2a protein, such functions of Ydj1p/Hsp70 chaperones also might be important and the lack of Ydj1p might result in irreversible aggregation. Normally, such denatured proteins are degraded by a ubiquitin-proteasome-dependent pathway, in which Ydj1p plays an important role for substrate recognition (37). Thus, the ydj1 mutation might also be critical for preventing degradation of the 10,000 × g-precipitable form of 2a protein.
ydj1 mutation inhibits negative-strand RNA synthesis.
Prior to negative-strand synthesis of BMV, the 1a protein associates with the ER membranes and recruits the 2a protein and viral RNA templates to the membrane-bound replication complex (11, 12, 44, 47). Here, we have shown that in ydj1i yeast, negative-strand synthesis is inhibited but 1a-dependent recruitment of 2a and RNA replication templates occurs with nearly the same efficiency as in wt cells (Fig. 5 and 6). Therefore, Ydj1p likely is necessary to convert the components of the BMV replication complex recruited to ER membranes into an active complex capable of negative-strand RNA synthesis.
Neither depletion nor addition of Ydj1p detectably affected the ability of detergent-solubilized BMV RdRp to synthesize negative-strand RNAs from exogenously added BMV RNA templates (Fig. 7). This detergent-solubilized BMV RdRp, however, shows anomalous characteristics such as lack of positive-strand RNA synthesis and lack of response to replication enhancer containing the box B sequence that is necessary for efficient negative-strand synthesis in vivo (19, 42, 49). These observations, together with the importance of membranes for the formation of BMV replication complex in vivo (38, 47) and inability to extract active RdRp from yeast expressing 1a and 2a but not replicating BMV-related RNA (42), suggest that the detergent-solubilized RdRp is derived from the active BMV replication complex preformed in vivo and does not reflect the process of forming the BMV replication complex in vivo. Therefore, the results obtained here with the detergent-solubilized RdRp do not necessarily deny the involvement of Ydj1p in the in vivo formation of the BMV RNA replication complex.
Another yeast mutation that inhibits BMV RNA replication, ole1w, was found to do so by reducing the level of unsaturated fatty acids, which is known to be a primary determinant of lipid fluidity and flexibility in yeast membranes (38). Like the ydj1 mutation, ole1w affected negative-strand RNA synthesis but not 1a-dependent recruitment of the 2a protein and RNA replication templates (38). This observation suggests that the activation of negative-strand synthesis occurring on ER membranes is a dynamic event that requires both the action of a Ydj1p-containing chaperone system and a specific status (e.g., fluidity and flexibility) of membranes. The contributions of YDJ1 and OLE1 to negative-strand RNA synthesis appear distinct, since parallel analysis of lipid extracts from wt and ydj1i yeast shows that the ydj1 mutation does not alter lipid composition (W. M. Lee, personal communication).
Activation of the hepadnavirus reverse transcriptase (RT) and subsequent nucleocapsid assembly and initiation of viral DNA synthesis require Hsp90 and its chaperone partners (22, 23, 24). Recently, Hu et al. succeeded in initiating hepadnavirus protein-primed reverse transcription using bacterially expressed recombinant RT and the purified molecular chaperones Hsp90, Hsp70, Hop, and Hsp40, a Ydj1p homolog (23). In this system, the Hsp90 chaperone complex binds to RT and, utilizing the energy produced by ATP hydrolysis, establishes and maintains the conformation of RT as competent for binding with the ɛ sequence on the pregenomic RNA, the template of reverse transcription. This chaperone-dependent binding of RT to ɛ initiates both the mutually dependent encapsidation of RT and pregenomic RNA and protein-primed reverse transcription. Once the initial protein-priming reaction is completed, Hsp90 is no longer necessary for DNA elongation (22). In parallel with this finding, Ydj1p, which functions as part of a chaperone complex, might be needed to bring the BMV replication complex into a conformation active for negative-strand synthesis. By further analogy with hepadnaviruses and in keeping with the induction by ydj1 mutation of a rapidly precipitating, potentially aggregated form of 2a, a prime point of interaction for this chaperone complex might be the 2a protein. Moreover, just as Hsp90 is needed for initial polymerase activation but not for hepadnavirus negative-strand synthesis, Ydj1p does not appear to be needed by a preformed, active BMV RdRp complex during negative-strand synthesis (Fig. 7). Thus, like the role of Hsp90 in hepadnavirus reverse transcription, the role of Ydj1p in BMV negative-strand synthesis appears to be in the formation rather than the continued function of the complex.
E. coli DnaJ, a close relative to Ydj1p, is involved in disassembly of macromolecular complexes. In the initiation of bacteriophage λ DNA replication, the DnaK, DnaJ, and GrpE chaperone system disassembles the λO-λP-DnaB complex formed at the replication origin of λ DNA. In the λO-λP-DnaB complex, the DnaB helicase is inactive. Upon disassembly of the complex, the DnaB helicase is activated and λ DNA replication starts (48). Similar molecular mechanisms are suggested to underlie the initiation of DNA replication by simian virus 40 and related viruses, where the J domains of the viral T antigens replace DnaJ (6, 33, 49). By analogy with such functions of DnaJ family proteins, the membrane-bound components of the preinitiation complex for BMV negative-strand synthesis might be subjected to at least partial disassembly by Ydj1p-containing molecular chaperone systems. For stringent selection of the RNA molecules to be replicated and for proper assembly of replication preinitiation complex, multiple accurate molecular recognition events, each releasing significant free energy, should be necessary. As a result, the preinitiation complex might be fairly stable and therefore difficult to rearrange to start replication. The free energy released upon ATP hydrolysis by molecular chaperones might be utilized to resolve these contradictory requirements.
Acknowledgments
We thank Elizabeth A. Craig, Avrom J. Caplan, Yoko Kimura, and Susan Lindquist for materials and discussions, Narushi Iizuka and Tetsuo Meshi for experimental advice, and Sunao Itahashi for excellent technical assistance. We used the facilities of the Biopolymer Analysis Laboratory in the Faculty of Agriculture, Hokkaido University, and the Research Center for Molecular Genetics at Hokkaido University.
This work was supported in part by grants from the Japan Society for the Promotion of Science to M.I. and by National Institutes of Health grant GM35072 to P.A. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 2.Ahola, T., and P. Ahlquist. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atencio, D. P., and M. P. Yaffe. 1992. Mas5, a yeast homolog of DnaJ involved in mitochondrial protein import. Mol. Cell. Biol. 12:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 5.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 7.Caplan, A. J., D. M. Cyr, and M. G. Douglas. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71:1143-1155. [DOI] [PubMed] [Google Scholar]
- 8.Caplan, A. J., and M. G. Douglas. 1991. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 114:609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplan, A. J., J. Tsai, P. J. Casey, and M. G. Douglas. 1992. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 267:18890-18895. [PubMed] [Google Scholar]
- 10.Cheetham, M. E., and A. J. Caplan. 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cyr, D. M., and M. G. Douglas. 1994. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J. Biol. Chem. 269:9798-9804. [PubMed] [Google Scholar]
- 14.Cyr, D. M., T. Langer, and M. G. Douglas. 1994. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19:176-181. [DOI] [PubMed] [Google Scholar]
- 15.Cyr, D. M., X. Lu, and M. D. Douglas. 1992. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 267:20927-20931. [PubMed] [Google Scholar]
- 16.Dey, B., A. J. Caplan, and F. Boschelli. 1996. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol. Biol. Cell 7:91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinant, S., M. Janda, P. A. Kroner, and P. Ahlquist. 1993. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 67:7181-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 21.Hinnenbusch, A. G., and S. W. Liebman. 1991. Protein synthesis in Saccharomyces, p. 627-735. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, J., D. Toft, D. Anselmo, and X. Wang. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa, M., J. Díez, M. Restrepo-Hartwig, and P. Ahlquist. 1997. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc. Natl. Acad. Sci. USA 94:13810-13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa, M., M. Janda, M. A. Krol, and P. Ahlquist. 1997. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 71:7781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa, M., P. Kroner, P. Ahlquist, and T. Meshi. 1991. Biological activities of hybrid RNAs generated by 3′-end exchanges between tobacco mosaic and brome mosaic viruses. J. Virol. 65:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 30.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao, C. C., and P. Ahlquist. 1992. Identification of the domains required for the direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66:7293-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, C. C., R. Quadt, R. P. Hershberger, and P. Ahlquist. 1992. Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J. Virol. 66:6322-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley, W. L., and C. Georgopoulos. 1997. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc. Natl. Acad. Sci. USA 94:3679-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura, Y., I. Yahara, and S. Lindquist. 1995. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 268:1362-1365. [DOI] [PubMed] [Google Scholar]
- 35.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai, M. M. C. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 37.Lee, D. H., M. Y. Sherman, and A. L. Goldberg. 1996. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, W.-M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273:27824-27830. [DOI] [PubMed] [Google Scholar]
- 40.O'Reilly, E. K., J. D. Paul, and C. C. Kao. 1997. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J. Virol. 71:7526-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Restrepo-Hartwig, M. A., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 46.Rüdiger, S., J. Schneider-Mergener, and B. Bukau. 2001. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20:1042-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 48.Skowyra, D., K. McKenney, and S. H. Wickner. 1995. Function of molecular chaperones in bacteriophage and plasmid DNA replication. Virology 6:43-51. [Google Scholar]
- 49.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickner, R. B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]