Abstract
Protein traffic from the cell surface or the trans-Golgi network reaches the lysosome via a series of endosomal compartments. One of the last steps in the endocytic pathway is the fusion of late endosomes with lysosomes. This process has been reconstituted in vitro and has been shown to require NSF, α and γ SNAP, and a Rab GTPase based on inhibition by Rab GDI. In Saccharomyces cerevisiae, fusion events to the lysosome-like vacuole are mediated by the syntaxin protein Vam3p, which is localized to the vacuolar membrane. In an effort to identify the molecular machinery that controls fusion events to the lysosome, we searched for mammalian homologues of Vam3p. One such candidate is syntaxin 7. Here we show that syntaxin 7 is concentrated in late endosomes and lysosomes. Coimmunoprecipitation experiments show that syntaxin 7 is associated with the endosomal v-SNARE Vamp 8, which partially colocalizes with syntaxin 7. Importantly, we show that syntaxin 7 is specifically required for the fusion of late endosomes with lysosomes in vitro, resulting in a hybrid organelle. Together, these data identify a SNARE complex that functions in the late endocytic system of animal cells.
INTRODUCTION
Endocytosed proteins that do not recycle to the plasma membrane or the trans-Golgi network are ultimately delivered to the lysosome. As one of the primary sites for protein degradation, lysosomes are also the recipients of phagocytosed material and autophagized proteins. There are ample data to support the view that all of these pathways converge in a late endocytic compartment before fusion with the lysosome (Futter et al., 1996; Mellman, 1996; Storrie and Desjardins, 1996; Bright et al., 1997). However, little is known about the molecular mechanisms that mediate this fusion process.
The process of fusion between late endosomes and lysosomes has been examined both in vivo and in vitro. This process is characterized by the formation of a hybrid organelle with intermediate properties of both compartments, implying that continued efflux of material from this hybrid is required for the ultimate reformation of dense lysosomes (Bright et al., 1997; Mullock et al., 1998; Luzio et al., 2000; Pryor et al., 2000). An alternative hypothesis is that late endosomes transfer their cargo to lysosomes by an abbreviated “kiss-and-run” scenario (Storrie and Desjardins, 1996). Both of these models, unlike the notion that lysosomes “mature” from endocytic compartments, rely on a highly specific fusion event with preexisting lysosomes (Mellman, 1996). Previous studies have identified a few molecular players involved in late endosome and lysosome fusion, such as NSF and a Rab GDI-sensitive GTPase. However, a better understanding of this overall process will require more detailed characterization of the proteins that mediate this specific fusion event (Mullock et al., 1998).
During the last few years, the SNARE hypothesis has provided a compelling mechanistic model for a number of fusion events throughout the secretory and endocytic pathways (Rothman and Warren, 1994; Rothman and Wieland, 1996), with particular combinations of SNAREs mediating distinct fusion events in vivo (Bennett, 1995). The SNARE proteins form a very stable parallel array of coiled-coil domains forming a four-helix bundle; it is thought that the formation of this complex may provide some of the energy that drives vesicle fusion (Hanson et al., 1997; Poirier et al., 1998; Sutton et al., 1998; Weber et al., 1998). Because both v-SNARE and t-SNARE proteins have been found in transport vesicles and target organelles, it can be more useful to categorize v- and t-SNAREs as R- and Q-SNAREs, respectively, according to their structural composition (Fasshauer et al., 1998; Sutton et al., 1998). Although the stoichiometry of the core complex may differ depending on the particular set of SNARE proteins (Carr et al., 1999; Ungermann et al., 1999), in general, v-SNAREs contribute a critical arginine residue in the interior of the bundle that is aligned with glutamine residues contributed by each of the three helices from the t-SNAREs to form a core complex (Sutton et al., 1998). What remains unclear about SNARE-mediated transport steps, however, is exactly which steps are mediated by some of the more recently described SNARE proteins as well as what regulates the specificity of these steps, especially throughout the endocytic pathway, where the role of vesicular transport is less certain.
In Saccharomyces cerevisiae, the process of vacuolar biogenesis parallels that of lysosomal biogenesis in mammalian cells (Bryant and Stevens, 1998). Many of the t-SNARE proteins that act at discrete trafficking steps along the mammalian secretory pathway display the greatest level of identity with the yeast t-SNARE that functions in an analogous step (i.e., Sed5p is most similar to syntaxin 5, and Sso1p/Sso2p are most similar to syntaxins 1–4) (Aalto et al., 1993; Banfield et al., 1994; Rowe et al., 1998). It is likely that similar parallels will be found in the endocytic pathway. The yeast syntaxin Vam3p has been localized to the yeast vacuole and is required for the delivery of vacuolar proteins that follow a variety of membrane trafficking pathways (Darsow et al., 1997; Piper et al., 1997; Wada et al., 1997). Vam3p is required for homotypic vacuole fusion (Nichols et al., 1997; Ungermann et al., 1998), fusion of endosomal compartments and prevacuolar autophagic organelles with the vacuole (Darsow et al., 1997), and fusion of a new type of transport intermediate that carries the membrane protein alkaline phosphatase to the vacuole (Piper et al., 1997; Rehling et al., 1999). Recently, a Vam3-related protein was identified in Arabidopsis thaliana that is likely to have similar functions to the yeast Vam3p, given its localization to the plant vacuole. Because of the prominent role of Vam3p in a variety of fusion events with the vacuole, we sought to identify a t-SNARE from mammalian cells that may functionally correspond to Vam3p. With this aim, we identified mammalian syntaxin 7 with the highest level of homology to S. cerevisiae and A. thaliana Vam3 proteins. Syntaxin 7 (Syn7) was originally proposed to participate in traffic from the trans-Golgi network to the endocytic system based on its sequence similarity to Pep12p (Wang et al., 1997); and partial colocalization of Syn7 with markers of early endosomes suggested a role in fusion events within the early endocytic system (Wong et al., 1998b). Our studies, however, show that Syn7 is concentrated in late endosomes and lysosomes and can be isolated in a complex with Vamp 8. Importantly, we find that Syn7 is required for the fusion of late endosomes with lysosomes with the use of an in vitro content-mixing assay. Together, these data suggest that Syn7 plays a pivotal role in the final steps of lysosomal biogenesis.
MATERIALS AND METHODS
Materials
Enzymes used in DNA manipulations were from New England Biolabs (Beverly, MA), Boehringer Mannheim Biochemicals (Indianapolis, IN), Bethesda Research Laboratories (Gaithersburg, MD), or United States Biochemical (Cleveland, OH). Radioactive materials were purchased from New England Nuclear (Boston, MA), and nitrocellulose membranes were from Micron Separations Inc. (Westborough, MA). All tissue culture reagents were from Life Technologies (Gaithersburg, MD). Bicinchoninic acid protein reagent was from Pierce (Rockford, IL). Glutathione–agarose beads were from Pharmacia (Uppsala, Sweden). Fixed Staphylococcus aureus cells (IgG Sorb) were purchased from the Enzyme Center (Walden, MA). All other chemicals were of high-purity commercial grade. Oregon Green– and Texas Red–conjugated goat anti-rabbit and anti-mouse secondary antibodies, biotinylated anti-rabbit antibodies, and Alexa 488–conjugated streptavidin were purchased from Molecular Probes (Eugene, OR). FITC- and Texas Red–conjugated donkey anti-rabbit and anti-goat secondary antibodies were purchased from Jackson Laboratories (West Grove, PA). HRP-conjugated goat anti-rabbit immunoglobulin G (IgG) and ECL detection kits were from Amersham (Arlington Heights, IL).
DNA and Plasmid Constructions
DNA manipulations and DNA-mediated transformation were performed by routine procedures (Sambrook et al., 1989). A BLAST search through the Merck/Washington University expressed sequence tag database revealed several putative ORFs that encoded proteins related to Vam3p (Altschul et al., 1990). We amplified one of these (H33185) with the oligonucleotides CCATTTCCAGAGTATCGGGTGGC and TCTATGCTCTCAATAACATCTCC. This PCR fragment was used to probe a mouse 3T3-L1 cDNA library as described previously (Tellam et al., 1997). A full-length clone was isolated and sequenced to reveal an ORF of 266 amino acids (GenBank accession number AFO56323). Sequence comparison analysis was performed with the DNASTAR (Madison, WI) MegAlign software with the use of the Clustal algorithm.
The plasmid pGSTSyn7a encoding a GST–Syn7 fusion protein was made by PCR amplifying codons 2–242 of the Syn7 ORF and subcloning the fragment into the BamHI–EcoRI sites of pGEX-3X with the use of the oligonucleotides CACAGGATCCAGTCTTACACTCCGGGGATTGG and CTCCAATTCTAAGTTTTCCTGGATTTGCGCTGA (Smith and Johnson, 1988). The plasmid pCWS002 encoding a GST–Vamp 8 protein was made by reverse transcription–PCR amplifying codons 1–76 of the mouse Vamp 8 ORF and subcloning the fragment into the BamHI–Xho sites of pGEX-4T3 with the use of the oligonucleotides CCGGGATCCATGGAGGAGGCCAGTGGG and CCGCTCGAGTTACATCTTCACATTCTTCCA. Northern analysis on whole RNA was conducted as described previously with the use of the entire cDNA as probe or with a glyceraldehyde-3-phosphate dehydrogenase cDNA probe as an internal control (Tellam et al., 1997).
An expression plasmid (pRCP316) that expressed a hemagglutinin (HA) epitope–tagged Syn7 was made by amplifying the complete ORF of mouse Syn7 with the oligonucleotides ATGTATCCTTACGACGTACCAGATTACGCATACCCCATATGATGTTCCTGACTATGCGTCTTACACTCCGGGGATTGGT and TCAGCCTTTCAGTCCCCATACGA and subcloning the resulting fragment into pCDNA-V5–6XHIS (Invitrogen, Carlsbad, CA). Both strands of the resulting construct were sequenced for orientation and fidelity of amplification. The resulting plasmid expressed the Syn7 ORF preceded by two tandem repeats of the HA epitope (YPYDVPDYAG).
A GST–syntaxin 13 (Syn13) fusion construct encoding amino acids 1–243 of rat Syn13 was kindly provided by Dr. Rohan Teasdale (Monash Medical Center, Melbourne, Australia).
Antibodies
Polyclonal anti-GST antibodies and polyclonal and monoclonal anti-Syn7 antibodies were raised against the GST–Syn7 fusion protein encoded by pGSTSyn7a. Anti-Vamp 8 antibodies were raised against the GST–Vamp 8 fusion protein encoded by pCWS002.
The bacterial fusion proteins were purified over glutathione–agarose and eluted with 25 mM glutathione as reported previously (Smith and Johnson, 1988). Two rabbit polyclonal antibodies (#1 and #2) and one goat polyclonal antibody to Syn7 were raised and were purified similarly. Rabbit immunizations were performed as described previously (Roberts et al., 1989). Serum from immunized animals was subjected to a series of affinity purification steps to yield specific antibodies to GST and Syn7. Serum was first passed over an Affigel column (Affigel 10 and Affigel 15, Bio-Rad, Hercules, CA) to which GST was attached. Anti-GST antibodies were eluted in 0.2 M glycine, pH 2.5, and dialyzed in three changes of PBS. The remaining serum that was depleted of anti-GST antibodies was then applied to another Affigel column to which the GST–Syn7 fusion protein was attached. Antibodies were eluted in 0.2 M glycine, pH 2.5, and dialyzed in three changes of PBS. Monovalent Fab fragments of rabbit anti-Syn7#2 IgG were prepared according to Coulter and Harris (1983) and then affinity purified like the intact antibodies. Antibodies specific to Vamp 8 were purified similarly. One antiserum raised against the C-terminal tail of the cation-independent mannose-6-phosphate receptor (CI-MPR) was a kind gift from Dr. Gwyn Gould (University of Glasgow, Glasgow, Scotland), and a second was as described previously (Reaves et al., 1996). Affinity-purified anti-Rab7 rabbit polyclonal antibodies were a kind gift of Marino Zerial (EMBL, Heidelberg, Germany). mAbs to the transferrin receptor were purchased from Zymed Laboratories (San Francisco, CA) and Chemicon (mAb 1451; Temecula, CA). Monoclonal anti-syntaxin 6 and anti-early endosome–associated protein 1 (EEA1) antibodies were purchased from Transduction Laboratories (Lexington, KY). mAbs to the HA epitope were purchased from BAbCO (Berkeley, CA). Polyclonal anti-syntaxin 6 antibodies were affinity purified over a GST–syntaxin 6 affinity column as described previously (Tellam et al., 1997). mAbs to rat lgp120 were as described previously (Grimaldi et al., 1987; Reaves et al., 1996). Rabbit polyclonal antibodies to rat lgp120 were a gift from Keitaro Kato, Yoshitake Tanaka, and Masaru Himeno (Kyushu University, Fukuoka, Japan) (Furuno et al., 1989). Anti-Syn13 antibodies were raised against the GST–Syn13 fusion protein or were a gift of Marino Zerial. Rabbit polyclonal antibodies to EEA1 were used as described previously (Mu et al. 1995). Anti-Syn13 antibodies were raised against the GST–Syn13 fusion protein (antiserum 4972) or were a kind gift from Marino Zerial.
For the production of the monoclonal anti-Syn7 antibody Syn7.1C3, hybridoma cell lines were produced and characterized as described previously (Brooks et al., 1997). Positive clones were screened for reactivity to the GST–Syn7 fusion protein by ELISA and assessed for specificity by screening for reactivity to GST–Syn13 and GST fusion proteins. Hybridoma culture supernatant was purified on a 0.1-ml protein G cartridge (Pharmacia) and eluted with 0.1 M NaH2PO4, pH 2.5, into 100 μl of 1 M Na2HPO4, pH 9.0, and then dialyzed against three changes of PBS.
Cell Culture
Madin-Darby canine kidney (MDCK) cells and normal rat kidney (NRK) cells were grown at 37°C in an atmosphere of 5% CO2 in DMEM supplemented with 10% FCS, 2 mM l-glutamine, 100 U/l penicillin, 100 mg/l streptomycin, and 0.1 mM MEM nonessential amino acid solution. Cells were grown to near confluence in either 150-mm culture dishes for membrane preparation and cryosection immunoelectron microscopy or on coverslips for immunofluorescence microscopy.
WIF-B cells were grown as described (Ihrke et al., 1993) with some modifications. Briefly, cells were grown in Cassio modified Ham's F12 medium (GIBCO/BRL, Grand Island, NY) supplemented with 10 μM hypoxanthine, 0.04 μM aminopterin, 1.6 μM thymidine, and 3.5% FCS (Hyclone, Logan, UT). For experiments, cells were plated on glass coverslips at 0.8–1.2 × 106 cells/cm2 and cultured until they had reached maximal polarization (10–14 d after plating).
For transfection studies, cells were grown until 50% confluent and washed three times in sterile PBS. DNA was transfected with Lipofectamine according to the manufacturer's directions (Life Technologies) for 10 h in serum-free DMEM. Transfection medium was replaced with DMEM containing 10% FCS, and cells were allowed to incubate for an additional 48 h. Cells were then cultured in 1 mg/ml geneticin for 10 d, after which colonies were isolated and screened for expression of HA epitope–tagged Syn7.
Cell Fractionation
Whole cell extracts and whole membrane fractions were prepared from rat tissues that were dissected and stored at −85°C. Tissues were minced in PBS containing PMSF, leupeptin, and α-tosyl-l-lysine chloromethyl ketone hydrochloride at 4°C and then homogenized by 25–30 strokes with a Teflon/glass Dounce. After a 1500 × g spin for 2 min, the supernatant fractions were solubilized in Laemmli sample buffer to make whole cell extracts.
Isolation of rat liver fractions enriched in late endosomes, hybrid organelles, or lysosomes was performed as described previously (Mullock et al., 1998). Briefly, endosome/lysosome hybrid organelles formed after the content-mixing assay incubation were isolated at the 20% Nycodenz/20% Ficoll interface of a step gradient. Electron-dense lysosomes were collected from a preparative 45%/20% Nycodenz step gradient. Late endosomes were isolated from a preparative 1–22% Ficoll gradient.
Immunofluorescence
Immunofluorescence on MDCK cells was performed with cells that were grown on glass coverslips until 50% confluent. Cells were fixed for 20 min in 2% paraformaldehyde in PBS at 25°C and permeabilized for 15 min in 0.2% Triton X-100 containing 50 mM glycine. Cells were washed in PBS containing 2% donkey serum and labeled with primary antibodies for 1 h at 25°C. After this incubation, the cells were washed three times in PBS for 5 min and then incubated for 30 min with conjugated secondary antibodies diluted at 1:250 in PBS containing 1% donkey serum. For double-labeling experiments with the use of rabbit anti-Syn7 antibodies in combination with a mouse mAb, Oregon Green–conjugated goat anti-rabbit and Texas Red–conjugated goat anti-mouse secondary antibodies were used. For double-labeling experiments with the use of goat anti-Syn7 antibodies in combination with a rabbit polyclonal antibody, FITC-conjugated donkey anti-goat and Texas Red–conjugated donkey anti-rabbit secondary antibodies were used. After this incubation, the cells were washed three times in PBS for 5 min and the coverslips were mounted onto slides in mounting medium (50% glycerol/1% N-propyl gallate in PBS).
As a test for the specificity of immunofluorescence labeling, parallel antibody solutions, containing a 1:300 dilution of rabbit or goat anti-Syn7 primary antibodies in PBS with 5% donkey serum, were incubated with no additions or 300 μg/ml purified GST–Syn13 fusion protein or 300 μg/ml GST–Syn7 fusion protein at room temperature for 2 h. Antibody solutions were then used to label parallel coverslips of MDCK cells. Photography was performed with the use of a set exposure and the same normalization settings.
For immunofluorescence localization in transfected NRK cells, cells were grown on coverslips until 50% confluent. Cells were washed three times in PBS at 25°C and then fixed in 100% methanol for 20 min at −20°C. Cells were then labeled with primary and secondary antibodies as described above.
Polarized WIF-B cells grown on coverslips were fixed for 2 min in 3% paraformaldehyde/0.05% glutaraldehyde in PBS in the presence of 0.2% sodium metabisulfite at 37°C, rinsed with PBS, and incubated with 0.5% sodium borohydride for 10 min at room temperature. Cells were incubated in 1% BSA/0.1% saponin in PBS for 30 min and then incubated for 1 h with primary antibodies diluted in 0.5% BSA/0.25% saponin in PBS. The rabbit anti-Syn13 antiserum (number 4972, 1:200) contained 300 μg/ml GST (preincubated for 30 min before adding to cells). Purified mouse IgG to Syn7 and affinity-purified anti-Vamp 8 were used at 10 μg/ml; mouse ascites fluid to lgp120 and mAb 1451 to the transferrin receptor were diluted 1:400 and 1:50, respectively; rabbit antiserum to lgp120 was diluted 1:300. After washes with PBS, the cells were incubated for 30 min with FITC- or Cy3-conjugated secondary antibodies (at 10 and 4 μg/ml, respectively). Cells were mounted in Prolong Antifade (Molecular Probes).
Images were captured with a Hamamatsu (Shizuoka, Japan) ORCA charge-coupled device camera mounted on an Olympus (Tokyo, Japan) BX-60 microscope equipped with a 60× oil objective (numerical aperture 1.4). In instances stated, a 4-μm z series was collected with a z-step of 0.1 μm (40 sections total) with a Ludl (Hawthorne, NY) z-stepper for each fluorophore. Image stacks were then subjected to a constrained iterative deconvolution algorithm (Agard et al., 1989) with the use of Microtome software (Vaytek, Fairfield, IA) and empirically determined point spread functions with the use of blue, green, and red fluorescent 0.2-μm microspheres (Molecular Probes). Paired sets of 10 images from each stack (1 μm thick) were combined by making maximum point projections and then overlaying the images with the use of Adobe Photoshop (Adobe Systems, Mountain View, CA). Alternatively, confocal images were taken with a Leica (Bucks Milton Keynes, UK) TCS SP system, a Zeiss (Thornwood, NY) LSM 510 laser scanning microscope, or a Bio-Rad laser scanning system, all equipped with 63× Plan-Apo objectives (numerical aperture 1.4). Images were collected at a resolution of 1024 × 1024 pixels. Adobe Photoshop software was used for image processing.
Immunogold Electron Microscopy on Isolated MDCK Membranes and Rat Liver Membranes
Preparation of membranes before fixation was performed at 0–4°C with the use of a procedure described previously (Martin et al., 1996). MDCK cells were washed three times in HES buffer (20 mM HEPES, 250 mM sucrose, 1 mM EDTA, pH 7.4), scraped in HES buffer (5 ml) containing protease inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin, 250 μM PMSF), and homogenized by 10 passes through a 22-gauge needle. A postnuclear supernatant was obtained by centrifugation at 9600 × g (SS-34 rotor, Sorvall Instruments, Wilmington, DE); it was then layered onto a 1.5 M sucrose cushion (containing 20 mM HEPES, pH 7.4, and 1 mM EDTA) and centrifuged at 154,000 × g (SW-41 rotor, Beckman Instruments, Fullerton, CA) for 1 h. The intracellular membrane fraction above the 1.5 M sucrose cushion was recovered and immediately fixed in a final concentration of 2% paraformaldehyde and stored at 4°C.
Formvar-coated carbon-stabilized grids were layered onto 10-μl drops of paraformaldehyde-fixed intracellular membranes for 10 min. Grids were incubated sequentially on 0.02 M glycine/PBS (four times) and 0.1% BSA/PBS. Grids were then incubated for 30 min with 5-μl drops of primary antibody diluted 1:50 in 1% BSA/PBS. After the first primary antibody incubation, the grids were washed in 0.1% BSA/PBS (four times) and then incubated for 20 min with 5-μl drops of colloidal gold conjugated to protein A (Electron Microscopy Sciences, Fort Washington, PA) diluted in 0.1% BSA/PBS. The grids were then washed in PBS (four times) and fixed with 1% glutaraldehyde for 5 min. When double labeling was performed, the grids were washed sequentially in 20 mM glycine/PBS (four times) and then returned to the 0.1% BSA/PBS step; the second primary antibody was then incubated, followed by incubation with a second protein A–gold conjugate and glutaraldehyde fixation. The grids were then washed in ultrapure H2O (seven times) and stained with uranyl acetate:methyl cellulose (1:9; 4% uranyl acetate in 0.15 M oxalic acid, pH 7–8, 2% methyl cellulose) for 10 min on ice. Grids were dried and visualized with the use of a transmission electron microscope. The order of primary antibody incubations in double-label experiments was alternated to determine if a particular antigen was compromised by glutaraldehyde treatment. We did not observe any affect of glutaraldehyde fixation on the antigens used, because the same level of labeling (number of gold particles per structure) was observed for all antibodies both before and after glutaraldehyde treatment. The specificity of double labeling was checked by omitting the second primary antibody and confirming the absence of the second gold conjugate. Random fields were quantitated in terms of the number of gold particles and the morphology of the labeled structure. Specifically for the analysis in Table 1, we quantitated the average number of Syn7-specific gold particles per distinct membrane structure. This was also performed for structures labeled for Rab7, CI-MPR, EEA1, and Vamp 8. To calculate the percentage of colocalization, we then quantitated the percentage of structures that were positive for Syn7 labeling that were also positive for one of the other markers and vice versa.
Table 1.
Quantitation of immunoelectron microscopy results on subcellular fractions prepared from MDCK cells
| Comparison | Structures counted | Number of gold particles/structure | Percent of structures labeled for Syn7 that were labeled for the indicated marker | Percent of structures labeled for the indicated marker that were labeled for Syn7 |
|---|---|---|---|---|
| Syntaxin 7 | 237 | 7.1 | 32 | 28 |
| EEA1 | 2.2 | |||
| Syntaxin 7 | 169 | 8.1 | 51 | 49 |
| VAMP 3 | 2.3 | |||
| Syntaxin 7 | 263 | 4.1 | 87 | 84 |
| CI-MPR | 3.9 | |||
| Syntaxin 7 | 266 | 3.5 | 84 | 82 |
| VAMP 8 | 3.6 |
Immunoelectron microscopy of the late endosome, hybrid, and lysosome fractions was performed as described previously (Mullock et al., 1998) with the use of the rabbit anti-Syn7 or anti-Vamp 8 antibody at a dilution of 1:50 followed by protein A conjugated to 15-nm colloidal gold (Department of Cell Biology, University of Utrecht, Utrecht, The Netherlands). The asialofetuin (ASF)–avidin content of late endosomes and hybrids was sequentially immunolabeled with 8-nm colloidal gold as described previously (Mullock et al., 1998). Quantitation of anti-Syn7 labeling and anti-Vamp 8 labeling per micrometer on the boundary membranes of organelles was performed with the use of intersections with a lattice overlay method (Griffiths, 1993). Late endosomes and hybrids were defined by their content of ASF–avidin, and random micrographs of the lysosome preparations were scored by their electron-dense morphology.
Electron Microscopy on Ultrathin Cryosections
MDCK cells were fixed in 8% paraformaldehyde in 0.1 M phosphate buffer, pH 7.35, for 1 h at 25°C. They were then washed with 0.2 M phosphate buffer, scraped from the culture dish, and pelleted at 10,000 rpm in a microfuge. Cells were resuspended in warm gelatin (10% in phosphate buffer) and repelleted at maximum speed in the microfuge. After cooling, the gelatin-embedded cells were infiltrated with polyvinylpyrrolidone–sucrose overnight at 4°C and then processed for frozen sectioning as described previously (Parton et al., 1997). Ultrathin frozen sections (60–80 nm) were labeled, stained, and viewed (JEOL 1010, Center for Microscopy and Microanalysis, University of Queensland) according to published techniques (Parton et al., 1997) with the use of gold-conjugated protein A.
Late Endosome–Lysosome Fusion Assays
Rat liver late endosomes loaded in vivo with avidin–ASF conjugate and rat liver lysosomes loaded in vivo with 125I-labeled biotinylated polymeric immunoglobulin A (125I-bpIgA) were resuspended separately in pig brain cytosol as described by Mullock et al. (1998). Antibody in a total volume of 200 μl was added to each membrane suspension to give a final volume of 700 μl. After 15 min on ice, 100-μl samples of the antibody-treated endosomes and lysosomes were used in the standard late endosome–lysosome fusion assay in a final volume of 240 μl for 10 min at 37°C (Mullock et al., 1998).
Immunoprecipitations
MDCK cells were washed three times in HES buffer, scraped in HES buffer (5 ml) containing protease inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin, 250 μM PMSF), and homogenized by 10 passes through a 22-gauge needle. A postnuclear supernatant was obtained by centrifuging the homogenate at 5000 × g for 5 min. One volume of PBS containing 2% Triton X-100 was added to the postnuclear supernatant, which was then centrifuged at 100,000 × g at 4°C (TLA-45 rotor, Beckman Instruments) for 40 min. The supernatant (protein concentration, 100 μg/ml) was then divided, and anti-Syn7 or anti-GST antibodies were added and incubated on ice for 6 h. Fifty microliters of a 50% slurry of IgG Sorb was then added followed by incubation for 45 min on ice. The immunoprecipitate was separated by centrifugation and washed three times with PBS containing 0.1% Triton X-100.
RESULTS
Syn7 Is Similar to Vam3p and Is Widely Expressed
After conducting a BLAST search of the Merck/Washington University expressed sequence tag database, several candidate mammalian homologues of the yeast vacuolar syntaxin Vam3p were identified. One expressed sequence tag that described a candidate cDNA (GenBank accession number H33185) was PCR amplified and used to screen a mouse cDNA library from which we recovered a clone encoding a 266-amino acid protein with a high level of identity to the human and rat sequences for Syn7, also known as TSL-5 (Bock and Scheller, 1997; Weimbs et al., 1997; Wong et al., 1998b). Of the non-mammalian syntaxins, Syn7 shows the highest level of identity (30%) with the Vam3p and Pep12p homologues from A. thaliana, AtVam3 and AtPep12, respectively (da Silva Conceicao et al. 1997; Sato et al. 1997; Sanderfoot et al. 1998). Overall, Syn7 shows the most homology (50%) with mammalian Syn13, a syntaxin confined mainly to the early endosome, indicating that these isoforms may have related functions in the endocytic pathway (Prekeris et al., 1998; McBride et al., 1999). Consistent with this is the observation that in A. thaliana AtVam3 shares 65% identity with AtPep12p. Furthermore in yeast, both Pep12p and Vam3p also occupy the endocytic system, and despite the fact that they indeed have distinct functional roles, high-level expression of one of these syntaxins can partially substitute for the function of the other (Darsow et al., 1997; Gotte and Gallwitz, 1997). Thus, by sequence comparisons alone, it would appear that Syn7 and Syn13 represent a pair of similar syntaxins that would have a role in the endocytic system as AtVam3 and AtPep12 have in A. thaliana and as Vam3 and Pep12p have in S. cerevisiae. Given that Pep12p and Syn13 have been shown to function in fusion events to the endosome, we decided to test whether Syn7, like Vam3p, participates in fusion events in the late endosomal/lysosomal system (Becherer et al. 1996; McBride et al., 1999).
As a first step in this analysis, we produced both rabbit and goat affinity-purified polyclonal antibodies with the use of a GST fusion protein comprising the entire cytosolic tail of Syn7. The polyclonal anti-Syn7 antibodies recognized a single ∼38-kDa protein that was expressed in all tissues examined (Figure 1). Syn7 levels were highest in brain and kidney. These levels correlated with the level of Syn7 mRNA within these tissues. The level of similarity between Syn7 and Syn13 presented the possibility that the anti-Syn7 antibodies could cross-react with Syn13. To ensure the specificity of the antibodies, we found that preincubation of either of the rabbit polyclonal anti-Syn7 antibodies or the goat anti-Syn7 antibody with a GST–Syn7 fusion protein but not a GST–Syn13 fusion protein abolished immunoblot reactivity. Similar results were obtained in immunolocalization experiments (see below). A mAb to Syn7 (Syn7.1C3) was also generated and recognized a single ∼38-kDa protein in MDCK cells. Characterization of Syn7.1C3 by ELISA revealed reactivity with the GST–Syn7 fusion protein but not with the GST–Syn13 fusion protein.
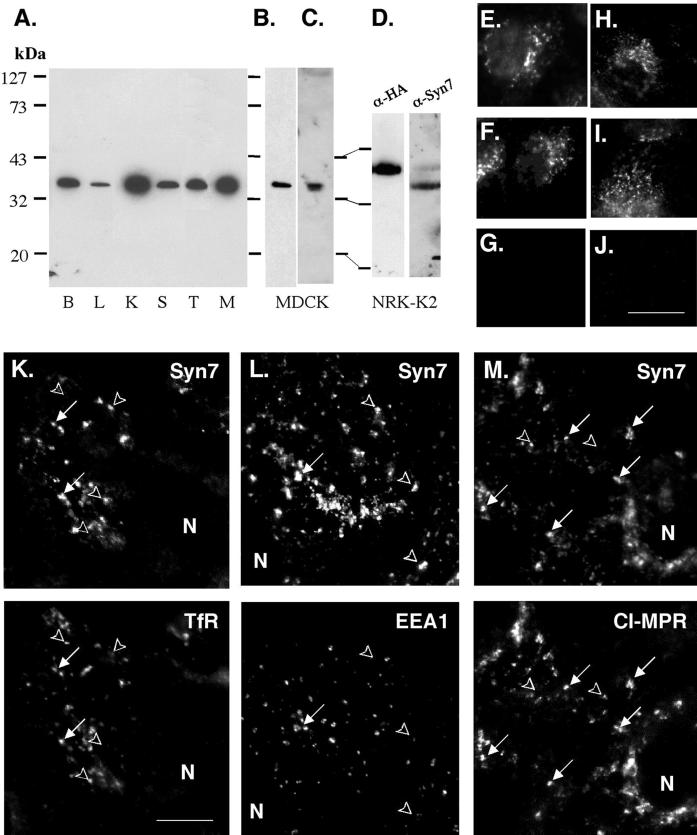
Figure 1.
Expression of Syn7. (A) Total membrane fractions prepared from rat brain (B), liver (L), kidney (K), spleen (S), testis (T), and MDCK (M) cells. Aliquots (10 μg of protein) were subjected to SDS-PAGE and immunoblotting with affinity-purified rabbit anti-Syn7 polyclonal antibody (rabbit anti-Syn7#1). Total membrane fraction from MDCK cells (10 μg of protein) was immunoblotted with an affinity-purified goat anti-Syn7 polyclonal antibody (B) or a separate rabbit antibody to Syn7 (rabbit anti-Syn7#2) (C). (D) Total membrane fraction (10 μg protein/lane) from a clone of NRK cells (NRK-K2) stably transfected with HA epitope–tagged Syn7 was subjected to immunoblot analysis with monoclonal anti-HA antibodies or rabbit anti-Syn7#1. (E–J) Rabbit anti-Syn7#1 (E–G) or goat anti-Syn7 antibodies (H–J) were incubated with no additions (E and H), 300 μg/ml GST–Syn13 (F and I), or 300 μg/ml GST–Syn7 (G and J) before labeling MDCK cells. Images were collected from a single focal plane and normalized to control. (K–M) Subconfluent MDCK cells were labeled for Syn7 with Alexa 488–conjugated secondary antibodies (top) in combination with antibodies specific for TfR (K), EEA1 (L), or CI-MPR (M) with Texas Red–conjugated secondary antibodies (bottom). Syn7 was labeled with the rabbit anti-Syn7#1 (K or L) or with the goat anti-Syn7 antibody (M). Cells were imaged by collecting a 4-μm-thick z-series (step, 0.1 μm) and subjecting the series to computed deconvolution before assembling a maximum point projection representing a 1-μm-thick plane. Arrows point to structures that are positive for two markers, and arrowheads point to structures that are positive for only one marker. N, nucleus. Bars, 15 μm for E–J; 5 μm for K–M.
Syn7 Is Localized to Late Endosomal/Lysosomal Structures by Immunofluorescence
To facilitate our localization studies of Syn7, we surveyed various cell types in culture for high levels of endogenous Syn7 expression. As shown in Figure 1A, high levels were found in MDCK cells. Thus, we initiated our immunofluorescence localization studies in MDCK cells with the use of both the rabbit and goat affinity-purified anti-Syn7 polyclonal antibodies (Figure 1, E–M). To ensure the specificity of the immunolabeling procedure, we labeled MDCK cells in the presence of either a GST–Syn7 fusion protein or a GST–Syn13 fusion protein. No change in the labeling pattern or the intensity of labeling was observed when the GST–Syn13 fusion protein was included during the primary antibody incubation; however, labeling was completely abolished with the inclusion of the GST–Syn7 fusion protein (Figure 1, E–J). In a series of double-labeling experiments (Figure 1, K–M), Syn7 labeling in MDCK cells was seen in intracellular structures that were most prominent in the perinuclear region but were also found throughout the cells. There was little overlap between Syn7 and syntaxin 6, a trans-Golgi network marker (Bock et al., 1997). The labeling pattern for Syn7 was quite distinct from that of the early endosome marker EEA1, which is generally concentrated in sorting endosomes in MDCK cells (Mu et al., 1995; Stenmark et al., 1996). Some overlap with Syn7 labeling was observed in double-labeling experiments with transferrin receptor (TfR), a more general marker of the recycling endocytic system. However, the extent of colocalization with TfR was poor because a considerable amount of labeling was observed in TfR-negative structures. In contrast, we found the highest level of colocalization was with the CI-MPR, which is concentrated in late endosomes in MDCK cells (Griffiths et al., 1988).
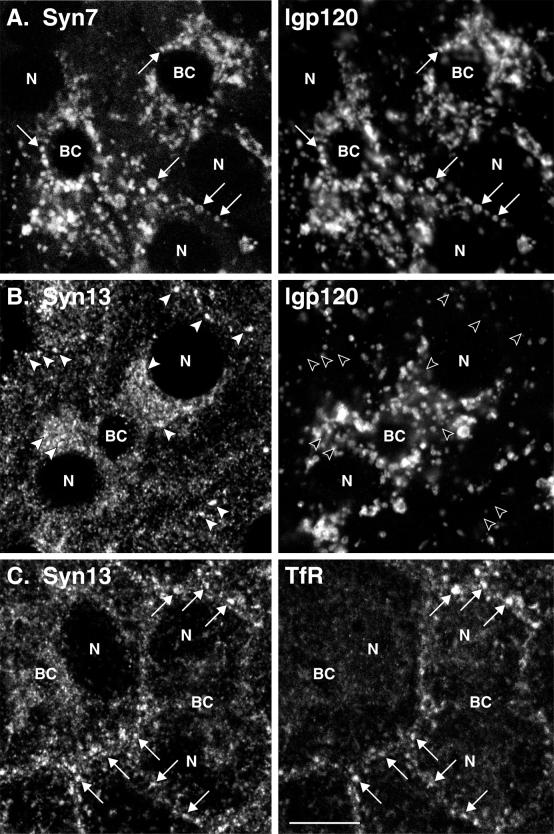
Because of the overlap of Syn7 with the CI-MPR in MDCK cells, we examined the distribution of Syn7 in relation to lgp120, a well-established marker of late endosomes and lysosomes (Lewis et al., 1985). Antibodies to lgp120 available to us did not cross-react with MDCK cells, and because we subsequently wanted to use a cell-free late endosome–lysosome fusion system from rat liver, we chose to examine Syn7 in WIF-B cells. WIF-B is a hepatic cell line that possesses many of the characteristic features of polarized hepatocytes, including the formation of the apical bile canalicular membrane (Ihrke et al., 1993; Shanks et al., 1994). We also took further steps to ensure the specificity of our labeling procedure with the use of a mAb raised against the cytosolic domain of Syn7 (Syn7.1C3). This mAb also recognizes a single 38-kDa protein in MDCK cell membrane fractions, and the binding of the antibody was specifically competed with GST–Syn7 fusion protein but not with GST–Syn13 fusion protein. Figure 2 shows that Syn7 was concentrated within the perinuclear region of WIF-B cells, well away from the periphery of the cell, where early endosomes are typically located. Double labeling for lgp120 showed a high level of colocalization in vacuolar structures. In contrast, Syn13 showed very little colocalization with lgp120 but extensive overlap with TfR, consistent with previous analyses indicating that Syn13 populates early endosomes and catalyzes early endosome homotypic fusion (Prekeris et al., 1998; McBride et al., 1999).
Figure 2.
Distribution of Syn7 and Syn13 in WIF-B cells by confocal microscopy. Polarized WIF-B cells were fixed and permeabilized and then double-labeled with antibodies to Syn7 and lgp120 (A), Syn13 and lgp120 (B), or Syn13 and TfR (C). Mouse mAbs were used to label Syn7 (A), lgp120 (B), and TfR (C) with rabbit polyclonal antibodies labeling lgp120 (A) and Syn13 (B and C). FITC-conjugated antibodies to mouse IgG (A, Syn7; B, lgp120; C, TfR) and Cy3-conjugated antibodies to rabbit IgG (A, lgp120; B and C, Syn13) were used as secondary antibodies. Optical sections in A and B are through the middle of the cells, whereas the section in C is slightly closer to the substratum to show more peripheral endosomes. Arrows point to structures that are positive for two markers, and arrowheads point to structures that are positive for only one marker. Note the good coincidence of Syn7 with lgp120-positive structures, i.e., lysosomes (A), and of Syn13 with TfR, i.e., early endosomes (C), whereas little overlap is seen between Syn13 and lgp120 (B). BC, bile canalicular-like space; N, nucleus. Bar, 10 μm.
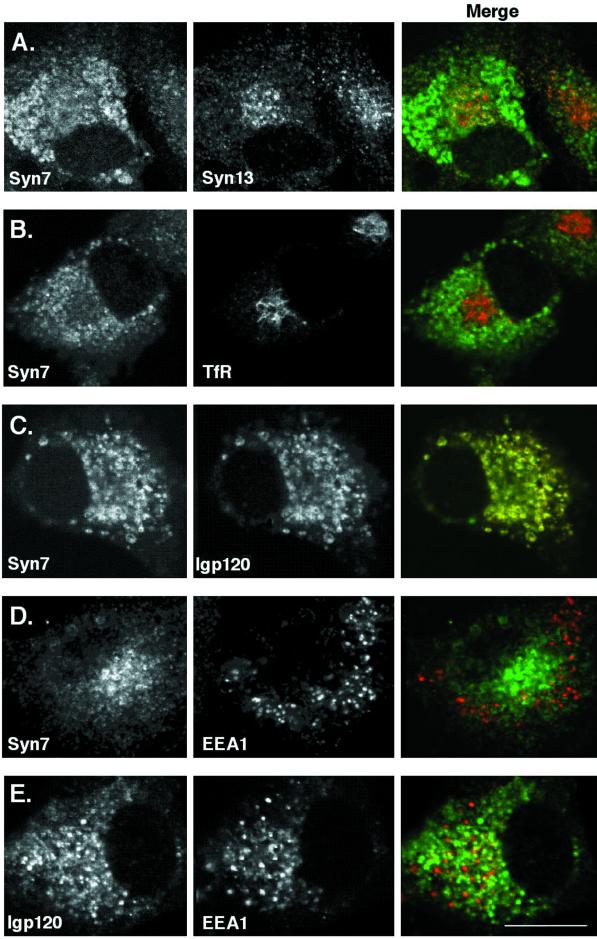
Although in the studies described above of MDCK and WIF-B cells we used polyclonal and monoclonal antibodies that do not cross-react with Syn13 or other syntaxins with molecular weights different from that of Syn7 (Figure 1), we could not completely eliminate the possibility that our antibodies cross-reacted with another protein in our immunofluorescence studies. Therefore, we chose to localize an epitope-tagged version of Syn7 in which we inserted two HA epitopes immediately after the initiation codon. The site for tag insertion has been used with a variety of other syntaxins and in those cases did not result in missorting of the syntaxin (Banfield et al., 1994; Advani et al., 1998; Carr et al., 1999). To analyze the distribution of epitope-tagged Syn7, we chose a cell line (NRK) in which the various compartments of the endocytic system can be distinguished morphologically and in which the localization and biogenesis of lysosomes have been characterized extensively (Bright et al., 1997). A stable clonal NRK cell line (NRK-K2) was isolated after transfection with a plasmid (pRCP316) encoding HA-tagged Syn7 and subsequent screening for G418-resistant cells. Western blot analysis of whole cell extract from NRK-K2 cells showed that the HA-tagged Syn7 was expressed as a single species with an apparent molecular mass ∼ 3 kDa larger than endogenous Syn7 (Figure 1D). Immunoblotting the same extract with anti-Syn7 antibody showed that the level of the epitope-tagged Syn7 was far less than that of the endogenous Syn7 (Figure 1D). As shown in Figure 3, the HA-Syn7 was distributed in large intracellular structures around the nucleus that showed significant overlap with lgp120. In contrast, very little overlap was observed between HA-Syn7 and TfR (which was found within a large tubular network). Furthermore, labeling of HA-Syn7 was distinct from that of EEA1 and Syn13, both of which were found in punctate structures throughout the cytoplasm. The observation that epitope-tagged Syn7 colocalized with lgp120 agrees well with the localization of endogenous Syn7 in MDCK and WIF-B cells.
Figure 3.
Localization of epitope-tagged Syn7 in NRK cells by confocal microscopy. Stably transfected NRK cells expressing an HA epitope–tagged Syn7 (NRK-K2 cells) were labeled for HA with the use of monoclonal anti-HA antibodies (A, C, and D, left) or rabbit anti-HA polyclonal antibodies (B, left) in combination with rabbit polyclonal antibodies to Syn13 (A, middle), mouse mAbs to TfR (B, middle), rabbit polyclonal antibodies to lgp120 (C, middle), or rabbit polyclonal antibodies to EEA1 (D, middle). E shows labeling with anti-lgp120 rabbit polyclonal antibodies (left) with anti-EEA1 mAbs (middle). Images were collected from a 0.5-μm focal plane by confocal microscopy. Note that variations in the distribution of Syn7 result from the different focal planes selected to show the localization of epitope-tagged Syn7 relative to each marker protein. Green/red merged images are shown on right. Bar, 10 μm.
Syn7 Is Localized to Late Endosomes and Lysosomes by Immunoelectron Microscopy
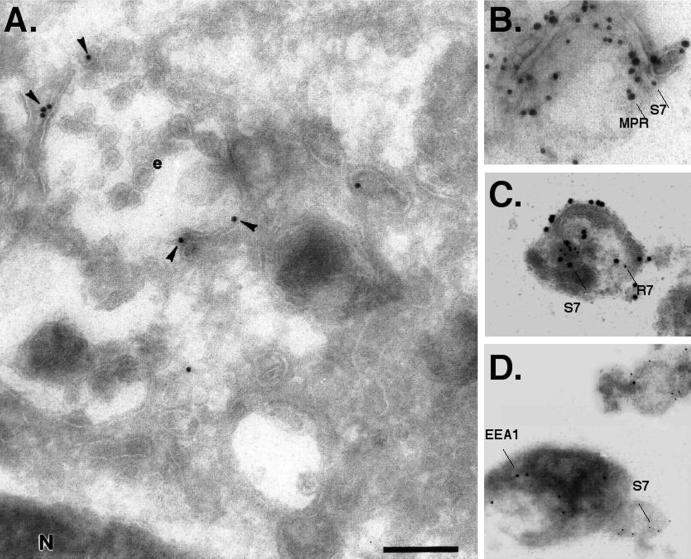
The colocalization of Syn7 with lgp120 by immunofluorescence in WIF-B and NRK cells indicated that a significant proportion of Syn7 is localized to late endosomes and lysosomes. Yet the lower levels of Syn7 coinciding with TfR-containing compartments by immunofluorescence (Figure 1K) in MDCK cells indicated that some Syn7 may also occupy earlier endosomal compartments. To characterize further the intracellular localization of Syn7 in MDCK cells, we examined ultrathin cryosections and isolated membranes by immunogold electron microscopy. Specific labeling for Syn7 was observed in the perinuclear region of the cell associated with multivesicular endosomes (Figure 4A). In addition, labeling was often observed on small tubulovesicular structures in close association with these multivesicular endosomes. However, despite the use of only the mild fixative paraformaldehyde, labeling on cryosections was invariably low. This proved problematic for convincing double-labeling experiments, so we adopted a procedure previously used to analyze GLUT-4–containing vesicles (Martin et al., 1996). A postnuclear supernatant was prepared from MDCK cells, fixed in paraformaldehyde, adsorbed to Formvar-coated grids, and labeled with antibodies to Syn7, CI-MPR, Rab7, and Vamp 3 (cellubrevin). Specific labeling for Syn7 was observed on large, electron-dense, multivesicular structures (Figure 4, B–D) that were similar to those structures labeled on cryosections. A large proportion of the structures that were positive for Syn7 labeling also showed labeling for the late endosome marker CI-MPR (Griffiths et al., 1988). In contrast, only a minor proportion of the structures that were positive for Syn7 showed labeling for EEA1 (Table 1), and most of the EEA1 labeling was restricted to small electron-lucent structures, confirming that the bulk of Syn7 and EEA1 occupy different compartments.
Figure 4.
Immunogold electron microscopic localization of Syn7 in MDCK cells. (A) Ultrathin cryosections were labeled with a specific affinity-purified antibody against Syn7 (anti-Syn7#1) followed by 15-nm protein A–gold. Specific labeling (arrowheads) was observed in the perinuclear region of the cell associated with multivesicular endosomes (e). (B–D) Intracellular MDCK membranes were fixed with paraformaldehyde and adsorbed to Formvar-coated grids. Membranes were then sequentially double labeled for Syn7 (15-nm protein A–gold; S7) and CI-MPR (20-nm protein A–gold; MPR) (B); Syn7 (20-nm protein A–gold; S7) and Rab7 (10-nm protein A–gold; R7) (C); or Syn7 (10-nm protein A–gold; S7) and EEA1 (15-nm protein A–gold) (D). Bar, 300 nm
We also examined the distribution of Syn7 in late endosomes and lysosomes isolated from rat liver (Figure 5, A and C) not only to confirm the localization of Syn7 that we observed in lgp120-positive structures in the hepatocyte WIF-B cell line but also because we used a cell-free late endosome–lysosome fusion reaction with the use of membranes derived from liver cells (see below). In addition, we examined the distribution of Syn7 in hybrid organelles formed after the in vitro late endosome–lysosome fusion assay (Figure 5B). Based on our previous pulse-chase studies with endocytosed ASF–avidin in whole livers, we prepared liver membrane fractions from rats that had been injected with ASF–avidin 6 min before killing, because this procedure results in the enrichment of ASF–avidin in multivesicular late endosomes (Mullock et al., 1994, 1998). Quantitation of ASF–avidin in these structures showed that there were 33 gold particles/μm2 compared with 0.5 gold particles/μm2 over other tissue components of the pellet, thus defining late endosomal structures. Lysosomes and late endosome–lysosome hybrids were identified in enriched fractions by their characteristic morphology. As shown in Figure 5, late endosomes, hybrids, and lysosomes showed significant labeling for Syn7, with the highest labeling seen on lysosomes (Figure 5D). Together, these data show that Syn7 is enriched in late endosomes and lysosomes. However, these data do not exclude the possibility that lower levels of Syn7 are distributed in earlier endocytic compartments. Therefore, we embarked on functional studies aimed at substantiating the proposed role of Syn7 in mediating fusion events to the late endosome.
Figure 5.
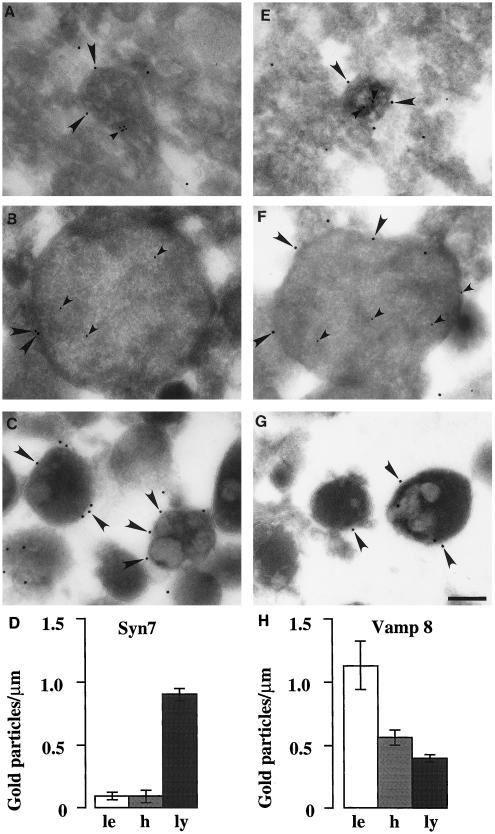
Immunogold electron microscopy of Syn7 and Vamp 8 associated with rat liver late endosomes, lysosomes, and hybrid organelles. Immunoelectron microscopy of Syn7 (A–C) labeling with the use of rabbit anti-Syn7#1 (15-nm gold; large arrowheads), Vamp 8 (E–G) labeling (15-nm gold; large arrowheads), and internalized ASF-avidin (A, B, E, and G) (8-nm gold; small arrowheads) associated with rat liver late endosomes (A and E), hybrid organelles (B and F), and lysosomal fractions (C and G). Late endosomes and hybrid organelles were identified by their content of ASF–avidin. Lysosomes were identified by their electron-dense morphology. Bar, 200 nm. (D and H) Quantitation of the level of Syn7 labeling (D) and Vamp 8 labeling (H) on the peripheral membrane of late endosomes (le; 30 μm scored), hybrid organelles isolated after late endosome–lysosome fusion in the cell-free fusion system (h; 65 μm scored), and lysosomes isolated after incubation with cytosol and ATP under the conditions of the cell-free fusion system (ly; 1000 μm scored). Error bars represent SEM for the number of organelles scored.
Antibodies to Syn7 Inhibit the Fusion of Late Endosomes with Lysosomes
Given the distribution of Syn7, we sought to determine whether Syn7 played a role as a t-SNARE that can coordinate specific fusion events between late endosomes and lysosomes. Previous studies have established an in vitro system that reconstitutes one of the last steps in lysosomal biogenesis, i.e., the fusion of late endosomes with lysosomes. This assay measures content mixing between distinct compartments containing either ASF–avidin or 125I-bpIgA by immunoprecipitating the avidin and quantitating the level of bound 125I-bpIgA. Labeled late endosomes are obtained from rat livers allowed to uptake ASF–avidin for 6 min, whereas labeled lysosomes are obtained by allowing uptake of 125I-bpIgA for 30 min. Fusion between these two compartments has been shown to require ATP and cytosol and is dependent on NSF and Rab proteins (Mullock et al., 1994, 1998).
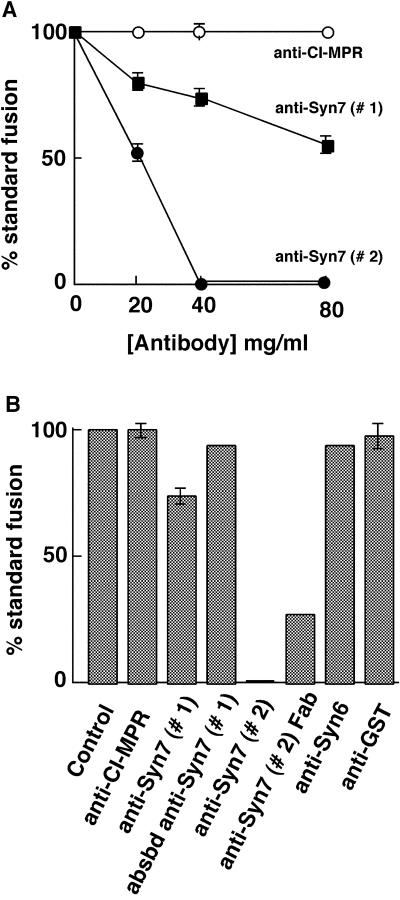
Both the late endosome and lysosome fractions were labeled with anti-Syn7 antibodies (Figure 5, A–C), and therefore both were treated with anti-Syn7 antibodies before the fusion reaction. We found that a 15-min preincubation at 4°C of the late endosomes and lysosomes with affinity-purified anti-Syn7 antibody significantly inhibited content mixing between these compartments after the reaction was initiated by combining fractions and incubating at 37°C. With the use of affinity-purified antibodies from two separate rabbit anti-Syn7 antisera (#1 and #2), the inhibition was dose-dependent and specific to anti-Syn7 antibodies (Figure 6A). Antibodies to the CI-MPR cytosolic tail had no effect at the same concentrations. In our initial experiments, we used the anti-Syn7 that was purified from the first rabbit we immunized (anti-Syn7#1). At the highest concentration used (80 μg/ml), we observed a 45% inhibition of fusion. However, increasing the antibody concentration in hopes of achieving complete inhibition was technically difficult. Therefore, we tested antibodies raised in additional animals. One such antibody was the goat anti-Syn7, which resulted in consistent and specific inhibition but which had a lower specific inhibitory activity than the affinity-purified rabbit anti-Syn7#1. Another rabbit antibody against Syn7 (anti-Syn7#2) proved to have an approximately fourfold higher level of specific inhibitory activity, yielding 48% inhibition at only 20 μg/ml and complete inhibition at and above 40 μg/ml. All three affinity-purified polyclonal antibody preparations were specific for Syn7 by immunoblotting. Neither an anti-syntaxin 6 antibody nor an anti-GST antibody purified in an identical manner nor anti-Syn7 antibodies that had been preincubated with the GST–Syn7 fusion protein affected the content-mixing assay (Figure 6B). Steric hindrance was not responsible for the inhibition by anti-Syn7 antibodies because monovalent Fab fragments prepared from the most inhibitory rabbit polyclonal antibody (anti-Syn7#2) also inhibited fusion by >70% at 45 μg/ml (Figure 6B).
Figure 6.
Syn7 antibodies specifically inhibit late endosome–lysosome fusion in vitro. (A) Titration experiment assaying content mixing between rat liver late endosomes and lysosomes. Effects of affinity-purified rabbit antibodies to Syn7#1 (closed squares) and Syn7#2 (closed circles) and rabbit antibodies to the cytosolic tail of the CI-MPR (open circles) were measured after treating the two fractions separately with the indicated concentrations of antibodies for 15 min on ice and keeping the antibodies present during the cell-free late endosome–lysosome fusion assay. Standard fusion (100%) is defined as the amount of fusion obtained after preincubation with buffer only, i.e., without the addition of antibodies. Data are presented as means ± SEM of duplicate samples from six separate experiments for anti-CI-MPR, five separate experiments for anti-Syn7#1 at 20 μg/ml, four separate experiments for anti-Syn7#1 at 40 μg/ml, three separate experiments for anti-Syn7#2 at 20 μg/ml, and two separate experiments for anti-Syn7#1 at 80 μg/ml. All other data presented are means of duplicate samples. (B) Summation of several experiments with the indicated antibodies to perturb the fusion assay between late endosomes and lysosomes. Before fusion, both late endosomes and lysosomes were preincubated with buffer only (Control) or the following antibodies at 40 μg/ml: anti-CI-MPR, affinity-purified antibodies to the cytosolic tail of the CI-MPR; anti-Syn7#1, affinity-purified antibodies to the cytosolic domain of Syn7 from serum #1; absbd anti-Syn7#1, preincubation with anti-Syn7#1 previously absorbed with GST–Syn7 (equivalent to anti-Syn7#1 in the assay at 40 μg/ml); anti-Syn7#2, affinity-purified antibodies to the cytosolic domain of Syn7 from serum #2; anti-syntaxin 6, affinity-purified antibodies to the cytosolic domain of syntaxin 6; and anti-GST, affinity-purified antibodies (40 μg/ml) to GST. Anti-Syn7(Fab), Fab fragments prepared from anti-Syn7#2 antibody were used at 45 μg/ml. Data presented are means of duplicate samples or means ± SEM in cases of three or more separate experiments.
Syn7 Complexes with the Endocytic v-(R)-SNARE Vamp 8
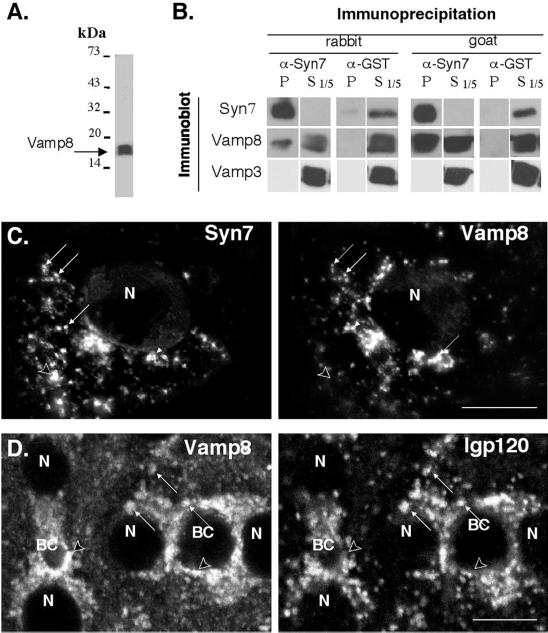
Because it is a syntaxin (Q-SNARE), we would predict that Syn7 would specifically complex with a subset of v-SNARE proteins (R-SNARE) to mediate fusion events with the late endosome and lysosome. Thus, in an effort to identify a putative v-SNARE partner for Syn7, we once again exploited the analogy between the yeast syntaxin, Vam3p, and its putative mammalian counterpart, Syn7. The work of Wickner and colleagues (Nichols et al., 1997; Ungerman et al., 1998, 1999) has established a role for Vam3p and the v-SNARE Nyv1p in fusion events within the late endocytic system of yeast, i.e., homotypic fusion of vacuoles. Furthermore, a significant proportion of these proteins are found in a complex, as demonstrated by coimmunoprecipitation experiments. Therefore, we searched the database for v-SNAREs that showed the highest level of identity with yeast Nyv1p. Although only low levels of identity were seen among the known mammalian v-SNAREs, the closest relative was Vamp 8, which has been localized to endocytic compartments (Advani et al., 1998; Wong et al., 1998b). Therefore, we determined whether Vamp 8 was associated with Syn7 by coimmunoprecipitation experiments. Figure 7A shows the specificity of the affinity-purified anti-Vamp 8 antibody generated to the cytosolic domain of Vamp 8. As reported previously, Vamp 8 migrates as an ∼17-kDa protein that is enriched in endosomal compartments (Wong et al., 1998a). Syn7 was immunoprecipitated from a Triton X-100–soluble fraction prepared from MDCK cells and then immunoblotted for Vamp 8 and Vamp 3 (cellubrevin) (Figure 7B). Immunoprecipitations performed with either the goat anti-Syn7 or the rabbit anti-Syn7#1 antibody resulted in a quantitative recovery of Syn7 in the immunoprecipitate. Blotting for candidate Vamps showed that 10–20% of the total Vamp 8 was associated with Syn7, whereas no detectable Vamp 3 was found associated with the immunoprecipitated Syn7.
Figure 7.
Immunofluorescence localization and coimmunoprecipitation of Vamp 8 with Syn7. (A) Specificity of Vamp 8 antibodies was demonstrated by subjecting whole cell extracts of MDCK cells to SDS-PAGE and immunoblotting with affinity-purified rabbit anti-Vamp 8 antibodies. (B) A Triton X-100–soluble fraction was prepared from a postnuclear supernatant from MDCK cells and allowed to incubate with the affinity-purified rabbit anti-Syn7 antibody or the rabbit anti-GST antibody (left panels) or the affinity-purified goat anti-Syn7 antibody and a matching goat anti-GST antibody (right panels). Fixed S. aureus cells were then added, and immunoprecipitates were washed, subjected to nonreduced SDS-PAGE, and immunoblotted for Syn7, Vamp 8, and Vamp 3. One-fifth of the corresponding supernatant from the immunoprecipitate was also immunoblotted. For immunoprecipitations with the rabbit anti-Syn7#1 antibody, the goat anti-Syn7 antibody was used for immunoblot analysis. For immunoprecipitations with the goat anti-Syn7 antibody, the rabbit anti-Syn7#1 antibody was used for immunoblot analysis. SDS-PAGE was performed under nonreducing conditions. (C) MDCK cells were labeled with goat anti-Syn7 (left) and rabbit anti-Vamp 8 (right) in combination with FITC-conjugated donkey anti-goat and Texas Red–conjugated donkey anti-rabbit secondary antibodies. Matched 1-μm-thick planes from a deconvoluted 4-μm-thick z-series are shown (D). Polarized WIF-B cells were fixed and permeabilized, double labeled with rabbit anti-Vamp 8 (left) and mouse anti-lgp120 (right), and imaged by confocal microscopy. Primary antibodies were followed by Cy3- and FITC-conjugated secondary antibodies, respectively. In C and D, arrows point to structures that are positive for both markers, and arrowheads indicate structures that contain only one marker. BC, bile canalicular-like space; N, nucleus. Bar, 10 μ m.
Consistent with our finding of a specific complex between Vamp 8 and Syn7, we also observed a significant level of overlap in the localization of Vamp 8 and Syn7 by immunofluorescence in MDCK cells (Figure 7C). This colocalization was confirmed by immunoelectron microscopy labeling of MDCK membranes (Table 1). Despite this colocalization, there were other Vamp 8–positive structures that were distinct from those positive for Syn7. One possibility is that Vamp 8 is restricted only to early endosomal compartments and that the overlap between Syn7 and Vamp 8 reflects the low level of Syn7 in early endosomal compartments. To determine whether any Vamp 8 occupied late endosomes or lysosomes, we also localized Vamp 8 by double-label immunofluorescence in WIF-B cells (Figure 7D) and by immunoelectron microscopy on isolated late endosome and lysosome fractions from rat liver (Figure 5, E–G). These experiments showed significant localization of Vamp 8 to lgp120-positive structures. Interestingly, by immunoelectron microscopy, Vamp 8 was enriched in late endosomes but far less so in lysosomes (Figure 5H). Thus, although Vamp 8 and Syn7 are localized to late endosome/lysosome compartments, the general distribution of each protein is different throughout the endocytic system.
DISCUSSION
Our effort to characterize the function of Syn7 was motivated by hopes of finding a functional homologue of the yeast Vam3p that would play a role in the late stages of lysosomal biogenesis. We present two major findings that support a role for Syn7 in lysosomal biogenesis. First, Syn7 is localized to late endosomes and lysosomes. Second, Syn7 is required for the fusion of late endosomes with lysosomes in vitro.
The mouse sequence we identify here is 93 and 94% identical to human and rat Syn7, respectively. Outside of the animal kingdom, mammalian Syn7 shows the highest level of identity (∼30%) with the plant Vam3p homologue AtVam3p, which like its proposed yeast counterpart is localized to the vacuole (Sato et al., 1997). Furthermore, Syn7 is expressed in all tissues examined, consistent with the role we hypothesize it fulfills in the endosomal system. By immunofluorescence microscopy, immunoelectron microscopy, and subcellular fractionation analysis, we find that significant levels of Syn7 occur in late endosomes and lysosomes and much less in EEA1-positive compartments.
As a potential partner v-SNARE for Syn7, we found that Vamp 8 partially colocalized with Syn7 and that a significant proportion of Vamp 8 is associated with Syn7 in immunoprecipitates from detergent-solubilized fractions prepared from MDCK cells. Whether Syn7 and Vamp 8 represent a SNARE pair that operates in vivo needs to be interpreted in light of recent experiments conducted in vitro that indicate that v-(R)-SNAREs can form complexes with many different t-(Q)-SNAREs in a manner that belies the specificity of the trafficking event they may catalyze in vivo (Yang et al., 1999). In contrast to these in vitro studies, we found a significant amount of Syn7 in a complex with Vamp 8 by immunoprecipitation from cell extracts, indicating that this complex exists in vivo. Syn7 was not observed in a complex with the v-SNARE Vamp 3 (cellubrevin), which in MDCK cells shows less colocalization with Syn7 than Vamp 8 (Table 1) and which has previously been localized to endosomes (McMahon et al., 1993; Chilcote et al., 1995; Daro et al., 1996). Thus, the specificity of the Syn7/Vamp 8 complex we observed diminishes the likelihood that the association of Syn7 and Vamp 8 is serendipitous as a result of their transit through the same compartment, because some Vamp 3 would be predicted to coprecipitate given the extent of colocalization we observed. Because of the specific association of Vamp 8 and Syn7 and the degree of colocalization we observed by immunofluorescence and by immunoelectron microscopy studies, we propose that Vamp 8 may be at least one of the v-SNARE partners that functionally interacts with Syn7 in vivo. This does not preclude Vamp 8 or Syn7 from interacting with other SNAREs to effect a variety of fusion events within the endocytic system. Further experiments are required to determine which transport steps this putative SNARE pair controls, because to date we have failed to show inhibition of fusion between late endosomes and lysosomes in the cell-free assay with the use of the single anti-Vamp 8 rabbit antiserum we have prepared.
Given the localization of Syn7 as well as the effect of anti-Syn7 antibodies and Fab fragments on the fusion of late endosomes and lysosomes in vitro, we believe that Syn7 participates in fusion events within the late endocytic pathway. This is in contrast to the function of the close homologue Syn13, which shares 50% identity with Syn7 and is also localized to the endocytic pathway. Syn13, however, is concentrated in early endosomes and participates with rab5 and EEA1 in the fusion of early endosomes (Prekeris et al., 1998; McBride et al., 1999). So far, no role for Syn7 has been found in early endosome fusion (McBride et al., 1999). Thus, Syn7 and Syn13, like their putative counterparts Vam3p and Pep12p, are likely to have distinct, nonoverlapping functions. Using an in vitro fusion reaction that measures content mixing between endosomes and lysosomes isolated from rat liver (Mullock et al., 1994, 1998), we were able to substantiate that Syn7 is likely to have a functional role in at least one fusion event with the lysosome in vivo. No vesicular carrier has been identified for transport from the late endosome to the lysosome in either mammalian or yeast cells. In contrast, it has been shown that this transport reaction is characterized by the fusion of large multivesicular endosomes with preexisting lysosomes (Futter et al., 1996; Mellman, 1996; Storrie and Desjardins, 1996; Mullock et al. 1998). In vitro, this fusion reaction results in the production of morphologically distinct late endosome–lysosome hybrids that can also be observed in vivo (Futter et al., 1996; Bright et al., 1997; Mullock et al., 1998). Previous studies have shown that this fusion reaction is dependent on NSF and α and γ SNAPs and is also sensitive to exogenous Rab GDI, a hallmark for the requirement of Rab GTPases (Mullock et al., 1998). These findings are consistent with the involvement of SNARE proteins in this fusion reaction, because it is well established that this cohort of proteins acts together to promote fusion (Lian et al., 1994; Sogaard et al., 1994; Luapshin and Waters, 1997). Syn7 appears to be one of the SNARE proteins in this fusion step, because antibodies directed to the cytosolic tail of Syn7 inhibited content mixing in a dose-dependent manner. The complete inhibition of content mixing observed with anti-Syn7#2 is consistent with the total inhibition of yeast vacuole fusion observed in a cell-free assay in the presence of anti-Vam3p (Nichols et al., 1997)
Wong and colleagues (1998b) reported that Syn7 was localized to early endosomes based on partial colocalization with the TfR. Although we find low levels of Syn7 in TfR-positive structures in MDCK cells, most of the labeling we observe is found coincident with markers of late endosomes and/or lysosomes. In the studies of Wong and colleagues, the TfR was labeled after the internalization of antibodies directed to the lumenal domain of the TfR, which, if in excess, may have also entered late endosomes and lysosomes and could have obscured detection of Syn7 in early compartments. Recent studies by Prekeris et al. (1999) that used immunogold labeling of cryosections indicated that Syn7 was restricted to clathrin-coated vesicles budding from early endosomes and was far more restricted in its localization pattern than the overlap with TfR that was reported by Wong et al. (1998b). Although the labeling observed in these immunoelectron microscopy studies was discrete and specific, the possibility remains that only a subset of the intracellular Syn7 molecules was detected under the fixation and labeling methods used. In our experiments, we saw a far more broad distribution of Syn7 by a variety of techniques. During the course of our immunoelectron microscopy experiments, however, we generated rabbit antibodies, a goat antibody, and a mouse mAb to Syn7 to use in immunogold labeling of cryosections. Although we observed specific labeling that was associated with late endosomal structures, we were unable to boost the overall level of immunogold labeling in sections, indicating that many of the Syn7 epitopes were obscured by this method despite our attempts with many different antibody preparations. Furthermore, when we labeled lysosomal fractions isolated from rat liver, we found the labeling efficiency to increase significantly when we incubated the fraction in cytosol and ATP before fixation and cryosectioning. Thus, the highly restricted localization of Syn7 in the studies of Prekeris and others may be due in part to the selective detection of a subpopulation of Syn7 that may reside in a specific protein complex (Prekeris et al., 1999). Our conclusion that Syn7 is distributed throughout the endocytic pathway and concentrated in late endosomes and lysosomes is based on a variety of immunofluorescence and immunoelectron microscopy localization studies that were performed in MDCK cells, NRK cells expressing epitope-tagged Syn7, and hepatocytes (e.g., rat liver and WIF-B cells) in which Syn7 was largely colocalized with markers of late endosomes and lysosomes. By electron microscopy, Syn7 was found on multivesicular endosomes that also labeled with antibodies to CI-MPR and Rab7 (Figure 4) or that contained internalized avidin-conjugated ASF (Figure 5). Importantly, we also found significant levels of Syn7 on the limiting membrane of lysosomes isolated from rat liver (Figure 5) at even higher levels than that observed for late endosomes also isolated from rat liver. Consistent with our localization results is recent work by Nakamura et al. (2000), who also found Syn7 localized to late endosomes and lysosomes in colabeling experiments with LAMP1 (the mouse homologue of rat lgp120). Furthermore, these authors provide evidence for a functional role for Syn7 in the late endocytic pathway by examining the effects of overexpressing the cytosolic domain of Syn7 to induce a dominant-negative effect. This resulted in a block in the transfer of endocytosed material from early endosomes to late endosomes. These results must be interpreted carefully, because the dominant-negative Syn7 works by titrating out Syn7-binding proteins that might function elsewhere. Also, soluble fragments of Syn7 may be able to bind promiscuously to inappropriate SNARE partners. Our results in vitro support a role for Syn7 per se in late endosome–lysosome fusion, and it will be interesting to determine exactly how Syn7 functions in vivo. In this regard, experiments aimed at assigning the exact function of Syn7 are reminiscent of a similar analysis of the role of Rab7. Experiments with the use of hyperactive Rab7 and biochemical in vitro fusion experiments have implied and/or shown a direct role for Rab7 and its yeast homologue Ypt7p in the fusion of lysosomes and vacuoles in animal and yeast cells, respectively (Haas et al., 1995; Bucci et al., 2000). However, analysis of dominant-negative Rab7 in transfected cells supports the conclusion that Rab7/Ypt7p participates in early to late endosome transport (Press et al., 1998). These results may mean that Rab7, or in our case Syn7, works at multiple steps in the late endocytic pathway or could mean that interruption of normal lysosomal fusion events results in indirect effects earlier in the endocytic pathway. At the very least, we believe that our results substantiate a role for Syn7 in the process of late endosome–lysosome fusion.
Given that Syn7 is found in both late endosomes and lysosomes, the question arises whether Syn7 catalyzes fusion as a t-SNARE on the lysosomal membrane or on the late endosome. Perhaps the best model for Syn7 function is exemplified by the yeast Vam3p, which has been shown to mediate homotypic fusion of yeast vacuoles (Nichols et al., 1997). This fusion reaction is dependent on NSF, Ypt7p (a Rab7 homologue), Vam3p, and Nyv1p (a v-SNARE). The best model is that Vam3p and the v-SNARE Nyv1p are not only present but are functionally active on both fusion partners (Nichols et al., 1997; Ungermann et al., 1998). Thus, Syn7 may be active on both late endosomes and lysosomes and catalyze fusion events from a number of different pathways that converge at late endosomal/lysosomal compartments. This would be consistent with the multifaceted role of Vam3p, which catalyzes not only fusion of endosomes with the vacuole but also fusion of autophagic vesicles with the vacuole and delivery of transport intermediates derived from the AP-3 transport pathway originating from the Golgi (Cowles et al., 1997; Darsow et al., 1997; Piper et al., 1997).
ACKNOWLEDGMENTS
The authors thank Judy Tellam for efforts in cloning Syn7 cDNA; the personnel at the University of Iowa Central Microscope facility, Nick Wade, and Kara Brown for their efforts in obtaining mAbs and polyclonal antibodies to syntaxin 7; Sally Gray for technical assistance; and Linda Pelleymounter for performing immunofluorescence studies. This work was supported by grants from the American Heart Association (9730275N), the National Health and Medical Research Council, the Medical Research Council, SmithKline Beecham, and the Australian Research Council. This work was also supported by a Human Frontiers Science Program grant to J.P.L., D.E.J., and R.C.P. The Center for Molecular and Cellular Biology is a Special Research Center for the Australian Research Council.
REFERENCES
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani RJ, Bae HR, Bock JB, Chao DS, Doung YC, Prekeris R, Yoo JS, Scheller RH. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273:10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- Agard DA, Yasushi H, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HR. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK. SNAREs and the specificity of transport vesicle targeting. Curr Opin Cell Biol. 1995;7:581–586. doi: 10.1016/0955-0674(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JB, Scheller RH. A fusion of new ideas. Nature. 1997;387:133–135. doi: 10.1038/387133a0. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright NA, Reaves BJ, Mullock BM, Luzio JP. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J Cell Sci. 1997;110:2027–2040. doi: 10.1242/jcs.110.17.2027. [DOI] [PubMed] [Google Scholar]
- Brooks DA, Bradford TM, Carlsson SV, Hopwood JJ. A membrane protein primarily associated with the lysosomal compartment. Biochim Biophys Acta. 1997;1327:162–170. doi: 10.1016/s0005-2736(97)00053-9. [DOI] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcote TJ, Galli T, Mundigl O, Edelmann L, McPherson PS, Takei K, De Camilli P. Cellubrevin and synaptobrevins: similar subcellular localization and biochemical properties in PC12 cells. J Cell Biol. 1995;129:219–231. doi: 10.1083/jcb.129.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter A, Harris R. Simplified preparation of rabbit Fab fragments. J Immunol Methods. 1983;59:199–203. doi: 10.1016/0022-1759(83)90031-5. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Daro E, van der Sluijs P, Galli T, Mellman I. Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc Natl Acad Sci USA. 1996;93:9559–9564. doi: 10.1073/pnas.93.18.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Conceicao A, Marty-Mazars D, Bassham DC, Sanderfoot AA, Marty F, Raikhel NV. The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell. 1997;4:571–582. [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D, Sutton B, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno KT, Ishikawa T, Akasaki K, Yano S, Tanaka Y, Yamaguchi Y, Tsuji H, Himeno M, Kato K. Morphological localization of a major lysosomal membrane protein glycoprotein in the endocytic membrane system. J Biochem. 1989;106:708–716. doi: 10.1093/oxfordjournals.jbchem.a122921. [DOI] [PubMed] [Google Scholar]
- Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte M, Gallwitz D. High expression of the yeast syntaxin-related Vam3 protein suppresses the protein transport defects of a pep12 null mutant. FEBS Lett. 1997;411:48–52. doi: 10.1016/s0014-5793(97)00575-9. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Fine Structure Immunocytochemistry. Berlin: Springer-Verlag; 1993. [Google Scholar]
- Griffiths G, Holflack B, Simons K, Mellman I, Kornfeld S. The mannose-6-phosphate receptor and the biogenesis of lysosomes. Cell. 1988;52:329–341. doi: 10.1016/s0092-8674(88)80026-6. [DOI] [PubMed] [Google Scholar]
- Grimaldi KA, Hutton JC, Siddle K. Production and characterization of monoclonal antibodies to insulin secretory granule membranes. Biochem J. 1987;245:557–566. doi: 10.1042/bj2450557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis V, Green SA, Marsh M, Vihko P, Helenius A, Mellman I. Glycoproteins of the lysosomal membrane. J Cell Biol. 1985;100:1839–1847. doi: 10.1083/jcb.100.6.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JP, Stone S, Jiang Y, Lyons P, Ferro Novick S. Ypt1p implicated in v-SNARE activation. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- Luapshin VV, Waters MG. t-SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Rous BA, Bright NA, Pryor PR, Mullock BM, Piper RC. Lysosome-endosome fusion and lysosome biogenesis. J Cell Sci. 2000;113:1515–1524. doi: 10.1242/jcs.113.9.1515. [DOI] [PubMed] [Google Scholar]
- Martin S, Tellam J, Livington C, Slot JW, Gould GW, James DE. The glucose transporter (GLUT-4) and vesicle-associated membrane protein-2 (VAMP-2) are segregated from recycling endosomes in insulin-sensitive cells. J Cell Biol. 1996;134:625–635. doi: 10.1083/jcb.134.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Ushkaryov YA, Edelman L, Link E, Binz T, Niemann H, Jahn R, Sudhof TC. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Mu F, Callaghan J, Steele-Mortimer O, Stenmark H, Parton R, Campbell P, McCluskey J, Yeo J, Tock E, Toh B. EEA1, an early endosome-associated protein: EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Mullock BM, Bright NA, Fearon CW, Gray SR, Luzio JP. Fusion of lysosomes with late-endosomes produces a hybrid organelle of intermediate density and is NSF dependent. J Cell Biol. 1998;140:591–601. doi: 10.1083/jcb.140.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullock BM, Perez JH, Kuwana T, Gray SR, Luzio JP. Lysosomes can fuse with a late endosomal compartment in a cell-free system from rat liver. J Cell Biol. 1994;126:1173–1182. doi: 10.1083/jcb.126.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Yamamoto A, Wada Y, Futai M. Syntaxin 7 mediates endocytic trafficking to late endosomes. J Biol Chem. 2000;275:6523–6529. doi: 10.1074/jbc.275.9.6523. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Parton RG, Way M, Zorzi N, Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole via a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Chen YA, Scheller RH. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R, Yang B, Oorschot V, Klumperman J, Scheller RH. Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol Biol Cell. 1999;10:3891–3908. doi: 10.1091/mbc.10.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP. The role of intra-organellar Ca2+ in late endosome-lysosome heterotypic fusion and in reformation of lysosomes from hybrid organelles. J Cell Biol. 2000;149:1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves BJ, Bright NA, Mullock BM, Luzio JP. The effect of wortmannin on the localization of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J Cell Sci. 1996;109:749–762. doi: 10.1242/jcs.109.4.749. [DOI] [PubMed] [Google Scholar]
- Rehling P, Darsow T, Katzmann DJ, Emr SD. Formation of AP-3 transport intermediates requires Vps41 function. Nat Cell Biol. 1999;6:346–353. doi: 10.1038/14037. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Pohlig G, Rothman JH, Stevens TH. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989;108:1363–1373. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Rowe T, Dascher C, Bannykh S, Plutner H, Balch W. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc Natl Acad Sci USA. 1998;95:9920–9925. doi: 10.1073/pnas.95.17.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato MH, Nakamura N, Ohsumi Y, Kouchi H, Kondo M, Hara NI, Nishimura M, Wada Y. The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J Biol Chem. 1997;272:24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- Shanks MR, Cassio D, Lecoq O, Hubbard AL. An improved polarized rat hepatoma hybrid cell line: generation and comparison with its hepatoma relatives and hepatocytes in vivo. J Cell Sci. 1994;107:813–825. doi: 10.1242/jcs.107.4.813. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sogaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh B, D'Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- Storrie B, Desjardins M. The biogenesis of lysosomes: is it a kiss and run continuous fusion and fission process? BioEssays. 1996;18:895–903. doi: 10.1002/bies.950181108. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Tellam JT, James DE, Stevens TH, Piper RC. Identification of a mammalian Golgi Sec1p-like protein, mVps45p. J Biol Chem. 1997;272:6187–6193. doi: 10.1074/jbc.272.10.6187. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Nichols B, Pelham H, Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol. 1998;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, von Mollard GF, Jensen ON, Margolis N, Stevens TH, Wickner W. Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J Cell Biol. 1999;145:1435–1442. doi: 10.1083/jcb.145.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Nakamura N, Ohsumi Y, Hirata A. Vam3p, a new member of syntaxin related protein, is required for vacuolar assembly in the yeast Saccharomyces cerevisiae. J Cell Sci. 1997;110:1299–1306. doi: 10.1242/jcs.110.11.1299. [DOI] [PubMed] [Google Scholar]
- Wang H, Frelin L, Pevsner J. Human syntaxin 7: a Pep12p/Vps6p homologue implicated in vesicle trafficking to lysosomes. Gene. 1997;199:39–48. doi: 10.1016/s0378-1119(97)00343-0. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofman K. A conserved domain is present in different families of vesicular fusion protein: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Xu Y, Zhang T, Hong W. Syntaxin 7, a novel syntaxin member associated with the early endosomal compartment. J Biol Chem. 1998a;273:375–380. doi: 10.1074/jbc.273.1.375. [DOI] [PubMed] [Google Scholar]
- Wong SH, Zhang T, Xu Y, Subramaniam VN, Griffiths G, Hong W. Endobrevin, a novel synaptobrevin/VAMP-like protein preferentially associated with the early endosome. Mol Biol Cell. 1998b;9:1549–1563. doi: 10.1091/mbc.9.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Gonzalez LJ, Prekeris R, Steegmaier M, Advani RJ, Scheller RH. SNARE interactions are not selective: implications for membrane fusion specificity. J Biol Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]