Abstract
Dramatic DNA reorganization and elimination processes occur during macronuclear differentiation in ciliates. In this study we analyzed whether cytosine methylation of specific sequences plays a functional role during DNA rearrangement. Three classes of sequences, macronuclear-destined sequences (MDSs, pCE7), members from a large family of transposon-like elements and micronuclear-specific sequences (pLJ01), differing in their structure and future destiny during nuclear differentiation, were studied in the micronucleus, the developing macronucleus and, when present, in the mature macronucleus. While the MDSs become processed to a 1.1 and 1.3 kb gene-sized macronuclear DNA molecule, the family of transposon-like elements represented by MaA81 becomes removed late in the course of polytene chromosome formation. The micronuclear-specific sequence pLJ01 is eliminated together with bulk micronuclear DNA during degradation of polytene chromosomes. No methylated cytosine could be detected in the vegetative macronucleus and no difference in methylation pattern was observed either between micronucleus and developing macronucleus in MDSs or in a micronuclear-specific sequence. However, a significant percentage of the cytosines contained in the transposon-like element becomes methylated de novo in the course of macronuclear differentiation. This is the first demonstration that cytosine methylation in specific sequences occurs during macronuclear differentiation and may provide a first step towards understanding epigenetic factors involved in DNA processing.
INTRODUCTION
In eukaryotic cells methylation of cytosine residues very frequently correlates with the silencing of genes (1) and formation of heterochromatin (2) and it seems to be essential for genomic imprinting and development in mammalian organisms (reviewed in 3,4). Once a specific methylation pattern is established it can be stably propagated by the action of maintenance methyltransferases (reviewed in 5). The methylation pattern is established by de novo methyltransferases while demethylation is achieved either by suppression of maintenance DNA methyltransferase followed by the passive loss of methyl groups during replications or by the action of specific demethylases (6). In vertebrate nuclei cytosine methylation occurs predominantly at the sequence CpG, although other sequence motifs in which cytosine methylation occurs are described (7). In addition to cytosine methylation the methylation of adenosine has been described in a variety of organisms including ciliates (8). Only recently cytosine methylation has been described to occur early during Drosophila embryogenesis using very sensitive detection techniques (7,9). Eukaryotic organisms in which cytosine methylation has not yet been found are yeast, Caenorhabditis (7) and the nuclei of stichotrichous ciliates.
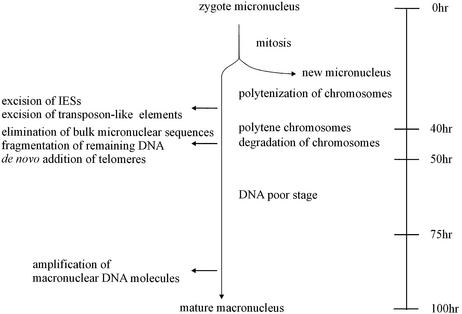
Vegetative cells of ciliated protozoa contain two types of nuclei: macronuclei and micronuclei. While the DNA-rich macronucleus is transcriptionally very active expressing all the RNAs required for vegetative growth, the diploid micronucleus seems to be transcriptionally inert and its main function only becomes obvious during sexual reproduction. In the course of this process of conjugation, haploid micronuclei are exchanged between sexual partners where they fuse with a remaining haploid micronucleus to form a diploid zygote nucleus. This nucleus divides mitotically and one of the daughter nuclei differentiates into a new micronucleus, whereas the other forms a new macronucleus while the old macronucleus degenerates. In spirotrichous ciliates, such as Euplotes, Oxytricha or Stylonychia, macronuclear differentiation is accompanied by massive DNA rearrangement and DNA elimination events (Fig. 1). In a first DNA synthesis phase, which eventually leads to the formation of polytene chromosomes, short non-coding DNA sequences (internal eliminated sequences, IESs) interrupting macronuclear- destined sequences (MDSs) and part of transposon-like sequences (10) are spliced off the DNA. As the polytene chromosomes become degraded, a large percentage of DNA is eliminated and in the course of this process the remaining DNA becomes fragmented into short DNA molecules (gene-sized pieces, gsp) ranging in size between ∼0.4 and 40 kb, to which telomeric sequences are added de novo. These short gene-sized DNA molecules carry the genes that will be transcribed in the vegetative macronucleus (11). So far, modification of nucleotides was only analyzed in the macronuclei and micronuclei of several ciliates using HPLC and methylation-dependent cleavage assays. Methylated cytosines have been detected in macronuclear DNA of Blepharisma (12) and Colpoda (13), whereas only methylated adenine was found in macronuclear DNA of most other ciliates (8,14–17).
Figure 1.
Schematic diagram of macronuclear development indicating major events during this process [modified after Prescott (11) and Kraut et al. (30)].
During macronuclear differentiation MDSs have to be discriminated from sequences to be eliminated. This elimination process can be regarded as the most extreme form of irreversible silencing. We therefore analyzed the DNA not only from vegetative nuclei but also from the developing macronucleus (macronuclear anlagen) with respect to methylation of cytosines using RP-HPLC. Moreover, specific sequences, such as macronuclear precursor sequences, micronuclear-specific sequences and a family of transposon-like elements, were analyzed in detail for cytosine methylation in the various nuclei using a bisulphate-based PCR strategy (18).
MATERIALS AND METHODS
Growth of Stylonychia lemnae cells, isolation of nuclei and DNA
Vegetative Stylonychia cells were cultivated as described earlier (19). Conjugations were set up under conditions described previously (19). Isolation of macronuclei, micronuclei and anlagen of different stages of development and preparation of DNA followed the protocols described previously (19,20).
RP-HPLC
For RP-HPLC, DNA (100 µg) was needle sheared and completely digested to nucleosides using Benzonase (Novagen, 300 U, 24 h), nuclease P1 (Roche, 10 U, 24 h) and alkaline phosphatase (Roche, 10 U, 3 h). The nucleosides were analyzed on a LIChrosorb RP-18 (7 µm) column (Merck) according to Gowher et al. (21). DNA was analyzed in the presence and absence of 5 µg methylcytidine (Sigma).
Bisulphite reaction and strand specific PCR
Bisulphite treatment was done according to the method described by Clark et al. (18) with some minor modifications. For each reaction 25 µg DNA isolated from the different nuclei was used in a volume of 100 µl. The genomic DNA was needle sheared, denatured in 0.3 M NaOH (45°C for 30 min). Sulphonation was done in 5 mM 0.1 M hydroquinone, 3.6 M sodium bisulphite, pH 5.0 (55°C for 6 h). Free bisulphite was removed using the Promega DNA clean-up-system. Alkali desulphonation was performed in 0.3 M NaOH (45°C for 30 min). The DNA was then ethanol precipitated and resuspended in 250 µl ddH2O. Aliquots of 5–10 µl were used in each PCR. PCR conditions were 15 min 94°C initial denaturation, 35 cycles of 45 s 94°C, 45 s 53°C, 45 s 72°C followed by a final extension of 15 min at 72°C to enable TA cloning using the pGEM-T easy system by Promega. A hotstart Taq polymerase (Genecraft) was used. In the first PCR, primer combination (a) was used and in the following, nested reaction primer combination (b) (see Table 1), with the exception of P13/P14 where the nested PCR was not necessary. Due to the structural instability of the bisulphite treated template (18) only short regions of the sequences of interest could be amplified in one reaction.
Table 1. Primers used for PCR analysis.
| Primer | Sequence | Size of PCR product |
|---|---|---|
| P1 | GATAAGGTGTTGGGGTTTAATTTG | |
| P2 | TCAAACAACAAAACACTTCATAAACA | 458 bp |
| P3a | TTGAGATTGAGGTGGGTTGA | |
| P4a | TCATTCCAATTAAACACTTCCTCTT | 532 bp |
| P3b | TTGGTAATTTTTGAATTTGGGAAT | |
| P4b | TTCAAAAACCTTATTACACCTTAACA | 279 bp |
| P5 | GAGGAAGTGTTTAATTGGAATGAAGT | |
| P6 | CAATTTAATCATCAAACCACCTTATCC | 913 bp |
| P7a | TTGTTTGGTGTAAAGATGAGAGGA | |
| P8a | CCATTAAACACTCAAAACTTTCAA | 425 bp |
| P7b | TTTTGGGTTTATTATAGGTGAAGAA | |
| P8b | CTATTAAATATAACCATATTAAAATCC | 288 bp |
| P9a | GTTTGAAAGATGAGATGTTTAGGTG | |
| P10a | TTTCCATAAATCTATCCAAAAACATT | 647 bp |
| P9b | AGATGTTTAGGTGGAAATGTTGAAG | |
| P10b | ATCCACCATAACCTATAATCCACTT | 586 bp |
| P11a | GAGGTATGAAGAAGAGTTTAAGAAAGTT | |
| P12a | CAATAATTAACCAAAACATCCCCTTATTAAC | 515 bp |
| P11b | GATTTTTAGATAAATATTGTAATTGTGTTAG | |
| P12b | AAAAAAATATCTAAAAATAACAATTTTCCTAC | 368 bp |
| P13 | TGAGTTTTGTGGTTTATATATTGTTTGAAG | |
| P14 | AAATCAAATTAACAACCAAAATTTCAATAAACTC | 628 bp |
Primer combination (a) was used in the first PCR, primer combination (b) are nested primers used in a second PCR. The length of the obtained fragment is indicated behind each primer pair.
RESULTS
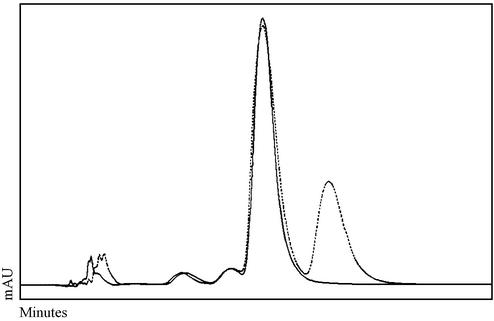
DNA was isolated from macronuclei, micronuclei and macronuclear anlagen of the stichotrichous ciliate Stylonychia (19,20), digested with nucleases and phosphatases and subjected to RP-HPLC analysis. A typical elution profile showing the positions of cytosine and methylated cytosine is shown in Figure 2. Methylated cytosine could not be detected in any case. Since this technique would not allow the detection of very low amounts (<0.1 µg methylcytosine/100 µg total DNA) of this nucleoside we decided to use the bisulphite-based PCR strategy to analyze exemplary sequences for the presence of methylated cytosines.
Figure 2.
RP-HPLC analysis of anlagen DNA in an early stage of polytenization. DNA (100 µg) was analyzed in the presence (broken line) and absence (continuous line) of 5 µg methylcytidine. Similar profiles were observed for micronuclear, macronuclear and anlagen DNA of different stages of development.
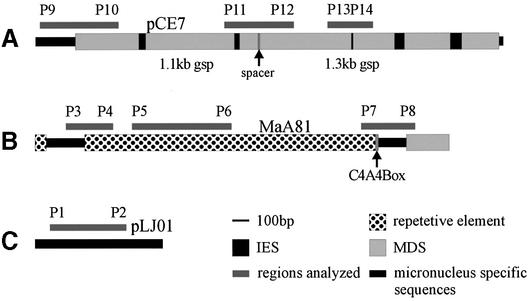
The following sequences with different destinies during macronuclear differentiation were further analyzed using PCR. The observed cytosine methylation pattern was compared in the micronucleus, the macronuclear anlagen and, if processed, also in the mature macronucleus (Fig. 3). (i) A cluster of MDSs (pCE7) that become processed in the mature macronucleus to a 1.1 and 1.3 kb gsp (22). In micronuclear DNA these two MDSs are separated by a 10 bp spacer region and interrupted by five IESs (Fig. 3A). (ii) Members (for which clone MaA81 is one example) of a family of transposon-like elements showing very high sequence identity (>90%) present in about 5000–7000 copies per haploid genome whose removal starts late during polytene chromosome formation but before fragmentation of DNA occurs. However, the exact mechanisms by which these elements are removed has not yet been determined and it is not clear whether all members of this family become excised during polytene chromosome formation or become eliminated during chromosome breakdown. This element consists of two long direct repeats flanking the truncated version of a macronuclear-specific sequence and a 16mer telomeric repeat (23,24) (Fig. 3B). (iii) A 800 bp micronuclear-specific DNA sequence (pLJ01) isolated from a micronuclear gene library (Fig. 3C). This sequence hybridizes to micronuclear DNA and to DNA isolated from late polytene chromosome stage but not to macronuclear DNA. Thus pLJ01 becomes eliminated together with bulk micronuclear DNA as part of the DNA elimination process. Its copy number was estimated to be below 20 copies per haploid genome (data not shown). pLJ01 shows no structural homology to transposon-like elements and most likely represents an intergenic DNA-spacer separating MDS clusters in the micronuclear genome. Six bisulphite conversions were performed with DNA isolated from the different nuclei. Sequence data were obtained from six to ten cloned PCR fragments from each set of sequences. Results from these analyses are summarized in Table 2. An alignment of the sequences of several cloned PCR products are provided as Supplementary Material.
Figure 3.
Sequences chosen for further analysis by PCR of bisulphite treated DNA (for explanation see text). Bars indicate the regions investigated. The position of the primer pairs for nested PCR are indicated by P1 to P12 (A)/(B) respectively (see Table 1). (A) pCE7, (B) MaA81, (C) pLJ01.
Table 2. Results of the bisulphite analysisa.
| Analyzed sequence | Macronuclear DNA | Micronuclear DNA | Anlagen DNA |
|---|---|---|---|
| pCE7 | |||
| P9b/P10a | b | 4% | 4% |
| P11b/P12b | b | – | – |
| P13/P14 | – | – | – |
| MaA81 | |||
| P3b/P4b | c | – | – |
| P5/P6 | c | – | 25% |
| P7b/P8b | c | – | – |
| pLJ01 | |||
| P1/P2 | c | – | – |
Given is the percentage of methylated cytosines within the obtained PCR fragments.
aFor sequence data see the Supplementary Material provided.
bNo PCR fragment can be obtained from macronuclear DNA.
cSequence not present in macronuclear DNA.
Although cytosine methylation was not detected in the analyzed sequences from macronuclear DNA, methylated cytosines were found in pCE7 in both the micronuclei and the macronuclear anlagen. In these sequences cytosine methylation was exclusively observed in the symmetric sequence motif CCWGG at the internal cytosine. This sequence motif is found seven times in pCE7 of which one motif is in the micronuclear-specific sequence upstream of the 1.1 kb MDS, four are found in its 5′-subtelomeric region, one in its 3′-subtelomeric region and one in the 5′-subtelomeric region of the 1.3 kb MDS. However, cytosines in this motif are not necessarily methylated. The modified nucleotides are only found in the micronuclear-specific sequence flanking the 1.1 kb gsp homologous sequence and in its 5′-subtelomeric region. No other cytosine methylation could be detected in pCE7. This methylation pattern was identical in different bisulphite conversions and PCRs (for sequence data see Supplementary Material). Also no cytosine methylation could be found in the micronuclear-specific sequence pLJ01. Most interestingly no cytosine methylation can be detected in the transposon-like element in the micronucleus but a significant percentage of cytosines becomes methylated (∼25%) in the analyzed region of this element (PCR product obtained with primers P5/P6) in the course of macronuclear differentiation. In all analyses the methylation pattern observed was identical and the nucleotide differences between different clones were <2% (for sequence data see Supplementary Material) demonstrating that probably all members of this family are modified early during macronuclear development. Interestingly methylation in these elements appeared clustered within 500 bp of these elements. In contrast to the methylation sites found in conjunction with the MDSs in the micronucleus and macronuclear anlagen, no specific sequence motif could be identified that is linked to cytosine methylation in the transposon-like element during macronuclear differentiation.
DISCUSSION
The methylation of cytosines has been attributed to be relevant in the silencing of genes, imprinting and the formation of heterochromatin. Ciliated protozoa, especially spirotrichous ciliates such as Euplotes, Oxytricha and Stylonychia, represent attractive models for studying programmed DNA rearrangement and elimination processes. Various reports approached the question whether cytosine methylation occurs in these cells. Methylated cytosines were shown to be present in macronuclear DNA of Blepharisma (12) and Colpoda (13). Only methylated adenine could be found in the macronucleus of most other species using standard chromatographic techniques (8,14–17). In these studies only DNA from the two nuclei occurring in the vegetative cell, macronucleus and micronucleus, were analyzed. Since a dramatic nuclear remodeling and differentiation process occurs only after sexual reproduction, we investigated the problem of cytosine methylation again and included DNA from various stages of the developing macronucleus. Since methylated cytosines could not be detected by RP-HPLC, the level of modified cytosines has to be <0.1 μg per 100 μg genomic DNA. We therefore used PCR as a more sensitive technique on a set of DNA sequences known to have different destinies during the process of DNA remodeling in S.lemnae: MDSs that become processed to a functional macronuclear DNA molecule, a family of transposon-like elements whose removal starts late during polytene chromosome formation and a micronuclear-specific sequence being eliminated with bulk micronuclear DNA after the breakdown of polytene chromosomes. In contrast to Colpoda (13) and Blepharisma (12) no modification of cytosine could be found in the mature macronucleus of Stylonychia whereas a low level of cytosine methylation occurring in the sequence motif CCWGG was found in the micronuclear-specific sequences upstream of one MDS and in its subtelomeric region. This methylation was identical in the micronucleus and the macronuclear anlagen suggesting that it is not involved in nuclear differentiation but may be relevant in the silencing of these sequences in the micronucleus and the developing macronucleus. CCWGG was already described as a prokaryotic methylation motif (25) and was recently found in mammalian cells possibly being involved in B cell lymphoma gene silencing (26). No cytosine methylation was found in the micronuclear-specific sequence pLJ01 eliminated with bulk micronuclear DNA in the micronucleus or the macronuclear anlagen. However, a dramatic difference in cytosine methylation extent is observed in the transposon-like element between micronuclei and developing macronuclei. While no cytosine is methylated in micronuclear DNA, cytosines are methylated de novo in this element during macronuclear differentiation. Excision of transposon-like elements has been extensively studied in Euplotes. Part of them are excised before breakdown of polytene chromosomes (10,27) in the form of heterochromatic chromatin rings (28) and evidence has been provided that chromatin configuration plays an important role in this excision process (29). Our data suggest that de novo methylation of cytosines is involved in the formation of heterochromatic regions, which could be a necessary prerequisite for correct removal of transposon-like elements during macronuclear differentiation.
In summary, we show for the first time a low level of cytosine methylation in micronuclear DNA of S.lemnae of the sequence motif CCWGG and demonstrate that the cytosines in a class of sequences, whose removal starts before elimination of bulk DNA, become de novo methylated during macronuclear differentiation. This implies two different DNA methylation systems in the cell. Maintenance methyltransferases are involved in the preservation of the methylation status in micronuclei. In this case methylation is observed exclusively in one sequence motif. A de novo methyltransferase will methylate cytosines in a very specific class of DNA sequences in the course of macronuclear differentiation. This may be a first step towards understanding epigenetic factors involved in programmed DNA reorganization.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the National Science Foundation (grant number 128-6114-1) and the Alfried von Bohlen and Halbach Foundation.
REFERENCES
- 1.Tate P.H. and Bird,A.P. (1993) Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev., 3, 226–231. [DOI] [PubMed] [Google Scholar]
- 2.Lewis J. and Bird,A. (1991) DNA methylation and chromatin structure. FEBS Lett., 285, 155–159. [DOI] [PubMed] [Google Scholar]
- 3.Feil R. and Khosla,S. (1999) Genomic imprinting in mammals: an interplay between chromatin and DNA methylation? Trends Genet., 15, 431–435. [DOI] [PubMed] [Google Scholar]
- 4.Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh C.L. (2000) Dynamics of DNA methylation pattern. Curr. Opin. Genet. Dev., 10, 224–228. [DOI] [PubMed] [Google Scholar]
- 6.Wolffe A.P., Jones,P.L. and Wade,P.A. (1999) DNA demethylation. Proc. Natl Acad. Sci. USA, 96, 5894–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyko F., Ramsahoye,B.H. and Jaenisch,R. (2000) DNA methylation in Drosophila melanogaster. Nature, 408, 538–540. [DOI] [PubMed] [Google Scholar]
- 8.Karrer K.M. and VanNuland,T.A. (1998) Position effect takes precedence over target sequence in determination of adenine methylation patterns in the nuclear genome of a eukaryote, Tetrahymena thermophila. Nucleic Acids Res., 26, 4566–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowher H., Leismann,O. and Jeltsch,A. (2000) DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J., 19, 6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frels J.S., Tebeau,C.M., Doktor,S.Z. and Jahn,C.L. (1996) Differential replication and DNA elimination in the polytene chromosomes of Euplotes crassus. Mol. Biol. Cell., 7, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prescott D.M. (1994) The DNA of ciliated protozoa. Microbiol. Rev., 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvini M., Durante,M., Citti,L. and Nobili,R. (1984) 5′-Methyl-cytosine in the macronuclear DNA of Blepharisma japonicum. Experientia, 40, 1401–1403. [DOI] [PubMed] [Google Scholar]
- 13.Palacios G., Martin-Gonzalez,A. and Gutierrez,J.C. (1994) Macronuclear DNA demethylation is involved in the encystment process of the ciliate Colpoda inflata. Cell Biol. Int., 18, 223–228. [DOI] [PubMed] [Google Scholar]
- 14.Ammermann D. and Steinbruck,G. (1981) Methylated bases in the DNA of the ciliate Stylonychia mytilus. Eur. J. Cell Biol., 24, 154–156. [PubMed] [Google Scholar]
- 15.Karrer K.M. and VanNuland,T.A. (2002) Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res., 30, 1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cummings D.J., Tait,A. and Goddard,J.M. (1974) Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta, 374, 1–11. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez J.C., Callejas,S., Borniquel,S. and Martin-Gonzalez,A. (2000) DNA methylation in ciliates: implications in differentiation processes. Int. Microbiol., 3, 139–146. [PubMed] [Google Scholar]
- 18.Clark S.J., Harrison,J., Paul,C.L. and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ammermann D., Steinbruck,G., von Berger,L. and Hennig,W. (1974) The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma, 45, 401–429. [DOI] [PubMed] [Google Scholar]
- 20.Elsevier S.M., Lipps,H.J. and Steinbruck,G. (1978) Histone genes in macronuclear DNA of the ciliate Stylonychia mytilus. Chromosoma, 69, 291–306. [DOI] [PubMed] [Google Scholar]
- 21.Gowher H., Ehrlich,K.C. and Jeltsch,A. (2001) DNA from Aspergillus flavus contains 5-methylcytosine. FEMS Microbiol. Lett., 205, 151–155. [DOI] [PubMed] [Google Scholar]
- 22.Eder C., Maercker,C., Meyer,J. and Lipps,H.J. (1993) The processing of macronuclear DNA sequences during macronuclear development of the hypotrichous ciliate Stylonychia lemnae. Int. J. Dev. Biol., 37, 473–477. [PubMed] [Google Scholar]
- 23.Stoll S., Zirlik,T., Maercker,C. and Lipps,H.J. (1993) The organization of internal telomeric repeats in the polytene chromosomes of the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res., 21, 1783–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maercker C., Stoll,S., Rosenkranz,K., Becker,E.M. and Lipps,H.J. (1997) Characterization of a family of repetitive sequences that is eliminated late during macronuclear development of the hypotrichous ciliate Stylonychia lemnae. Dev. Genet., 21, 201–211. [DOI] [PubMed] [Google Scholar]
- 25.Lieb M. and Bhagwat,A.S. (1996) Very short patch repair: reducing the cost of cytosine methylation. Mol. Microbiol., 20, 467–473. [DOI] [PubMed] [Google Scholar]
- 26.Malone C.S., Miner,M.D., Doerr,J.R., Jackson,J.P., Jacobsen,S.E., Wall,R. and Teitell,M. (2001) CmC(A/T)GG DNA methylation in mature B cell lymphoma gene silencing. Proc. Natl Acad. Sci. USA, 98, 10404–10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klobutcher L.A., Turner,L.R. and LaPlante,J. (1993) Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev., 7, 84–94. [DOI] [PubMed] [Google Scholar]
- 28.Meyer G.F. and Lipps,H.J. (1980) Chromatin elimination in the hypotrichous ciliate Stylonychia mytilus. Chromosoma, 77, 285–297. [DOI] [PubMed] [Google Scholar]
- 29.Jahn C.L. (1999) Differentiation of chromatin during DNA elimination in Euplotes crassus. Mol. Biol. Cell., 10, 4217–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraut H., Lipps,H.J. and Prescott,D.M. (1986) The genome of hypotrichous ciliates. Int. Rev. Cytol., 99, 1–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.