Abstract
Photoactive yellow protein (E-PYP) is a blue light photoreceptor, implicated in a negative phototactic response in Ectothiorhodospira halophila, that also serves as a model for the Per–Arnt–Sim superfamily of signaling molecules. Because no biological signaling partner for E-PYP has been identified, it has not been possible to correlate any of its photocycle intermediates with a relevant signaling state. However, the PYP domain (Ppr-PYP) from the sensor histidine kinase Ppr in Rhodospirillum centenum, which regulates the catalytic activity of Ppr by blue light absorption, may allow such issues to be addressed. Here we report the crystal structure of Ppr-PYP at 2 Å resolution. This domain has the same absorption spectrum and similar photocycle kinetics as full length Ppr, but a blue-shifted absorbance and considerably slower photocycle than E-PYP. Although the overall fold of Ppr-PYP resembles that of E-PYP, a novel conformation of the β4–β5 loop results in inaccessibility of Met-100, thought to catalyze chromophore reisomerization, to the chromophore. This conformation also exposes a highly conserved molecular surface that could interact with downstream signaling partners. Other structural differences in the α3–α4 and β4–β5 loops are consistent with these regions playing significant roles in the control of photocycle dynamics and, by comparison to other sensory Per–Arnt–Sim domains, in signal transduction. Because of its direct linkage to a measurable biological output, Ppr-PYP serves as an excellent system for understanding how changes in photocycle dynamics affect signaling by PYPs.
Photoactive yellow proteins (PYPs) are 14-kDa water-soluble blue-light photoreceptors thought to mediate negative phototactic responses in purple bacteria (1). PYPs are members of the Per–Arnt–Sim (PAS) domain superfamily, an α/β structure found through all kingdoms of life, for which PYP from Ectothiorhodospira halophila (E-PYP) serves as the structural prototype (2). PAS domains are found in signaling and sensory proteins and are often involved in mediating protein–protein interactions. In bacteria they can act as sensors of light, redox potential, or oxygen concentration in the sensor histidine kinases of two-component signal transduction systems (3). The PAS domain is extremely versatile: it can accommodate different ligands such as heme in the oxygen sensor FixL (4) or flavin mononucleotide (5) and flavin adenine dinucleotide (6, 7) in the subclass of PAS domains known as light-oxygen-voltage domains. PAS domains can relay signals originating at these chemically diverse ligands to different classes of effector domains, such as histidine or serine/threonine kinases. In PYPs, a conserved cysteine residue is covalently linked to a 4-hydroxycinnamic acid chromophore (8), which, on the absorption of a blue light photon, undergoes a fully reversible photocycle characterized by trans–cis isomerization about its C2–C3 double bond. This photocycle is associated with structural transitions in the protein (9–13) that presumably are associated with differential levels of biological signaling activity to yet-to-be identified downstream regulatory partners.
Because of its favorable spectroscopic properties and putative biological role, E-PYP has become a well-studied system for understanding how light absorption results in biological signaling. In the dark or ground state, the deprotonated 4-hydroxycinnamic acid chromophore is completely shielded from solvent in the chromophore binding pocket and is stabilized in the trans configuration by participation of its phenolate oxygen in a distal hydrogen bonding network consisting of Tyr-42, Glu-46 and Thr-50. After excitation by blue light, the ground state, whose maximal absorption is at 446 nm, converts through a series of red-shifted intermediates I0, I0‡, and I1, in picoseconds, hundreds of picoseconds, and nanoseconds respectively (14), before decaying to a blue-shifted intermediate I2, which completes the photocycle by reverting to the ground state in hundreds of milliseconds (15). Because it is the longest-lived state in the photocycle, I2 is considered to be the “signaling state” of E-PYP and is associated with structural heterogeneity (12), partial protein unfolding (16), and transient exposure of hydrophobic residues (17, 18). The photocycle and its intermediates have been studied by a number of crystallographic (9–11, 19, 20) and spectroscopic methods (12, 14, 15, 21–24). Although these studies have contributed a great deal to understanding the photocycle dynamics of E-PYP, a unified picture of the photocycle and its intermediates has yet to emerge; sometimes contradictory results have been obtained by using these different methodologies, and it has not been possible to correlate any of its spectroscopic states with a biologically relevant signaling state.
The identification of the novel photoreceptor Ppr from Rhodospirillum centenum, which has a PYP domain linked to bacteriophytochrome and histidine kinase domains (25), may enable us to understand how PYPs are able to generate and transmit biological signals to effector domains and how such signaling activity is associated with photocycle dynamics. In Ppr, absorption of blue light by its PYP domain results in inhibition of its histidine kinase activity. A R. centenum Ppr knockout showed no defects in phototaxis but had lower levels of expression of chalcone synthase, an enzyme involved in photoprotection through flavonoid biosynthesis (25). Although the PYP domain of Ppr (Ppr-PYP) is 45% identical in sequence to E-PYP and has similar or identical residues to all those suggested to be critical to absorption spectra and photocycle dynamics in E-PYP (26, 27), Ppr-PYP has a considerably slower photocycle than E-PYP (25). Furthermore, as Ppr-PYP controls the activity of a histidine kinase, its study affords not only the opportunity to understand the signaling mechanism of PYPs, but of the entire class of PAS-domain-regulated sensor histidine kinases, which are ubiquitous in bacteria (28).
Here we report a room temperature crystal structure of Ppr-PYP at 2.0-Å resolution. We identify critical differences in conformation that are responsible for the slow photocycle kinetics of Ppr-PYP and regions of conformational variability that correspond to those important to signal transduction in other sensory PAS domains. We propose a model for signaling by PYP that links structural changes associated with its photocycle to a biological signaling state.
Materials and Methods
Protein Expression and Purification.
From a plasmid encoding full-length Ppr (25), PCR amplification was performed by using N- and C-terminal primers encoding residues 1–129. The PCR product was ligated into the TOPO TrcHis vector (Invitrogen) to generate a construct of Ppr-PYP with an N-terminal hexahistidine tag and enterokinase cut site. This construct was transformed into Escherichia coli TOP10F cells (Invitrogen), which were grown to an OD600 of 0.5 at 37°C, 225 rpm in a shaker, before induction with 1 mM IPTG. After growth for a further 4 h, the cells were spun down and lysed. The chromophore was then reconstituted with the apoprotein as described (29). After centrifugation, the supernatant containing Ppr-PYP was applied to a chelating fast flow Sepharose resin charged with Ni2+ (Amersham Pharmacia), followed by dialysis against 10 mM Tris (pH 8.0) and anion exchange chromatography using high performance Q Sepharose (Amersham Pharmacia). After incubation with bovine enterokinase (Boehringer Mannheim) and a second round of anion exchange chromatography, purity was assessed optically (OD434/OD280) and by SDS/PAGE. Electrospray ionization mass spectrometry revealed that the purified component consisted of residues 5–129.
Crystallization and Data Collection.
Crystals were grown by the hanging drop vapor diffusion method in which 5 μl drops consisting of 1 μl of 12 mg/ml protein solution and 4 μl of a reservoir solution of 36% polyethylene glycol 6000, 10 mM N-(2-acetamido)-2-aminoethanesulfonic acid) (pH 7.0) were equilibrated to that solution. Crystals grew in bunches of rods (200 μm × 50 μm × 30 μm), which were split with a sharp blade and individually mounted in capillaries. Monochromatic oscillation data were collected at room temperature at BioCARS beamline 14BM-C at the Advanced Photon Source, Argonne, IL. (Room temperature was used to allow for time-resolved crystallographic studies of Ppr-PYP.) The crystals initially diffracted to >1.7-Å resolution, but because of radiation damage, the resolution limit was ≈2.0 Å after ≈80° of data collection. Data from three crystals were indexed and then scaled together by using hklsuite (30).
Structure Determination.
Molecular replacement was performed by using epmr (31) with a search model of residues 23–124 of E-PYP (PDB ID code 2phy) in which all nonidentical residues were replaced by alanine and the 4-hydroxycinnamic acid chromophore was removed. A successful molecular replacement solution for three molecules resulted in the reappearance of density consistent with the chromophore. Refinement was carried out by using cns (32). After initial stages in which 3-fold noncrystallographic symmetry constraints were applied, the three molecules were refined independently with rounds of simulated annealing, conjugate gradient, and individual B factor minimizations followed by identification of water molecules. Strong chromophore geometry restraints were based on a structure of ground state E-PYP at 1.0-Å resolution collected at room temperature (S. Anderson, personal communication). Model building was performed in o (33) and xtalview (34). Figures were prepared with ribbons (35) and grasp (36). The sequence numbering for E-PYP will be used when referring to corresponding residues in Ppr-PYP.
Absorption Spectroscopy.
Absorption spectra of Ppr-PYP in 10 mM Tris, pH 8.0 were recorded on a Perkin–Elmer Lambda 6 UV/Vis spectrophotometer. An absorption spectrum of wild-type E-PYP under similar conditions was provided by S. Anderson. Spectroscopy of crystals and solution was performed by using a previously described apparatus (37, 38) with a xenon light monitoring beam and a diode array detector (Ocean Optics, Dunedin, FL). Data could not be recorded below ≈370 nm because of absorption by glass components in this system.
Least-Squares Superpositions and Surface Area Calculations.
Pairwise least-squares superpositions of Cα atomic coordinates were performed by using the CCP4 program lsqkab (39). Because chain B was the most complete model of the three chains of Ppr-PYP (see Results and Discussion), it was selected for comparison with the ground state structure of E-PYP. Deviations between E-PYP and Ppr-PYP were considered statistically significant if they were greater than the sum of the average rms deviation (rmsd) between the three chains of Ppr-PYP plus two standard deviations of that value (0.96 Å), a very conservative cutoff. Solvent accessible surface area calculations were performed by using the CCP4 program areaimol (39), using a solvent sphere radius of 1.6 Å.
Results and Discussion
Crystal Structure of Ppr-PYP.
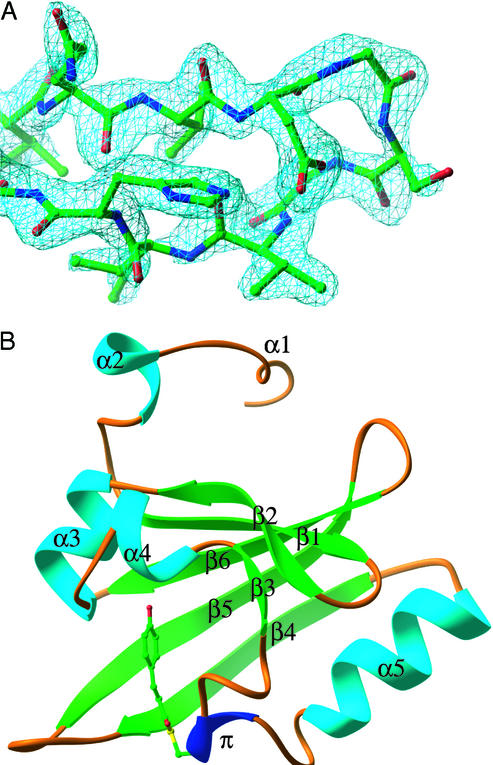
Crystals of Ppr-PYP grew in space group P21 with three molecules in the asymmetric unit, which we refer to as chains A, B, and C. P21 crystals of Ppr-PYP and P63 crystals of E-PYP each have ≈30% solvent content; however, the crystal packing constraints are looser in Ppr-PYP, because only 43% of its solvent-accessible surface area is buried in packing interfaces compared with 55% in E-PYP. The N termini of all three chains and the β4–β5 loop of the C chain, which are flexible during E-PYP's photocycle (12), are largely disordered. The moderately high values of R factor, B values, and rmsd bond lengths (Table 1) can be attributed to data collection at room temperature, the merging of data from multiple crystals, and the inability to model portions of the molecule. The quality of the final model is illustrated in a simulated annealing composite omit map of the β1–β2 turn of chain B (Fig. 1A).
Table 1.
Data and refinement statistics
| Resolution range, Å | 40–2.0 (2.04–2.00) |
| 〈I/σ(I)〉 | 23.2 (3.2) |
| Rmerge | 0.080 (0.379) |
| Unique reflections | 19,047 |
| Completeness, % | 93.2 (78.9) |
| Redundancy | 4.2 |
| Space group | P21 |
| a, b, c, Å | 34.56, 143.07, 36.45 |
| α, β, γ, ° | 90, 117.96, 90 |
| No. of monomers in the asymmetric unit | 3 |
| Rwork, % | 24.5 |
| Rfree, % | 26.9 |
| B, Å2 | 43 |
| rmsd bond lengths, Å | 0.015 |
| rmsd angles, ° | 1.54 |
| Water molecules | 114 |
| Ramachandran plot, % | |
| Most favorable | 89.2 |
| Allowed | 10.8 |
| Not allowed | 0 |
Last resolution shell statistics are shown in parentheses. The test set was composed of 10% of all reflections, and Rfree was calculated with the formula for Rcryst using only test set reflections. Rmerge = ∑hkl(|Ihkl − 〈I〉|/∑hklIhkl). Rcryst = ∑hkl (∥Fobs| − |Fcalc∥/∑hkl|Fobs|).
Figure 1.
Crystal structure of Ppr-PYP. (A) A simulated annealing composite omit map at 2.0σ level of the β1–β2 turn of chain B. (B) Chain B of Ppr-PYP with secondary structure labeled according to ref. 20.
The structure of chain B of Ppr-PYP is shown in Fig. 1B. Ppr-PYP maintains the conserved structural elements of PYPs (25, 40) with high conservation in those residues forming its two hydrophobic cores and the chromophore binding pocket (20, 25, 40) and low homology in the two-helix N-terminal cap, which has been identified as a region of enhanced flexibility in an E-PYP photostationary state (21). All PYPs contain residues Cys-69, required for chromophore attachment, and Arg-52, which acts as a gate to the chromophore binding pocket. In the distal hydrogen-bonding network, Tyr-42 and Glu-46 are conserved in all PYPs, but not Thr-50, which is a Ser in Ppr-PYP. Residue Met-100, thought to catalyze chromophore reisomerization in E-PYP (27), is not highly conserved in PYPs, but is found in Ppr-PYP.
Structural Variability in Ppr-PYP and E-PYP.
Least-squares pairwise superpositions of the three chains of Ppr-PYP with one another and of chain B with E-PYP allows the identification of those regions with the greatest conformational variability in Ppr-PYP and between Ppr-PYP and E-PYP, respectively. A plot of the Cα rmsd by residue and a listing of the mean Cα rmsds between these chains after superposition are shown in Fig. 2A. The three chains of Ppr-PYP are very similar to one another: none of the deviations, excluding those in the N termini and the β4–β5 loop in chain C, are statistically significant (see Materials and Methods). A superposition of Ppr-PYP with E-PYP shows much more pronounced differences (Fig. 2B). In addition to the highly variable N-terminal cap, three regions of Ppr-PYP have statistically significant differences in their coordinates from E-PYP: the α3-α4 region containing the conserved Arg-52, the α5–β4 loop, and the β4–β5 loop containing Met-100. All of these regions are flexible in E-PYP (10, 22, 41); the first and third are adjacent to the chromophore.
Figure 2.
Differences between PYP molecules. (A) Cα rmsds after pairwise superpositions of the A and B chains (blue; mean rmsd, 0.28 Å), C and B chains (red; mean rmsd, 0.34 Å), A and C chains (green; mean rmsd, 0.42 Å), and E-PYP and B chain (cyan; mean rmsd, 0.80 Å). Those deviations >0.96 Å (thin black line) are considered to be statistically significant. (B) Superposition of B chain of Ppr-PYP (red) and E-PYP (blue). The N and C termini, the Cys-69 required for chromophore attachment, and the three loops that have statistically significant differences between E-PYP and Ppr-PYP are labeled.
Structural changes in the β4–β5 and α3–α4 loops would be expected to affect photocycle dynamics because of the presence of critical residues in those regions (26, 27). As in E-PYP, the β4–β5 loop is flexible in Ppr-PYP, evidenced by high B factors in this region in chains A and B and its disorder in chain C. The disorder of the β4–β5 loop of chain C is caused by a weak packing interaction between it and the N-terminal helices of chain A, which are similarly disordered. This interaction is maintained between the β4–β5 loops of chains A and B and the N termini of symmetry-related chains B and C, respectively. In chains A and B, the β4–β5 loop of Ppr-PYP adopts a novel conformation (Fig. 3A), unlike any reported structure of E-PYP (20, 22, 41–43). Large differences between the dihedral angles of residues 97–100 of Ppr-PYP and E-PYP make the β4–β5 loop of Ppr-PYP a type III tight turn. In E-PYP, Tyr-98 takes a position that fully limits solvent accessibility to the chromophore, but in Ppr-PYP, Phe-98 has flipped up and taken a position closer to the aromatic ring of the chromophore, a position occupied by Met-100 in E-PYP (Fig. 3B). This limits the access of Met-100 to the chromophore and also results in increased solvent accessibility of the carbonyl oxygen of the chromophore, which in the crystal forms a packing interface in chains A and B. In the α3–α4 loop, Gly-47 and Gly-51 have been suggested to play roles in the control of photocycle dynamics in E-PYP (43). Gly-47 is replaced by a Ser in Ppr-PYP, and Ppr-PYP's loop conformation is more similar to that of a Gly-47-Ser mutant of E-PYP than to wild-type E-PYP (43). The largest structural shift in the α3–α4 loop is at the highly conserved Pro-54, though unlike the β4–β5 loop, there are no significant differences in dihedral angles between Ppr-PYP and E-PYP (data not shown).
Figure 3.
Conformation of the β4–β5 loop. Views from below (A) and the side (B) of the β4–β5 loop of Ppr-PYP (red) and E-PYP (blue), after superposition as in Fig. 2B.
The β4–β5 loop conformation of Ppr-PYP would affect the photocycle dynamics considerably, as residue Met-100 has been proposed to play a role in catalyzing chromophore reisomerization. Mutations of this residue slow the rate of decay from I2 to the ground state by two to three orders of magnitude (27, 44). Barring the access of Met-100 to the chromophore and changing the stability of the chromophore binding pocket by radically altering the conformation of the β4–β5 loop would be expected to slow the photocycle considerably. Although associated with smaller kinetic effects than the β4–β5 loop, the α3–α4 loop conformation would also contribute to the slower recovery kinetics of Ppr-PYP because the conformation observed here results in a four-fold slowing of the rate of I2 to ground state decay in a Gly47Ser E-PYP mutant. A combination of both of these effects likely contributes to the slower photocycle kinetics of Ppr-PYP.
The Chromophore Binding Pocket.
The ground state absorption spectrum of Ppr-PYP is similar to that of full-length Ppr with a maximal absorption at 434 nm (25), a 12 nm blue shift relative to, but maintaining the same qualitative form as, the wild-type E-PYP spectrum (Fig. 4A). It does not have an extra absorption peak at 350 nm, such as those observed in E-PYP Met-100 mutants, arising from a population of protonated trans chromophore because of destabilization of the chromophore binding pocket (27, 44). Ppr-PYP also maintains the slow photocycle kinetics of full-length Ppr relative to E-PYP in solution (data not shown). The absorption spectrum of Ppr-PYP in a crystalline state is very similar to that of solution, with a maximum at 431 nm (Fig. 4B), consistent with the chromophore-binding pocket adopting a similar conformation in solution and in the crystal. Fig. 5 shows the chromophore binding pockets of Ppr-PYP and E-PYP after a least squares superposition of their chromophores. There are no statistically significant differences between their distal hydrogen bonding networks at this resolution. However, two hydrogen bonds that could affect photocycle dynamics are present in E-PYP but absent in Ppr-PYP: one between the hydroxyl of Tyr-94 and the backbone carbonyl oxygen of Cys-69 and another between the side chain of Arg-52 and the backbone carbonyl of Phe-98. As noted above, the position taken by Met-100 in E-PYP is occupied by Phe-98 in Ppr-PYP, a polarizable aromatic side chain that would change the local dielectric environment. Also, hydrogen bonding by water to the proximal carbonyl oxygen, which is solvent-accessible in Ppr-PYP (see above), could also contribute to a blue shift in absorption because of resonance stabilization between quinonic and phenolic forms of the chromophore (19). However, it is difficult to correlate the structural changes observed in this structure with the specific and nonspecific protein environmental effects that affect chromophore absorption in PYPs (45, 46).
Figure 4.
Absorption spectra of Ppr-PYP and E-PYP. (A) Absorption spectra of Ppr-PYP (dash) and E-PYP (line) in solution. (B) Absorption spectra of Ppr-PYP in solution (dash) and in P21 crystals (line).
Figure 5.
Chromophore binding pockets. Chain B of Ppr-PYP (red) and E-PYP (blue) after a least-squares superposition of their hydroxycinnamic acid chromophores.
Conformational Substates and Signaling in PYPs.
E-PYP should also be capable of adopting the conformation of the β4–β5 loop found in Ppr-PYP because the sequence similarity between the two is high in this region and replacement of Phe-98 by Tyr in Ppr-PYP does not result in any steric clashes. This conformation exposes ≈80 Å2 of additional solvent accessible surface area relative to E-PYP (Fig. 6), an area sufficient to bind Nile Red, a hydrophobic dye, to a region adjacent to the chromophore in the I2 state of E-PYP (18). Furthermore, a hydrogen bond, which is present in ground state E-PYP between the side chain of Arg-52 and the backbone carbonyl oxygen of residue 98 (Fig. 5), is not present in Ppr-PYP and in the I2 state (10), which would destabilize the ground state E-PYP conformation and favor a loop conformation such as that observed in Ppr-PYP. Both lines of evidence are consistent with the conformational substate favored in the ground state of Ppr-PYP being more “I2-like” than the ground state of E-PYP. This finding is also consistent with the slower photocycle kinetics of Ppr-PYP compared with E-PYP, as energetic stabilization of I2 relative to the ground state would result in slower thermal reisomerization of the chromophore. We believe that the structure observed here is a partially activated conformational substate that Ppr-PYP is capable of adopting some of the time in solution.
Figure 6.
Molecular surfaces of E-PYP (A) and Ppr-PYP (B). (Left) Residues color-coded by electrostatic potential with red denoting negative and blue denoting positive potential (36). Secondary structure elements are labeled according to Fig. 1. (Right) Residues identical in all PYPs (40) are colored green.
The surface exposed in this substate is not unusual in terms of electrostatic potential (Fig. 6 Left) or hydrophobicity (data not shown), but is highly conserved in all PYPs (Fig. 6 Right). This surface is dissected by the β4–β5 loop in E-PYP (Fig. 6A Right) but is fully contiguous in Ppr-PYP (Fig. 6B Right), allowing the PYP to interact with signaling partners through a more extended, conserved surface. Possible signaling partners include histidine kinases such as that found in Ppr, and other effector domains that are commonly controlled by PAS domains (3). A similar role has been proposed for this surface in E-PYP (2) and in the light-oxygen-voltage domains (47), a subset of photosensory PAS domains that bind flavins and control a wide variety of effector domains.
Comparison of Ppr-PYP with other sensory PAS domains identifies additional structural features in common in this superfamily. The three regions of Ppr-PYP that show statistically significant displacements in Cα coordinates from E-PYP correspond to null mutants of the Aer redox sensor, a PAS domain that binds flavin adenine dinucleotide (FAD). Mutations that retain the ability to bind FAD but are unable to signal map to the α3–α4, α5–β4, and β4–β5 loops of Ppr-PYP (48), suggesting that the structural variability observed in these loops is related to biological activity. Null mutations of Aer that lie in the α3–α4 and β4–β5 regions are consistent with the formation of a biologically-relevant molecular surface by those loops. The α5–β4 loop of E-PYP corresponds to the FG loop of the oxygen-sensing domain of the sensor histidine kinase FixL, where mutations result in a protein that can still bind heme but is incapable of sensing oxygen (49). This loop has also been proposed to mediate an interaction between the PAS domain and serine/threonine kinase of the human protein PAS kinase (50). The similarities to other PAS domains suggest that there is some conservation of mechanism in this diverse family of sensory proteins. Such conservation likely involves the importance of these loops in controlling the dynamics of the protein and its activation to a competent signaling state.
These findings are consistent with a model for signaling in PYPs in which, on absorption of blue light, Arg-52 is pushed out of the chromophore-binding pocket (10, 11), and the hydrogen bond with Tyr-98 is broken. This allows the β4–β5 loop to adopt an altered conformation in I2 and expose a new molecular surface that exhibits altered interaction with a signaling partner. This could lead to higher or lower affinity in Ppr-PYP for the Ppr histidine kinase, resulting in a modulation of its activity. Although experiments to test the biological significance of mutations in these critical regions are not possible in E-PYP, they can be performed by using the full-length Ppr sensor histidine kinase. Ppr can therefore serve as an excellent system for bridging research on the photocycle dynamics of PYPs to that on the mechanism of PAS-mediated signal transduction.
Acknowledgments
We thank Zeyu Jiang and Carl Bauer for supplying a plasmid encoding full-length Ppr to Marius Schmidt of this laboratory, Vukica Srajer for help with the crystal microspectrophotometer; Spencer Anderson for a structure of E-PYP; and Xiaojing Yang, Carl Correll, Sean Crosson, and Jason Key for helpful advice. This work was supported by National Institutes of Health Grant GM36452 (to K.M.) and a Medical Scientist Training Program fellowship (to S.R.). The BioCARS facility at the Advanced Photon Source is supported by National Institutes of Health Grant RR07707 (to K.M.).
Abbreviations
- PYP
photoactive yellow protein
- E-PYP
PYP from Ectothiorhodospira halophila
- Ppr-PYP
PYP from the Ppr sensor histidine kinase in Rhodospirillum centenum
- PAS
Per–Arnt–Sim domain superfamily
- rmsd
rms deviation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1MZU).
References
- 1.Sprenger W W, Hoff W D, Armitage J P, Hellingwerf K J. J Bacteriol. 1993;175:3096–3104. doi: 10.1128/jb.175.10.3096-3104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellequer J L, Wager-Smith K A, Kay S A, Getzoff E D. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor B L, Zhulin I B. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilles-Gonzalez M A, Gonzalez G, Perutz M F, Kiger L, Marden M C, Poyart C. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 5.Christie J M, Reymond P, Powell G K, Bernasconi P, Raibekas A A, Liscum E, Briggs W R. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 6.Froehlich A C, Liu Y, Loros J J, Dunlap J C. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 7.He Q, Cheng P, Yang Y, Wang L, Gardner K H, Liu Y. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- 8.Baca M, Borgstahl G E, Boissinot M, Burke P M, Williams D R, Slater K A, Getzoff E D. Biochemistry. 1994;33:14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 9.Perman B, Srajer V, Ren Z, Teng T, Pradervand C, Ursby T, Bourgeois D, Schotte F, Wulff M, Kort R, Hellingwerf K, Moffat K. Science. 1998;279:1946–1950. doi: 10.1126/science.279.5358.1946. [DOI] [PubMed] [Google Scholar]
- 10.Genick U K, Borgstahl G E, Ng K, Ren Z, Pradervand C, Burke P M, Srajer V, Teng T Y, Schildkamp W, McRee D E, Moffat K, Getzoff E D. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 11.Ren Z, Perman B, Srajer V, Teng T Y, Pradervand C, Bourgeois D, Schotte F, Ursby T, Kort R, Wulff M, Moffat K. Biochemistry. 2001;40:13788–13801. doi: 10.1021/bi0107142. [DOI] [PubMed] [Google Scholar]
- 12.Rubinstenn G, Vuister G W, Mulder F A, Dux P E, Boelens R, Hellingwerf K J, Kaptein R. Nat Struct Biol. 1998;5:568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 13.Lee B C, Pandit A, Croonquist P A, Hoff W D. Proc Natl Acad Sci USA. 2001;98:9062–9067. doi: 10.1073/pnas.111153598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ujj L, Devanathan S, Meyer T E, Cusanovich M A, Tollin G, Atkinson G H. Biophys J. 1998;75:406–412. doi: 10.1016/S0006-3495(98)77525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoff W D, van Stokkum I H, van Ramesdonk H J, van Brederode M E, Brouwer A M, Fitch J C, Meyer T E, van Grondelle R, Hellingwerf K J. Biophys J. 1994;67:1691–1705. doi: 10.1016/S0006-3495(94)80643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Brederode M E, Hoff W D, Van Stokkum I H, Groot M L, Hellingwerf K J. Biophys J. 1996;71:365–380. doi: 10.1016/S0006-3495(96)79234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salamon Z, Meyer T E, Tollin G. Biophys J. 1995;68:648–654. doi: 10.1016/S0006-3495(95)80225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendriks J, Gensch T, Hviid L, van Der Horst M A, Hellingwerf K J, van Thor J J. Biophys J. 2002;82:1632–1643. doi: 10.1016/S0006-3495(02)75514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genick U K, Soltis S M, Kuhn P, Canestrelli I L, Getzoff E D. Nature. 1998;392:206–209. doi: 10.1038/32462. [DOI] [PubMed] [Google Scholar]
- 20.Borgstahl G E, Williams D R, Getzoff E D. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 21.Craven C J, Derix N M, Hendriks J, Boelens R, Hellingwerf K J, Kaptein R. Biochemistry. 2000;39:14392–14399. doi: 10.1021/bi001628p. [DOI] [PubMed] [Google Scholar]
- 22.Dux P, Rubinstenn G, Vuister G W, Boelens R, Mulder F A, Hard K, Hoff W D, Kroon A R, Crielaard W, Hellingwerf K J, Kaptein R. Biochemistry. 1998;37:12689–12699. doi: 10.1021/bi9806652. [DOI] [PubMed] [Google Scholar]
- 23.Brudler R, Rammelsberg R, Woo T T, Getzoff E D, Gerwert K. Nat Struct Biol. 2001;8:265–270. doi: 10.1038/85021. [DOI] [PubMed] [Google Scholar]
- 24.Takeshita K, Imamoto Y, Kataoka M, Tokunaga F, Terazima M. Biochemistry. 2002;41:3037–3048. doi: 10.1021/bi0110600. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Z, Swem L R, Rushing B G, Devanathan S, Tollin G, Bauer C E. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- 26.Genick U K, Devanathan S, Meyer T E, Canestrelli I L, Williams E, Cusanovich M A, Tollin G, Getzoff E D. Biochemistry. 1997;36:8–14. doi: 10.1021/bi9622884. [DOI] [PubMed] [Google Scholar]
- 27.Devanathan S, Genick U K, Canestrelli I L, Meyer T E, Cusanovich M A, Getzoff E D, Tollin G. Biochemistry. 1998;37:11563–11568. doi: 10.1021/bi9803776. [DOI] [PubMed] [Google Scholar]
- 28.Galperin M Y, Nikolskaya A N, Koonin E V. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 29.Imamoto Y, Ito T, Kataoka M, Tokunaga F. FEBS Lett. 1995;374:157–160. doi: 10.1016/0014-5793(95)01096-w. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Kissinger C R, Gehlhaar D K, Fogel D B. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 32.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Jones T A, Bergdoll M, Kjeldgaard M. In: Crystallographic and Modeling Methods in Molecular Design. Bugg C, Ealick S, editors. New York: Springer; 1990. pp. 189–195. [Google Scholar]
- 34.McRee D E. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 35.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 36.Nicholls A, Bharadwaj R, Honig B. Biophys J. 1993;64:A166. [Google Scholar]
- 37.Chen Y, Srajer V, Ng K, Legrand A, Moffat K. Rev Sci Instrum. 1994;65:1506–1511. [Google Scholar]
- 38.Ng K, Getzoff E D, Moffat K. Biochemistry. 1995;34:879–890. doi: 10.1021/bi00003a022. [DOI] [PubMed] [Google Scholar]
- 39.Bailey S. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 40.Kort R, Hoff W D, Van West M, Kroon A R, Hoffer S M, Vlieg K H, Crielaand W, Van Beeumen J J, Hellingwerf K J. EMBO J. 1996;15:3209–3218. [PMC free article] [PubMed] [Google Scholar]
- 41.van Aalten D M, Crielaard W, Hellingwerf K J, Joshua-Tor L. Protein Sci. 2000;9:64–72. doi: 10.1110/ps.9.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brudler R, Meyer T E, Genick U K, Devanathan S, Woo T T, Millar D P, Gerwert K, Cusanovich M A, Tollin G, Getzoff E D. Biochemistry. 2000;39:13478–13486. doi: 10.1021/bi0009946. [DOI] [PubMed] [Google Scholar]
- 43.van Aalten D M, Haker A, Hendriks J, Hellingwerf K J, Joshua-Tor L, Crielaard W. J Biol Chem. 2002;277:6463–6468. doi: 10.1074/jbc.M109313200. [DOI] [PubMed] [Google Scholar]
- 44.Kumauchi M, Hamada N, Sasaki J, Tokunaga F. J Biochem (Tokyo) 2002;132:205–210. doi: 10.1093/oxfordjournals.jbchem.a003211. [DOI] [PubMed] [Google Scholar]
- 45.Kroon A R, Hoff W D, Fennema H P, Gijzen J, Koomen G J, Verhoeven J W, Crielaard W, Hellingwerf K J. J Biol Chem. 1996;271:31949–31956. doi: 10.1074/jbc.271.50.31949. [DOI] [PubMed] [Google Scholar]
- 46.Yoda M, Houjou H, Inoue Y, Sakurai M. J Phys Chem B. 2001;105:9887–9895. [Google Scholar]
- 47.Crosson S, Rajagopal S, Moffat K. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 48.Repik A, Rebbapragada A, Johnson M S, Haznedar J O, Zhulin I B, Taylor B L. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukai M, Nakamura K, Nakamura H, Iizuka T, Shiro Y. Biochemistry. 2000;39:13810–13816. doi: 10.1021/bi001184x. [DOI] [PubMed] [Google Scholar]
- 50.Amezcua C A, Harper S M, Rutter J, Gardner K H. Structure (London) 2002;10:1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]