Abstract
The myocyte enhancer factor-2 (MEF2) transcription factor regulates muscle development and calcium-dependent gene expression. MEF2 activity is repressed by class II histone deacetylases (HDACs), which dissociate from MEF2 when phosphorylated on two serine residues in response to calcium signaling. To explore the potential importance of MEF2/HDAC interactions in the heart, we generated transgenic mice expressing a signal-resistant form of HDAC5 under cardiac-specific and doxycycline-inducible regulation. Transgene expression resulted in sudden death in male mice accompanied by loss and morphologic changes of cardiac mitochondria and down-regulation of mitochondrial enzymes. The transcriptional coactivator PGC-1α, a master regulator of mitochondrial biogenesis and fatty acid oxidation, was also down-regulated in response to HDAC5 expression. Examination of the PGC-1α promoter revealed two MEF2-binding sites that mediate transcriptional activation by MEF2 and repression by HDAC5. These findings identify PGC-1α as a key target of the MEF2/HDAC regulatory pathway and demonstrate this pathway's importance in maintenance of cardiac mitochondrial function.
Acetylation of nucleosomal histones represents a central mechanism for regulation of gene expression. Histone acetylation relaxes chromatin, allowing access of the transcriptional machinery to specific regions of DNA. Control of this process is tightly regulated and can affect individual genes in a specific and directed fashion (1). Acetylation of histones by histone acetyltransferases is associated with activation of gene expression. Conversely, gene repression is achieved by histone deacetylases (HDACs) (2).
HDACs in mammalian cells comprise three distinct classes on the basis of protein structure and homology to yeast HDACs. Class I and III HDACs are ubiquitously expressed, whereas class II HDACs are highly enriched in striated muscles and brain (2). Class II HDACs (HDAC4, 5, 7, and 9) repress transcriptional activity of myocyte enhancer factor-2 (MEF2) transcription factors, which are also highly expressed in muscle and brain (3). This repression is relieved by phosphorylation of HDACs on two conserved serine residues, creating docking sites for the 14–3-3 chaperone protein, which mediates translocation of class II HDACs out of the nucleus (4). Mutation of these serine residues to alanines in HDAC5 prevents 14–3-3 binding and export to the cytosol (4–6). This signal-resistant HDAC mutant, which we refer to here as HDAC5S/A, acts as a dominant negative regulator of MEF2 signaling, because it binds and represses MEF2 factors but cannot be exported from the nucleus. Because histone acetyltransferases (HATs) and HDACs compete for the same binding site on MEF2, class II HDACs may also inhibit binding of HATs to MEF2 and subsequent histone acetylation (7). Expression of HDAC5S/A in neonatal rat cardiomyocytes prevents agonist-induced hypertrophy and activation of the fetal cardiac gene program (8). The notion that class II HDACs may counteract hypertrophic signals is supported by the recent demonstration that HDAC9 mutant mice show an exaggerated response to hypertrophic stimuli (8).
To further investigate the importance of HDAC5 in cardiac function, we generated transgenic mice harboring a doxycycline (DOX)-inducible cardiac-specific transgene encoding the HDAC5S/A signal-resistant mutant. Because this mutant is able to efficiently inhibit MEF2 transactivation (4), we hypothesized that it would allow us to more clearly identify MEF2/HDAC targets free from complications of unknown kinases and phosphatases that may alter the activity of endogenous HDAC5. Expression of HDAC5S/A in the adult heart resulted in sudden death of male mice accompanied by gross aberrations in mitochondrial architecture and down-regulation of mitochondrial enzymes and the transcriptional coactivator peroxisome proliferator-activated receptor (PPAR) γ coactivator 1α (PGC-1α). PGC-1α coactivates numerous transcription factors involved in metabolism, including MEF2, and cardiac-specific overexpression of PGC-1α is sufficient to induce mitochondrial biogenesis (9–11). We show that two MEF2-binding sites in the PGC-1α upstream region mediate transcriptional responsiveness to MEF2 and repression by HDAC5, identifying the MEF2/HDAC pathway as a key regulator of the PGC-1α gene and mitochondrial biogenesis in the heart.
Methods
Generation of Transgenic Animals.
We cloned the cDNA for human HDAC5 containing serine-to-alanine mutations at amino acids 259 and 498 into the tetracycline (tet)-responsive pUHG-10 vector to generate the tetHDAC5S/A vector (4). This vector was injected into B6C3F1 mouse oocytes and implanted into surrogate female ICR mice. Transgenic offspring were bred to α-myosin heavy chain (MHC)-tet transactivator transgenic mice while receiving 200 μg/ml DOX in water (12). Animals were maintained on DOX as needed. All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center.
Histology and Microscopy.
For light microscopy, hearts were fixed overnight in 10% formalin, then embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin/eosin. Alternatively, sections were immunostained with anti-FLAG primary antibody (20 μg/ml; Sigma–Aldrich) and goat anti-mouse fluorescein isothiocyanate conjugated secondary antibody (1:1,000; Vector Laboratories, Burlingame, CA). For transmission electron microscopy, hearts were fixed overnight in 3% glutaraldehyde in PBS at 4°C, then postfixed in 1% OsO4 and dehydrated in an ethanol series. Samples were then embedded in Spurr resin (Ted Pella, Redding, CA), stained with uranyl acetate and lead citrate, and sectioned at 80 nm.
Western Blot Analysis.
Protein was isolated from hearts by homogenization in lysis buffer (PBS, 0.1% Triton X-100/1 mM EDTA, pH 7.4) containing Complete protease inhibitors (Roche Diagnostics). After electrophoresis and blotting onto Sequi-blot poly(vinylidene difluoride) membranes (Bio-Rad), blots were probed with anti-FLAG antibodies and horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biosciences). Signal was detected with Western blotting Luminol Reagent (Santa Cruz Biotechnology), followed by exposure of blots to BioMax film (Kodak).
Cell Culture and Luciferase Assays.
COS cells were maintained in 10% FBS in DMEM. Fugene 6 (Roche Diagnostics) was used for all transfections. The first 3.1 kb of genomic sequence upstream of the ATG translational start codon for mouse PGC-1α was amplified from BAC RPCI23–260A10 (CHORI-BacPac, Oakland, CA; www.chori.org) by high-fidelity PCR and TA cloned into the pCRII-TOPO vector (upstream primer: 5′-GGGGTACCCCATTTGGGAATCCTCTATACAAAGTTG-3′; downstream primer: 5′-GAAGATCTTCCCAGCTCCCGAATGACG-3′), then subcloned into the pGL3 basic luciferase reporter vector (Promega). A QuikChange kit (Stratagene) was used for site-directed mutagenesis of this reporter with primers to introduce several A/T to C/G mutations in the two putative MEF2-binding sites (MEF2 site 1-distal: 5′-AAGACAGAGAGAAAATTAACCATGGAAACTGCCTGGGGAG-3′; MEF2 site 2-proximal: 5′-ATGGTGCTTTATAAATTAGGTCTAGATGCATAGGGACTTT-3′; mutations are underlined). Luciferase assays were performed on COS cell lysates by using a kit (Promega). Cardiomyocytes were isolated from neonatal rat pups as described (8). After isolation, cells were grown overnight and then infected in serum-free medium with adenoviruses encoding either cytomegalovirus (CMV)-GFP or CMV-HDAC5S/A at a multiplicity of infection of 50/cell or were left uninfected (8). After infection, cells were incubated in serum-free medium for 24 h then harvested for RNA isolation.
Electrophoretic Gel Mobility-Shift Assay.
Double-stranded oligomers were labeled with [32P]dCTP by using Klenow DNA polymerase. Labeled oligomers were incubated with extracts from COS cells transfected with either GFP or MEF2C expression plasmids, and gel mobility-shift assays were performed as described (13). DNA–protein complexes were resolved on 6% acrylamide gels and exposed to BioMax film.
RNA Analysis.
Total cardiac RNA was isolated from mice by using Trizol (Invitrogen). A 20-μg aliquot representative of three animals per sample group was then analyzed on Affymetrix U74Av2 microarrays. A subset of differentially expressed RNAs was further characterized by semiquantitative RT-PCR. Briefly, 2 μg of RNA from each sample was used to generate cDNA by using a SuperScript First-Strand Synthesis kit (Invitrogen). PCR was carried out by using primers specific to each gene of interest, with product labeling by [32P]dCTP. Cycling parameters were determined for each product to ensure linearity of response, and reactions lacking reverse transcriptase were carried out as negative controls. PCR products were resolved on 6% acrylamide native gels and exposed to BioMax film. A similar protocol was followed for semiquantitative RT-PCR of rat neonatal cardiomyocytes, and for DNA isolated by chromatin immunoprecipitation (ChIP).
ChIP.
After adenoviral infection, neonatal rat cardiomyocytes were prepared for ChIP assays by using a commercial kit and antiacetyl-histone H3 antibodies (Upstate Biotechnology, Lake Placid, NY). After ChIP, DNA was purified by phenol/chloroform extraction and PCR carried out as described above.
Results
Generation of Transgenic HDAC5S/A Mice.
Our initial goal was to express the signal-resistant HDAC5 protein, HDAC5S/A, in the heart and to determine the consequences on cardiac gene expression. Attempts to generate mice transgenic for HDAC5S/A under control of the α-MHC promoter were unsuccessful, suggesting that constitutive high-level expression of this HDAC5 mutant caused lethality (T. A. McKinsey and E.N.O., unpublished results). We therefore incorporated a tet-off inducible system in which transgene expression is inhibited in the presence of the tet analog, DOX, but is induced upon withdrawal of the drug (12). FLAG-tagged HDAC5S/A was cloned into a tet-responsive expression vector to generate a tetHDAC5 mouse line. This line was crossed with transgenic mice harboring a tet transactivator (tTA) gene under control of the α-MHC promoter (α-MHC-tTA mice) to provide cardiac-specific expression (12).
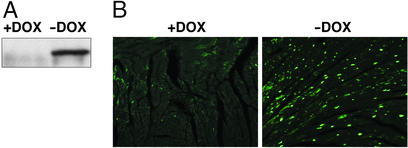
We confirmed expression of HDAC5S/A by Western blot analysis of total protein from cardiac extracts in the presence or absence of DOX. No transgene expression was detected in the presence of DOX (Fig. 1A) or in noncardiac tissues such as liver (data not shown). Maximal transgene expression was attained within 5 days after DOX withdrawal. Immunohistochemistry of cardiac sections using anti-FLAG antibody revealed intense staining of cardiac nuclei after withdrawal of DOX (Fig. 1B). Microarray analysis of total RNA from mice after DOX withdrawal revealed a 3.5-fold increase in HDAC5 expression, but no changes in expression levels of HDAC1, 2, 3, or 6 (see below).
Figure 1.
Characterization of HDAC5S/A expression in DOX-inducible HDAC5S/A mice. (A) Western blotting of equal amounts of total cardiac protein of α-MHC-tTA/tetHDAC5S/A double transgenic mice before or after DOX withdrawal. (B) Immunohistochemical staining for FLAG-HDAC5S/A of cardiac sections from double transgenic mice before or after DOX withdrawal.
Cardiac Sudden Death and Mitochondrial Defects in HDAC5S/A Mice.
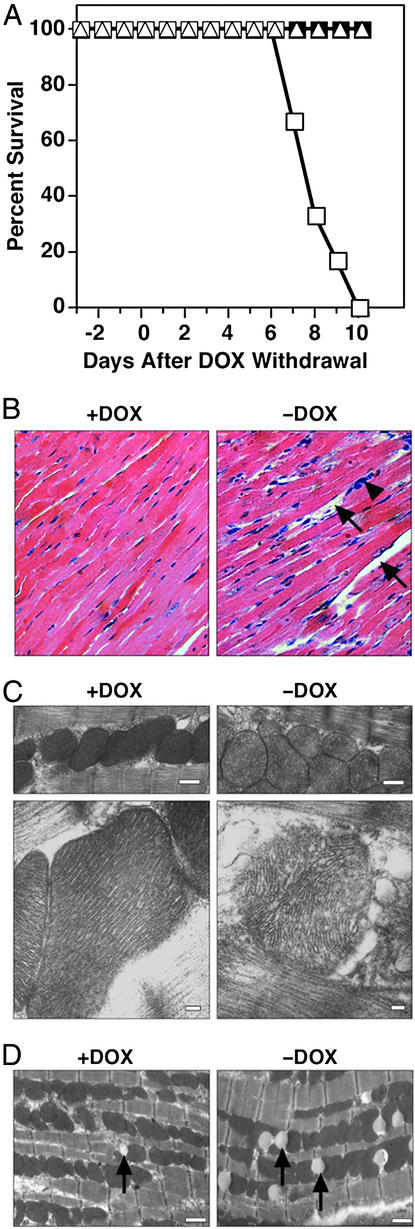
Mice harboring both the α-MHC-tTA and tetHDAC5S/A transgenes were normal when maintained on DOX. However, withdrawal of DOX resulted in death of 100% of male mice within 7–10 days (Fig. 2A). In contrast, female transgenic mice survived ≈30 days after DOX withdrawal, indicating gender specificity in response to HDAC5S/A. This sexual dichotomy is particularly interesting in light of the phenotype of PPARα-deficient mice. Inhibition of mitochondrial fatty acid import by treatment of PPARα−/− mice with etomoxir resulted in the death of 100% of male mice, compared with 25% of females, but estrogen treatment of males normalized mortality to that of the females (14). The mechanism underlying increased longevity in the females in our study is currently under investigation. Two days before death, male mice become extremely lethargic, ceasing to groom themselves or respond to handling. Heart rate declined to ≈230 beats per minute in unanesthetized animals, compared with >600 in normal littermates as measured by ECG recording. There was no change in heart-to-body weight ratio at the time of death of male transgenic mice removed from DOX.
Figure 2.
Cardiac and mitochondrial abnormalities in HDAC5S/A mice. (A) Survival curve for male and female HDAC5S/A mice before and after withdrawal of DOX on day 0. Transgene activation results in death of 100% of male mice expressing HDAC5S/A within 10 days (n = 6 males or females per group). Squares, males; triangles, females; filled symbols, +DOX; open symbols, −DOX. (B) Hematoxylin/eosin-stained cardiac sections from double-transgenic mice before or after DOX withdrawal. Arrows denote areas of cardiomyocyte death or dropout. The arrowhead indicates inflammatory cell infiltration. Magnification is ×40. (C) Transmission electron micrographs of mitochondria in double transgenic mice before and after DOX withdrawal. (Bar = 500 nm in Upper and 100 nm in Lower.) (D) Transmission electron micrographs of cardiac sections from control mice (Left) or from transgenic mice after 5 days of DOX withdrawal. (Bar = 1 μm.) Arrows denote lipid bodies.
Histological examination of cardiac tissue sections revealed signs of cellular necrosis and inflammation in the hearts of male mice after removal of DOX (Fig. 2B). Hearts of female mice at the time of death showed high levels of collagen deposition and exhibited dilated cardiomyopathy (data not shown), suggesting a different process of pathogenesis from that in the males.
Further examination of cardiac tissue sections from male mice by transmission electron microscopy revealed dramatic changes in mitochondrial morphology (Fig. 2C). The mitochondrial cristae were highly disturbed, and large areas of blebbing could be seen in mice 8 days after DOX withdrawal. These changes led to a loss of electron-dense staining of the mitochondria that was easily visible at lower magnifications (Fig. 2C). Mitochondria from these mice appeared swollen and were >60% larger than controls (average mitochondrial area: 0.76 ± 0.42 μm2 versus 0.47 ± 0.24 μm2, respectively, P < 0.0001; n = 680–820). Hearts from animals without DOX also contained significantly fewer mitochondria (86 ± 24 mitochondria per field versus 137 ± 26 in controls, P < 0.01; field size = 264 μm2; n = 6 fields).
Transmission electron micrographs also revealed an unusual pattern of lipid body formation in hearts of transgenic mice after DOX withdrawal. Relatively few lipid bodies were seen in hearts from control animals, but numbers increased dramatically at 5 days after DOX withdrawal (Fig. 2D). At this stage, mitochondrial morphology appeared normal. The increase in lipid bodies was reversed by 8 days after DOX withdrawal (2.8 ± 1.5 lipid bodies per field in controls versus 10.7 ± 4.5 at 5 days, P < 0.05, versus 1.7 ± 1.0 at 8 days; field size = 264 μm2; n = 6–12 fields).
Abnormalities in Cardiac Gene Expression in HDAC5S/A Mice.
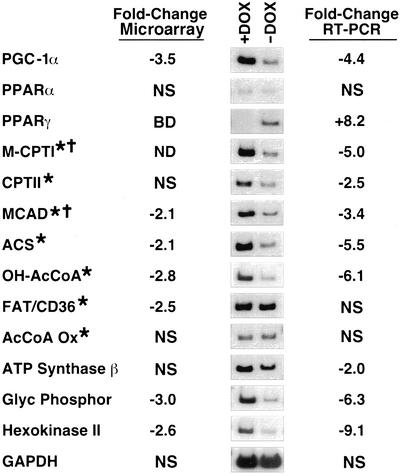
The mitochondrial abnormalities and behavior of mice removed from DOX were consistent with a loss of cardiac energy reserves and heart failure as the likely cause of death. To further investigate the basis for this phenotype, we isolated total cardiac RNA from mice removed from DOX for 8 days and from mice receiving DOX and analyzed gene expression levels using an Affymetrix U74Av2 microarray (Fig. 3). Expression of a subset of genes was also examined by semiquantitative RT-PCR. Of particular interest were changes in the expression levels of numerous genes involved in the mitochondrial fatty acid oxidation pathway.
Figure 3.
Inhibition of expression of PGC-1α and metabolic enzymes after DOX withdrawal. Total RNA from hearts of HDAC5S/A mice was analyzed by microarray or by semiquantitative RT-PCR. The fold changes in gene expression for a panel of enzymes involved in cardiac energy metabolism pathways are presented in the chart (+DOX mice vs. −DOX), as well as images of bands obtained for each transcript by RT-PCR. NS, no significant change; BD, below detection limit; ND, not determined; M-CPTI, muscle-type carnitine palmitoyltransferase I; CPTII, carnitine palmitoyltransferase II; MCAD, medium chain acyl-CoA dehydrogenase; ACS, acyl-CoA synthetase; OH-AcCoA, OH-long-chain acyl-CoA dehydrogenase; FAT/CD36, fatty acid translocase; AcCoA Ox, acyl-CoA oxidase; Glyc Phosphor, glycogen phosphorylase. Asterisks denote direct targets of PPARα regulation; crosses denote genes regulated by PGC-1α.
PGC-1α, a critical regulator of mitochondrial biogenesis and function, was dramatically down-regulated in mice after withdrawal of DOX, consistent with the mitochondrial pathology observed. We also examined expression of PPARα and PPARγ, nuclear receptors that regulate expression of enzymes involved in fatty acid oxidation and adipogenesis, respectively. Expression of PPARα was unchanged, but expression of PPARγ, which is usually expressed in the heart at very low levels, was up-regulated on DOX withdrawal. This may result from excessive levels of unoxidized fatty acids present in the heart due to inhibition of the fatty acid oxidation pathway (see below), because increases in fatty acid levels may up-regulate PPARγ expression (15–17). The up-regulation of PPARγ may be responsible for the loss of lipid bodies observed between 5 and 8 days of transgene induction (Fig. 2D), because the PPARγ agonist troglitazone caused a similar loss of lipid bodies in a rat steatosis model, although the mechanism of this process is unclear (18).
The carnitine palmitoyltransferases M-CPT-I and CPT-II are responsible for mitochondrial import of fatty acids and represent critical rate-limiting steps in the mitochondrial fatty acid oxidation pathway (9). Transcripts encoding both enzymes were down-regulated in mice removed from DOX (Fig. 3). Other key enzymes of this pathway were also dramatically down-regulated including medium chain acyl-CoA dehydrogenase (MCAD), acyl-CoA synthetase, and OH-long-chain acyl-CoA dehydrogenase. The capacity for fatty acid oxidation is therefore likely to be highly reduced in these animals, which may contribute to the accumulation of lipid bodies observed after 5 days of gene induction (Fig. 2D). Expression of these fatty acid oxidation genes is regulated by PPARα (9). Because expression levels of PPARα did not change in these animals, however, the down-regulation of these genes may be due to impaired coactivation by PGC-1α. In fact, expression of M-CPT-I and MCAD is coactivated by PGC-1α (19). Although we cannot rule out the possibility of posttranslational modifications of PPARα being responsible for this effect, some transcriptional targets of PPARα, including the fatty acid translocase/CD36 and acyl-CoA oxidase genes, showed no changes in expression level, suggesting that the down-regulation is not due to changes in PPARα activity directly (Fig. 3).
Various other metabolic enzymes involved in energy generation were down-regulated upon DOX withdrawal, including ATP synthase-β, which is involved directly in ATP synthesis in the mitochondria, glycogen phosphorylase, the rate-limiting enzyme comprising the first step of the glycogenolysis pathway, and hexokinase II, one of the first enzymes in the glycolytic pathway (20). Microarray analysis also revealed down-regulation of the MEF2 transcriptional targets creatine kinase (3.5-fold) and GLUT4 (3-fold), the primary glucose transporter in the myocardium and a target of PGC-1α coactivation (11, 21). Together, these findings indicate that overexpression of a mutant HDAC, which represses MEF2 activity, negatively impacts the major pathways of cardiac energy production. Because PGC-1α acts as a master regulator of mitochondrial biogenesis and gene expression, it was a logical target for the effects of HDAC5S/A on the heart.
Regulation of PGC-1α Expression by MEF2.
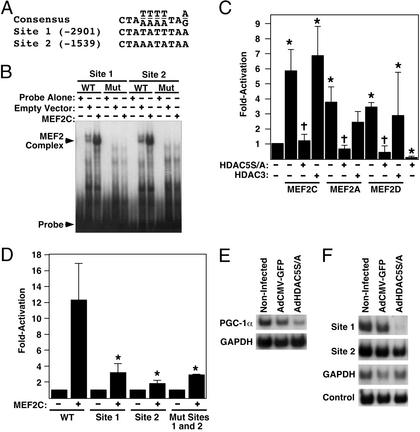
Class II HDACs repress the activity of MEF2 transcription factors. We therefore examined the 5′ flanking region of the mouse PGC-1α gene and identified two putative MEF2-binding sites with conformity to the consensus MEF2 site [CTA(A/T)4TAG/A] (Fig. 4A).
Figure 4.
Regulation of the PGC-1α promoter by MEF2 and HDAC5. (A) Sequence of putative MEF2 transcription factor-binding sites in the 3.1-kb proximal promoter of the mouse PGC-1α gene, compared with the consensus-binding sequence. Numbers in brackets refer to location in base pairs relative to the ATG start codon for PGC-1α. (B) Electrophoretic mobility-shift assay for binding of MEF2C to the putative MEF2 sites of the 3.1-kb proximal PGC-1α promoter. WT, wild-type oligomer; Mut, mutant oligomer with A/T to C/G conversion. (C) Activation of PGC-1α-promoter–luciferase reporter by MEF2 factors and attenuation by HDAC5S/A in COS cells. Asterisks denote P < 0.05 vs. reporter alone; crosses denote P < 0.05 vs. same sample in absence of HDAC5S/A. Error bars represent standard deviation of the mean for three experiments. (D) Effects of MEF2-binding site mutations in the PGC-1α promoter on transcriptional activation by MEF2C. Asterisks denote P < 0.01 vs. wild-type reporter plus MEF2C. Error bars represent standard deviation of the mean for three experiments. (E) Semiquantitative RT-PCR of PGC-1α expression in noninfected neonatal rat cardiomyocytes compared with cells infected with AdCMV-GFP or AdCMV-HDAC5S/A. (F) Immunoprecipitation of acetylated histone H3/DNA complexes from noninfected neonatal rat cardiomyocytes or cells infected with AdCMV-GFP or AdCMV-HDAC5S/A. Immunoprecipitated DNA was subjected to 32P-PCR under nonsaturating conditions using primers for PGC-1α promoter MEF2-binding sites or GAPDH. An aliquot of nonimmunoprecipitated DNA was used as an input control.
Gel-shift analysis of these sites using extracts from MEF2C-transfected COS cells revealed that they bound MEF2C (Fig. 4B). Mutated MEF2 sites with A/T to C/G conversions showed no complex formation, demonstrating the specificity of binding.
We then examined whether MEF2 factors were able to activate the PGC-1α promoter using a luciferase reporter fused to the 3.1-kb PGC-1α promoter region. All three MEF2 factors tested (MEF2C, -A, and -D) significantly activated the PGC-1α promoter (Fig. 4C). Mutation of either of the two MEF2 sites significantly reduced transactivation by MEF2C (Fig. 4D), and mutation of both sites had no further inhibitory effect. Together, these findings demonstrate that both of the MEF2-binding sites are necessary, but neither is sufficient, for transactivation of the PGC-1α promoter.
Repression of PGC-1α Expression by HDAC5S/A.
On the basis of the ability of MEF2 to activate the PGC-1α promoter, we speculated that HDAC5 would repress this promoter, since HDAC5 is recruited by MEF2. Indeed, PGC-1α promoter activation could be completely attenuated by expression of HDAC5S/A, but not by the class I HDAC, HDAC3 (Fig. 4C), demonstrating a specific effect of HDAC5. HDAC5S/A also suppressed expression of the PGC-1α reporter in the absence of exogenous MEF2, possibly due to inhibition of endogenous MEF2 factors present in COS cells. Reporter assays using the homologous human PGC-1α promoter region show similar activation and inhibition of the promoter by MEF2 and HDAC5S/A, respectively (data not shown).
To determine whether HDAC5S/A was capable of inhibiting expression of the endogenous PGC-1α gene, we infected neonatal rat cardiomyocytes in vitro with an adenovirus encoding HDAC5S/A (AdHDAC5S/A) and examined expression of PGC-1α by semiquantitative RT-PCR (8). Infection with AdHDAC5S/A significantly inhibited PGC-1α expression, whereas infection with a control adenovirus expressing GFP (AdCMV-GFP) had no effect (Fig. 4E). We conclude that endogenous PGC-1α gene expression is modulated by HDAC5.
Histone Deacetylation at an Essential MEF2 Site Correlates with Repression of PGC-1α.
We examined whether the inhibition of PGC-1α gene expression by HDAC5 was due to deacetylation of histones in the region of the essential MEF2-binding sites within the PGC-1α promoter by performing ChIP analysis. Acetylation of histones at the distal MEF2-binding site (−2901) was dramatically decreased in cardiomyocytes infected with AdHDAC5S/A compared with noninfected or AdCMV-GFP-infected cardiomyocytes (Fig. 4F). No changes were observed in acetylation of histones at the other PGC-1α MEF2 site or at the GAPDH promoter. Nonimmunoprecipitated input DNA was used as a control.
It might be expected that because the proximal MEF2 site at −1539 was required for transactivation of the luciferase reporter gene (Fig. 4D), deacetylation of histones in this region would be observed. One possible explanation for the lack of deacetylation may be that the proximal site is required for assembly of a transcriptional complex, but HDACs themselves are not recruited to this region. Alternatively, dominant-acting histone acetyltransferases may be recruited to the second site but not the more distal site at −2901.
Discussion
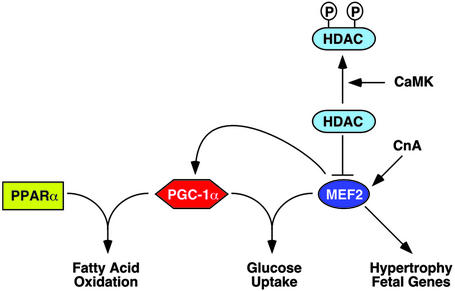
The results of this study suggest a model of high-level control of mitochondrial energy production by MEF2 and HDAC regulation of PGC-1α expression (Fig. 5). This model provides a means of fine-tuning PGC-1α levels in response to changing energy demands and predicts a role for MEF2 in matching increased energy demand during myocyte hypertrophy to increased energy production in the mitochondria.
Figure 5.
Model for interactions between MEF2/HDAC5 and PGC-1α. PGC-1α and PPARα cooperate to activate genes encoding enzymes involved in cardiac fatty oxidation. PGC-1α and MEF2C have been demonstrated to regulate glucose uptake in muscle by controlling expression of the GLUT4 glucose transporter, the main glucose uptake mechanism in muscle. MEF2 also regulates fetal gene programs during muscle hypertrophy. CnA, calcineurin.
Previous studies showed that the calcineurin and calcium/calmodulin-dependent protein kinase (CaMK)-signaling pathways induce the transformation of fast glycolytic skeletal muscle fibers to slow oxidative fibers, accompanied by the up-regulation of PGC-1α (22, 23). Overexpression of PGC-1α in skeletal muscle also results in increased slow fiber number and expression of slow fiber metabolic and contractile genes, which are MEF2-dependent (24). Given that calcineurin and CaMK signaling stimulate MEF2 activity, the present study suggests that MEF2 links these signaling pathways to PGC-1α transcription, providing a potential mechanism for the increase in mitochondrial number and PGC-1α expression observed in response to CaMK signaling in skeletal muscle (23). CaMK stimulates phosphorylation of class II HDACs, leading to their export from the nucleus (4). Our data suggest that inactivation of HDACs by CaMK or related kinases would derepress PGC-1α expression, resulting in increased mitochondrial biogenesis.
Overexpression of PGC-1α in the heart enhances mitochondrial biogenesis (10). Conversely, as shown here, down-regulation of PGC-1α expression results in a loss of cardiac mitochondria. We interpret the severe cardiac abnormalities in HDAC5S/A transgenic mice to reflect the extreme potency of this mutant HDAC in suppressing MEF2 activity. Although our initial interest was to determine whether the signal-resistant mutant of HDAC5 could block cardiac hypertrophy in vivo as it does in vitro (8), the cardiac abnormalities evoked by overexpression of HDAC5S/A suggest that there is likely to be an essential level of MEF2 activity required for cardiac homeostasis, such that suppression of this activity results in cardiac demise. Consistent with this notion, mice lacking MEF2A exhibit a deficiency of mitochondria and high postnatal mortality (25). It is tempting to speculate that the phenotype of HDAC5S/A mice reflects the combined loss of function of all MEF2 factors in the adult heart, although we cannot rule out the possible involvement of other transcription factors.
It is possible that the mitochondrial changes observed in the HDAC5S/A over-expressing animals are not directly due to repression of PGC-1α expression, because MEF2 has numerous target genes. Indeed, transcripts encoding a variety of MEF2 target genes were down-regulated by microarray analysis after DOX withdrawal, including β-enolase, phosphoglycerate mutase, and contractile proteins such as skeletal α-actin and α-MHC (results not shown). However, the in vitro reporter and ChIP assays, combined with the rapid repression (within 24 h) of PGC-1α expression in isolated cardiomyocytes after AdHDAC5S/A infection, strongly suggest a direct effect of HDAC5S/A on PGC-1α expression.
It remains to be seen whether HDAC repression of PGC-1α expression plays a role in cardiac decompensation and failure in human disease progression. Heart failure is often marked by mitochondrial dysfunction, and children with genetic mitochondrial diseases commonly present with cardiovascular deficiencies, indicating that precise balance is required between mitochondrial energy production and cardiac energy demand (26, 27). The rapid progression of heart failure in HDAC5S/A mice suggests that depletion of mitochondrial energy stores may play a critical role in this process. A potential model may involve activation of MEF2 factors during adaptive hypertrophy, resulting in activation of fetal gene programs, activation of PGC-1α-dependent fatty acid metabolism and mitochondrial growth, and an increase in work capacity. However, if this activation is prolonged, or if other stresses act on the heart, other mechanisms may activate HDACs, repressing MEF2 activation of PGC-1α expression and negatively impacting fatty acid metabolism and cardiac energy production. This would help explain the observations of a shift from fatty acid oxidation toward glucose utilization in heart failure and fatty acid oxidation enzyme down-regulation in failing human hearts (28, 29). Should HDACs prove to play a role in these pathways, HDAC inhibitors may be efficacious in treatment of cardiac failure by derepressing PGC-1α expression, thereby increasing fatty acid oxidation and energy production.
Acknowledgments
We thank A. Tizenor for graphics; T. McKinsey and C. Zhang for vectors and valuable discussion; J. Shelton and D. Bellotto for histology; and B. Conklin (University of California, San Francisco) and H. Bujard (University of Heidelberg) for the pUHG-10 vector. We also thank R. Bassel-Duby, O. Nakagawa, and D. Garry for critical reading of the manuscript. E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Foundation, and the Texas Advanced Technology Program. M.P.C. was the recipient of a postdoctoral fellowship from the Canadian Institutes for Health Research.
Abbreviations
- HDAC
histone deacetylase
- MEF2
myocyte enhancer factor-2
- HDAC5S/A
HDAC5 double serine-to-alanine mutant
- PPAR
peroxisome proliferator-activated receptor
- PGC-1α
PPARγ coactivator 1α
- DOX
doxycycline, ChIP, chromatin immunoprecipitation
- MHC
myosin heavy chain
- tet
tetracycline
References
- 1.Eberharter A, Becker P B. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKinsey T A, Zhang C L, Olson E N. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 3.Black B L, Olson E N. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 4.McKinsey T A, Zhang C L, Lu J, Olson E N. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grozinger C M, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKinsey T A, Zhang C L, Olson E N. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinsey T A, Zhang C L, Olson E N. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C L, McKinsey T A, Chang S, Antos C L, Hill J A, Olson E N. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehman J J, Kelly D P. Clin Exp Pharmacol Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 10.Lehman J J, Barger P M, Kovacs A, Saffitz J E, Medeiros D M, Kelly D P. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michael L F, Wu Z, Cheatham R B, Puigserver P, Adelmant G, Lehman J J, Kelly D P, Spiegelman B M. Proc Natl Acad Sci USA. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Z, Redfern C S, Fishman G I. Circ Res. 1996;79:691–697. doi: 10.1161/01.res.79.4.691. [DOI] [PubMed] [Google Scholar]
- 13.McFadden D G, Charite J, Richardson J A, Srivastava D, Firulli A B, Olson E N. Development (Cambridge, UK) 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 14.Djouadi F, Weinheimer C J, Saffitz J E, Pitchford C, Bastin J, Gonzalez F J, Kelly D P. J Clin Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patane G, Anello M, Piro S, Vigneri R, Purrello F, Rabuazzo A M. Diabetes. 2002;51:2749–2756. doi: 10.2337/diabetes.51.9.2749. [DOI] [PubMed] [Google Scholar]
- 16.Meadus W J, MacInnis R, Dugan M E. J Mol Endocrinol. 2002;28:79–86. doi: 10.1677/jme.0.0280079. [DOI] [PubMed] [Google Scholar]
- 17.Fabris R, Nisoli E, Lombardi A M, Tonello C, Serra R, Granzotto M, Cusin I, Rohner-Jeanrenaud F, Federspil G, Carruba M O, et al. Diabetes. 2001;50:601–608. doi: 10.2337/diabetes.50.3.601. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y T, Grayburn P, Karim A, Shimabukuro M, Higa M, Baetens D, Orci L, Unger R H. Proc Natl Acad Sci USA. 2000;97:1784–1789. doi: 10.1073/pnas.97.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vega R B, Huss J M, Kelly D P. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Depre C, Rider M H, Hue L. Eur J Biochem. 1998;258:277–290. doi: 10.1046/j.1432-1327.1998.2580277.x. [DOI] [PubMed] [Google Scholar]
- 21.Mora S, Pessin J E. J Biol Chem. 2000;275:16323–16328. doi: 10.1074/jbc.M910259199. [DOI] [PubMed] [Google Scholar]
- 22.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Kanatous S B, Thurmond F A, Gallardo T, Isotani E, Bassel-Duby R, Williams R S. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 24.Lin J, Wu H, Tarr P T, Zhang C Y, Wu Z, Boss O, Michael L F, Puigserver P, Isotani E, Olson E N, et al. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 25.Naya F J, Black B L, Wu H, Bassel-Duby R, Richardson J A, Hill J A, Olson E N. Nat Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 26.Guertl B, Noehammer C, Hoefler G. Int J Exp Pathol. 2000;81:349–372. doi: 10.1046/j.1365-2613.2000.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanley W C, Chandler M P. Heart Fail Rev. 2002;7:115–130. doi: 10.1023/a:1015320423577. [DOI] [PubMed] [Google Scholar]
- 28.Recchia F A, McConnell P I, Bernstein R D, Vogel T R, Xu X, Hintze T H. Circ Res. 1998;83:969–979. doi: 10.1161/01.res.83.10.969. [DOI] [PubMed] [Google Scholar]
- 29.Sack M N, Rader T A, Park S, Bastin J, McCune S A, Kelly D P. Circulation (Cambridge, UK) 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]