Abstract
The nature and extent of interindividual and temporal variation in gene expression patterns in specific cells and tissues is an important and relatively unexplored issue in human biology. We surveyed variation in gene expression patterns in peripheral blood from 75 healthy volunteers by using cDNA microarrays. Characterization of the variation in gene expression in healthy tissue is an essential foundation for the recognition and interpretation of the changes in these patterns associated with infections and other diseases, and peripheral blood was selected because it is a uniquely accessible tissue in which to examine this variation in patients or healthy volunteers in a clinical setting. Specific features of interindividual variation in gene expression patterns in peripheral blood could be traced to variation in the relative proportions of specific blood cell subsets; other features were correlated with gender, age, and the time of day at which the sample was taken. An analysis of multiple sequential samples from the same individuals allowed us to discern donor-specific patterns of gene expression. These data help to define human individuality and provide a database with which disease-associated gene expression patterns can be compared.
Numerous studies have described efforts to map and characterize variations in human gene expression patterns associated with differences in cell and tissue type, physiological processes, and disease. In vitro experiments have examined aspects of physiological regulation of human gene expression programs, including the identification of genes periodically expressed in the cell cycle (1), the response of human cells to various stimuli (2, 3), and the dissection of signaling pathways (4, 5). Profiles of gene expression patterns are also helping to define the complex biological processes associated with both health and disease in vivo. Considerable progress has already been made in clinical cancer research, using systematic analysis of gene expression patterns to define tumor subtypes, identify molecular markers, and investigate new therapies (6–8). The insights made possible by genome-scale assessment of gene expression in cancer have provided oncologists with a greater understanding of tumor biology and the potential for better patient care and prognosis. Experiments investigating host response to infection with in vitro models have revealed insights into mechanisms of pathogenesis (9), and, as with studies of cancer, have highlighted the potential for application of microarray technology to the study of infection in vivo.
The extent, nature, and sources of variation in gene expression among healthy individuals is a fundamental, yet largely unexplored, aspect of human biology (10). Future investigations of human gene expression programs associated with disease, and their potential application to detection and diagnosis, will depend on an understanding of their normal variation within and between individuals, over time, and with age, gender, and other aspects of the human condition.
Peripheral blood is an accessible source of cells with which to investigate these questions. Moreover, circulating leukocytes can be viewed as scouts, continuously maintaining a vigilant and comprehensive surveillance of the body for signs of infection or other threats. The gene expression responses of circulating leukocytes can potentially provide an early warning of the threats they discover. Thus, peripheral leukocytes have the potential to be used diagnostically as surrogates for direct sampling of sites of infection or other disease processes, including cancer, autoimmune, genetic, and metabolic disorders.
We carried out a survey of the variation in gene expression patterns in the blood of healthy individuals, by using cDNA microarrays. Our results revealed a surprising consistency in these patterns, but also evidence of distinct patterns of interindividual and temporal variation. Some features of variation in the expression patterns were associated with differences in the cellular composition of the blood sample, with gender, age, and time of day. These results expand our understanding of human variation and hematological physiology and provide a genome-scale molecular portrait of a healthy human tissue.
Materials and Methods
Patient Information and Blood Samples.
Blood samples from 75 apparently healthy human donors were obtained after informed consent and treated anonymously throughout the analysis. Volunteer blood donors from the United States averaged 36.5 (±14.8) years of age, and males and females were similarly represented (40 male, 35 female). Samples were also obtained from seven apparently healthy volunteer donors in Kathmandu, Nepal (25–30 years of age, four females and three males). Complete blood counts were determined at the Stanford University Hospital Clinical Laboratory by automated procedures. Measured parameters included total white count, differential counts for neutrophils, lymphocytes, monocytes, eosinophils, and basophils, red blood cell count, platelet count, hemoglobin, hematocrit, and erythrocyte indices [mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, and red cell distribution width (RDW)]. Time of blood draw was recorded, as was the self-reported health status and medication use of each subject, by using a standardized questionnaire. Peripheral blood mononuclear cells (PBMCs) from 8 ml of blood were isolated by using Vacutainer cell preparation tubes with sodium citrate (Becton Dickinson) and stored in Trizol reagent (Life Technologies, Rockville, MD) at −80°C before RNA extraction. Total RNA was isolated from 2.5 ml of whole blood with the PaxGene Blood RNA System (PreAnalytiX, Hombrechtikon, Switzerland) within 24 h. Whole blood and PBMC total RNA was linearly amplified as described (11).
Microarray Procedures.
Microarray methods followed closely those described in a previous study (6). More detailed information including data selection and manipulation methods, as well as searchable figures and all raw microarray data can be found at http://genome-www.stanford.edu/normalblood.
Results and Discussion
Cellular and Physiological Themes in Gene Expression Variation in Whole Blood.
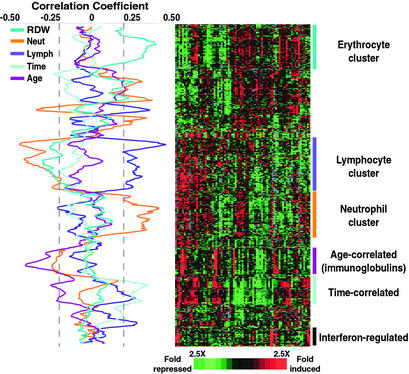
To explore and characterize the variation in gene expression in a complex tissue, in a large and diverse group of healthy human subjects, 77 whole-blood RNA samples from 75 individuals were analyzed by comparative hybridization to DNA microarrays containing 37,632 elements, representing ≈18,000 different genes (12). The patterns of variation revealed distinct features that can be associated with variation in cellular composition, physiological responses, and interindividual variation. Fig. 1 provides an overview of the gene expression profiles of these blood samples, focusing on the subset of genes whose expression levels were most variable between individuals.
Figure 1.
Variation in gene expression patterns in human blood. Whole blood was drawn from 75 healthy volunteers. Two volunteers donated samples on two occasions, resulting in a total of 77 samples. Genes with at least a 2.0-fold change in level of expression from the mean in at least five (of 77) samples are shown. The expression pattern of the corresponding ≈370 genes is displayed in hierarchical cluster format where rows represent genes (unique cDNA elements) and columns represent experimental samples. Each expression measurement represents the normalized ratio of fluorescence from the hybridized experimental sample to a reference sample (see Supporting Materials and Methods for details). Missing or excluded data are represented by gray squares. Correlation coefficients were calculated between the gene expression of every gene and each parameter (neutrophil count, lymphocyte count, time of blood draw, age, and RDW) across the 77 samples. The correlation values are plotted as moving averages of 11 genes (along the vertical axis). Gene expression data were randomly permuted 10,000 times; the correlation coefficients derived from each permutation of the data were compared with that from the actual data. The P value for each gene was calculated as the fraction of the permutations that resulted in a correlation coefficient as high as was observed in the actual data. A dashed line indicates the lowest value of correlation coefficient significant (P < 0.05) for each parameter. Supplemental data and enhanced versions of the figures, including searchable clusters and raw microarray data, can be found at http://genome-www.stanford.edu/normalblood.
We suspected that a significant fraction of the observed variation reflected variation in the relative proportions of the different peripheral blood cell types. In an attempt to identify genes most closely associated with specific subsets of cells and other independently determined characteristics of the samples, we calculated correlation coefficients for the association of each gene's expression pattern with complete blood count parameters, age of donor, and time of blood draw. The resulting correlation curves, plotted as moving averages (window size = 11 genes), are displayed in Fig. 1 Left.
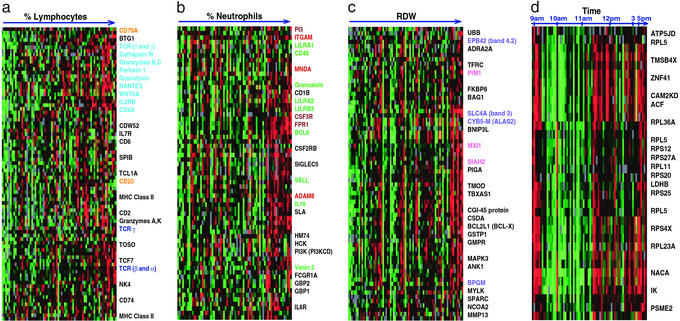
Fifty-five unique genes (15% of the 370 genes with the most variable expression) were members of a “lymphocyte-associated” cluster (Fig. 2a). The correlation coefficient between the average expression level of these genes and the absolute number of lymphocytes was 0.45. Many genes in this cluster have previously been reported to be expressed specifically in T or B cells (e.g., CD20, CD2, CD79A, Spi-B transcription factor). Cytotoxic T lymphocyte (CTL)-associated genes were particularly apparent; CD8, IL-2RB, RANTES, granulysin, and perforin transcripts, all characteristic of CTLs, varied in parallel across these samples.
Figure 2.
Gene clusters associated with measured complete blood count parameters and time. The lymphocyte (a), neutrophil (b), reticulocyte (c), and time-correlated (d) clusters. Samples were reordered from Fig. 1 according to the scale shown across the top, and genes were hierarchically clustered. Colored text indicates the following: genes specific to B cells (orange), cytotoxic T lymphocytes or natural killer cells (light blue), T cells (dark blue), erythrocytes (blue), neutrophils (brown), myeloid cells (red), ubiquitously expressed (green), and Myc-regulated (pink).
Somewhat surprisingly, the pattern of variation in expression of Ig genes was only weakly correlated with lymphocyte count and was distinct from the expression patterns of other genes characteristically expressed in lymphocytes in general or B cells in particular. CD20 expression correlated well with relative lymphocyte count, and the variation in expression of other genes characteristically expressed by B cells (CD19, CD22, CD72, BTK), although not of sufficient amplitude for inclusion in this set of variable genes, paralleled the expression patterns of CD20 and other lymphocyte-associated genes (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Ig gene expression was also considerably more variable among these samples than expression of the CD20 cluster of B cell genes. This finding could be explained by variation in the percentage of mature B and plasma cells among the total circulating B cell population. Alternatively, variation in Ig expression may reflect regulation in response to environmental cues and physiological conditions or baseline genetic variation in Ig expression.
Genes known to be expressed in neutrophils, the most abundant leukocytes in the samples, also demonstrated significant variability in expression among healthy blood donors (Fig. 2b). The correlation coefficient between the average transcript levels of the 52 unique genes in this cluster and the absolute number of neutrophils in the 77 samples was 0.42. Based on previous reports of their expression patterns, the genes in this cluster could be grouped into three increasingly specific families: (i) those ubiquitously expressed in many types of circulating immune cells; (ii) those expressed by cells of the myeloid lineage; and (iii) those specific to granulocytes. BCL6, vanin 2, IL-16, and selectin L are among the genes expressed by many immune cell subsets, but which correlated most closely with neutrophil count in our dataset. Integrin alpha M and myeloid cell nuclear differentiation antigen are expressed primarily in cells of the myeloid lineage (granulocytes and monocytes), whereas formyl peptide receptor 1 and colony stimulating factor 3 receptor have been shown to be specifically expressed by granulocytes (13).
We found a significant correlation between RDW, a measure of variability in red blood cell size, and the expression pattern of a cluster of 57 genes (Fig. 2c). Elevation of the RDW often reflects a higher percentage of reticulocytes in the circulating blood (14). Some of the genes in this group encode proteins related to erythrocyte structure and maturation. EPB42 (band 4.2) and SLC4A1 (band 3) are erythrocyte membrane proteins. BCL2L1 (BCL-X), BNIP3L, and BAG1 are BCL2-associated proteins involved erythropoietin's known suppression of apoptosis during erythropoiesis (15). Other genes correlated with RDW are involved in the heme biosynthetic pathway (ALAS2) and in regulating hemoglobin oxygen affinity (BPGM) (16, 17).
To identify additional genes whose expression is characteristic of specific cell types, we collected from each of 11 donors separate samples for analysis of gene expression in both PBMC and whole blood. The PBMC fraction does not include several cell types present in the whole-blood samples: neutrophils, eosinophils, platelets, reticulocytes, and red blood cells. A comparative analysis revealed ≈2,000 genes with at least a 2-fold average difference in expression between whole blood and PBMCs, most of which appear to reflect the difference in cellular composition (http://genome-www.stanford.edu/normal blood). As expected, transcripts of genes associated with neutrophils (FPR1, PI3, HM74, grancalcin), eosinophils (IL-5RA), and reticulocytes and red blood cells (hemoglobin, EPB42, ANK1, spectrin) were more relatively more abundant in whole blood than in the PBMC fraction. Conversely, many of the genes with higher levels of expression in PBMCs varied in parallel with lymphocyte count in the whole-blood samples (e.g., PTP4A2, MHC class II, transcription factor 7, T cell receptor interacting molecule). Numerous transcripts encoding translation initiation and elongation factors, ribosomal proteins, and splicing factors were enriched in the PBMC fraction of blood, perhaps reflecting the greater importance of new transcription and protein synthesis in physiological responses of lymphocytes and monocytes as compared with granulocytes. A gene expression “signature” characteristically induced by prolonged handling of human cells and tissues (including FOS, early lymphocyte activation markers CD83 and CD69, tumor necrosis factor α-induced protein 3, and dual specificity phosphatase 2; C. Perou, personal communication) suggests that the handling required for isolation of the mononuclear cell fraction from whole blood may incite changes in gene expression.
Variations Associated with Age and Gender.
An unexpected and interesting result was the clear negative correlation between Ig gene expression and donor age (Fig. 1, P < 0.03). The Ig λ and κ light chain and γ 3 and μ heavy chain genes all exhibited this association, whereas the J-chain gene showed a weaker inverse correlation with age. Although there is evidence in mice that aging has an effect on B cell longevity and generation (18), little is known about the effects of aging on Ig expression in humans. Decreased serum IgM and IgG levels have been associated with increased age in humans (19). The relationship between the expression patterns seen in PBMCs and lymphoid organs remains unexplored.
Gender is a major determinant of variation in physiology, morphology, and disease susceptibility in humans. Many immunological and inflammatory diseases have a striking gender bias in incidence and severity (20, 21). We used the significance analysis of microarrays (SAM) procedure to search for genes whose expression differed significantly between healthy male and female blood donors (see Supporting Materials and Methods, which are published as supporting information on the PNAS web site) (22). The genes whose expression was most sex-specific were located on the X or Y chromosome, but we also found autosomal genes with a significant gender bias in expression level (Table 1, which is published as supporting information on the PNAS web site). The majority of these were more highly expressed in females, including several transcripts whose expression was also correlated with neutrophil count (e.g., SLA, TNFRSF1B, IFN-GR2, IFITM3), although the neutrophil count itself was not correlated with gender. IFN-γ has long been suspected as playing a role in the sexual dimorphism of autoimmunity, but the specific mechanisms behind these gender-specific differences remain an enigma (20, 23). We found a significant gender bias in the expression of IFN-GR2, the β chain of the IFN-γ receptor complex, which has been implicated as a potentially important factor in determining the number of functional receptor complexes that transduce IFN-γ signals, and hence, the difference between a proliferative or apoptotic response to IFN-γ (24). IFITM3, another IFN-responsive gene, was also more highly expressed in the blood of females than males. Other genes demonstrating a gender bias in their expression included some related to cell cycle control, proliferation, and apoptosis (ITM2B, BTG3, ARHGDIB) and not obviously correlated with any cell type. EIF1A, a translation initiation factor whose expression was correlated with lymphocyte count, was also more highly expressed in females.
Temporal Variation in Whole-Blood Gene Expression.
Although all of the samples were drawn between 8 a.m. and 5 p.m., and 85% were taken between 9 a.m. and 1 p.m., we identified a group of genes whose expression in whole-blood samples varied significantly (Fig. 1, P < 0.05) with the time of day that the sample was drawn (Fig. 2d). A remarkable fraction of these genes (35%) encode ribosomal proteins. Other researchers have observed gene expression signatures indicative of circadian clock regulation in the mouse. Storch et al. (25) recently reported diurnal expression differences in genes encoding several ribosomal proteins in the liver and heart of mice. The RPS4X mouse ortholog of the RPS4X (ribosomal protein 4, X-linked) gene, one of the genes with time-of-day associated variation in our dataset, was reported to show circadian regulation in the mouse. Panda et al. (26) found that genes involved in protein synthesis comprised the largest group of coordinately regulated, time-dependent transcripts in the suprachiasmatic nucleus of the mouse hypothalamus, including genes encoding ribosomal proteins, NACA (nascent-polypeptide-associated complex α polypeptide), and genes involved in protein folding and proteasome-mediated degradation. Interestingly, NACA and a proteasome activator subunit (PSME2) were also among the 30 time-of-day dependent genes we observed in human whole blood. These results thus provide further evidence that systemic regulation of genes responsible for protein synthesis and control of protein degradation, perhaps reflecting circadian cycles of cell growth and/or nutrient availability, may be a fundamental feature of the diurnal cycle in mammals.
Variation in IFN-Regulated Genes.
Significant individual variation in many genes had no identifiable correlation with differences in the composition of the blood samples or characteristics of their donors. One of the most prominent groups of such genes comprised 15 genes known to be regulated by IFN (Fig. 1). These transcripts changed 10-fold, on average, from maximum to minimum, although we observed little variation in IFN-γ expression (<3-fold). Previous experiments have reported minimal interindividual variation in IFN-γ production compared with other cytokines and suggested that the level of production of this cytokine is characteristic of an individual (27). The gene expression pattern we observed among healthy blood donors could indicate inherent interindividual variability in the basal activity of this system or in the response to IFN or related stimuli, as suggested by additional results described further below. Subclinical or recent infection in a fraction of donors could also account for some of this variability.
Individuality in Gene Expression Patterns.
The observed variation in gene expression patterns reflects both physiological variation and intrinsic interindividual variation. To identify genes whose variation in expression in these samples is most likely caused by intrinsic interindividual differences, we analyzed variation in gene expression in PBMCs from 16 individuals whose blood was sampled on multiple days. Importantly, when the samples were clustered based on their patterns of expression of ≈600 genes whose expression varied by a minimum of 2.5-fold from the mean in at least three of the 48 specimens, the multiple samples from the same donor did not consistently cluster together (Fig. 6c, which is published as supporting information on the PNAS web site). Thus, intrinsic individual differences are not the dominant source of variation in gene expression among these samples. To focus specifically on these intrinsic variations, we calculated, for each of the ≈25,000 genes in the primary dataset, the ratio of the mean squared pairwise difference in that gene's transcript levels between individuals, to the mean squared pairwise difference in the gene's transcript levels between multiple samples from the same individual (the “intrinsic score”). When we clustered the PBMC samples based on expression of the 340 genes with the highest intrinsic score (those >2 SD from the mean), the multiple samples from each individual clustered together as nearest neighbors for all but three subjects (Figs. 3a and 6a). These intrinsic differences in expression patterns are likely caused by differences in genotype, although they might also reflect epigenetic or environmental factors. For example, hierarchical analysis of expression patterns clearly distinguished specimens from males and females. Most of the gender-specific differences were directly attributable to differences in sex chromosomes; when genes located on the Y or inactive X chromosome were removed from the intrinsic gene list (≈20 of 340 genes), we did not observe male-female segregation of specimens (Fig. 6b).
Figure 3.
Intrinsic interindividual differences in gene expression patterns. (a) Global gene expression was measured in PBMCs from 16 individuals who were each sampled more than once over the course of 1 month. Also included are three patients sampled only once (gray branches). Genes displayed are those with the greatest variation in intrinsic individual differences in expression, i.e., those with intrinsic scores >2 SD from the mean (see text). The expression pattern of the corresponding ≈340 genes is displayed in hierarchical cluster format. Gender is represented by colored branches: blue for male and pink for female. Blue and pink brackets represent samples from the same patient that clustered together. The other colored branches represent repeated samples from single subjects that did not cluster together. (b) DDX17 gene expression (log2) for up to six samples collected over the course of a month for 20 patients. Data are expressed as the difference from the mean expression level for the entire dataset.
Several of the genes that appeared to have the greatest intrinsic individual variation in their expression in PBMCs are also known to have highly polymorphic sequences, supporting the notion that the variation in expression of many of these genes may reflect underlying allelic variation in their regulatory sequences. For example, classical MHC class II genes were well represented among the genes with the highest intrinsic scores, and HLA-DQ (α and β chains) and HLA-DR (β only) are among the most polymorphic known genes. TAP2, the gene encoding one of the transporters associated with antigen processing for class I presentation, was also highly individual-specific in its expression pattern.
We also identified significant interindividual variation in expression of genes that have not previously been characterized as polymorphic. DDX17 (also referred to as RH70), a member of the dead-box protein family, had the highest intrinsic score and was markedly variable in expression in different individuals, but showed little variation over time in any single individual (Fig. 3b). Moreover, its pattern of variation in expression was not closely correlated with any other gene, suggesting that the variation in its expression may be caused by variation in its promotor or enhancer sequences. The DDX17 protein was recently reported to function as an RNA helicase with a role in pre-mRNA splicing, raising the interesting possibility that the variation in its expression might lead in turn to individual variants in mRNA splicing (28). The DDX17 gene is located within a highly polymorphic region of chromosome 22 that contains many genes associated with inherited and acquired disease (29, 30).
BRCA1 mutations are associated with a sharply increased risk of breast and ovarian cancer (31). This gene was generally more highly expressed in PBMCs from females than in PBMCs from males, and also varied in expression among female subjects. These data suggest the intriguing possibility of sexual dimorphism in BRCA1 expression and that expression polymorphism of BRCA1 in leukocytes, and perhaps other cells, may be relatively common.
We found significant intrinsic, interindividual variation in the prion protein gene (PRNP) expression in PBMCs. It is worth noting that three polymorphisms in the regulatory region of PRNP have been reported to be more common in sporadic Creutzfeld–Jakob disease (CJD) patients than in controls (32). The possibility that intrinsic differences in levels of PRNP expression, detectable in PBMCs, might influence susceptibility to CJD is an interesting question for further investigation.
One of the most striking examples of individuality in expression patterns was a cluster of six IFN-regulated genes (OAS3, MTAP44, INADL, MX1, GS3686, and IFIT1), suggesting intrinsic differences in IFN-responsive gene expression. Donor-specific differences in expression of IFN-responsive genes were a prominent feature in gene expression programs in PBMCs responding to bacteria (9). These findings support our previous hypothesis that the variability we observed in expression of IFN-regulated genes was caused by intrinsic variability in the response to IFN and highlights the potential importance of these inherent differences in disease susceptibility. In a recently published study, a mutation in OAS1, an IFN-response gene, was associated with enhanced susceptibility to West Nile virus infection in mice (33). Similarly, failure to produce the Mx protein, encoded by an IFN-response gene, was associated with increased susceptibility to influenza virus in mice (34). The basis of these variations, and their implications for susceptibility to infection and autoimmune disease in humans, will be important questions for further investigation.
Variation in Gene Expression in Health and Disease.
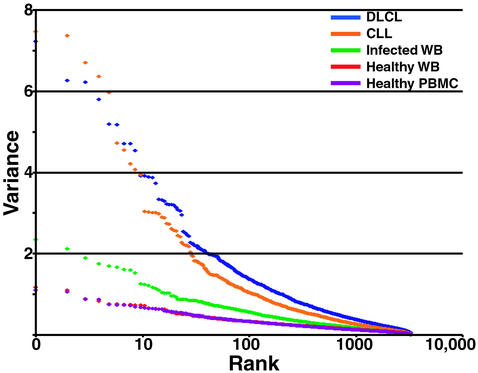
The gene expression responses of circulating leukocytes may provide a basis for detection and diagnosis of infections and other diseases (35). An essential premise for development of diagnostic tools based on expression patterns in blood is that the temporal and individual variation in healthy subjects should be clearly distinguishable from that seen in the diseases and disorders we want to diagnose. We compared the variation in global gene expression patterns in whole blood and PBMCs from diverse normal subjects with the variation in expression patterns in biopsy samples of diffuse large B-cell lymphomas (DLCL) and leukophoresis samples of chronic lymphocytic leukemia (CLL). Both of these malignancies derive from lymphoid cells and therefore comprise a related and relatively homogeneous series of cancer samples for comparison of variability in expression in similar human cell types. The CLL samples consisted of purified CD19+ B cells from peripheral blood, and the DLCL samples consisted of a bulk lymph node biopsy (6). A series of whole-blood samples obtained from patients at the time of presentation with a fever of at least 38°C, in whom a bacterial infection was subsequently diagnosed, were analyzed as a pilot study of the utility of blood gene expression profiles in infectious disease diagnosis (S.J.P., unpublished data). To eliminate the effect of sample size differences, datasets were limited to the same number of samples and array elements in each experimental group, as constrained by the dataset with the smallest number of samples (DLCL with 45 samples) and elements (CLL with 3,826 elements).
The variation in gene expression patterns observed in the blood of healthy subjects was strikingly smaller than the variation observed among samples from subjects with either of the cancers or in blood from patients with a bacterial infection (Fig. 4). Thus, the alterations in global gene expression associated with these disruptions of healthy human biology are significantly greater than the background variation in normal gene expression, supporting the potential value of DNA microarray profiling of whole blood in identifying disease signatures. The relative lack of variation in global gene expression patterns in the blood samples from healthy individuals highlights the homeostasis of cellular composition and the physiology of the diverse cells in this tissue.
Figure 4.
Variation in gene expression in health and disease. Global gene expression was measured in 45 samples from each of five studies. See text for details on each sample set. Of the well-measured microarray elements, 3,826 were randomly selected from each group and variance was calculated for each. Variance was plotted in rank order (highest to lowest) for the genes in each of the specimen groups. DLCL, diffuse large B-cell lymphomas; CLL, chronic lymphocytic leukemia; WB, whole blood.
Conclusion
We conducted a large-scale survey of the variation in gene expression in a single complex tissue (blood) in healthy subjects. These data help to define the extent and nature of the normal variability in gene expression in human blood and also provide insight into the factors that contribute to this variation. Importantly, although the variability among healthy individuals was sufficient to reveal many distinct systematic features, it was far more restricted than the variability observed in disease states affecting related tissues. Our ability to recognize systematic features in the patterns of variation in gene expression in human blood, together with the relatively limited scope and magnitude of normal variation, provides strong support for the feasibility of using gene expression patterns in peripheral blood as a basis for detection and diagnosis of disease in human patients.
Supplementary Material
Acknowledgments
We thank the volunteers who donated blood for this study. We thank the members of the Brown and Relman laboratories for helpful discussions, and Chih Long Liu for assistance with the permutation analyses. We gratefully acknowledge support for this work from the Defense Advanced Research Planning Agency (Grant N65236-99-1-5428 to P.O.B. and D.A.R.), the National Cancer Institute (to P.O.B.), the National Institute of Allergy and Infectious Diseases/National Institutes of Health (Grant AI39587 to D.A.R.), the Ellison Medical Foundation (Grant ID-SS-0103-01 to D.A.R.), and the Howard Hughes Medical Institute. P.O.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- PBMC
peripheral blood mononuclear cell
- RDW
red cell distribution width
References
- 1.Whitfield M L, Sherlock G, Saldanha A J, Murray J I, Ball C A, Alexander K E, Matese J C, Perou C M, Hurt M M, Brown P O, Botstein D. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 3.Belcher C E, Drenkow J, Kehoe B, Gingeras T R, McNamara N, Lemjabbar H, Basbaum C, Relman D A. Proc Natl Acad Sci USA. 2000;97:13847–13852. doi: 10.1073/pnas.230262797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diehn M, Alizadeh A, Rando O J, Liu C L, Stankunas K, Botstein D, Crabtree G R, Brown P. Proc Natl Acad Sci USA. 2002;99:11796–11801. doi: 10.1073/pnas.092284399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fambrough D, McClure K, Kazlauskas A, Lander E S. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 6.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 7.Welsh J B, Zarrinkar P P, Sapinoso L M, Kern S G, Behling C A, Monk B J, Lockhart D J, Burger R A, Hampton G M. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang B D, Swift M E, Shen M, Fang J, Broude E V, Roninson I B. Proc Natl Acad Sci USA. 2002;99:389–394. doi: 10.1073/pnas.012602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldrick J C, Alizadeh A A, Diehn M, Dudoit S, Liu C L, Belcher C E, Botstein D, Staudt L M, Brown P O, Relman D A. Proc Natl Acad Sci USA. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H, Yuan W, Velculescu V E, Vogelstein B, Kinzler K W. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 11.Wang E, Miller L D, Ohnmacht G A, Liu E T, Marincola F M. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 12.Alizadeh A, Eisen M, Davis R E, Ma C, Sabet H, Tran T, Powell J I, Yang L, Marti G E, Moore D T, et al. Cold Spring Harbor Symp Quant Biol. 1999;64:71–78. doi: 10.1101/sqb.1999.64.71. [DOI] [PubMed] [Google Scholar]
- 13.Itoh K, Okubo K, Utiyama H, Hirano T, Yoshii J, Matsubara K. Blood. 1998;92:1432–1441. [PubMed] [Google Scholar]
- 14.Roberts G T, El Badawi S B. Am J Clin Pathol. 1985;83:222–226. doi: 10.1093/ajcp/83.2.222. [DOI] [PubMed] [Google Scholar]
- 15.Silva M, Grillot D, Benito A, Richard C, Nunez G, Fernandez-Luna J L. Blood. 1996;88:1576–1582. [PubMed] [Google Scholar]
- 16.Sadlon T J, Dell'Oso T, Surinya K H, May B K. Int J Biochem Cell Biol. 1999;31:1153–1167. doi: 10.1016/s1357-2725(99)00073-4. [DOI] [PubMed] [Google Scholar]
- 17.Fujita T, Suzuki K, Tada T, Yoshihara Y, Hamaoka R, Uchida K, Matuo Y, Sasaki T, Hanafusa T, Taniguchi N. J Biochem (Tokyo) 1998;124:1237–1244. doi: 10.1093/oxfordjournals.jbchem.a022243. [DOI] [PubMed] [Google Scholar]
- 18.Kline G H, Hayden T A, Klinman N R. J Immunol. 1999;162:3342–3349. [PubMed] [Google Scholar]
- 19.Challacombe S J, Percival R S, Marsh P D. Oral Microbiol Immunol. 1995;10:202–207. doi: 10.1111/j.1399-302x.1995.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 20.Verthelyi D. Int Immunopharmacol. 2001;1:983–993. doi: 10.1016/s1567-5769(01)00044-3. [DOI] [PubMed] [Google Scholar]
- 21.Cutolo M, Sulli A, Seriolo B, Accardo S, Masi A T. Clin Exp Rheumatol. 1995;13:217–226. [PubMed] [Google Scholar]
- 22.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarvetnick N, Fox H S. Mol Biol Med. 1990;7:323–331. [PubMed] [Google Scholar]
- 24.Bernabei P, Coccia E M, Rigamonti L, Bosticardo M, Forni G, Pestka S, Krause C D, Battistini A, Novelli F. J Leukocyte Biol. 2001;70:950–960. [PubMed] [Google Scholar]
- 25.Storch K, Lipan O, Leykin I, Viswanathan N, Davis F, Wong W, Weitz C. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 26.Panda S, Antoch M P, Miller B H, Su A I, Schook A B, Straume M, Schultz P G, Kay S A, Takahashi J S, Hogenesch J B. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 27.Yaqoob P, Newsholme E A, Calder P C. Cytokine. 1999;11:600–605. doi: 10.1006/cyto.1998.0471. [DOI] [PubMed] [Google Scholar]
- 28.Lee C G. J Biol Chem. 2002;277:39679–39683. doi: 10.1074/jbc.C200337200. [DOI] [PubMed] [Google Scholar]
- 29.Brown M A, Edwards S, Hoyle E, Campbell S, Laval S, Daly A K, Pile K D, Calin A, Ebringer A, Weeks D E, Wordsworth B P. Hum Mol Genet. 2000;9:1563–1566. doi: 10.1093/hmg/9.11.1563. [DOI] [PubMed] [Google Scholar]
- 30.Castells A, Gusella J F, Ramesh V, Rustgi A K. Cancer Res. 2000;60:2836–2839. [PubMed] [Google Scholar]
- 31.Rebbeck T R. Environ Mol Mutagen. 2002;39:228–234. doi: 10.1002/em.10050. [DOI] [PubMed] [Google Scholar]
- 32.McCormack J E, Baybutt H N, Everington D, Will R G, Ironside J W, Manson J C. Gene. 2002;288:139–146. doi: 10.1016/s0378-1119(02)00466-3. [DOI] [PubMed] [Google Scholar]
- 33.Mashimo T, Lucas M, Simon-Chazottes D, Frenkiel M P, Montagutelli X, Ceccaldi P E, Deubel V, Guenet J L, Despres P. Proc Natl Acad Sci USA. 2002;99:11311–11316. doi: 10.1073/pnas.172195399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staeheli P, Grob R, Meier E, Sutcliffe J G, Haller O. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown P O, Hartwell L. Nat Genet. 1998;18:91–93. doi: 10.1038/ng0298-91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.