Abstract
The presence of HIV-1 in latently infected, resting CD4+ T cells has been clearly demonstrated in infected individuals; however, the extent of viral expression and the underlying mechanisms of the persistence of HIV-1 in this viral reservoir have not been fully delineated. Here, we show that resting CD4+ T cells from the majority of viremic patients are capable of producing cell-free HIV-1 spontaneously ex vivo. The levels of HIV-1 released by resting CD4+ T cells were not significantly reduced in the presence of inhibitors of cellular proliferation and viral replication. However, resting CD4+ T cells from the majority of aviremic patients failed to produce virions, despite levels of HIV-1 proviral DNA and cell-associated HIV-1 RNA comparable to viremic patients. The DNA microarray analysis demonstrated that a number of genes involving transcription regulation, RNA processing and modification, and protein trafficking and vesicle transport were significantly upregulated in resting CD4+ T cells of viremic patients compared to those of aviremic patients. These results suggest that active viral replication has a significant impact on the physiologic state of resting CD4+ T cells in infected viremic patients and, in turn, allows release of HIV-1 without exogenous activation stimuli. In addition, given that no quantifiable virions were produced by the latent viral reservoir in the majority of aviremic patients despite the presence of cell-associated HIV-1 RNA, evidence for transcription of HIV-1 RNA in resting CD4+ T cells of aviremic patients should not necessarily be taken as direct evidence for ongoing viral replication during effective therapy.
Keywords: HIV‖latency‖viremia‖DNA microarray
The use of highly active antiretroviral therapy (HAART) in HIV-1-infected individuals has dramatically changed the clinical outcome in many infected individuals and has led to a substantial decline in the incidence of AIDS and in AIDS-related mortality (1). However, the presence of latently infected, resting CD4+ T cells has been consistently demonstrated in the majority of HIV-1-infected individuals receiving HAART in whom plasma viremia has been successfully suppressed for prolonged periods of time (2–4). The persistence of this latently infected viral reservoir has emerged as the major obstacle in preventing the eradication of HIV-1 (2–6). Previous studies have suggested that this viral reservoir does not spontaneously release cell-free HIV-1 ex vivo in the absence of activating stimuli, as measured by p24 ELISA (7, 8). However, the presence of replication-competent virus (2–4), HIV-1 proviral DNA (2), and cell-associated HIV-1 RNA (9–13) has been subsequently demonstrated in the resting CD4+ T compartment in the majority of infected individuals whose detectable viremia has been successfully suppressed by HAART. Although the above observations clearly indicate that HIV-1 persists in infected individuals in the absence of any measurable plasma viremia, they do not conclusively demonstrate that latently infected, resting CD4+ T cells contribute to low levels of on-going viral replication or that they produce HIV-1 in the absence of activating stimuli. In particular, several recent studies have demonstrated the presence of cell-associated HIV-1 RNA in resting CD4+ T cells in patients whose plasma viremia fell to below detectable levels while receiving HAART and have directly linked such findings to evidence for on-going viral replication within the latent viral reservoir (9–13). However, considering that transcription of HIV-1 RNA from integrated HIV-1 DNA is not a target of current anti-HIV-1 drug regimens, it is possible that the detection of cell-associated HIV-1 RNA in patients whose viremia has been suppressed while receiving HAART results from basal cellular transcription activities that may or may not lead to production and secretion of HIV-1 virions. In light of this possibility, direct quantitation of cell-free HIV-1 produced by highly purified, resting CD4+ T cells in the absence of activating stimuli along with examination of the molecular physiology of resting CD4+ T cells from viremic and aviremic infected individuals may serve as a means to prove or disprove that there is on-going viral replication within the latent viral reservoir and the circumstances under which this replication may or may not occur. The present study addresses the above questions.
Materials and Methods
Study Patients.
Eleven viremic and 12 aviremic HIV-1-infected individuals were studied. The clinical profiles of the patients are shown in Table 1. Some viremic and all aviremic patients were receiving various HAART regimens containing at least one protease inhibitor and/or one nonnucleoside reverse transcriptase inhibitor in addition to two reverse transcriptase inhibitors of HIV-1. Leukopheresis procedures were conducted in accordance with protocols approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Table 1.
Profiles of HIV-1-infected patients
| Patient | Antiretroviral therapy at the time of study* | Duration of therapy, month | CD4 count, per μl | CD8 count, per μl | Plasma viral load, copies per ml† |
|---|---|---|---|---|---|
| 1 | None | NA | 551 | 577 | 1,160 |
| 2 | None | NA | 484 | 847 | 8,039 |
| 3 | HAART | 1.0 | 761 | 1437 | 8,722 |
| 4 | HAART | 5.0 | 354 | 553 | 592 |
| 5 | HAART | 0.5 | 1153 | 891 | 853 |
| 6 | None | NA | 557 | 571 | 10,991 |
| 7 | None | NA | 1168 | 935 | 10,132 |
| 8 | None | NA | 794 | 1446 | 2,539 |
| 9 | None | NA | 905 | 1430 | 2,756 |
| 10 | HAART | 1.0 | 452 | 909 | 4,129 |
| 11 | HAART | 10.0 | 682 | 1437 | 837 |
| 12 | HAART | 16.0 | 364 | 434 | <50 |
| 13 | HAART | 7.5 | 662 | 536 | <50 |
| 14 | HAART | 15.0 | 717 | 1315 | <50 |
| 15 | HAART | 7.0 | 723 | 890 | <50 |
| 16 | HAART | 18.0 | 377 | 468 | <50 |
| 17 | HAART | 13.0 | 622 | 903 | <50 |
| 18 | HAART | 50.0 | 474 | 876 | <50 |
| 19 | HAART | 34.5 | 341 | 861 | <50 |
| 20 | HAART | 13.5 | 519 | 948 | <50 |
| 21 | HAART | 40.0 | 589 | 574 | <50 |
| 22 | HAART | 10.0 | 339 | 597 | <50 |
| 23 | HAART | 34.5 | 347 | 944 | <50 |
NA, not applicable.
HAART contained at least one protease inhibitor and/or one nonnucleoside reverse transcriptase inhibitor plus two reverse transcriptase inhibitors of HIV-1.
Measured by ultrasensitive bDNA assay with a detection limit of 50 copies per ml of plasma.
Isolation of Resting CD4+ T Cells.
Peripheral blood mononuclear cells (PBMC) were obtained from leukopheresis by Ficoll-Hypaque density gradient centrifugation. Resting CD4+ T cells were isolated from PBMC of HIV-1-infected individuals by using a column-based cell separation technique (StemCell Technologies, Vancouver), as described (14). The purity of isolated resting CD4+ T cells was generally >99.8%, as measured by fluorescence activated cell sorter (FACS) analysis.
Cultures and Quantitation of Cell-Free HIV-1 Virions.
Freshly isolated resting CD4+ T cells were incubated with media consisting of RPMI medium 1640 supplemented with 10% (vol/vol) FCS, penicillin-streptomycin, and l-glutamine ± cyclosporin A (1 μg/ml), HAART (4 μM AZT, 5 μM 3TC, and 10 μM Indinavir), actinomycin D (0.1 μg/ml), or anti-CD3 antibody for 1–3 days at a density of 2 × 106 cells per ml. At days 1 and 3, culture supernatants were harvested, and cell-free HIV-1 was quantified by using Amplicor HIV-1 Monitor Test (Roche Diagnostics; detection limit: 50 copies per ml) according to the manufacturer's instructions. In some experiments, cells were treated with pronase (20 mg/ml) before incubation to rule out detection of cell-bound HIV-1 virions that would later be released into the supernatant.
Quantitative Real-Time PCR for Measurements of HIV-1 DNA and Cell-Associated, Unspliced HIV-1 RNA.
To determine the frequency of resting CD4+ T cells carrying HIV-1 provirus in infected individuals, real-time PCR was carried out on genomic DNA isolated from 1–2 × 106 purified resting CD4+ T cells by using the Puregene DNA isolation kit according to the manufacturer's specifications (Gentra Systems). One microgram of DNA was then used as template for real-time PCR in an iCycler (Bio-Rad). The amplification reaction was carried out in triplicate by using 0.5 μM primers, 0.2 μM fluorescent probe, 0.8 mM dNTPs, 5 mM MgCl2, and 2.5 units of Platinum Taq Polymerase (Life Technologies, Rockville, MD) in 50 μl of total volume. The following primers and probe were used: 5′-GGTCTCTCTGGTTAGACCAGAT-3′ (5′ primer) and 5′-CTGCTAGAGATTTTCCACACTG-3′ (3′ primer), along with the fluorescent probe 5′-6FAM-AGTAGTGTGTGCCCGTCTGTT-TAMRA-3′. PCR conditions consisted of a denaturation step at 95°C for 3 min followed by 45 cycles of 15 sec at 95°C and 1 min at 58°C. Serially diluted ACH-2 DNA was also subjected to the above PCR to obtain standard curves. For detection of the cell-associated, unspliced form of HIV-1 RNA, total cellular RNA was isolated from 2–5 × 106 resting CD4+ T cells by using TRIzol reagent, according to the manufacturer's specifications (Invitrogen). Synthesis of cDNA and real-time PCR were performed in an iCycler by using TaqMan EZ RT-PCR Kit (Applied Biosystems), according to the manufacturer's specifications. PCR conditions consisted of denaturation at 95°C for 3 min followed by 45 cycles of 15 sec at 95°C and 1 min at 60°C. The following primers and probe were used: 5′-TCTCTAGCAGTGGCGCCC-GAACA-3′ (5′ primer), 5′-TCTCCTTCTAGCCTCCGCTAG-TC-3′ (3′ primer), and 5′-6FAM-CAAGCCGAGTCCTGCGT-CGAGAG-TAMRA-3′ (probe). Serially diluted in vitro trans-cribed HIV-1 RNA was also subjected to the above PCR to obtain standard curves.
Oligonucleotide Arrays.
Total RNA from freshly isolated resting CD4+ T cells (3–5 × 106) was extracted by using the TRIzol method (Invitrogen), according to the manufacturer's instruction. A total of 5 μg of RNA was used per each microarray analysis. First- and second-strand DNA synthesis reactions were performed by using the Superscript Choice System (Invitrogen) followed by in vitro transcription (Enzo Diagnostics) using biotin-labeled dNTPs. The resulting biotin-labeled cRNA was quantitated and analyzed for purity on a 2% agarose gel. cRNA samples were then fragmented and prepared for hybridization to Affymetrix Human Genome U95A oligonucleotide arrays according to protocols specified by the manufacturer (Affymetrix). A significant analysis of microarrays (SAM; ref. 15) algorithm was used to determine the genes that were significantly up-regulated after extensive prefiltering processes. The expression values and normalization were derived from DChip Software by using the model-based algorithm “Model-Based Expression Index (MBEI)” (16). The list of differentially expressed genes in resting CD4+ T cells of four HIV-negative donors, five aviremic, and five aviremic patients was first identified as the genes having <5% false discovery rate using the multiclass (pooled variance) method of SAM followed by 1,000 random permutations of the data. For cluster analysis, individual gene expression values were Z-scored across all samples. Genes were grouped by K-means clustering, and samples were grouped by hierarchical clustering (Spotfire Decisionsite For Functional Genomics).
Statistical Analysis.
Levels of cell-free HIV-1 virions in the supernatants of each culture condition within the viremic patient group were compared by Student's t test for paired samples. Levels of virions produced by anti-CD3 antibody-stimulated cells and levels of HIV-1 proviral DNA and cell-associated HIV-1 RNA in resting CD4+ T cells of the two patient groups were compared by Student's two-sample t test. The data were log-normally distributed and were logged for analysis. The Bonferroni method was used to adjust P values for multiple testing.
Results
Quantitation of Cell-Free HIV-1 Virions Released by Latently Infected, Resting CD4+ T Cells from Viremic and Aviremic Patients.
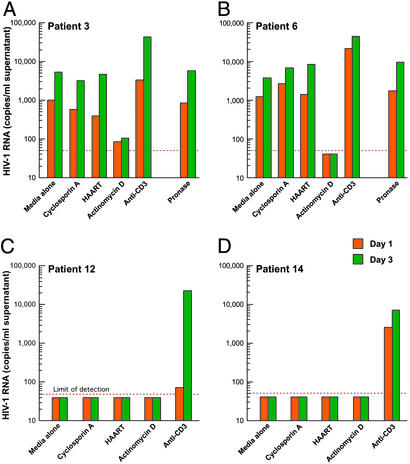
To investigate whether the latent viral reservoir is capable of producing cell-free HIV-1, 11 viremic and 12 aviremic HIV-1-infected individuals were studied. The term “aviremia” is defined as level of plasma viremia that is below the limit of detection measured by ultrasensitive bDNA assay (50 copies of HIV-1 RNA per ml of plasma). The clinical profiles of the patients are shown in Table 1. Resting CD4+ T cells were isolated from PBMC of HIV-1-infected individuals by using a column-based cell separation technique (StemCell Technologies), as described (14). The purity of isolated CD4+ T cells lacking activation markers (CD25−/CD69−/HLA-DR−) was consistently >99.8%, as measured by FACS analysis. Highly purified resting CD4+ T cells were incubated ex vivo for a short period in the absence of activating stimuli with or without inhibitors of cellular proliferation and viral replication. Cell-free supernatants collected from each culture at day 1 and 3 were subjected to Amplicor HIV-1 Monitor Test (detection limit: 50 copies of HIV-1 RNA per ml) for quantitation of HIV-1. As shown in Fig. 1, resting CD4+ T cells from representative viremic patients 3 and 6 released measurable amounts of cell-free HIV-1 in the absence of activating stimuli after 24-h incubation (Fig. 1 A and B). Given that, during the short culture period, purified resting CD4+ T cells maintained a viability of >99%, as determined by trypan blue exclusion, it is highly unlikely that detection of cell-free HIV-1 in the supernatants was caused by release of HIV-1 RNA upon cell death. In addition, the high-speed centrifugation performed in the ultrasensitive version of Amplicor HIV-1 Monitor Test precludes the precipitation of naked HIV-1 RNA. The levels of HIV-1 released by resting CD4+ T cells were not significantly affected by the presence of inhibitors of cellular proliferation (cyclosporin A) and viral replication (in vitro HAART); however, as expected, actinomycin D, an inhibitor of both cellular and viral RNA synthesis, significantly reduced the number of virions released by the latent viral reservoir. Of note, the viability of resting CD4+ T cells that were incubated with actinomycin D overnight was greater than 90%, as determined by trypan blue exclusion. To rule out detection of cell-bound HIV-1 virions that were later released into the culture supernatants, resting CD4+ T cells were pretreated with pronase before incubation ex vivo (Fig. 1 A and B). Given that pronase treatment did not abrogate production of HIV-1 virions, and that further incubation (up to 3 days in culture) of resting CD4+ T cells resulted in higher levels of cell-free HIV-1 without expression of activation markers (data not shown), these results clearly demonstrate that the latent viral reservoir in viremic patients is capable of producing HIV-1 in the absence of activating stimuli. In contrast, resting CD4+ T cells from representative aviremic patients 12 and 14, who were receiving HAART, did not produce cell-free HIV-1 in the absence or presence of inhibitors of cellular proliferation and viral replication (Fig. 1 C and D). When resting CD4+ T cells were stimulated by anti-CD3 antibody as a positive control, HIV-1 were detected in the culture supernatants in both patients, confirming that these patients carried an inducible latent viral reservoir that was capable of producing HIV-1 when appropriately stimulated.
Figure 1.
Quantitation of cell-free HIV-1 virions released by latently infected, resting CD4+ T cells from representative viremic (A and B) and aviremic (C and D) patients. Cell-free supernatants collected from each culture at days 1 and 3 were subjected to Amplicor HIV-1 test (detection limit: 50 copies of HIV-1 RNA per ml) for quantitation of HIV-1 virions.
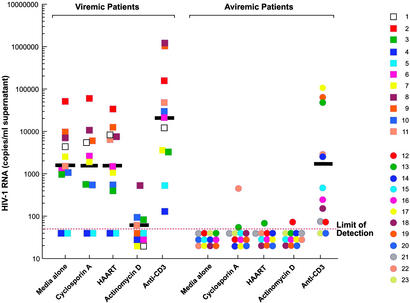
Resting CD4+ T cells from 11 viremic and 12 aviremic patients were subjected to the above culture conditions followed by quantitation of cell-free HIV-1 in the culture supernatants at day 1. As shown in Fig. 2, cell-free HIV-1 produced by resting CD4+ T cells were detected in 9 of 11 viremic patients (82%) in the absence of activating stimuli. Given that, on average, only 0.02% of activated CD4+ T cells harbor integrated HIV-1 DNA (8), and that the vast majority of these cells carry defective virus, it is highly unlikely that cell-free virions released by purified resting CD4+ T cells originated from a small number of CD4+ T cells expressing activation markers. Of note, a population of resting CD4+ T cells obtained by FACS (based on both size and phenotype, >99.9% purity) from a viremic patient also produced a substantial level of cell-free virions (data not shown). The levels of HIV-1 in the culture supernatants were not reduced by cyclosporin A (P > 0.5) or in vitro HAART (P > 0.5); however, actinomycin D significantly decreased the number of virions produced by resting CD4+ T cells (P = 0.003). Resting CD4+ T cells from the majority of aviremic patients (9 of 12 patients, 75%) did not produce any cell-free HIV-1, and those that did produce virus did so at very low levels. Inducible HIV-1 virions were detected in 10 of 12 (83%) patients when cells were stimulated with anti-CD3 antibody. Of note, the levels of virus that were released into the supernatants after stimulation of resting CD4+ T cells by anti-CD3 antibody were not significantly different between the two patient groups (P = 0.06), suggesting that the latent viral reservoir in all patients that were examined carried comparable levels of inducible HIV-1.
Figure 2.
Levels of cell-free HIV-1 virions released by latently infected, resting CD4+ T cells from viremic and aviremic patients. Cell-free supernatants from each culture harvested at day 1 were subjected to Amplicor HIV-1 test (detection limit: 50 copies of HIV-1 RNA per ml). The geometric mean values are shown as black bars.
Frequency of Latently Infected, Resting CD4+ T Cells Carrying HIV-1 Proviral DNA and Cell-Associated, Unspliced HIV-1 RNA from Viremic and Aviremic Patients.
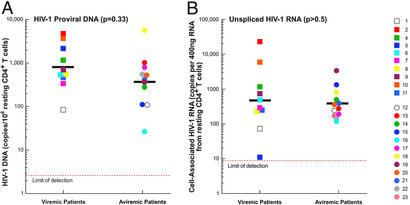
To rule out the possibility that the inability of resting CD4+ T cells to produce virus in the majority of aviremic patients was caused by the absence of HIV-1 in these resting cells, the frequencies of resting CD4+ T cells carrying HIV-1 proviral DNA and cell-associated, unspliced HIV-1 RNA were determined in viremic and aviremic individuals. Freshly isolated, resting CD4+ T cells were subjected to real-time PCR for detection of HIV-1 proviral DNA and cell-associated HIV-1 RNA. The geometric mean frequencies of resting CD4+ T cells carrying HIV-1 proviral DNA from viremic (870 copies per 106 resting CD4+ T cells) vs. aviremic (380 copies per 106 resting CD4+ T cells) patients were not statistically different (P = 0.33; Fig. 3A). In addition, the geometric mean frequencies of resting CD4+ T cells carrying cell-associated, unspliced HIV-1 RNA from viremic (487 copies per 400 ng of RNA from resting CD4+ T cells) vs. aviremic (382 copies per 400 ng of RNA from resting CD4+ T cells) patients were not statistically different (P > 0. 5; Fig. 3B). These results strongly suggest that the absence of virion production by resting CD4+ T cells from the majority of aviremic patients was not caused by the lack of intracellular HIV-1 present in this latent viral reservoir.
Figure 3.
Frequency of latently infected, resting CD4+ T cells carrying HIV-1 proviral DNA (A) and cell-associated, unspliced HIV-1 RNA (B) from viremic and aviremic patients. Freshly isolated, resting CD4+ T cells were subjected to real-time PCR for detection of HIV-1 proviral DNA and cell-associated HIV-1 RNA. The geometric mean values are shown as black bars.
DNA Microarray Analysis of Resting CD4+ T Cells from Viremic and Aviremic Patients and Healthy HIV-Negative Donors.
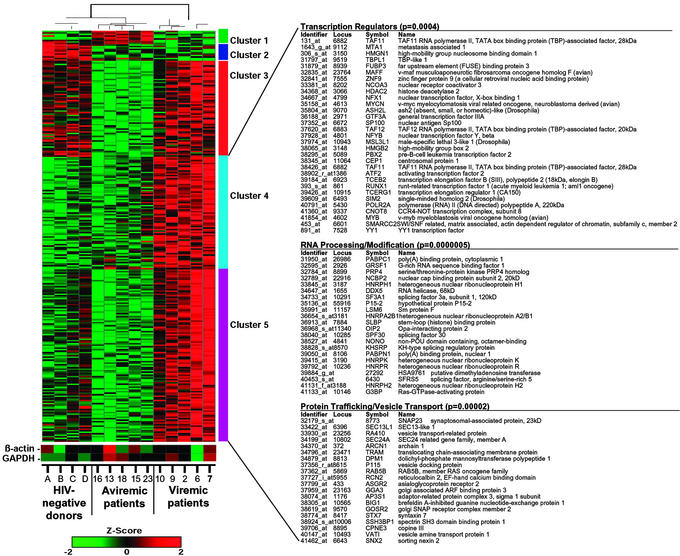
In an attempt to delineate the mechanism(s) by which the latent viral reservoir of viremic patients supported production of cell-free HIV-1 whereas that of aviremic patients did not, we performed a DNA microarray analysis by using total RNA isolated from fresh resting CD4+ T cells of five viremic, five aviremic patients, and four healthy HIV-negative donors. By using Affymetrix Human Genome U95A oligonucleotide arrays consisting of probes encompassing 12,600 genes and a significant analysis of microarrays (SAM) algorithm (15), 535 differentially expressed genes were identified. The corresponding genes and samples from the above donors were grouped by using K-means clustering and hierarchical clustering, respectively (Fig. 4). The hierarchical clustering analysis demonstrated that the transcriptional profile of resting CD4+ T cells of aviremic patients is more closely related to that of HIV-negative donors but more distantly related to that of viremic patients (Fig. 4). Clusters 4 and 5 indicate a set of 370 host genes that were significantly up-regulated in resting CD4+ T cells of viremic patients relative to those of aviremic patients and HIV-negative donors. Statistical analysis (Fisher exact test) identified genes associated with transcription regulators (P = 0.0004), RNA processing/modification (P = 0.0000005), and protein trafficking/vesicle transport (P = 0.00002) as the top three functional categories enriched with genes from clusters 4 and 5 relative to the frequency of all annotated genes on the oligonucleotide arrays. To rule out that a small number of contaminating activated CD4+ T cells may have contributed to the patterns of up-regulated host gene expression found in the cells of viremic patients, we conducted an independent DNA microarray analysis in which increasing percentages of activated CD4+ T cells were added to a fixed number of resting CD4+ T cells. It took severalfold more than 1% of contaminating activated CD4+ T cells to give rise to any detectable levels of activation-related gene expression (data not shown). In addition, the majority of the up-regulated genes in activated CD4+ T cells was related to cell cycle regulation/progression and cellular proliferation; only 2.9% of these genes (4 of 139) were also shown to be up-regulated in resting CD4+ T cells of viremic patients relative to those of aviremic patients and HIV-negative donors. Taken together, these data strongly suggest that the up-regulated genes seen in the resting CD4+ T cell population of viremic patients did not originate from contaminating activated CD4+ T cells.
Figure 4.
Clustering of differentially expressed genes in resting CD4+ T cells of viremic patients, aviremic patients, and healthy HIV-negative donors. Transcription profiles of resting cells from five viremic patients, five aviremic patients, and four HIV-negative donors were examined by using high-density microarrays. Levels of gene expression were assayed on Affymetrix U95A chips, and 535 differentially expressed genes were identified. Genes were grouped by using K-means clustering, and samples were grouped by hierarchical clustering. Clusters 4 and 5 indicate a set of 370 genes that were up-regulated in viremic patients relative to both aviremic and healthy individuals. Statistical analysis (Fisher exact test) of the genes in clusters 4 and 5 determined that the top three functional categories, transcription regulators, RNA processing/modification, and protein trafficking/vesicle, were overrepresented in this list of genes relative to all annotated genes on the chip. Differences in relative levels of gene expression (Z-score) are indicated in color, where red indicates up-regulation and green indicates down-regulation. Some genes were listed twice because certain probes recognize multiple regions of a single gene. Levels of expression of housekeeping genes are also shown.
The levels of gene expression in the above descriptive categories also strongly correlated with the plasma viral loads of the patients we examined (r > 0.90, P < 0.0004, data not shown). These results suggest that active viral replication in vivo may lead directly or indirectly to up-regulation of a number of host genes in the resting CD4+ T cell compartment, including the latent viral reservoir, which, in turn, may provide enough metabolic energy for completion of viral assembly and release of cell-free HIV-1 in the absence of expression of cell-surface activation markers.
Discussion
The persistence of HIV-1 in latently infected, resting CD4+ T cells has been clearly demonstrated in infected individuals receiving HAART and in whom plasma viremia has fallen below the limit of detection (2–4). Given that this viral reservoir has posed one of the major obstacles to the achievement of eradication of HIV-1 in infected individuals receiving clinically successful HAART, intensive investigation has been aimed at further elucidating the role of this latently infected reservoir in the pathogenesis of HIV-1 disease. In particular, contrary to previous speculations that the latent viral reservoir does not spontaneously release cell-free HIV-1 in the absence of exogenous activating stimuli (7, 8), recent studies have demonstrated that resting CD4+ T cells lacking cell-surface activation markers support viral replication in simian immunodeficiency virus (SIV)-infected monkeys (17) as well as productive infection of resting CD4+ T cells in the microenvironment of lymphoid tissues ex vivo (18). In the present study, we clearly demonstrate that the latent viral reservoir of resting CD4+ T cells of viremic patients supports production of cell-free HIV-1 in the absence of activating stimuli, and that inhibitors of cellular proliferation and antiretroviral drugs do not effectively block this process. We also demonstrate that the latent viral reservoir of resting CD4+ T cells of the vast majority of aviremic patients receiving HAART, despite the fact that they seem to be phenotypically identical to resting CD4+ T cells of viremic patients by classical activation markers, do not produce detectable levels of cell-free HIV-1. Of note, resting CD4+ T cells from 2 of 11 infected individuals who had been receiving HAART for a relatively short period (≤1 month) with detectable plasma viremia continued to spontaneously produce relatively low levels of cell-free HIV-1 (<1,055 copies of HIV-1 RNA per ml) in the absence of activating stimuli. This observation corroborates the study in which sustained expression of SIV RNA in CD4+ T cells lacking any activation markers was demonstrated in infected monkeys shortly after initiation of HAART in which undetectable levels of plasma viremia may not have been achieved (17). Given that resting CD4+ T cells from viremic patients spontaneously produced detectable levels of cell-free HIV-1, whereas those from aviremic patients did not, despite the presence of comparable levels of HIV-1 proviral DNA, cell-associated HIV-1 RNA, and inducible virus upon cellular stimulation, we speculated that the latently infected resting CD4+ T cell reservoir in patients with active viral replication may differ physiologically from that of patients in whom plasma viremia is maximally suppressed by effective HAART.
It has been clear for some time that HIV disease is characterized by aberrant immune activation, and that this state of cellular activation drives viral replication. In turn, active viral replication and the presence of plasma viremia directly or indirectly induces and maintains a state of enhanced cellular activation, creating a vicious cycle that has an important impact on the pathogenesis of HIV-1 (19, 20). Although active viral replication has recently been implicated in increased proliferation and death of CD4+ T cells of HIV-1-infected individuals (21, 22), it has been unclear whether the presence of viremia impacts the latently infected viral reservoir that phenotypically seems to be in the resting state, as determined by classicalmarkers of cellular activation. In this regard, the DNA microarray analysis in the present study demonstrates clearly that the presence of active viral replication, manifested by detectable plasma viremia, has a significant impact on the latent viral reservoir of resting CD4+ T cells and is associated with the enhancement of expression of certain host genes in this cellular compartment despite the absence of expression of classical cell-surface activation markers. Some of the genes that were up-regulated in resting CD4+ T cells of viremic patients relative to those of aviremic patients and HIV-negative donors include those involved in early events of signal transduction (PIK3CD, PIK4CB, Lck, PAK2, MAPK1, and MAP3K11) and several transcription factors that modulate levels of HIV transcription (CMYB, YY1, TFCP2 and RUNX1). Lck signaling events can be linked to RNA processing and translation via the Lck-mediated phosphorylation of heterogeneous nuclear ribonucleoprotein K, a multifunctional protein that acts at multiple tiers of gene expression including transcription, translation, and RNA processing (23). The levels of four other heterogeneous nuclear ribonucleoproteins, including HNRPA2B1, HNRPH1, HNRPH2, and HNRPR, were also up-regulated in resting CD4+ T cells of viremic patients relative to those of aviremic and HIV-negative donors. HNRPR interacts with poly(A)-binding protein 1 (PABP1) to stabilize mRNA and stimulate cap-dependent and independent translation initiation in the cytoplasm (24). PABP1 (cytoplasmic form) and PABP2 (nuclear form), which are involved in the mRNA trafficking, translation, and stability, were expressed at higher levels in viremic patients relative to those of aviremic and HIV-negative donors. Several recent studies have implicated ubiquitination and the endosome-sorting pathway in the transport of retroviral proteins, including HIV Gag, to the budding site of the plasma membrane (25–28). Numerous genes specifically up-regulated in viremic patients were found to be involved in the various steps of protein/vesicle transport including ER and Golgi proteins (ARCN1, DPM1, EXT2, EXTL3, FUT8, PPGB, RCN2, SSH3BP1), vesicle coat-associated proteins (AP3S1, GGA3, GOSR2, KIAA0905, P115, RA410, RAB5B, SEC24A, TRAM, VAT1), endo/exocytosis related proteins (ASGR2, BIG1, KIAA0254, KIAA0528, KIAA1067, PIK4CB, SNAP23, SNX2, STX7), and ubiquitination-related proteins (APC10, NCUBE1, SMURF2, TRIP12, UBE2D2, UREB1, USP14, VDU1), strongly suggesting that enhanced activities in secretory pathways of resting CD4+ T cells of viremic patients may aid in the release of viral particles in the absence of exogenous activation stimuli. Taken together, the up-regulation of these host genes in the resting CD4+ T cells from viremic patients may provide a favorable environment and sufficient metabolic energy for completion of the HIV-1 replication cycle and release of cell-free virions through enhancement of cellular transcriptional machinery and facilitation of protein trafficking and vesicle transport. However, in the setting of effective antiviral therapy and, thus, in the absence of the immune activation caused by active viral replication, latently infected, resting CD4+ T cells may lack sufficient levels of appropriate cellular gene expression to support production of HIV-1 virions. Thus, the presence of active virus replication, in addition to its well documented deleterious effects on the infected host, may have subtle, but important, effects on the propagation and evolution of the resting CD4+ T cell reservoir, in the absence of obvious phenotypic changes in this cellular compartment. Finally, given that this study clearly demonstrates that the latent viral reservoir from the majority of aviremic patients did not produce quantifiable cell-free virions despite the presence of cell-associated HIV-1 RNA, transcription of HIV-1 RNA in resting CD4+ T cells, which cannot be blocked by currently available antiviral drugs, should not necessarily be taken as evidence for ongoing, clinically relevant viral replication during effective therapy.
Acknowledgments
We thank Claudia Cicala for helpful discussions and Joseph Adelsberger and Catherine Watkins for technical assistance. We also thank the patients for their participation in this study as well as the staff of the National Institute of Allergy and Infectious Diseases HIV Clinic for their invaluable assistance in the execution of this study.
Abbreviation
- HAART
highly active antiretroviral therapy
References
- 1.Centers for Disease Control and Prevention. HIV/AIDS Surv Rep. 2000;12:1–44. [Google Scholar]
- 2.Chun T-W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, et al. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 4.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 5.Ho D D. Science. 1998;280:1866–1867. doi: 10.1126/science.280.5371.1866. [DOI] [PubMed] [Google Scholar]
- 6.Chun T-W, Fauci A S. Proc Natl Acad Sci USA. 1999;96:10958–10961. doi: 10.1073/pnas.96.20.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun T-W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano R F. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 8.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, et al. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 9.Natarajan V, Bosche M, Metcalf J A, Ward D J, Lane H C, Kovacs J A. Lancet. 1999;353:119–120. doi: 10.1016/s0140-6736(05)76156-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, et al. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 11.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 12.Patterson B K, McCallister S, Schutz M, Siegel J N, Shults K, Flener Z, Landay A. AIDS. 2001;15:1635–1641. doi: 10.1097/00002030-200109070-00005. [DOI] [PubMed] [Google Scholar]
- 13.Zhu T, Muthui D, Holte S, Nickle D, Feng F, Brodie S, Hwangbo Y, Mullins J I, Corey L. J Virol. 2002;76:707–716. doi: 10.1128/JVI.76.2.707-716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun T-W, Engel D, Berrey M M, Shea T, Corey L, Fauci A S. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Wong W H. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus K A, Reimann K A, Reinhart T A, Rogan M, Cavert W, Miller C J, et al. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 18.Eckstein D A, Penn M L, Korin Y D, Scripture-Adams D D, Zack J A, Kreisberg J F, Roederer M, Sherman M P, Chin P S, Goldsmith M A. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 19.Ascher M S, Sheppard H W. Clin Exp Immunol. 1988;73:165–167. [PMC free article] [PubMed] [Google Scholar]
- 20.Fauci A S. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 21.Mohri H, Perelson A S, Tung K, Ribeiro R M, Ramratnam B, Markowitz M, Kost R, Hurley A, Weinberger L, Cesar D, et al. J Exp Med. 2001;194:1277–1287. doi: 10.1084/jem.194.9.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovacs J A, Lempicki R A, Sidorov I A, Adelsberger J W, Herpin B, Metcalf J A, Sereti I, Polis M A, Davey R T, Tavel J, et al. J Exp Med. 2001;194:1731–1741. doi: 10.1084/jem.194.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrowski J, Schullery D S, Denisenko O N, Higaki Y, Watts J, Aebersold R, Stempka L, Gschwendt M, Bomsztyk K. J Biol Chem. 2000;275:3619–3628. doi: 10.1074/jbc.275.5.3619. [DOI] [PubMed] [Google Scholar]
- 24.Grosset C, Chen C Y, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu A B. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 25.VerPlank L, Bouamr F, LaGrassa T J, Agresta B, Kikonyogo A, Leis J, Carter C A. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez O D, Nolan G P. Immunity. 2001;15:687–690. doi: 10.1016/s1074-7613(01)00238-2. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Serrano J, Zang T, Bieniasz P D. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 28.Carter C A. Trends Microbiol. 2002;10:203–205. doi: 10.1016/s0966-842x(02)02350-8. [DOI] [PubMed] [Google Scholar]