Abstract
To better understand the function of the conserved C terminus of the cystic fibrosis (CF) transmembrane conductance regulator, we studied constructs containing deletions in the C-terminal tail. When expressed in well differentiated CF airway epithelia, each construct localized predominantly to the apical membrane and generated transepithelial Cl− current. The results suggested that neither the C-terminal PSD-95/Discs-large/ZO-1 (PDZ)-interacting motif nor other C-terminal sequences were absolutely required for apical expression in airway epithelia. Surprisingly, deleting an acidic cluster near the C terminus reduced both channel opening rate and transepithelial Cl− transport, indicating that it influences channel gating. These results may help explain the relative paucity of CF-associated mutations in the C terminus.
Conservation of sequence at the C terminus of cystic fibrosis (CF) transmembrane conductance regulator (CFTR) suggests this domain may serve a significant function (1–3). Clinical observations in humans support this conclusion. Deletion of 26 C-terminal residues by the S1455X mutation was associated with elevated sweat Cl− concentrations, but not other manifestations of CF (4), whereas deletion of the last 70 residues by the Q1412X mutation caused CF (C. J. Taylor, personal communication). Although earlier studies have shown anion transport in channels missing portions of the C terminus (2, 5), several studies have identified sequences and regions of the C terminus that influence the localization, stability, and function of CFTR. The most extensively studied region has been the extreme C terminus, which contains the PSD-95/discs-large/ZO-1 (PDZ)-interacting motif DTRL (single-letter amino acid code) (Fig. 1A). This sequence binds several proteins that contain PDZ domains, including NHERF (also known as EBP50), CAP70, and CAL (6–10). Interactions between the DTRL sequence and PDZ proteins may influence CFTR in several ways. An interaction with NHERF or other PDZ proteins may be required for the normal apical localization of CFTR (11–14). Interactions between the Golgi-associated CAL and the CFTR PDZ-interacting motif favored retention of CFTR within the cell (10). And interactions between the DTRL sequence and either NHERF or CAP70 may increase Cl− channel activity (9, 15).
Figure 1.
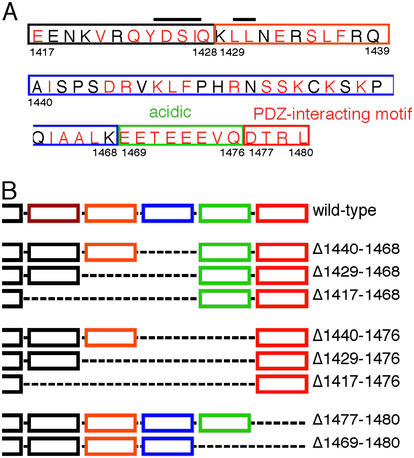
The CFTR C terminus. (A) Sequence from 1,417 to 1,480. Residues that are identical or conserved among species are shown in red; nonconserved are in black. The tyrosine-based and di-leucine-based endocytosis motifs are overlined. The PDZ-interacting motif is boxed in red, the acidic cluster in green, and internal deletions in other colors. (B) Schematic representation of the constructs studied.
Other C-terminal sequences also play a role. A tyrosine-based internalization motif (Fig. 1A) interacts with the endocytic adaptor complex AP-2 and mediates entry via clathrin-coated vesicles (3). Mutating this motif reduced CFTR endocytosis and increased cell surface expression (16, 17). In addition, a di-leucine-based motif may contribute to endocytosis (18). The C terminus also influences protein stability. Deleting 70 or more C-terminal residues accelerated CFTR turnover by destabilizing the mature plasma membrane protein and enhancing its proteasome-dependent degradation (19–21). In contrast, deleting 26 or 41 residues did not accelerate turnover. Some C-terminal deletions also interfered with normal protein biosynthesis; e.g., a hydrophobic patch at residues 1,413–1,416 was essential for normal CFTR biosynthesis (18, 21, 22).
Although sequences in the CFTR C terminus may confer a number of important properties, the consequences of deleting these sequences on protein localization and function remain uncertain, especially in epithelia that normally express CFTR. Therefore, in this study we examined the contribution of the C terminus to CFTR function in airway epithelia where the loss of CFTR is the major cause of CF morbidity and mortality (23). We were also interested in using C-terminal truncations to develop gene transfer vectors as a potential treatment for CF airway disease. Although adeno-associated virus vectors offer several potential advantages for CF gene therapy (24), they are limited by the small size of transgene that can be accommodated. A study testing insert size suggested that 4,100–4,900 bp is the optimal genome size for packaging (25). In comparison, the coding sequence of full-length CFTR is 4,450 bp (1), and inclusion of the adenovirus-associated virus inverted terminal repeats, promoter-enhancer elements, and untranslated regions can easily exceed the adenovirus-associated virus-packaging capacity. One approach to circumventing this limitation is to shorten all inserted elements, including selective deletion of the CFTR-coding sequence (5, 26, 27). Therefore, we also examined C-terminal deletions with an eye toward developing constructs that might be useful for gene transfer to the CF airway.
Experimental Procedures
Construction of CFTR Variants.
CFTR deletion variants were made by PCR deletion mutagenesis (QuikChange Mutagenesis, Stratagene) as described (27). Deletions were confirmed by sequencing. Variants are named by the residues that are deleted; e.g., in Δ1,440–1,468, residues between and including residues 1,440 and 1,468 are deleted. We produced adenovirus vectors encoding each construct driven by a cytomegalovirus promoter/enhancer. An identical adenovirus expressing GFP served as a negative control.
Well Differentiated CF Airway Epithelia and Other Cells.
Primary cultures of human airway epithelia were obtained from CF bronchus and cultured at the air-liquid interface as described (28). Epithelia were used at least 14 days after seeding when they developed a well-differentiated, pseudostratified epithelium consisting of ciliated, goblet, and other nonciliated cells. Epithelia were infected with an adenovirus multiplicity of infection of 200 by using 5 mM EGTA in H2O applied to the apical surface to transiently disrupt the tight junctions (27). For patch-clamp studies, variants were expressed in HeLa cells by using vaccinia virus-T7 RNA polymerase expression systems (29) or the adenovirus constructs. For biochemical studies, H441 cells were used as described (30).
Protein Biochemistry.
To confirm protein size and phosphorylation, HeLa cells were infected with a 200 multiplicity of infection of adenovirus vector for 45 min. Cells were lysed 18–24 h later, CFTR-immunoprecipitated with anti-CFTR [13-1 (anti-R domain), R & D Systems], and phosphorylated with [γ-32P]ATP and the catalytic subunit of protein kinase A as described (31). For pulse–chase studies, H441 cells were methionine starved and labeled with [35S]methionine for 30 min, and pulse–chase studies were carried out as described (31). Proteins were separated on 6% SDS/PAGE, and counts in bands B and C were quantitated by phosphorimaging.
Immunocytochemistry.
Three days after gene transfer, epithelia were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, blocked with 5% normal goat serum (Jackson ImmunoResearch) in SuperBlock (Pierce), and stained with anti-CFTR (13-1) and anti-ZO-1 [polyclonal (Zymed) or monoclonal (Transduction Laboratories, Lexington, KY)] primary Abs. Epithelia were then stained with Alexa Fluor-conjugated secondary Abs (goat anti-mouse A488 or A568 and goat anti-rabbit A568, Molecular Probes). As controls, epithelia were also infected with adenovirus expressing GFP and GFP fused to the coxsackie and adenovirus receptor (32). Epithelia were examined by confocal laser scanning microscopy.
Patch–Clamp Studies.
Excised, inside-out membrane patches were studied as described (33). The pipette (extracellular) solution contained (in mM) 140 N-methyl-d-glucamine, 2 MgCl2, 5 CaCl2, 100 l-aspartic acid, and 10 Hepes (pH 7.3) with HCl. The bath (intracellular) solution contained (in mM) 140 N-methyl-d-glucamine, 3 MgCl2, 1 cesium EGTA (CsEGTA), and 10 Hepes (pH 7.3) with HCl. Experiments were performed at 23–26°C. Channels were studied in the presence of 80 units/ml protein kinase A catalytic subunit (Promega) and 1 mM MgATP on the cytoplasmic surface. Holding voltage was −100 mV. Data were low-pass filtered at 700 Hz with an eight-pole Bessel filter and digitized at 7 kHz. Single channel analysis was performed as described (33) with a burst delimiter of 20 ms. Events ≤3 ms duration were ignored. To test statistical significance, we used a one-way ANOVA in combination with a multiple comparison procedure (Dunnett′s method) (sigmastat statistical software; SPSS Science Software, Erkrath, Germany). P < 0.05 was considered statistically significant.
Ussing Chamber Studies.
Three days after gene transfer, short-circuit current (Isc) was measured in symmetrical solutions containing (in mM): 135 NaCl, 1.2 MgCl2, 1.2 CaCl2, 2.4 K2PO4, 0.6 KH2PO4, 5 dextrose, and 5 Hepes (pH 7.4), as described (27). After measuring baseline current, we sequentially added the following agents (28): (i) Mucosal amiloride (10−4 M) to inhibit apical Na+ channels, hyperpolarize the apical membrane, and thereby generate a driving force for a Cl− secretory Isc. (ii) Mucosal 4,4′-diisothiocyanotostilbene-2,2′-disulfonic acid (DIDS, 10−4 M) to inhibit DIDS-sensitive apical Cl− channels. (iii) The cAMP agonists mucosal forskolin (10−5 M) and 3-isobutyl-2-methylxanthine (IBMX, 10−4 M) to activate CFTR. (iv) Then, we added submucosal bumetanide (10−4 M) to inhibit basolateral Cl− cotransport. Under these conditions, bumetanide-sensitive Isc provides a good assessment of CFTR-dependent transepithelial Cl− transport (28).
Results
C-Terminal Deletion Constructs.
Because earlier studies indicated that deletions extending N-terminal of residue 1,416 destabilized the protein or interfered with its biosynthesis (18–21), we studied the C terminus between residues 1,417 and 1,480. We focused on five parts of this sequence (Fig. 1A): (i) The extreme C terminus (1,477–1,480). This sequence is a PDZ-interacting motif. (ii) An acidic cluster (1,469–1,476). This sequence has a very negative charge and is highly conserved across species (2, 3). (iii) Residues 1,440–1,468. These residues are only moderately conserved across species and are not found in other ABC transporters (22). (iv) Residues 1,429–1,439. This sequence contains a di-leucine-based endocytosis motif (18). (v) Residues 1,417–1,428. This cluster contains the YDSI endocytosis motif (16, 17). We generated constructs that deleted combinations of these five regions (Fig. 1B). Infecting HeLa cells with each of the constructs produced approximately equivalent amounts of protein of the predicted size; each was recognized by CFTR Abs and phosphorylated in vitro by protein kinase A (not shown).
Biosynthesis and Stability.
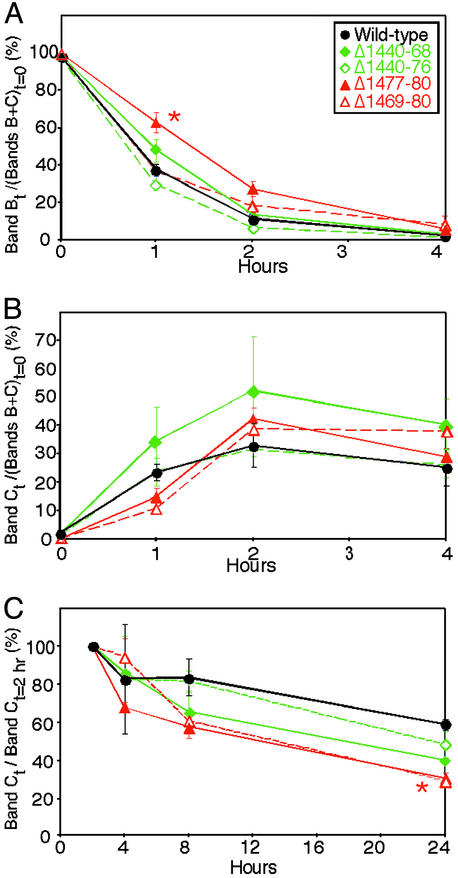
Previous work showed that C-terminal deletions of <65–70 residues did not disrupt CFTR biosynthesis (2, 18–22). To test the effect of shorter and internal deletions, we metabolically labeled several variants and used a pulse–chase analysis to track protein maturation and degradation in the airway cell line H441. The immature band B form of the variant proteins disappeared at approximately the same rate as WT; an exception was Δ1,477–1,480, which showed a delayed disappearance at 1 h (Fig. 2A). The mature band C protein appeared at approximately the same rate for all of the constructs (Fig. 2B). However 24 h after labeling, the amount of Δ1,477–1,480 and Δ1,469–1,480 was reduced compared to WT (Fig. 2C). These results suggest that loss of the PDZ-interacting motif may have altered stability of the mature protein.
Figure 2.
Pulse–chase analysis of WT and variant CFTR. (A) Disappearance of band B. Data are ratio of counts in band B at time t vs. counts in bands B plus C immediately after the pulse (t = 0). *, Δ1,477–1,480 was different from WT at 1 h, P < 0.05. (B) Appearance of band C. Data are ratio of counts in band C at time t vs. counts in bands B plus C at t = 0. (C) Time-dependent disappearance of the mature band C. Data are shown relative to counts 2 h after the pulse to ensure that the majority of the protein had been converted to the mature band C form. *, Δ1,477–1,480 and Δ1,469–1,480 were different from WT at 24 h, P < 0.05. Data are mean ± SEM. n = 3–4.
Immunolocalization.
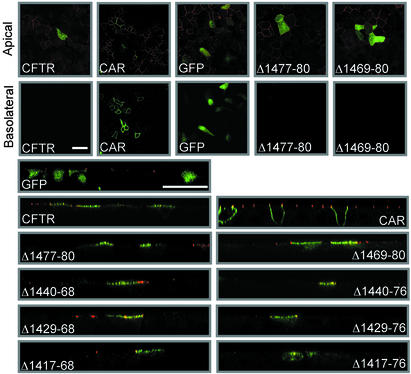
In airway epithelia, CFTR localizes at the apical membrane (34). To learn whether the C-terminal deletions altered apical targeting, we studied well-differentiated primary cultures of CF airway epithelia (28). To provide a landmark for the boundary between apical and basolateral membranes, we immunostained the tight junction-associated protein ZO-1. As controls, we expressed GFP, which showed a diffuse cytoplasmic expression pattern, and GFP fused to the coxsackie and adenovirus receptor (32), which was expressed in the basolateral membrane (Fig. 3).
Figure 3.
Immunostaining of differentiated CF airway epithelia expressing WT CFTR and the deletion constructs. Immunostaining of CFTR is green, and fluorescence for GFP and GFP-tagged coxsackie and adenovirus receptor (CAR) is green. Immunostaining for ZO-1 is in red. We used a mouse monoclonal anti-ZO-1 Ab in combination with the GFP-tagged CAR and a rabbit polyclonal anti-ZO-1 Ab in the other panels. The anti-ZO-1 mAb gave brighter and more specific immunostaining then the polyclonal Ab. Figures in the top row are X–Y confocal images taken at the level of apical membrane. The second row shows X–Y images taken through the basolateral membrane. Panels in the lower part of the figure are X–Z images with apical membrane at the top and filter support at the bottom. (Bars = 25 μm.)
With each C-terminal deletion, the majority of CFTR immunostaining appeared in the apical membrane. When only the PDZ-interacting motif was removed (Δ1,477–1,480), the protein localized at the apical membrane in a pattern that appeared identical to WT CFTR (Fig. 3). Internal deletions beginning at residues 1,440, 1,429, or 1,417 and ending at residue 1,468 did not change this pattern. When in addition to these deletions, we also removed the acidic cluster, the majority of the protein still localized to the apical surface, but staining also revealed some intracellular CFTR. None of the variants showed a pattern consistent with a basolateral membrane localization. These data suggest that no one portion of the C terminus is required for apical expression in airway epithelia. However the acidic cluster may influence one or more of the many mechanisms that determine the relative distribution of apical vs. intracellular protein.
Single-Channel Properties.
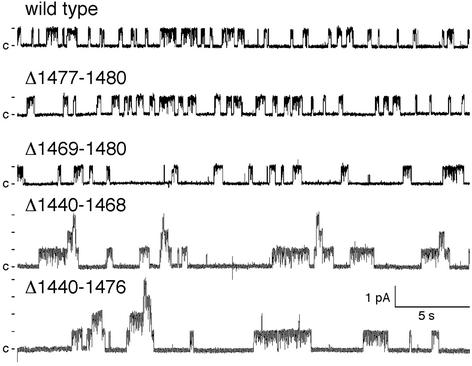
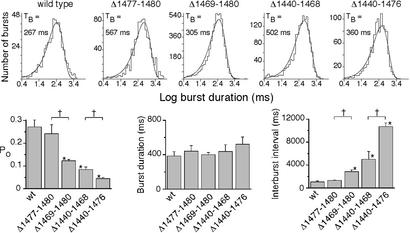
Despite the potential importance of the C terminus, few studies have examined the single-channel function of C-terminal deletions. A study in planar lipid bilayers reported little effect on open-state probability (Po) of D1425X or shorter deletions (22). In addition, whole-cell patch-clamp studies of BHK-21 and IB3–1 cells reported that the S1455X mutation had no effect on current, but the Q1412X mutation reduced current by 90% (4, 19). In cell-attached patches, the F1437X deletion was reported to increase open and closed times (5). We found that removing the four C-terminal residues (Δ1,477–1,480) did not alter Po (Figs. 4 and 5). However, the additional removal of the acidic cluster (Δ1,469–1,480) reduced Po by one-half. A longer deletion that retained the acidic cluster and the extreme C terminus (Δ1,440–1,468) reduced Po to one-third that of WT. When we combined that deletion with removal of the acidic cluster (Δ1,440–1,476), Po was again reduced by one-half. None of the deletions altered burst duration (Fig. 5 A and B) or single-channel conductance (not shown). Instead, prolongation of the closed interburst interval caused the fall in Po. Thus both an internal region contained within residues 1,440–1,468 and the acidic cluster (1,469–1,476) were involved in channel opening.
Figure 4.
Representative current traces from WT CFTR and indicated variants in excised, inside-out membrane patches. For purposes of illustration, the traces were low-pass filtered at 100 Hz. Patches contained one channel for WT and Δ1,477–1,480, two for Δ1,469–1,480, four for Δ1,440–1,468, and five for Δ1,440–1,476.
Figure 5.
Influence of C-terminal deletions on CFTR gating. (Upper) Burst duration histograms obtained from experiments shown in Fig. 4. (Lower) Single-channel properties from multiple patches. Data are means ± SEM from three to five patches. *, Value significantly different from WT (P < 0.05). +, Bracketed values are different from each other (P < 0.05). TB indicates mean burst duration.
Transepithelial Cl− Transport.
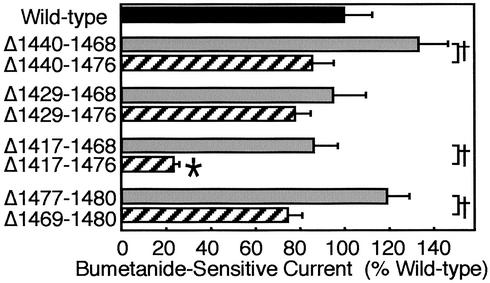
We expressed WT and variant CFTR constructs in airway epithelia, a site where CFTR is normally expressed. To avoid interference from endogenous WT CFTR, we used CF epithelia. We measured bumetanide-sensitive Isc, which represents CFTR-dependent transepithelial Cl− transport under the conditions used. All variants generated Cl− current (Fig. 6). Because we tested the constructs in CF epithelia obtained from multiple donors, we normalized Isc generated by the variants to that of epithelia expressing GFP (the negative control) or WT CFTR (the positive control). We made three general observations with the internal deletions. First, the magnitude of current generated by the variants was similar to that of WT CFTR, with the exception of Δ1,417–1,476. Second, deleting the acidic cluster tended to reduce the Cl− current compared to constructs in which it was retained; e.g., compare Δ1,440–1,468 to Δ1,440–1,476, and so on. Third, in general the larger the internal deletion, the less the Cl− current. Taken together, these data indicate that, with the possible exception of Δ1,417–1,476, the internal deletions did not completely disrupt biosynthetic processing, apical localization, or Cl− channel function. They also point to the acidic cluster as a sequence with previously unrecognized importance for CFTR function.
Figure 6.
Bumetanide-sensitive Isc in CF airway epithelia expressing constructs shown in Fig. 1. Epithelia were treated as described in Experimental Procedures. Data are change in current produced by bumetanide addition, corrected for current in GFP-expressing epithelia and normalized to current generated by WT CFTR. Data are mean ± SEM. n = 6–12 for each construct. *, P < 0.05 (by ANOVA) compared to WT; +, P < 0.05 (by unpaired t test) for the indicated pairs. Bumetanide-sensitive Isc for WT CFTR was 10.5 ± 0.5 μA/cm2.
In addition to the internal deletions, we studied deletions of the PDZ-interacting motif alone (Δ1,477–1,480) or together with the acidic cluster (Δ1,469–1,480) (Fig. 6). The Δ1,477–1,480 construct generated as much Cl− current as WT. This result suggests that loss of the PDZ-interacting motif did not redistribute CFTR from a predominantly apical location to a mixed apical and basolateral location; otherwise the Cl− current would have fallen. However, Δ1,469–1,480 generated less current than Δ1,477–1,480, suggesting that the acidic cluster influenced current.
Discussion
Several recent studies have examined the contribution of the C terminus to CFTR biosynthesis, membrane targeting, and function. In assessing the role played by the C terminus, our study has several advantages. We studied CFTR variants in airway epithelia, a tissue where CFTR is normally expressed in the apical membrane. Protein trafficking and membrane localization can be cell-type specific. For example, in Fischer rat thyroid epithelia, CFTR localizes to both apical and basolateral membranes (35), and in Madin–Darby canine kidney (MDCK) epithelia the ratio of apical:basolateral CFTR is estimated at 2:1 to 7.5:1 (11, 14). We also studied CFTR with no epitope tags that might alter its biology. Our study also had limitations. We expressed CFTR at a much higher level than endogenous CFTR, which is expressed at very low levels in airway epithelia (34, 36). However, if overexpression had altered targeting, we would have expected to see more intracellular or basolateral protein instead of the selective apical staining we observed. Another limitation is that our studies of protein biosynthesis and our single-channel studies were not performed in differentiated airway epithelia.
Preserved Biosynthesis of CFTR with C-Terminal Deletions.
Our data in H441 cells suggest that the internal and C-terminal deletions had minor effects on protein biosynthesis. However, deleting the PDZ-interacting motif accelerated degradation of the mature protein; this small effect presumably reflects loss of an interaction with a PDZ domain protein. These findings are consistent with previous reports that maturation and degradation of C-terminally truncated CFTR (S1455X and D1425X) is similar to WT in COS and BHK cells (19–21). In MDCK cells, removing the last three amino acids reduced the half-life of GFP-CFTR in the apical membrane (14). Earlier studies showed that mutating the phenylalanine-based (1,424–1,427) and di-leucine-based (1,430–1,431) motifs reduced endocytosis (17, 18). Our functional studies suggest that any alteration in biosynthesis, stability, or endocytosis had a relatively small effect in airway epithelia. Alternatively, opposing influences may have counterbalanced each other, e.g., reduced single-channel activity may have counterbalanced the deletion of endocytosis motifs. Addressing these possibilities may require time course studies with labeling of apical and basolateral proteins; such studies in primary cultures of airway epithelia are not practical at this time.
Targeting of CFTR with C-Terminal Deletions to the Apical Membrane.
Deleting the PDZ-interacting motif did not alter the apical localization of CFTR as detected by immunocytochemistry. Although removing the acidic cluster occasionally led to some intracellular staining, most immunostaining remained apical. None of the deletions produced immunostaining in a basolateral membrane pattern. The transepithelial transport studies agreed well with an apical localization. Under the conditions we used, Cl− is transported from basolateral to apical surface if CFTR is expressed apically. Deleting residues 1,477–1,480 did not reduce Isc. If disrupting a PDZ domain interaction had shifted the channel to the basolateral membrane exclusively, Cl− secretory current would have been abolished. If CFTR had redistributed equally to apical and basolateral membranes, Isc should have been cut in one-half. Thus, both immunostaining and functional data suggest that the C terminus, and hence an interaction with NHERF, CAP70, CAL, or some other PDZ protein (6–10) is not absolutely required for apical localization. These results suggest that other portions of the protein also contribute to its apical targeting, at least in primary cultures of differentiated airway epithelia.
Previous work examined CFTR with GFP fused to the N terminus; when GFP-CFTR was expressed in canine kidney epithelia (MDCK), the PDZ-interacting motif was required for apical localization (11–14). In addition, GFP fused to a peptide containing residues 1,370–1,480 localized apically in MDCK cells (13), although residues other than those at the C terminus also contributed to apical targeting. Those data contrast with our results. However, in pig kidney epithelia (LLCPK1), CFTR with a C-terminal VSV-G epitope, which disrupts the PDZ-interacting motif, was targeted to the apical membrane (37). Deleting the C terminus also produced different functional effects in MDCK cells, where it abolished transepithelial Cl− transport (12), and in airway epithelia, where it had no effect on Cl− current. There are several potential explanations for these differences in apical targeting. As noted above, different cell types might target CFTR to different sites and/or use different cellular targeting processes. In addition, epitopes fused to proteins might influence localization or processing. Moreover, as described above, the level of protein expression might influence localization. Future studies may be able to shed light on these and other alternative possibilities.
Retention of Transepithelial Cl− Current in CFTR with C-Terminal Deletions.
Transepithelial Cl− current is influenced by single-channel conductance, Po, and the number of CFTR channels at the apical and basolateral membranes. Despite the reduced Po caused by some deletions, the number of CFTR Cl− channels at the apical membrane was sufficient to generate WT levels of current with all but the shortest construct. A caveat in interpreting these studies is that basolateral membrane transport processes also influence transepithelial transport under the conditions we used. Thus if basolateral transport was the rate-limiting step in Cl− secretion, we could have missed the functional consequences of small changes in the number or activity of apical CFTR channels.
Role of the Acidic Cluster in Controlling CFTR Gating.
Our data revealed an unexpected role for the acidic cluster in regulating CFTR gating; removing this sequence reduced Po and CFTR-dependent Isc. Although the responsible mechanisms are not known, they appear different from effects previously attributed to the C and N termini. For example, previous data suggested that binding the CFTR PDZ-interacting motif to CAP70 or NHERF increased in Po (9, 15). However, that interaction influenced burst duration, whereas deleting the acidic cluster prolonged the interburst interval and left burst duration unaltered. Moreover, deleting the acidic cluster reduced Po in both the presence and the absence of the DTRL sequence. The CFTR N terminus has also been implicated in gating through an interaction with the R domain (38). Mutation of acidic residues in a putative N-terminal α-helix reduced Po, primarily by shortening burst duration (39), rather than by prolonging the interburst interval. Nevertheless, the relationship between the regulation of gating by the C and N termini is interesting and warrants further investigation.
Clusters of acidic residues have been observed in many proteins. In some cases they influence protein expression on the cell surface, an effect that can be mediated by binding to the cytosolic protein PACS-1 (40). Serines and threonines in acidic clusters are also targets for phosphorylation by casein kinase II. For example, in the cation-independent mannose 6-phosphate receptor, casein kinase II phosphorylation of an acidic cluster di-leucine sequence enhances its interaction with GGA adapter proteins (41). In our studies, the acidic cluster was not required for apical targeting, although when it was removed we noted an increase in intracellular protein. Thus, we do not know how the acidic cluster regulates channel gating, but we speculate that this cluster might bind the highly charged R domain via an electrostatic interaction. The effect of the acidic cluster on opening rate (interburst interval) but not burst duration is consistent with the way that other R domain interventions influence gating (42).
Implications for CF.
Because a shortened CFTR transgene might be of value for CF gene transfer, one aim of this work was to determine whether parts of the C terminus could be deleted without altering the normal properties of CFTR. Our data suggest that the acidic cluster should probably be retained because its deletion inhibited channel activity and transepithelial Cl− transport and appeared to increase the amount of intracellular protein. Likewise, the PDZ-interacting motif should probably be retained because its deletion slightly increased the disappearance rate of the mature protein. Moreover, although our data suggest that deleting this region did not alter apical expression in airway epithelia, the work of others has indicated that it can be involved in apical targeting. Deletions between residues 1,417 and 1,468 might be considered for a shortened CFTR transgene because some showed normal transepithelial Cl− current. However, they decreased single-channel function. Thus, in contrast to some regions of the R domain that could be deleted without altering function (27), the C-terminal deletions we studied tended to alter some properties of the protein.
Our finding that many C-terminal deletions retain function in airway epithelia may also help explain clinical observations. There is a relative paucity of mutations in the C terminus (see www.genet.sickkids.on.ca). The S1455X was associated with an elevated sweat Cl− concentration, but not airway or pancreatic disease (4), and the 1476X deletion was found in a person with congenital bilateral absence of the vas deferens (personal communication, M. Claustres, Montpellier, France). In contrast, the much longer Q1412X deletion appeared to cause CF. Nonsense variations in this region are also rare and their relationship to CF remains uncertain (see www.genet.sickkids.on.ca.).
Acknowledgments
We thank Evan Sivesind, Pary Weber, Tamara Nesselhauf, Janice Launspach, Tom Moninger, and Theresa Mayhew for excellent assistance. We thank Allan Berger for help with the statistical analysis. C.R. is supported by a grant of the Deutsche Forschungsgemeinschaft (Ra 682/5–1). M.J.W. is an Investigator of the Howard Hughes Medical Institute. We appreciate valuable assistance from the University of Iowa In Vitro Models and Cell Culture Core [supported by the National Heart, Lung and Blood Institute (NHLBI), the Cystic Fibrosis Foundation (CFF), and the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK, DK54759)], the University of Iowa Gene Transfer Vector Core (supported by the Roy J. Carver Charitable Trust, the NHLBI, CFF, and NIDDK DK54759), the DNA Core Facility (supported by DK25295), and the University of Iowa Central Microscopy Research Facility. This work was supported by the NHLBI, the CFF (RDP and ENGLH9850), and the Howard Hughes Medical Institute.
Abbreviations
- CF
cystic fibrosis
- CFTR
CF transmembrane conductance regulator
- Isc
short-circuit current
- Po
open-state probability
- PDZ
PSD-95/discs-large/ZO-1
- MDCK
Madin–Darby canine kidney
References
- 1.Riordan J R, Rommens J M, Kerem B S, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J L, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Rich D P, Gregory R J, Cheng S H, Smith A E, Welsh M J. Recept Channels. 1993;1:221–232. [PubMed] [Google Scholar]
- 3.Weixel K M, Bradbury N A. J Biol Chem. 2000;275:3655–3660. doi: 10.1074/jbc.275.5.3655. [DOI] [PubMed] [Google Scholar]
- 4.Mickle J E, Macek M J, Fulmer-Smentek S B, Egan M M, Schwiebert E, Guggino W, Moss R, Cutting G R. Hum Mol Genet. 1998;7:729–735. doi: 10.1093/hmg/7.4.729. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Wang D, Fischer H, Fan P D, Widdicombe J H, Kan Y W, Dong J Y. Proc Natl Acad Sci USA. 1998;95:10158–10163. doi: 10.1073/pnas.95.17.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall R A, Ostedgaard L S, Premont R T, Blitzer J T, Rahman N, Welsh M J, Lefkowitz R J. Proc Natl Acad Sci USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Raab R W, Schatz P J, Guggino W B, Li M. FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- 8.Short D B, Trotter K W, Reczek D, Kreda S M, Bretscher A, Boucher R C J, Stutts M J, Milgram S L. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Yue H, Derin R B, Guggino W B, Li M. Cell. 2000;103:169–179. doi: 10.1016/s0092-8674(00)00096-9. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Moyer B D, Milewski M, Loffing J, Ikeda M, Mickle J E, Cutting G R, Li M, Stanton B A, Guggino W B. J Biol Chem. 2002;277:3520–3529. doi: 10.1074/jbc.M110177200. [DOI] [PubMed] [Google Scholar]
- 11.Moyer B D, Denton J, Karlson K H, Reynolds D, Wang S, Mickle J E, Milewski M, Cutting G R, Guggino W B, Li M, et al. J Clin Invest. 1999;104:1353–1361. doi: 10.1172/JCI7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moyer B D, Duhaime M, Shaw C, Denton J, Reynolds D, Karlson K H, Pfeiffer J, Wang S, Mickle J E, Milewski M, et al. J Biol Chem. 2000;275:27069–27074. doi: 10.1074/jbc.M004951200. [DOI] [PubMed] [Google Scholar]
- 13.Milewski M I, Mickle J E, Forrest J K, Stafford D M, Moyer B D, Cheng J, Guggino W B, Stanton B A, Cutting G R. J Cell Sci. 2001;114:719–726. doi: 10.1242/jcs.114.4.719. [DOI] [PubMed] [Google Scholar]
- 14.Swiatecka-Urban A, Duhaime M, Coutermarsh B, Karlson K H, Collawn J, Milewski M, Cutting G R, Guggino W B, Langford G, Stanton B A. J Biol Chem. 2002;277:40099–40105. doi: 10.1074/jbc.M206964200. [DOI] [PubMed] [Google Scholar]
- 15.Raghuram V, Mak D D, Foskett J K. Proc Natl Acad Sci USA. 2001;98:1300–1305. doi: 10.1073/pnas.031538898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prince L S, Peter K, Hatton S R, Zaliauskiene L, Cotlin L F, Clancy J P, Marchase R B, Collawn J F. J Biol Chem. 1999;274:3602–3609. doi: 10.1074/jbc.274.6.3602. [DOI] [PubMed] [Google Scholar]
- 17.Peter K, Varga K, Bebok Z, McNicholas-Bevensee C M, Schwiebert L, Sorscher E J, Schwiebert E M, Collawn J F. J Biol Chem. 2002;277:49952–49957. doi: 10.1074/jbc.M209275200. [DOI] [PubMed] [Google Scholar]
- 18.Hu W, Howard M, Lukacs G L. Biochem J. 2001;354:561–572. doi: 10.1042/0264-6021:3540561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haardt M, Benharouga M, Lechardeur D, Kartner N, Lukacs G L. J Biol Chem. 1999;274:21873–21877. doi: 10.1074/jbc.274.31.21873. [DOI] [PubMed] [Google Scholar]
- 20.Benharouga M, Haardt M, Kartner N, Lukacs G L. J Cell Biol. 2001;153:957–970. doi: 10.1083/jcb.153.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentzsch M, Riordan J R. J Biol Chem. 2001;276:1291–1298. doi: 10.1074/jbc.M003672200. [DOI] [PubMed] [Google Scholar]
- 22.Gentzsch M, Aleksandrov A, Aleksandrov L, Riordan J R. Biochem J. 2002;366:541–548. doi: 10.1042/BJ20020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsh M J, Ramsey B W, Accurso F, Cutting G R. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, Childs B, Vogelstein B, editors. New York: McGraw–Hill; 2001. pp. 5121–5189. [Google Scholar]
- 24.Flotte T R. Curr Opin Mol Ther. 1999;1:510–516. [PubMed] [Google Scholar]
- 25.Dong J Y, Fan P D, Frizzell R A. Hum Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 26.Flotte T R, Afione S A, Solow R, Drumm M L, Markakis D, Guggino W B, Zeitlin P L, Carter B J. J Biol Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- 27.Ostedgaard L S, Zabner J, Vermeer D W, Rokhlina T, Karp P H, Stecenko A A, Randak C, Welsh M J. Proc Natl Acad Sci USA. 2002;99:3093–3098. doi: 10.1073/pnas.261714599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karp P H, Moninger T, Weber S P, Nesselhauf T S, Launspach J, Zabner J, Welsh M J. In: Epithelial Cell Culture Protocols. Wise C, editor. Vol. 188. Totowa, NJ: Humana; 2002. pp. 115–137. [DOI] [PubMed] [Google Scholar]
- 29.Sheppard D N, Ostedgaard L S, Winter M C, Welsh M J. EMBO J. 1995;14:876–883. doi: 10.1002/j.1460-2075.1995.tb07069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas C P, Auerbach S, Stokes J B, Volk K A. Am J Physiol. 1998;274:C1312–C1323. doi: 10.1152/ajpcell.1998.274.5.C1312. [DOI] [PubMed] [Google Scholar]
- 31.Ostedgaard L S, Zeiher B, Welsh M J. J Cell Sci. 1999;112:2091–2098. doi: 10.1242/jcs.112.13.2091. [DOI] [PubMed] [Google Scholar]
- 32. Ashbourne-Excoffon, K. J., Moninger, T. & Zabner, J. (2003) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 33.Carson M R, Travis S M, Welsh M J. J Biol Chem. 1995;270:1711–1717. doi: 10.1074/jbc.270.4.1711. [DOI] [PubMed] [Google Scholar]
- 34.Engelhardt J F, Zepeda M, Cohn J A, Yankaskas J R, Wilson J M. J Clin Invest. 1994;93:737–749. doi: 10.1172/JCI117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheppard D N, Carson M R, Ostedgaard L S, Denning G M, Welsh M J. Am J Physiol. 1994;266:L405–L413. doi: 10.1152/ajplung.1994.266.4.L405. [DOI] [PubMed] [Google Scholar]
- 36.Trapnell B C, Chu C S, Paakko P K, Banks T C, Yoshimura K, Ferrans V J, Chernick M S, Crystal R G. Proc Natl Acad Sci USA. 1991;88:6565–6569. doi: 10.1073/pnas.88.15.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa de Beauregard M A, Edelman A, Chesnoy-Marchais D, Tondelier D, Lapillonne A, El Marjou F, Robine S, Louvard D. Eur J Cell Biol. 2000;79:795–802. doi: 10.1078/0171-9335-00116. [DOI] [PubMed] [Google Scholar]
- 38.Naren A P, Cormet-Boyaka E, Fu J, Villain M, Blalock J E, Quick M W, Kirk K L. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- 39.Fu J, Ji H L, Naren A P, Kirk K L. J Physiol (London) 2001;536:459–470. doi: 10.1111/j.1469-7793.2001.0459c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crump C M, Xiang Y, Thomas L, Gu F, Austin C, Tooze S A, Thomas G. EMBO J. 2001;20:2191–2201. doi: 10.1093/emboj/20.9.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra S, Puertollano R, Kato Y, Bonifacino J S, Hurley J H. Nature. 2002;415:933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 42.Ostedgaard L S, Baldursson O, Welsh M J. J Biol Chem. 2001;276:7689–7692. doi: 10.1074/jbc.R100001200. [DOI] [PubMed] [Google Scholar]