Abstract
Glutamate transport is central to neurotransmitter functions in the brain. Impaired glutamate transport induces neurotoxicity associated with numerous pathological processes, including stroke/ischemia, temporal lobe epilepsy, Alzheimer's disease, amyotrophic lateral sclerosis, Huntington's disease, HIV-1-associated dementia, and growth of malignant gliomas. Excitatory amino acid transporter-2 (EAAT2) is a major glutamate transporter in the brain expressed primarily in astrocytes. We presently describe the cloning and characterization of the human EAAT2 promoter, demonstrating elevated expression in astrocytes. Regulators of EAAT2 transport, both positive and negative, alter EAAT2 transcription, promoter activity, mRNA, and protein. These findings imply that transcriptional processes can regulate EAAT2 expression. Moreover, they raise the intriguing possibility that the EAAT2 promoter may be useful for targeting gene expression in the brain and for identifying molecules capable of modulating glutamate transport that could potentially inhibit, ameliorate, or prevent various neurodegenerative diseases.
Keywords: sequential progressive genomic scanning‖nuclear run-on assays‖promoter reporter assays‖mRNA and protein expression
The amino acid glutamate is the major excitatory neurotransmitter in the mammalian CNS (1). Although compulsory for normal neuronal function and neurotransmission, this excitatory amino acid can accumulate in the extracellular fluid of the CNS, as a consequence of pathologic changes in the brain, thereby promoting neuronal damage and brain injury by a process termed “excitotoxicity” (2, 3). It is well established that the concentration of extracellular glutamate in the CNS is controlled by Na+-dependent transport systems present in astrocytes and neurons and that glutamate taken up by astrocytes is subsequently metabolized by glutamine synthase (1). In this context, glutamate transport represents an important mechanism for maintaining low levels of this neurotransmitter in the extracellular milieu to promote synaptic signaling and to restrict potential neurotoxicity resulting from the excitotoxic property of glutamate (1, 3).
Five excitatory amino acid transporter (EAAT) cDNAs have been identified and cloned (EAAT1–5) (4–6). EAAT2, also identified in the rodent as glutamate transporter-1 (GLT-1), and EAAT1, also known as GLAST (4–6), are the major glutamate transporters in the CNS, accounting for the majority of total glutamate uptake in the brain (7). Although EAAT2 protein is predominantly expressed in astrocytes, neuronal expression of EAAT2 has been observed during development (8), ischemic insult (9), and in neurons in culture (10). Controversy exists relative to the areas in the brain where the EAAT2 protein is expressed, i.e., two semiquantitative studies suggest uniform expression with minimal variations in different brain regions (1, 11), whereas others suggest greater expression (8- to 10-fold) in the forebrain than in the cerebellum (12, 13). Reductions in EAAT2 protein expression have been documented as a function of neuropathology resulting from ischemia (14), in temporal lobe epilepsy (15), Alzheimer's disease (16), Huntington's disease (3), and amyotrophic lateral sclerosis (17, 18). A potential role has also been proposed for astrocyte glutamate transport in HIV-1-related dementia (19). Moreover, malignant gliomas secrete glutamate, which may contribute to tumor expansion (20). These findings emphasize the importance of glutamate transport and the EAAT2 transporter in astrocytes to normal brain function and their association with multiple pathologic changes in the brain.
The mechanisms underlying regulation of EAAT2 and other glutamate transporters are not well defined, but they potentially involve transcriptional and/or posttranscriptional processes (1). To provide insights into the process of glutamate transport and its normal regulation and association with disease, a sequential progressive genomic scanning cloning approach was used to identify and isolate the human EAAT2 promoter. We have characterized regions in the promoter controlling expression in astrocytes and defined biochemical pathways involved in modulating human EAAT2 promoter expression and mRNA and protein levels. These studies document the importance of transcriptional processes in regulating EAAT2 expression and consequently EAAT2 protein levels in human astrocytes.
Materials and Methods
Primary Cell Cultures, Cell Lines, and Reagents.
Primary normal human fetal astrocytes (PHFA) were isolated from second trimester (gestational age 16–19 weeks) human fetal brains and cultured as described (21–23). Early passage primary human mammary epithelial and prostate epithelial cells were obtained (Clonetics, San Diego) and cultured as described (24, 25). Simian virus 40-immortalized normal human foreskin melanocyte cells (FM516-SV) and HO-1 human melanoma cells were cultured as described (25, 26). HCN-2, HCN-1A, DU-145, MCF-7, Colo 205, and PANC-1 cells were from the American Type Culture Collection and maintained as described (25). A normal cerebellum cell line was established and grown as described (27). Culture media and cells were tested for mycoplasma contamination with the Mycoplasma PCR ELISA kit (Roche Molecular Biochemicals) and only negative cultures were used. Epidermal growth factor (EGF), transforming growth factor α (TGF-α), and tumor necrosis factor α (TNF-α) were from Invitrogen; dibutyryl cAMP (dBcAMP), bromo-cAMP, AG1478, pyrrolidinedithiocarbamate (PDTC), and PD98059 were from Sigma; and KT5720 and wortmanin were from Calbiochem.
EAAT2 Promoter (EAAT2-Prom) Isolation.
Nylon filters containing a human genomic bacterial artificial chromosome (BAC) library were screened by using a PCR-amplified 32P-labeled exon 2 EAAT2 (105–605 bp) probe. This screening identified three BAC clones, FBAC-4434 BAC library, plate nos. 354j11, 362h20, and 433n05 (Incyte Genomics, Palo Alto, CA), which contained the EAAT2 second exon with a large intron preceding this sequence. The three BACs were sequenced with T3 and T7 primers to determine the sequences in the proximity of the vector enabling rescreening of the library. This sequencing information permitted the generation of an intervening sequence probe that extended ≈50 kb into the first intron and resulted in the identification of three additional clones. Southern blotting analysis revealed that these three BACs contained the first EAAT2 exon. SacII digestion (2.5 kb) of the BAC clones generated fragments containing the first exon of EAAT2 and the 5′ upstream region. This fragment, designated as p-2426, contained the putative EAAT2-Prom region and part of the first exon.
Primer Extension Analysis and Nuclear Run-On Assays.
Primer extension assays were performed as described (28). A primer with the sequence 5′-TAATCCGCGTCCCGGCTCTCCACGGCGCGCGA-3′ complementary to the 5′ UTR of the EAAT2 cDNA was used for this assay. Nuclear run-on assays were performed as described (29).
Construction of EAAT2-Prom Deletion Mutants and Luciferase Assays.
5′ Deletion mutants of the EAAT2-Prom were made with exonuclease III digestion by using the Erase-A-Base system (Promega) as described for the PEG-3-Prom (28). The FL-EAAT2-Prom and EAAT2-Prom deletion mutants were cloned into the pGL3-basic luciferase reporter vector (Promega), and luciferase reporter assays were performed as described with slight modifications (28, 30). Instead of using Lipofectamine, which was toxic to PHFA, the calcium phosphate precipitation technique was used (31).
Northern and Western Blotting Assays.
Total cellular RNA was isolated by the guanidinium/phenol extraction method, and Northern blotting was performed as described (25, 28, 30). Western blotting assays were performed as described (24).
Results
Cloning of the Human EAAT2-Prom Using the Sequential Progressive Genomic Scanning Cloning Approach and Identification of the EAAT2 Transcription Start Site.
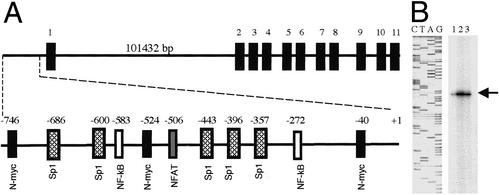
A previous study of EAAT2 structure concluded that the human EAAT2 gene region is composed of 11 exons spanning >50 kb of genomic DNA (32). However, despite the paramount importance of EAAT2 regulation in normal brain function and its potential involvement in multiple neuropathologies, the structure of the EAAT2-Prom or its role in controlling EAAT2 expression remain unknown. Our present studies provide a possible explanation for the difficulties encountered in cloning the EAAT2-Prom. We reanalyzed the EAAT2 genomic region and found that the previously proposed structure of the EAAT2 gene (32) is not correct relative to the 5′ region. Current information in GenBank (accession no. Z32517) contains only a partial sequence of exon 1, consisting of 105 bp. Exon 1 is separated from exon 2 by an intron of ≈100 kb (Fig. 1A). This structure and the small size of the previously identified exon 1 fragment (105 bp) prevented us from using a simple genomic walking approach or a single BAC library screening approach for identifying the putative 5′ region containing the EAAT2 promoter. To clone the EAAT2 promoter, we used a sequential progressive genomic scanning cloning strategy in which nylon filters containing a human genomic BAC library were initially screened by using a PCR-amplified [α-32P]dCTP-labeled EAAT2 exon 2 probe. This screening identified clones containing exon 2 with a large intron preceding this sequence. Additional screening using probes containing part of the sequence of intron 1 identified three clones that contained the sequence of exon 1 and ≈2.5 kb of the 5′ upstream region. Sequence analysis of this putative EAAT2-Prom region revealed that it contains five Sp1 sites and GC-rich repeats, but no TATA box (Fig. 1A). A similar genomic structure is found in the promoter of the ASCT1 gene, which also lacks well-defined cis elements while containing five Sp1 sites and GC-rich repeats (commonly found in early growth response genes, such as those in the EGF family and jun D) (33). Bioinformatic analysis of the promoter region revealed a number of potential regulatory transcription factor-binding elements that may contribute to EAAT2 expression and its regulation, including NFAT, NF-κB, and N-myc (Fig. 1A).
Figure 1.
Schematic of the human EAAT2 gene including intron–exon structure and its promoter (A) and primer extension analysis (B). (A) Exons are indicated by number above bold boxes; location of various binding sites for transcription factors is indicated above the approximate location in the EAAT2-Prom. The size of the first intron is indicated. (B) Defining the transcriptional start site of the EAAT2 cDNA. Lanes 1–3 contain different concentrations of labeled probe (1, 104 cpm; 2, ≈105 cpm; 3, ≈5 × 104 cpm).
To determine the transcriptional initiation site of the EAAT2 gene, a labeled antisense primer was hybridized to total RNA from PHFA, and the extension products were separated on a sequencing gel (Fig. 1B). This process demonstrated that the major transcript is being initiated from an adenosine residue located 283 bp upstream of the ATG start codon. Accordingly, this base was designated as +1 bp and extended the 5′ end of the previously cloned EAAT2 cDNA by 194 bp. These results confirm that the first exon contains 299 bp (the originally reported sequence of 105 bp and an additional 194 bp now identified by primer extension analysis) (Fig. 1).
Human EAAT2-Prom Activity and Deletion Analysis of the EAAT2-Prom.
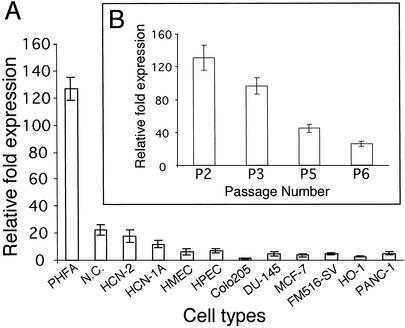
EAAT2 is expressed in brain-derived cells, mainly astrocytes (7, 34). Experiments were performed to confirm EAAT2-Prom activity in normal human astrocytes and to determine expression levels in other cell types (Fig. 2A). Primary early passage and established human normal and tumor cell lines were cotransfected with a putative full-length ≈2.5-kb EAAT2-Prom (p-2426/+44, SacII fragment; cloned into the pGL3-basic vector) (Promega) driving expression of the firefly luciferase (LUC) gene and a pSV-β-galactosidase expression plasmid. Relative-fold activity in the different cell types was determined by normalizing the LUC activity to β-galactosidase expression (Fig. 2A). Highest expression was consistently seen in early passage (nos. 1–3) PHFA. EAAT2-Prom activity was ≈6- to 10-fold higher in PHFA than in the other cell types analyzed (Fig. 2A). With repeated passage, EAAT2-Prom activity decreased ≈3- to 5-fold in PHFA cells by passage no. 5 or no. 6, respectively (Fig. 2B). The EAAT2-Prom displayed reduced activity versus early passage PHFA cells, in a normal adult human cerebellum culture (NC) and in human cortical neuronal cultures (HCN-2, HCN-1A) (Fig. 2A). Low-level EAAT2-Prom activity was also apparent in additional human cells, including human mammary epithelial cells (passage no. 5), human prostate epithelial cells (passage no. 4), FM516-SV, MCF-7, DU-145, PANC-1, HO-1, and Colo 205. Elevated EAAT2-Prom activity was found in one of six gliomas (data not shown). In summary, based on the cells currently studied, the highest activity of the EATT2-Prom was apparent in astrocytes, specifically early passage PHFA. Analysis of EAAT2 expression in multiple tissue Northern blots containing RNA from different regions of the adult human brain indicate that EAAT2-Prom activity may differ in different populations of astrocytes, with highest expression in the cerebral cortex (data not shown).
Figure 2.
Relative expression of the EAAT2-Prom in PHFA and various normal and tumor cell lines. (A) Relative fold expression of a FL-EAAT2-Prom-LUC construct was determined as described (28, 30), and activity versus transfection with a pSV-β-galactosidase plasmid, to equalize for differences in transfection efficiency, was determined (28, 30). (B) Effect of passage number on EAAT2-Prom-LUC activity in PHFA cells.
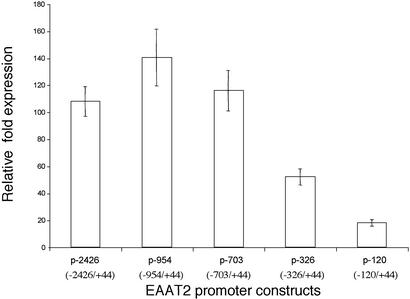
To identify cis-acting elements important for expression of EAAT2, a series of 5′ deletion mutants were constructed and evaluated in PHFA (Fig. 3). Deletion of the most distal region from −2426 to −703 did not alter EAAT2-Prom activity. However, a −703 to −326 deletion reduced promoter activity by ≈1.7-fold versus the putative FL (p-2426) or the p-703 deletion mutant (Fig. 3). Sequence analysis of this deleted region revealed five Sp1 and one NF-κB, N-myc, and NFAT transcription factor-binding site (Fig. 1A). Deletion of the region −326 to −120 further reduced the activity of the EAAT2-Prom by >2-fold (Fig. 3). These results suggest that a putative transcription regulatory element(s) present in this region contributes to basal EAAT2-Prom activity in PHFA.
Figure 3.
Deletion analysis of the EAAT2-Prom. 5′ deletions of the EAAT2-Prom were constructed, and relative fold luciferase activity versus β-galactosidase activity in PHFA was determined as in Fig. 2.
Positive and Negative Regulation of Human EAAT2 Transcription, Promoter Activity, and mRNA Levels in PHFA.
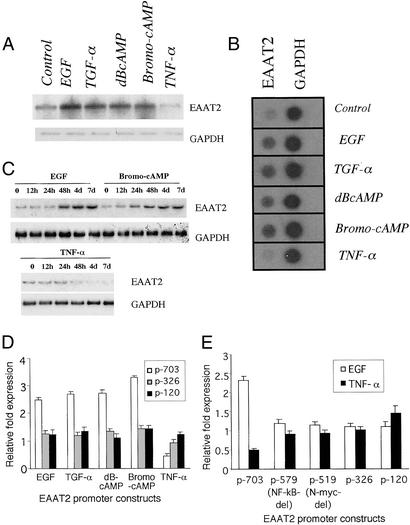
EGF, TGF-α, and dBcAMP enhance GLT-1 expression in rat astrocytes (35, 36). Consistent with observations in rat astrocytes, 7-day treatment with EGF, TGF-α, and two analogs of cAMP, dBcAMP, and bromo-cAMP stimulated EAAT2 mRNA expression in PHFA, whereas TNF-α decreased expression (Fig. 4A). EGF up-regulated EAAT2 mRNA to the highest extent at 48 h, and bromo-cAMP enhanced EAAT2 mRNA expression by 24 h (maximum at 48 h), whereas TNF-α decreased expression by 48 h (Fig. 4C). To examine whether stimulation of EAAT2 expression involves transcriptional changes, nuclear run-on assays were performed. The relative rate of transcription of EAAT2 RNA, as compared with the housekeeping gene GAPDH, was elevated in PHFA after treatment with EGF, TGF-α, dBcAMP, and bromo-cAMP and decreased with TNF-α treatment (Fig. 4B). These data confirm that these regulators of EAAT2 glutamate transporter function exert their effect on steady-state mRNA by altering EAAT2 transcription in PHFA.
Figure 4.
Effect of modulators of EAAT2 activity on EAAT2 expression and EAAT2-Prom deletion mutant activity in PHFA. (A) Northern analysis of EAAT2 and GAPDH mRNA expression after treatment with EGF (30 ng/ml), TGF-α (30 ng/ml), dBcAMP (200 μM), bromo-cAMP (250 μM), or TNF-α (200 units/ml) for 7 days. (B) Nuclear run-on assays determining relative rate of EAAT2 and GAPDH transcription as a function of 7-day treatment with the same concentration of the indicated agents. (C) Time-course mRNA expression of EAAT2 and GAPDH after treatment with EGF, bromo-cAMP, or TNF-α (same concentrations as in A). (D) Effect of deletion mutations in the EAAT2-Prom on basal and modulator-regulated expression. Relative fold expression of the p-703, p-326, and p-120 in untreated and 4-day compound-treated PHFA. (E) Effect of defined mutations in the EAAT2-Prom on basal, EGF, and TNF-α treatment. PHFA were treated for 4 days, and results represent relative fold expression.
To examine further the relationship between EAAT2 and treatment with the various glutamate transporter modulators, transient transfection assays were performed in PHFA with various EAAT2-Prom deletion constructs (Fig. 4 D and E). Four-day treatment of PFHA with EGF, TGF-α, dBcAMP, and bromo-cAMP resulted in ≈1.5- to 3.0-fold up-regulation of EAAT2-Prom activity. Deletion of the region between −2426 and −703 in the EAAT2-Prom did not significantly decrease stimulation by these agents (data not shown). On the other hand, deletion of the region between −703 and −326 in the EAAT2-Prom reduced up-regulation by these agents to a level approximating that found in uninduced PHFA. A further deletion of the EAAT2-Prom sequences between −326 and −120 did not reduce EAAT2-Prom activity as compared with control after treatment with the stimulating agents (Fig. 4D). These data suggest that putative transcription regulatory motifs in the human EAAT2-Prom between −703 and −326 are determinants of elevated EAAT2-Prom activity in PHFA after treatment with the various stimulating agents. These studies emphasize that specific regions of the EAAT2-Prom (located predominantly between −703 to −326) contain important cis-regulatory elements that enhance promoter activity after exposure to EGF, TGF-α, dBcAMP, and bromo-cAMP. In the case of TNF-α, deletion between −326 and −120 of the EAAT2-Prom resulted in promoter activity similar to that found in control untreated PHFA (Fig. 4 D and E).
Several recognized transcription regulatory motifs exist between p-703 and p-120 of the EAAT2-Prom, including NF-κB (−583 and −272), N-myc (−524), and NFAT (−506) (Fig. 1A). A mutant was constructed at p-579 that deletes the NF-κB site at −583. Transfection of p-579 into PHFA resulted in a reduction in basal activity as compared with a p-703 construct (Fig. 4E). In addition, the p-579 mutant lost responsiveness to the inducers, which did not decrease further relative to control in additional mutants lacking N-myc (p-519), NFAT (p-326), and three SP1 sites (p-326) or the second NF-κB site (p-120). In the case of TNF-α, only p-703 showed responsiveness, whereas the other constructs, including p-579, did not show TNF-α-mediated down-regulation of the EAAT2-Prom activity. These results argue that basal activity, as well as EGF-, TGF-α-, dBcAMP- and bromo-cAMP-enhanced activity and TNF-α inhibitory activity of the EAAT2-Prom is regulated by the NF-κB site at position −583. These findings provide a potential explanation of how both stimulators and inhibitors of EAAT2-Prom activity may function to differentially alter EAAT2 gene transcription.
Biochemical Basis for Positive and Negative Regulation of EAAT2 Expression in PHFA.
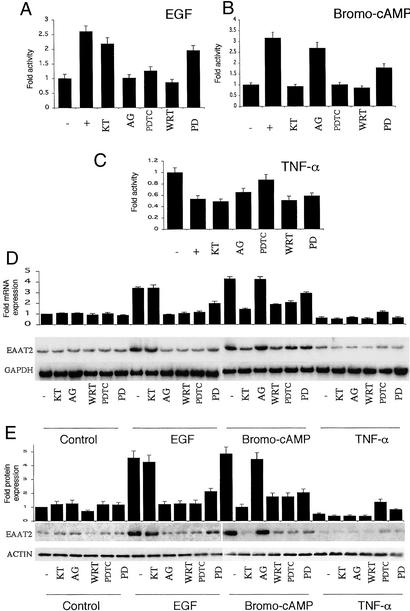
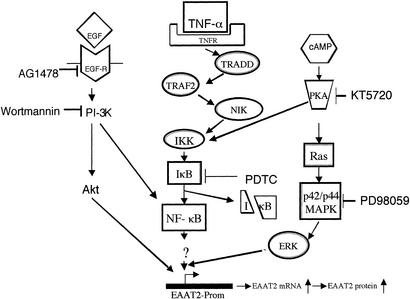
To define the biochemical pathways relevant to the regulation of EAAT2 expression in PHFA resulting from the different treatment protocols, we used a pharmacological approach. This process involved the use of well-characterized pathway-specific inhibitors and determining effects on EAAT2 promoter activity, mRNA levels, and protein levels (Fig. 5). The inhibitors included KT5720 [a protein kinase A (PKA) inhibitor], AG1478 (a tyrosine kinase inhibitor), wortmanin [a phosphatidylinositol 3-kinase (PI-3K) inhibitor], LY294002 (a PI-3K inhibitor) (data not shown), PDTC (an inhibitor of NF-κB activation), and PD98059 (a mitogen-activated kinase inhibitor) (MEK1/MEK2) (36). In the case of EGF (or TGF-α, data not shown), the enhancement of EAAT2-Prom activity and EAAT2 mRNA and protein levels was abolished by AG1478, PDTC, wortmanin, and LY294002, significantly inhibited but not extinguished by PD98059, and unaffected by KT5720 (Fig. 5 A, D, and E). In the case of bromo-cAMP (or dBcAMP, data not shown), the enhancement of EAAT2 expression was eliminated by KT5720, PDTC, wortmanin, and LY294002, partially inhibited by PD98059, and unaffected by AG1478 (Fig. 5 B, D, and E). These findings demonstrate that the various stimulators affect EAAT2 expression through several pathways involving related biochemical changes, both similar and distinct from each other (Fig. 6). Enhancement of human EAAT2 expression by the different stimulators was inhibited by blocking NF-κB activation and PI-3K stimulation and partially inhibited by altering mitogen-activated protein kinase (MEK1/MEK2) activation. In contrast, EGF and TGF-α enhancement of EAAT2 expression involved tyrosine kinase activation and occurred in a PKA-independent manner, whereas stimulation by bromo-cAMP and dBcAMP depended on the PKA pathway but was independent of tyrosine kinase activation (Fig. 6).
Figure 5.
Effect of pharmacological inhibitors on EAAT2-Prom activity, and mRNA and protein expression in PHFA after various treatment protocols. (A–C) EAAT2-Prom activity in PHFA either untreated (−) or treated with EGF (30 ng/ml) (A), bromo-cAMP (250 μM) (B), or TNF-α (200 units/ml) (C) in the absence (−) or presence (+) of KT5720 (5 μM), AG1478 (1 μM), PDTC (100 μM), wortmanin (WRT) (100 nM), or PD98049 (PD) (50 μM). The duration of the assay was 4 days. (D) Effect of the various treatment protocols on EAAT2 and GAPDH mRNA levels. (Upper) Relative EAAT2 RNA expression versus GAPDH expression compared with control untreated or treated samples based on scanning of autoradiograms. (Lower) Actual Northern blots. (E) Effect of the various treatment protocols on EAAT2 and ACTIN protein levels. (Upper) Relative EAAT2 protein expression versus ACTIN expression compared with control untreated or treated samples based on scanning of autoradiograms. Individual experiments were performed three times with triplicate samples, and SD from the mean is presented.
Figure 6.
Schematic of pathways and inhibitors effecting EAAT2-Prom activity. EGF-R, EGF receptor; TNFR, TNF-α receptor; TRADD, TNF receptor-1-associated death domain protein; TRAF2, TNF receptor-associated factor 2; NIK, NF-κB-induced kinase; IκK, I-κB kinase; IκB, inhibitor of NF-κB; ERK, extracellular signal-regulated kinase.
In contrast to the stimulatory effects of EGF, TGF-α, Bromo-cAMP, and dBcAMP on EAAT2 expression, TNF-α decreased EAAT2 expression in PHFA (Figs. 4 and 5). Cotreatment of PHFA with TNF-α and the various pharmacological inhibitors demonstrated that blocking activation of the NF-κB pathway with PDTC was able to restore EAAT2 promoter activity and mRNA and protein levels to that observed in untreated PHFA. A partial restoration of normal levels of EAAT2 protein was also found after treatment with TNF-α in combination with PD98059. The importance of NF-κB activation in the TNF-α inhibitory effect on EAAT2 expression was further documented by using an IκB mutant, which specifically inhibits NF-κB activation (data not shown).
Discussion
Although glutamate excitotoxicity is implicated in numerous CNS abnormalities, including pathological changes associated with head trauma and stroke, and diseases such as amyotrophic lateral sclerosis, Huntington disease, Alzheimer's disease, epilepsy, HIV-1-associated dementia, and immune-mediated damage in multiple sclerosis (3, 14–19, 37–39), the mechanism by which the brain (astrocytes) regulates glutamate transport and prevents glutamate damage to neurons remains to be defined. The present study provides insights into the mechanism by which human astrocytes regulate EAAT2 gene expression and consequently glutamate transport. Evidence is provided indicating that EAAT2 expression, occurring normally in astrocytes and as a consequence of treatment with physiological regulators of glutamate transport, operates through transcriptional processes, thereby resulting in changes in EAAT2 mRNA and protein. Based on these considerations, the EAAT2-Prom may prove amenable for developing high-throughput assays to identify small molecule modulators of glutamate transport. It is possible that these new reagents could afford protection from abnormal physiological alterations in glutamate transport resulting from trauma and stroke as well as those occurring during various disease-related pathological changes in the CNS. These agents could also represent a way of modulating glutamate transport in malignant glioma cells, concomitantly affecting their growth and ability to damage neurons (20). Further studies are required to explore these possibilities.
Traditionally, the astrocyte was considered to play a minor role in neuronal function and in directing overall activities in the brain, providing only a maintenance role in regulating brain homeostasis (2, 34). However, recent studies challenge these assumptions and suggest that rather than being an innocuous bystander, the astrocyte may play a crucial role in regulating neuronal activity and signal transmission; as emphasized previously, deficiencies in these functions may contribute to neurodegeneration (34, 40–42). One way astrocytes wield their effects on neuronal function is through the EAAT2 transporter and its capacity to maintain stimulatory, but nontoxic low levels of free intrasynaptic L-glutamate in the area adjacent to neurons (1–3). Abnormalities in this process result in the accumulation of extracellular glutamate in synaptic clefts and overexcitation and death of neurons in the process of glutamate excitotoxicity (1–3). Additional functions of astrocytes include stimulation of the number of synapses and an enhancement of synaptic efficiency by altering presynaptic and postsynaptic functions in vitro (43, 44). Astrocytes also display excitation properties similar to neurons, including the appearance of functional neuronal nicotinic acetylcholine receptors and Ca2+-dependent glutamate release (44–46). These traits of astrocytes permit intracellular signaling between astrocytes and neurons and may even assist in modulating neuronal signal transmission (44–46). Based on these significant properties of astrocytes and their interrelationship with neurons, factors that adversely affect astrocytic functions can be anticipated to directly influence neuronal function and survival.
Studies designed to comprehend the biochemical processes regulating glutamate transport have focused on rat astrocytes as a model system (36, 47, 48). These investigations indicate that multiple and converging signal transduction pathways affecting astrocyte maturation regulate rodent GLT-1 expression, as monitored by elevations in mRNA and protein levels and consequently glutamate transport (36, 47, 48). In the present study, we now document that similar signaling cascades are involved in regulating human EAAT2 expression in PHFA, and this regulation occurs at a transcriptional level. Expression of EAAT2 mRNA and protein is temporally up-regulated in PHFA cells after treatment with EGF, TGF-α, and cAMP analogs (dBcAMP and bromo-cAMP). Using a series of pharmacological inhibitors of defined biochemical pathways (Fig. 6), we have documented the role of multiple signaling events that impinge on EAAT2-Prom activity in PHFA (Figs. 4 and 5). In the case of EGF and TGF-α, signaling through the EGF receptor and activation of PI-3K and NF-κB are primary mediators of elevated EAAT2 expression. In the case of dBcAMP and bromo-cAMP, signaling through PKA is a major mediator of activity, and regulation of EAAT2 expression is also exerted by PI-3K and NF-κB. Cocultivation of neurons, or neuronal conditioned medium, with rat astrocytes stimulates GLT-1 expression (36). Similarly, rat neuronal conditioned medium also enhances human EAAT2 expression in PHFA (data not shown), suggesting that the human model is behaving in an analogous manner as the rodent astrocytes and factors regulating activity are not species specific. Because the rat GLT-1-Prom was not available and because actinomycin D (which inhibits transcription) was toxic, it was not previously possible to determine the mechanism, i.e., activation of gene transcription or increase in mRNA stability, involved in the increase in mRNA in rat astrocytes after treatment with EGF, TGF-α, and dBcAMP (36). The present studies using nuclear run-on and promoter-based reporter assays demonstrate that these modulators of rat GLT-1 expression can exert their effects in PHFA by altering transcription of the EAAT2 gene. We further document that the mechanism underlying basal, enhanced, and TNF-α-reduced EAAT2-Prom activity in PHFA involves the transcription factor NF-κB (Fig. 4 D and E).
TNF-α inhibits glutamate uptake by PHFA (49). This inhibition of glutamate transport by TNF-α was dose-dependent and very specific, as neutralizing antibody to TNF-α abolished this inhibition and a mAb that is an agonist at the 55-kDa TNF receptor induced inhibition (49). Infection of PHFA by HIV-1 or exposure of the cells to gp120 induces rapid and sustained inhibition of glutamate uptake by astrocytes, and this effect correlates with a decrease in the expression of EAAT2 protein and RNA (D.J.V., unpublished data). Consistent with this effect, exposure of PHFA to HIV-1 or gp120 decreases EAAT2-Prom activity in these cells (Z.-z.S., D.J.V., and P.B.F., unpublished data). These findings suggest that HIV-1, gp120, and other neuropathogenic agents can alter specific signaling pathways in astrocytes in a way that may impair important physiological functions of these cells in neuronal signal transmission and response to brain injury. The mechanism by which HIV-1, gp120, and TNF-α inhibit glutamate uptake in astrocytes is not clear. Our present studies provide insights into the mechanism underlying this process. TNF-α inhibited EAAT2 RNA transcription (nuclear run-on) and promoter activity and decreased the levels of EAAT2 mRNA and protein in PHFA cells (Figs. 4 and 5). This effect could be reversed by simultaneous treatment with PDTC, an inhibitor of NF-κB activation (Fig. 5). Moreover, the stimulatory effect of EGF, TGF-α, dBcAMP, and bromo-cAMP were also inhibited by PDTC, suggesting that NF-κB activation may be a primary contributor, acting both positively and negatively depending on the agent administered, in regulating EAAT2 expression in PHFA. Analysis of specific EAAT2-Prom deletion mutants confirms this possibility and provides further evidence for the importance of NF-κB in regulating EAAT2 expression in PHFA. These results support the hypothesis that TNF-α decreases EAAT2 activity in PHFA by decreasing transcription through NF-κB, steady-state mRNA, and protein levels of EAAT2.
In summary, regulation of extracellular glutamate levels in the brain is critical for normal brain function and is implicated in numerous neurological diseases. Our data provide a perspective into how EAAT2 expression is regulated in human astrocytes. Transcriptional processes control EAAT2 expression in astrocytes as confirmed with agents that can physiologically regulate glutamate transport in astroglial cells. Evidence is presented defining specific regions of the EAAT2-Prom and biochemical pathways, both involving NF-κB, that converge in either elevating or decreasing EAAT2 expression (Fig. 6). These studies suggest that it may now be possible to develop sensitive cell culture-based assays for identifying small molecules and compounds that can regulate glutamate transport in astrocytes. Although further studies are required, specific small molecules and compounds may provide new and effective approaches for preventing and treating diverse pathological changes in the brain.
Acknowledgments
We thank Drs. A. Ghorpade and H. Gendelman for PHFA. This work was supported by National Institutes of Health Grant NS31492, the Samuel Waxman Cancer Research Foundation, and the Chernow Endowment.
Abbreviations
- EAAT
excitatory amino acid transporter
- EAAT2-Prom
EAAT2 promoter
- PHFA
primary normal human fetal astrocytes
- PDTC
pyrrolidinedithiocarbamate
- PKA
protein kinase A
- EGF
epidermal growth factor
- TGF-α
transforming growth factor α
- TNF-α
tumor necrosis factor α
- BAC
bacterial artificial chromosome
- dBcAMP
dibutyryl cAMP
- PI-3K
phosphatidylinositol 3-kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF510107).
References
- 1.Robinson M B. Neurochem Int. 1999;33:479–491. doi: 10.1016/s0197-0186(98)00055-2. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls D, Attwell D. Trends Pharmacol Sci. 1990;11:462–468. doi: 10.1016/0165-6147(90)90129-v. [DOI] [PubMed] [Google Scholar]
- 3.Lipton S A, Rosenberg P A. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- 4.Arriza J L, Fairman W A, Wadiche J I, Murdoch G H, Kavanaugh M P, Amara S G. J Neurosci. 1994;14:5559–5569. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairman W A, Vandenberg R J, Arriza J L, Kavanaugh M P, Amara S G. Nature. 1995;375:599–603. doi: 10.1038/375599a0. [DOI] [PubMed] [Google Scholar]
- 6.Arriza J L, Eliasof S, Kavanaugh M P, Amara S G. Proc Natl Acad Sci USA. 1997;94:4155–4160. doi: 10.1073/pnas.94.8.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, et al. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- 8.Yamada K, Watanabe M, Shibata T, Nagashima M, Tanaka K, Inoue Y. J Neurosci. 1998;18:5706–5713. doi: 10.1523/JNEUROSCI.18-15-05706.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin L J, Brambrink A M, Lehmann C, Portera-Cailliau C, Koehler R, Rothstein J, Traystman R J. Ann Neurol. 1997;42:3335–3348. doi: 10.1002/ana.410420310. [DOI] [PubMed] [Google Scholar]
- 10.Brooks-Kayal A R, Munir M, Jin H, Robinson M B. Neurochem Int. 1998;33:95–100. doi: 10.1016/s0197-0186(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 11.Rothstein J D, Martin L, Levey A I, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl R W. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 12.Lehre K P, Levy L M, Ottersen O P, Storm-Mathisen J, Danbolt N C. J Neurosci. 1995;15:1835–1853. doi: 10.1523/JNEUROSCI.15-03-01835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milton I D, Banner S J, Ince P G, Piggott N H, Fray A E, Thatcher N, Horne C H, Shaw P J. Mol Brain Res. 1997;52:17–31. doi: 10.1016/s0169-328x(97)00233-7. [DOI] [PubMed] [Google Scholar]
- 14.Torp R, Lekieffre D, Levy L M, Haug F M, Danbolt N C, Meldrum B S, Ottersen O P. Exp Brain Res. 1995;103:51–58. doi: 10.1007/BF00241964. [DOI] [PubMed] [Google Scholar]
- 15.Mathern G W, Mendoza D, Lozada A, Pretorius J K, Dehnes Y, Danbolt N C, Nelson N, Leite J P, Chimelli L, Born D E, et al. Neurology. 1999;52:453–472. doi: 10.1212/wnl.52.3.453. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Mallory M, Alford M, Tanaka S, Masliah E. J Neuropathol Exp Neurol. 1997;56:901–911. doi: 10.1097/00005072-199708000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Bruijn L I, Becher M W, Lee M K, Anderson K L, Jenkins N A, Copeland N G, Sisodia S S, Rothstein J D, Borchelt D R, Price D L, et al. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 18.Lin C L, Bristol L A, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein J D. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 19.Kaul M, Garden G A, Lipton S A. Nature. 2001;410:988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- 20.Takano T, Lin J J-C, Arcuino G, Gao Q, Yang J, Nedergaard M. Nat Med. 2001;7:1010–1015. doi: 10.1038/nm0901-1010. [DOI] [PubMed] [Google Scholar]
- 21.Bencheikh M, Bentsman G, Sarkissian N, Canki M, Volsky D J. J Neurovirol. 1999;5:115–124. doi: 10.3109/13550289909021993. [DOI] [PubMed] [Google Scholar]
- 22.Canki M, Thai J N, Chao W, Ghorpade A, Potash M J, Volsky D J. J Virol. 2001;75:7925–7933. doi: 10.1128/JVI.75.17.7925-7933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Z-z, Kang D-c, Chen Y, Pekarskaya O, Chao W, Volsky D J, Fisher P B. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- 24.Su Z-z, Madireddi M T, Lin J J, Young C S H, Kitada S, Reed J C, Goldstein N I, Fisher P B. Proc Natl Acad Sci USA. 1998;95:14400–14405. doi: 10.1073/pnas.95.24.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang E Y, Madireddi M T, Gopalkrishnan R V, Leszczyniecka M, Su Z-z, Lebedeva I V, Kang D-c, Jiang H, Lin J J, Alexandre D, et al. Oncogene. 2001;20:7051–7063. doi: 10.1038/sj.onc.1204897. [DOI] [PubMed] [Google Scholar]
- 26.Lebedeva I V, Su Z-z, Chang Y, Kitada S, Reed J C, Fisher P B. Oncogene. 2002;21:708–718. doi: 10.1038/sj.onc.1205116. [DOI] [PubMed] [Google Scholar]
- 27.Guarini L, Temponi M, Bruce J N, Bollon A P, Duigou G J, Moulton T A, Ferrone S, Fisher P B. Int J Cancer. 1990;46:1041–1047. doi: 10.1002/ijc.2910460616. [DOI] [PubMed] [Google Scholar]
- 28.Su Z-z, Shi Y, Fisher P B. Oncogene. 2000;19:3411–3421. doi: 10.1038/sj.onc.1203666. [DOI] [PubMed] [Google Scholar]
- 29.Su Z-z, Shi Y, Fisher P B. Proc Natl Acad Sci USA. 1997;94:9125–9130. doi: 10.1073/pnas.94.17.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su Z-z, Shi Y, Friedman R, Qiao L, McKinstry R, Hinman D, Dent P, Fisher P B. Nucleic Acids Res. 2001;29:1661–1671. doi: 10.1093/nar/29.8.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babiss L E, Liaw W S, Zimmer S G, Godman G C, Ginsberg H S, Fisher P B. Proc Natl Acad Sci USA. 1986;83:2167–2171. doi: 10.1073/pnas.83.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer T, Munch C, Knappenberger B, Liebau S, Volkel H, Ludolph A C. Neurosci Lett. 1998;241:68–70. doi: 10.1016/s0304-3940(97)00973-7. [DOI] [PubMed] [Google Scholar]
- 33.Gegelashvili G, Schousboe A. Mol Pharmacol. 1997;52:6–15. doi: 10.1124/mol.52.1.6. [DOI] [PubMed] [Google Scholar]
- 34.Anderson C M, Swanson R A. Glia. 2000;32:1–14. [PubMed] [Google Scholar]
- 35.Swanson R A, Liu J, Miller J W, Rothstein J D, Farrell K, Stein B A, Longuemare M C. J Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zelenaia O, Schlag B D, Gochenauer G E, Ganel R, Song W, Beesley J S, Grinspan J B, Rothstein J D, Robinson M B. Mol Pharamacol. 2000;57:667–678. doi: 10.1124/mol.57.4.667. [DOI] [PubMed] [Google Scholar]
- 37.McDonald J W, Althomsons S P, Hyrc K L, Choi D W, Goldberg M P. Nat Med. 1998;4:291–297. doi: 10.1038/nm0398-291. [DOI] [PubMed] [Google Scholar]
- 38.Meldrum B S. J Nutr. 2000;130:1007S–1015S. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- 39.Smith T, Groom A, Zhu B, Turski L. Nat Med. 2000;6:62–66. doi: 10.1038/71548. [DOI] [PubMed] [Google Scholar]
- 40.Carmignoto G. Prog Neurobiol. 2000;62:561–581. doi: 10.1016/s0301-0082(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 41.Haydon P G. Nat Rev Neurosci. 2001;2:185–193. doi: 10.1038/35058528. [DOI] [PubMed] [Google Scholar]
- 42.Trotti D, Rolfs A, Danbolt N C, Brown R H, Jr, Hediger M A. Nat Neurosci. 1999;2:427–433. doi: 10.1038/8091. [DOI] [PubMed] [Google Scholar]
- 43.Oliet S H, Piet R, Poulain D A. Science. 2001;292:872–873. doi: 10.1126/science.1059162. [DOI] [PubMed] [Google Scholar]
- 44.Ullian E M, Sapperstein S K, Christopherson K S, Barres B A. Science. 2001;291:569–570. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- 45.Iino M, Goto K, Kakegawa W, Okado H, Sudo M, Ishiuchi S, Miwa A, Takayasu Y, Saito I, Tsuzuki K, et al. Science. 2001;292:926–929. doi: 10.1126/science.1058827. [DOI] [PubMed] [Google Scholar]
- 46.Sharma G, Vijayaraghavan S. Proc Natl Acad Sci USA. 2001;98:4148–4153. doi: 10.1073/pnas.071540198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gegelashvili G, Danbolt N C, Schousboe A. J Neurochem. 1997;69:2612–2615. doi: 10.1046/j.1471-4159.1997.69062612.x. [DOI] [PubMed] [Google Scholar]
- 48.Schlag B D, Vondrasek J R, Munir M, Kalandadze A, Zelenaia O A, Rothstein J D, Robinson M B. Mol Pharmacol. 1998;53:355–369. doi: 10.1124/mol.53.3.355. [DOI] [PubMed] [Google Scholar]
- 49.Fine S M, Angel R A, Perry S W, Epstein L G, Rothstein J D, Dewhurst S, Gelbard H A. J Biol Chem. 1996;271:15303–15306. doi: 10.1074/jbc.271.26.15303. [DOI] [PubMed] [Google Scholar]