Abstract
Streptococcus pneumoniae is one of the leading causes of invasive bacterial disease worldwide. Fragments of the cell wall and the cytolytic toxin pneumolysin have been shown to contribute substantially to inflammatory damage, although the interactions between pneumococcal components and host-cell structures have not been elucidated completely. Results of a previous study indicated that cell-wall components of pneumococci are recognized by Toll-like receptor (TLR)2 but suggested that pneumolysin induces inflammatory events independently of this receptor. In this study we tested the hypothesis that pneumolysin interacts with surface proteins of the TLR family other than TLR2. We found that pneumolysin stimulates tumor necrosis factor-α and IL-6 release in wild-type macrophages but not in macrophages from mice with a targeted deletion of the cytoplasmic TLR-adapter molecule myeloid differentiation factor 88, suggesting the involvement of the TLRs in pneumolysin recognition. Purified pneumolysin synergistically activated macrophage responses together with preparations of pneumococcal cell walls or staphylococcal peptidoglycan, which are known to activate TLR2. Furthermore, when compared with wild-type macrophages, macrophages from mice that carry a spontaneous mutation in TLR4 (P712H) were hyporesponsive to both pneumolysin alone and the combination of pneumolysin with pneumococcal cell walls. Finally, these TLR4-mutant mice were significantly more susceptible to lethal infection after intranasal colonization with pneumolysin-positive pneumococci than were control mice. We conclude that the interaction of pneumolysin with TLR4 is critically involved in the innate immune response to pneumococcus.
Infections by Streptococcus pneumoniae (pneumococcus) are among the leading causes of mortality from infectious diseases, claiming ≈10 million lives yearly worldwide including at least 1 million children in developing countries (1). In the United States, pneumococcus causes 50,000 cases of bacteremia and 3,000 cases of meningitis annually (2, 3). Faced with one of the most frequent nasopharyngeal colonizers of children, the host immune response can be assumed to play an important role in preventing the progression from colonization to invasive pneumococcal disease. At the same time, the morbidity and mortality associated with pneumococcal infections have also been ascribed to the host response. Indeed, the induction of cytokines by cellular components of pneumococci has been demonstrated to cause important damage to the host, particularly in experimental models of CNS infection (4, 5). Furthermore, interference with the inflammatory response to CNS invasion by this pathogen has been shown to attenuate the neurologic damage observed in experimentally infected rodents with pneumococcal meningitis (6).
There is abundant evidence that endotoxin, or lipopolysaccharide (LPS), is the predominant mediator of inflammatory responses to Gram-negative bacteria through its interaction with Toll-like receptor (TLR)4 (6–9) and MD2 (10–12). In contrast, despite the propensity of certain Gram-positive bacteria to cause the sepsis syndrome, an inflammatory mediator in Gram-positive bacteria that is equivalent in potency to LPS is yet to be identified. Several components of Gram-positive bacteria (peptidoglycan of Staphylococcus aureus, pneumococcal whole cells, and lipoteichoic acid) have been demonstrated to induce inflammatory responses, predominantly via TLR2 (13–18). These molecules, however, seem to have much weaker proinflammatory potential than LPS. The precise mechanisms through which the clinical presentation of severe Gram-positive infections such as pneumococcal septicemia can mimic that seen in typical Gram-negative shock or endotoxemia therefore remain to be explained.
Pneumolysin is a product of S. pneumoniae that contributes to virulence in experimental infection models and has numerous inflammatory effects on eukaryotic cells (19–23). This toxin, similar to other members of the cholesterol-dependent cytolysin family, is thought to bind to cholesterol in cell membranes, forming oligomers and creating transmembrane pores (24). Pneumolysin also interferes with specific functions of cells of the immune system such as the respiratory burst in polymorphonuclear leukocytes, chemotaxis, and bactericidal activity (19, 25). Pneumolysin can activate the classical pathway of complement (26), contributing to its proinflammatory characteristics. Furthermore, pneumolysin induces the release of tumor necrosis factor α (TNF-α) and nitric oxide from human polymorphonuclear leukocytes (21) and to be a potent inducer of apoptosis in cells of the CNS (27).
Because these inflammatory and proapoptotic properties have been commonly associated with molecules that interact with TLRs, we investigated whether pneumolysin engages and signals through a TLR. We found that the inflammatory response of macrophages to pneumolysin depends on TLR4. Furthermore, mutant mice that lack functional TLR4 are significantly more susceptible to invasive disease and death after exposure to pneumolysin-producing pneumococci when compared with control mice. Our results implicate the interaction of pneumolysin with TLR4 as an important protective component of the host response to pneumococcus.
Materials and Methods
Reagents.
PBS, Ham's F-12, RPMI medium 1640, DMEM, and trypsin-EDTA were from BioWhittaker. Low-endotoxin FBS was from HyClone. Ciprofloxacin was a gift from Miles Pharmaceuticals (West Haven, CT). Hygromycin B was purchased from Calbiochem, and G418 was from GIBCO/BRL. Native pneumolysin was purified according to published methods (28). PdT is a mutant of pneumolysin with undetectable cytotoxicity and no complement-activating properties by virtue of three amino acid substitutions (Trp-433 → Phe, Asp-385 → Asn, and Cys-428 → Gly) and was purified as described (22). Heat-treated LPS, pneumolysin, and PdT were prepared by heating aqueous stock suspensions in PBS (for LPS) and 50% glycerol/water (vol/vol) (for pneumolysin and PdT) in a boiling-water bath for 60 min. Protein-free LPS from Escherichia coli K235 was a gift from S. Vogel (University of Maryland, Baltimore), and soluble peptidoglycan from S. aureus was donated by Roman Dziarski (Indiana University, Indianapolis). The peptidoglycan preparation was found to be free of LPS contamination by the use of a Chinese hamster ovary (CHO)-reporter cell line [3E10 (29)] that is sensitive to LPS contamination as low as 10 pg/ml. Endotoxin-neutralizing protein-coated beads were purchased from Associates of Cape Cod. The synthetic lipoprotein analogs, consisting of palmitylated N-acyl-S-diacylglyceryl cysteine and a few COOH-terminal amino acids (Pam3Cys-Ser-Lys-4), were obtained from G. Jung (University of Tübingen, Tübingen, Germany).
Cell Lines.
The CHO/CD14.ELAM.tac reporter cell line (clone 3E10) is stably transfected with human CD14 and expresses inducible human CD25 under the transcriptional control of the nuclear factor κB (NF-κB)-dependent human E-selectin promoter (29). The LPS nonresponder CHO/CD14 cell line, clone 7.19, carries a point mutation resulting in a C95Y amino acid substitution for MD2 (11, 29). Human embryonic kidney (HEK) cell lines that are stably transfected with CD14, TLR2, and TLR4 have been described (30).
DNA Isolation and Manipulation.
Plasmid DNA was isolated by using either the Qiagen (Valencia, CA) midiprep or miniprep kit according to manufacturer recommendations. Chromosomal DNA from S. pneumoniae was isolated as described (31). Restriction endonuclease digestions, DNA ligations, agarose gel electrophoresis, and Southern hybridizations (ECL, Amersham Pharmacia) were performed by using standard techniques.
Bacterial Strains.
S. pneumoniae serotype-2 strain D39 and its isogenic pneumolysin-negative mutant PLN-A were as described (20). Strains were grown in modified chemically defined medium (32). D39 and PLN-A were grown without shaking at 37°C with 5% CO2 to midlog phase (A650, 0.3), harvested by centrifugation, killed in 70% ethanol (vol/vol) for 1 h on ice, washed again, and resuspended in LPS-free PBS. To ensure that the bacterial preparations represented equal amounts of bacteria, colony-forming units (cfu) were determined before killing, and dry weights of the final preparations were measured.
S. pneumoniae type-3 strain WU2 (33) and WU2-PLA, a pneumolysin-negative isogenic mutant of WU2, were used for animal experiments. The pneumolysin-negative mutant was constructed by insertion-duplication mutagenesis. Briefly, a 240-bp region within the pneumolysin gene (plyA) was amplified by using primers Ply601BamHI (5′-GATCGGATCCGCTAAGTGGCATCAAGAT-3′) and Ply841KpnI (5′-GATCGGTACCAGTATCTTGAAACACATC-3′) and ligated into pGEM-T cloning vector (Promega). The 240-bp pneumolysin fragment was released from pGEM-T by KpnI/BamHI restriction digestion, ligated into KpnI/BamHI-digested suicide vector pJY4164 (34), and transformed into E. coli strain DH5α. The resulting plasmid pJY4164-ply was transformed into S. pneumoniae strain WU2 by using competence-stimulating peptide 2 (as described in ref. 35; synthesized by the Biopolymer Facility at Harvard University) at a concentration of 100 ng/ml by using protocols as described (36). Colonies that were resistant to erythromycin (0.3 μg/ml) were screened for pneumolysin production by measuring the hemolytic activity of supernatants from overnight cultures in Todd–Hewitt medium supplemented with 0.5% yeast extract. Candidate mutants that lacked any hemolytic activity then were assessed by PCR and Southern hybridization to confirm that the pneumolysin gene had been disrupted. One clone, WU2-PLA, was subsequently selected for animal experiments. There was no detectable difference in growth rate between WU2-PLA and the parent strain. For bioluminescence experiments, strain A66.1 Xen 10 (a gift of Carolyn Bellinger-Kawahara, Xenogen, Alameda, CA) was used. This strain was derived from strain A66.1 (serotype 3) and stably transformed with a Gram-positive lux transposon as described (37).
Isolation of Peritoneal Macrophages.
Myeloid differentiation factor 88 (MyD88)−/− (38) and TLR4−/− (7) mice were a gift from S. Akira (Osaka). C57BL/6 mice were from Charles River Breeding Laboratories. C3H/HeOuJ and C3H/HeJ were from The Jackson Laboratory. Five- to 8-week-old mice were injected i.p. with 2.5 ml of 3% thioglycollate (Remel, Lenexa, KS). After 3 days, peritoneal lavage with 10 ml of DMEM with 10% FBS and 10 μg of ciprofloxacin per ml was performed on killed mice. Cells were washed, counted with a hemocytometer, and plated to the appropriate density in tissue-culture dishes (1 × 105 cells per well for 96-well plates, 1.5 × 106 cells per well for six-well plates). After 72 h, nonadherent cells were removed by washing with medium, and adherent cells were stimulated as described below.
Electrophoretic Mobility-Shift Assay (EMSA).
Nuclear translocation of NF-κB was determined by EMSA as follows. Three days after seeding on six-well dishes, peritoneal-derived macrophages were stimulated for 1 h with LPS (10 ng/ml), pneumolysin (10 μg/ml), Pam3Cys-Ser-Lys-4 (5 μg/ml), or medium alone. Nuclear extracts were isolated and analyzed for binding to a 32P-labeled NF-κB-specific oligonucleotide by EMSA as described (39).
Measurement of TNF-α and IL-6 Secretion from Peritoneal Macrophages and IL-8 from HEK Cells.
Peritoneal exudate cells or HEK cells were seeded at a density of 105 cells per well in 96-well plates in DMEM with 10% FBS. Three days later, cells were washed with medium and stimulated with the indicated preparations for 16 h. Supernatants then were collected and stored up to 7 days at −80°C until assayed for TNF-α, IL-6 (for murine macrophages), or IL-8 (for HEK cells) by ELISA for murine or human cytokines (R & D Systems), respectively. In these experiments, all conditions were performed in triplicate; the results are expressed as the means and SD of the three determinations. Results shown are from an experiment that is representative of three experiments.
Flow-Cytometry Analysis.
Cells were plated at a density of 7 × 104 per well in 24-well plates. The next day, cells were stimulated as indicated in Ham's F-12 medium with 10% FBS (for a total volume of 0.5 ml per well) and incubated for 18 h. Cells then were labeled with FITC-labeled anti-human CD25 (IL-2R) mAb (Becton Dickinson) in PBS with 1% FBS on ice. The cells were analyzed by flow cytometry by using a FACScan instrument (Becton Dickinson), and data were analyzed by using CELLQUEST software (Becton Dickinson).
Animal Models of Pneumococcal Nasopharyngeal Colonization and Sepsis.
C3H/HeOuJ and C3H/HeJ male mice 5–8 weeks of age were obtained from The Jackson Laboratory. C3H/HeJ mice carry a missense mutation of the TLR4 gene (P712H), which renders them hyporesponsive to LPS (40). For sepsis experiments, mice were restrained gently without anesthesia and inoculated intranasally with 10 μl of either strain WU2 (pneumolysin-positive type-3 strain) or WU2-PLA (pneumolysin-negative isogenic mutant of WU2) at an inoculum of 7 × 107 cfu. Mice then were observed daily for 5 days. For nasopharyngeal colonization experiments with bioluminescent bacteria, mice were gently restrained without anesthesia and inoculated intranasally with 10 μl (corresponding to 7 × 107 cfu) of A66.1 Xen 10. Twenty-four hours after inoculation, and on days 2 and 3, analysis of photon emission was determined from the ventral image of mice under Avertin (Sigma–Aldrich) anesthesia in an IVIS charge-coupled device camera coupled to the LIVINGIMAGE software package (both from Xenogen). As shown (37), the bioluminescent signal in mice correlates well with the number of cfu of pneumococci recovered even after several days in vivo.
Results
Ethanol-Killed but Not Heat-Killed Pneumococci Stimulate HEK Cells Transfected with TLR4.
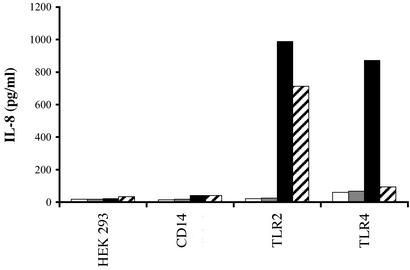
In previous experiments (ref. 41 and data not shown), we noted that ethanol- but not heat-killed whole-cell pneumococcal preparations administered intranasally with an adjuvant (cholera toxin) were immunogenic and protected rats against sepsis. These findings raised the possibility that heat treatment but not ethanol treatment had inactivated an important immunogenic component(s) of whole pneumococci. We explored whether the differences in immunogenicity of these mucosally applied bacteria could be related to a differential stimulatory effect on cells transfected with human TLRs. As shown in Fig. 1, exposure to ethanol-killed pneumococci led to higher release of IL-8 in HEK cells transfected with TLR4 compared with heat-killed pneumococci. In contrast, the response of TLR2-transfected HEK cells to ethanol- or heat-killed pneumococci was similar. These results suggested the presence of a heat-labile, ethanol-stable, TLR4-dependent ligand in whole-cell preparations of pneumococci. LPS contamination seemed an unlikely explanation for this selective TLR4-dependent response, because LPS would have been expected to resist heat treatment. Furthermore, as shown in the Fig. 1, in the absence of CD14 the TLR4-transfected HEK cells are LPS-hyporesponsive. Our experiments strongly suggested the existence of a heat-labile TLR4 ligand in pneumococcus.
Figure 1.
Differential induction of IL-8 secretion in transfected HEK cells by ethanol-killed vs. heat-killed S. pneumoniae. HEK 293 cells stably transfected with CD14, TLR2, or TLR4 were stimulated over 18 h with medium (white bars), 10 ng/ml LPS (gray bars), ethanol-killed pneumococcus (black bars), and heat-killed pneumococcus (hatched bars).
Stimulation with WT but Not Pneumolysin-Negative Pneumococci Leads to NF-κB Translocation in CHO Cells.
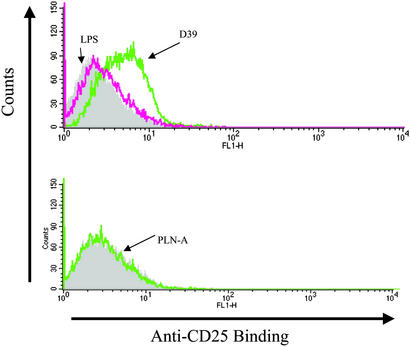
An important inflammatory stimulus in pneumococcus is the cholesterol-dependent cytolysin, pneumolysin, which has been shown by previous investigators to be heat-labile (42). Pneumolysin is a strong agonist of macrophages, stimulating the secretion of TNF-α as well as nitric oxide (21, 23). The formation of these inflammatory mediators depends, in part, on the nuclear translocation and activation of the transcription factor NF-κB. Therefore, we asked whether the observed response to pneumolysin was associated with NF-κB activation. We used clone 7.19, a CHO/CD14 cell line that contains a stably integrated NF-κB reporter plasmid (pELAM.tac) that is unresponsive to LPS because of a mutation in MD2 (C95Y) (11, 29); similar to all CHO cells, clone 7.19 also lacks TLR2 (43). The 7.19 cells were stimulated with ethanol-killed D39 or the isogenic pneumolysin-negative mutant known as PLN-A. As shown in Fig. 2, whereas stimulation with a concentration of killed PLN-A corresponding to 108 cfu/ml did not stimulate LPS nonresponder 7.19 cells, stimulation with the same concentration of killed WT strain, D39, resulted in NF-κB activation. A 10-fold higher concentration of PLN-A also failed to stimulate the 7.19 cells (data not shown). These results indicated that pneumolysin from pneumococcus could induce NF-κB translocation through a TLR2- and MD2-independent signaling pathway, thus differentiating pneumolysin recognition from other bacterial products including peptidoglycan and LPS.
Figure 2.
Pneumolysin induces NF-κB translocation in CHO cells. Clone 7.19, a CHO/CD14 cell line that contains an ELAM.tac (CD25) reporter plasmid, is LPS-nonresponsive because of a point mutation in MD2 (C95Y). This cell line was stimulated with ethanol-killed D39 (corresponding to 108 cfu/ml) or PLN-A (corresponding to 108 cfu/ml) over 18 h. After stimulation, cells were stained with anti-CD25 mAb and analyzed by flow microfluorometry.
The Secretion of IL-6 in Response to Pneumolysin Depends on the Presence of the TLR Adapter Molecule MyD88.
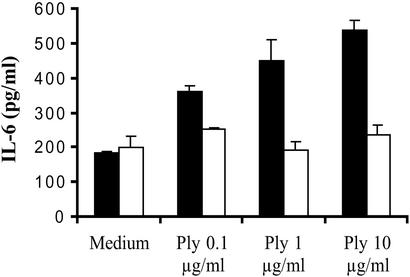
MyD88 is a 35-kDa protein that has been shown to operate in the human TLR signaling pathway (38). MyD88 is required for full signaling in the TLR pathways in mammals by linking other proteins of the signal-transduction pathway to the receptor complex such as the serine/threonine-kinase family of IL-1 receptor-associated kinase (IRAK). We compared the response of WT and MyD88-null peritoneal-derived macrophages to native pneumolysin. In these experiments, 96-well tissue-culture plates were seeded with peritoneal-derived macrophages of either C57BL/6 or MyD88−/− mice. As shown in Fig. 3, stimulation of WT macrophages with increasing concentrations of purified recombinant pneumolysin resulted in increased IL-6 secretion, whereas MyD88−/− macrophages failed to respond. These results indicated that signaling by pneumolysin requires MyD88 and that the secretion of proinflammatory cytokines in response to this toxin depends on one or multiple TLRs. On the basis of these data and the results obtained with the two preparations of killed pneumococci (ethanol- vs. heat-killed), we considered the hypothesis that signaling by pneumolysin requires the presence of functional TLR4.
Figure 3.
The inflammatory activity of pneumolysin is MyD88-dependent. Peritoneal macrophages from C57BL/6 (WT, black) and MyD88−/− (white) mice were stimulated with medium or indicated concentrations of pneumolysin (Ply) over 18 h.
Pneumolysin Induces Proinflammatory Responses in Macrophages in a TLR4-Dependent Manner in Synergy with Peptidoglycan or Whole Pneumococcal Cell Walls.
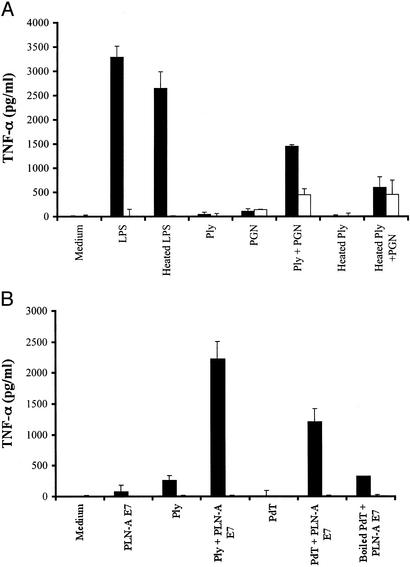
The inflammatory activity of pneumolysin was compared in peritoneal macrophages obtained from C3H/HeOuJ and HeJ mice. As shown in Fig. 4A, stimulation of macrophages with pneumolysin alone (1 μg/ml) resulted in an inflammatory response only in macrophages from TLR4-sufficient mice, whereas the response to staphylococcal peptidoglycan [a TLR2 ligand (13)] was similar between control and TLR4-deficient mice. Furthermore, a synergistic response with peptidoglycan or killed whole pneumococcal cells from a pneumolysin-negative strain of pneumococcus (PLN-A) was observed only in TLR4-bearing macrophages (Fig. 4). Because the recombinant pneumolysin preparation we used was purified from E. coli, we wished to exclude any effect of potentially contaminating LPS. First, we assessed the heat lability of the proinflammatory effect of pneumolysin, because LPS is heat-stable. The TLR4-dependent inflammatory response was lost when the pneumolysin was heated (100°C, 60 min). In contrast, heat treatment did not affect LPS-stimulated TNF-α secretion from C3H/HeOuJ macrophages. Furthermore, the inflammatory activity of pneumolysin was preserved after incubation with endotoxin-neutralizing protein-coated beads, which abrogated the response to 10 ng of LPS per ml (data not shown). Taken together, these results provide strong evidence that pneumolysin interacts with TLR4 and that this interaction is not due to LPS contamination.
Figure 4.
The inflammatory activity of pneumolysin is TLR4-dependent, heat-labile, and augmented by costimulation with TLR2 ligands (peptidoglycan or killed whole pneumococcal cells PLN-A). Peritoneal macrophages from C3H/HeOuJ (WT, black) and C3H/HeJ [TLR4 mutants (P712H), white] mice were stimulated with 10 ng/ml LPS, 1 μg/ml pneumolysin (Ply), heated pneumolysin with or without staphylococcal peptidoglycan (PGN, 1 μg/ml) (A), killed pneumococcal cells [PLN-A, concentrations of 107 (E7) cells per ml], pneumolysin, or PdT (a nonhemolytic mutant of pneumolysin, 1 μg/ml) with or without PLN-A E7, heated PdT with PLN-A E7 (B).
Next we assessed whether the TLR4-dependent inflammatory response to pneumolysin depended on the cytolytic properties of the molecule. PdT is a mutant of pneumolysin that lacks cytolytic and also complement-activating properties by virtue of three amino acid substitutions. As shown in Fig. 4B, the inflammatory activity of PdT is TLR4-dependent, heat-labile, and synergistic with that of killed, pneumolysin-negative pneumococcal cells. These data indicate that the TLR4-dependent proinflammatory activity of pneumolysin is independent of cytolytic activity.
Stimulation of Peritoneal Macrophages from WT but Not TLR4−/− Mice with Pneumolysin Leads to NF-κB Translocation.
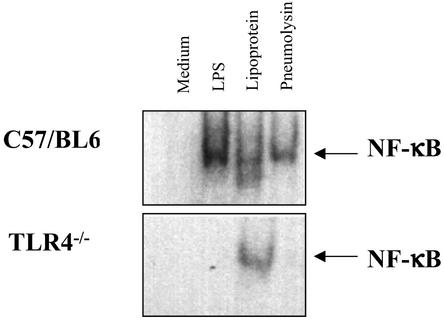
Peritoneal macrophages were also assessed for the induction of NF-κB translocation by pneumolysin with the EMSA. Peritoneal macrophages derived from WT C57BL/6 mice or TLR4−/− mice (7) were compared. Fig. 5 shows that, in response to a 1-h stimulation, macrophages derived from WT and TLR4−/− mice demonstrated NF-κB translocation in response to a TLR2 ligand (Pam3Cys-Ser-Lys-4). In contrast, only WT mice cells responded to exposure to LPS or pneumolysin. Thus, similar to LPS, pneumolysin-induced NF-κB translocation in macrophages requires TLR4.
Figure 5.
Nuclear translocation of NF-κB due to pneumolysin is TLR4-dependent. Peritoneal macrophages from C57BL/6 or TLR4−/− mice were stimulated for 1 h with medium, 10 ng/ml LPS, 5 μg/ml lipoprotein (Pam3Cys-Ser-Lys-4), or 10 μg/ml pneumolysin, and then nuclear extracts were prepared and analyzed by EMSA.
Mice with a Defined Mutation of TLR4 (P712H) Are Hypersusceptible to Pneumococcal Invasive Disease After Nasopharyngeal Challenge with Pneumolysin-Producing Pneumococci.
We tested the effects of TLR4 expression in an in vivo model of pneumococcal nasopharyngeal infection in mice. We hypothesized that, if pneumolysin signals through TLR4, mice with a mutation in the TLR4 gene would be more susceptible to sepsis and death after exposure to pneumolysin-producing pneumococci. To this end, we compared the mortality rates of C3H/HeJ and C3H/HeOuJ mice after intranasal exposure to WU2 (pneumolysin-producing) and its pneumolysin-negative derivative, strain WU2-PLA. Strain WU2 is a laboratory-passaged type-3 strain that is highly virulent in mice when injected i.p. or administered intranasally in anesthetized mice. In contrast, in unanesthetized mice, intranasal inoculation of WU2 in WT mice rarely leads to invasive disease or death, presumably because the organism is confined to the upper respiratory tract and does not progress to the bloodstream, lung, or other organs. Five- to 8-week-old C3H/HeJ and C3H/HeOuJ mice were challenged with 7 × 107 cfu WU2 or WU2-PLA (pneumolysin-negative isogenic derivative of WU2). Table 1 shows that strain WU2-PLA had low virulence in both mouse strains with comparable mortality rates. In contrast, after exposure to strain WU2, C3H/HeJ mice had a significantly increased mortality compared with C3H/HeOuJ mice [9 of 16 (56.2%) vs. 2 of 16 (12.5%); P = 0.023). These results indicate that mice lacking functional TLR4 are significantly more likely to develop lethal systemic infection than are TLR4-sufficient mice after nasopharyngeal challenge with pneumolysin-producing pneumococci. Taken together with the findings of in vitro studies, the results of animal-challenge experiments support an important role for TLR4 in the inflammatory response to pneumolysin and in host defense against pneumococcal infection.
Table 1.
Mice with a defined mutation of TLR4 are highly susceptible to sepsis due to pneumolysin-producing pneumococci
| Challenge strain | No. dead/total (%)
|
|
|---|---|---|
| C3H/HeJ mice | C3H/HeOuJ mice | |
| WU2 | 9/16 (56.3)* | 2/16 (12.5)* |
| WU2-PLA | 2/10 (20.0) | 1/10 (10.0) |
Mice were challenged intranasally with 7 × 107 cfu (in 10 μl) of either WU2 (pneumolysin-producing type 3 strain) or WU2-PLA (pneumolysin-negative isogenic mutant), and mortality was monitored over 5 days.
P = 0.023 by Fisher's exact test.
Mice with a Defined Mutation of TLR4 Are More Susceptible to Pneumococcal Colonization After Nasopharyngeal Challenge.
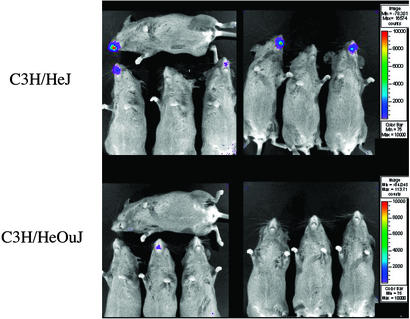
A possible explanation for the increased virulence of pneumolysin-producing pneumococci in TLR4-deficient mice is that these animals are more likely to be heavily colonized after exposure to pneumococci in the absence of a robust proinflammatory response to pneumolysin. To test this hypothesis, we evaluated the effects of TLR4 expression in an in vivo model of pneumococcal nasopharyngeal colonization in mice. To monitor nasopharyngeal colonization in mice, we used a bioluminescent pneumococcal strain, A66.1 Xen 10, engineered as described (37). The expression of the luciferase gene in this strain allows for real-time in vivo imaging of nasopharyngeal colonization in mice. As shown in Fig. 6 (obtained 24 h after intranasal inoculation), C3H/HeJ mice had a much higher bacterial nasopharyngeal burden (as determined by photon emission) than C3H/HeOuJ mice. This difference was notable also on days 2 and 3 after inoculation. The density of nasopharyngeal colonization, as determined by photon emission, also predicted mortality, because all the animals in which bioluminescent bacteria could be detected on day 1 subsequently developed bacteremia and died. As a confirmation of the results obtained with strain WU2, by 96 h after infection the mortality in C3H/HeJ mice significantly exceeded that of C3H/HeOuJ mice (11 of 20 vs. 3 of 17, respectively; P = 0.04).
Figure 6.
Mice lacking functional TLR4 are more susceptible to pneumococcal nasopharyngeal colonization. Emitted photons by luciferase-tagged S. pneumoniae in the nasopharynx from anesthetized C3H/HeJ (Upper) and C3H/HeOuJ (Lower) mice were detected by a charge-coupled camera after intranasal inoculation with 7 × 107 cfu of a type-3 pneumococcus (strain A66.1 Xen 10). Results are from 24 h after inoculation and represent the course of colonization in these animals. The animals with bioluminescence at 24 h (five C3H/HeJ and one C3H/HeOuJ mice in these pictures) subsequently developed bacteremia and died.
Discussion
With the exception of some horse-specific type-3 strains, S. pneumoniae is exclusively a human pathogen. The pneumococcus is extraordinarily well adapted to its natural host. Epidemiologic studies have demonstrated that with some variation due to geographic location, exposure to daycare centers, and age, 40–60% of infants and toddlers can be colonized with pneumococci at any one time (44, 45). In this context, the relatively low incidence of pneumococcal invasive disease [for all ages, 15–30 cases per 100,000 populations, and 150 cases per 100,000 children under 2 years (46, 47)] is further evidence of the exquisite adaptation of the colonizing pathogen to the human. Invasive pneumococcal disease is therefore a relatively rare consequence of colonization.
The natural sequence of events leading to pneumococcal invasive disease begins in the nasopharynx, where pneumococci have been shown to survive in the host for several months (44). In a few individuals, usually within 4 weeks after initial exposure, colonization leads to invasion and the subsequent development of bacteremia, meningitis, and/or sepsis. The underlying reasons why certain individuals develop invasive disease after colonization, whereas most do not, are not clear. Although anticapsular antibodies certainly play a role in protection against colonization and disease (48), our study strongly suggests that innate immune mechanisms, and in particular TLR4, play an important role in preventing colonizing bacteria from invading the host.
In the present study, we demonstrate the relationship between a cholesterol-dependent cytolysin released by virtually all clinical isolates of pneumococci and TLR4, the signal-transducing portion of the LPS receptor. Furthermore, mice lacking functional TLR4 are significantly more susceptible to invasive disease after colonization with a virulent, pneumolysin-producing type-3 strain of S. pneumoniae.
There are several important implications of these results. First, it has long been assumed that the ability of cholesterol-dependent cytolysins to cause inflammatory responses in cells was related to the formation of pores in cholesterol-rich membranes (49, 50). With respect to pneumolysin, careful virulence studies using derivatives with specific mutations that abolish the hemolytic and/or complement-activating properties suggested that other properties of the toxin also contribute to virulence (51, 52). We report here that a specific TLR participates in the proinflammatory response to pneumolysin by macrophages and hypothesize that the interaction of pneumolysin with this TLR is likely to be a specific one. This hypothesis is supported further by our data showing that PdT, a noncytolytic mutant of pneumolysin, similarly stimulates macrophages in a TLR4-dependent fashion. In other words, it seems unlikely that the pore-forming capabilities of pneumolysin are solely responsible for this inflammatory activity.
In addition, the interaction of pneumolysin with phagocytic cells seems to be synergistically potentiated by costimulation with staphylococcal peptidoglycan or killed, pneumolysin-negative pneumococcal cells, both of which activate TLR2 (13). The similarity of the sepsis syndrome induced by Gram-negative organisms and pneumococcus may be explained by the involvement of the same signal-transduction receptor, TLR4, in combination with TLR2-mediated stimulation by cell-wall components of lysed pneumococci.
Third, our finding that the response to live pneumococci is mediated by TLR4 further challenges the generally accepted paradigm that TLR4 is a receptor with primary ligands that are products of Gram-negative bacteria. Investigators have demonstrated the interaction between viral or mycobacterial components and TLR4 (30, 53), but the relative contribution of this interaction to pathogenesis was less clear than that of Gram-negative bacterial LPS. With respect to pneumococcus, investigators explored the possibility that C3H/HeJ mice may have differing susceptibilities to pneumococcal infections compared with C3H/HeOuJ mice by using a peritoneal challenge model in mice but saw no difference between the two mouse strains (51). In contrast, using an intranasal challenge model, we have shown that mice lacking functional TLR4 are significantly more prone to systemic infection after nasal inoculation with virulent, pneumolysin-producing pneumococci. Both the data obtained from peritoneal macrophages and in vivo challenge with a pneumolysin-negative mutant of pneumococcus support the hypothesis that pneumolysin plays an important role in innate immune recognition. As suggested by the bioluminescence experiments, the increased susceptibility of C3H/HeJ mice to pneumococcus may be viewed as the consequence of the absence of TLR4-mediated sentinel activity of phagocytic leukocytes that normally contain and help clear colonizing pneumococci in the nasopharynx.
The importance of TLR2 in the recognition of Gram-positive bacteria, primarily through cell-wall and membrane components such as peptidoglycan, lipoteichoic acid, and lipoproteins, has been well demonstrated. Most recently, Echchannaoui et al. (54) reported that TLR2-deficient mice were more susceptible to death after intracerebral injection with virulent S. pneumoniae compared with WT mice. Although these data are consistent with evidence implicating TLR2 in the recognition of Gram-positive bacteria, the protective effect of TLR2 was relatively modest, because it conferred only slightly prolonged survival after direct intracerebral inoculation. In contrast, in our mucosal challenge model, mice lacking TLR4 are profoundly compromised in their ability to resist pneumococcal infection. As suggested by the experiments using bioluminescent pneumococci, mice lacking functional TLR4 appear unable to control the proliferation of pneumococci in the nasopharynx, which subsequently leads to invasive disease and sepsis.
We conclude that TLR4 mediates innate immune responses to pneumococci through its interaction with one of the major virulence factors of the organism, the cholesterol-dependent cytolysin pneumolysin. Given these results, we suggest that the relatively rare occurrence of invasive disease after asymptomatic colonization with pneumococcus may be due to the robust inflammatory response to pneumococcus. This response is mediated, at least in part, by a synergistic TLR4-dependent response to pneumolysin.
Acknowledgments
We thank Drs. Porter Anderson and David Ludwig for helpful suggestions and discussions as well as their review of the manuscript. This work was funded in part by the Meningitis Research Foundation and The Milton Fund (to R.M.), Deutsche Forschungsgemeinschaft Grant He 3127/1-1 (to P.H.), and National Institutes of Health Grants 1 K08 AI51526-01 (to R.M.) and R01 GM54060, GM63244, and AI38515 (to D.T.G.).
Abbreviations
- LPS
lipopolysaccharide
- TLR
Toll-like receptor
- TNF-α
tumor necrosis factor α
- CHO
Chinese hamster ovary
- NF-κB
nuclear factor κB
- HEK
human embryonic kidney
- cfu
colony-forming units
- MyD88
myeloid differentiation factor 88
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.World Health Organization. Wkly Epidemiol Rec. 1998;73:187–188. [Google Scholar]
- 2.Zangwill K M, Vadheim C M, Vannier A M, Hemenway L S, Greenberg D P, Ward J I. J Infect Dis. 1996;174:752–759. doi: 10.1093/infdis/174.4.752. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Morbid Mortal Wkly Rep. 1997;46:1–24. [Google Scholar]
- 4.Tuomanen E, Tomasz A, Hengstler B, Zak O. J Infect Dis. 1985;151:535–540. doi: 10.1093/infdis/151.3.535. [DOI] [PubMed] [Google Scholar]
- 5.Tuomanen E I. Ann NY Acad Sci. 1996;797:42–52. doi: 10.1111/j.1749-6632.1996.tb52948.x. [DOI] [PubMed] [Google Scholar]
- 6.Koedel U, Bayerlein I, Paul R, Sporer B, Pfister H W. J Infect Dis. 2000;182:1437–1445. doi: 10.1086/315877. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 8.Chow J C, Young D W, Golenbock D T, Christ W J, Gusovsky F. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 9.da Silva Correia J, Soldau K, Christen U, Tobias P S, Ulevitch R J. J Biol Chem. 2001;276:21129–21135. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 10.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shchromm A B, Lien E, Henneke P, Chow J C, Yoshimura A, Heine H, Latz E, Monks B G, Schwartz D A, Miyake K, Golenbock D T. J Exp Med. 2001;194:79–88. doi: 10.1084/jem.194.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 14.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Dziarski R, Kirschning C J, Muzio M, Gupta D. Infect Immun. 2001;69:2270–2276. doi: 10.1128/IAI.69.4.2270-2276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michelsen K S, Aicher A, Mohaupt M, Hartung T, Dimmeler S, Kirschning C J, Schumann R R. J Biol Chem. 2001;276:25680–25686. doi: 10.1074/jbc.M011615200. [DOI] [PubMed] [Google Scholar]
- 18.Morath S, Stadelmaier A, Geyer A, Schmidt R R, Hartung T. J Exp Med. 2002;195:1635–1640. doi: 10.1084/jem.20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson M K, Boese-Marrazzo D, Pierce W A., Jr Infect Immun. 1981;34:171–176. doi: 10.1128/iai.34.1.171-176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry A M, Yother J, Briles D E, Hansman D, Paton J C. Infect Immun. 1989;57:2037–2042. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houldsworth S, Andrew P W, Mitchell T J. Infect Immun. 1994;62:1501–1503. doi: 10.1128/iai.62.4.1501-1503.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry A M, Alexander J E, Mitchell T J, Andrew P W, Hansman D, Paton J C. Infect Immun. 1995;63:1969–1974. doi: 10.1128/iai.63.5.1969-1974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun J S, Novak R, Gao G, Murray P J, Shenep J L. Infect Immun. 1999;67:3750–3756. doi: 10.1128/iai.67.8.3750-3756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M K, Geoffroy C, Alouf J E. Infect Immun. 1980;27:97–101. doi: 10.1128/iai.27.1.97-101.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paton J C, Ferrante A. Infect Immun. 1983;41:1212–1216. doi: 10.1128/iai.41.3.1212-1216.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton J C, Rowan-Kelly B, Ferrante A. Infect Immun. 1984;43:1085–1087. doi: 10.1128/iai.43.3.1085-1087.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun J S, Sublett J E, Freyer D, Mitchell T J, Cleveland J L, Tuomanen E I, Weber J R. J Clin Invest. 2002;109:19–27. doi: 10.1172/JCI12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paton J C, Lock R A, Lee C J, Li J P, Berry A M, Mitchell T J, Andrew P W, Hansman D, Boulnois G J. Infect Immun. 1991;59:2297–2304. doi: 10.1128/iai.59.7.2297-2304.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delude R L, Yoshimura A, Ingalls R R, Golenbock D T. J Immunol. 1998;161:3001–3009. [PubMed] [Google Scholar]
- 30.Kurt-Jones E A, Popova L, Kwinn L, Haynes L M, Jones L P, Tripp R A, Walsh E E, Freeman M W, Golenbock D T, Anderson L J, Finberg R W. Nat Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 31.Bricker A L, Camilli A. FEMS Microbiol Lett. 1999;172:131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- 32.Paoletti L C, Ross R A, Johnson K D. Infect Immun. 1996;64:1220–1226. doi: 10.1128/iai.64.4.1220-1226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briles D E, Nahm M, Schroer K, Davie J, Baker P, Kearney J, Barletta R. J Exp Med. 1981;153:694–705. doi: 10.1084/jem.153.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yother J, Handsome G L, Briles D E. J Bacteriol. 1992;174:610–618. doi: 10.1128/jb.174.2.610-618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison D A, Baker M F. Nature. 1979;282:215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- 36.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein L S, Piccoli L, Simon D, Morrison D A. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francis K P, Yu J, Bellinger-Kawahara C, Joh D, Hawkinson M J, Xiao G, Purchio T F, Caparon M G, Lipsitch M, Contag P R. Infect Immun. 2001;69:3350–3358. doi: 10.1128/IAI.69.5.3350-3358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 39.Delude R, Fenton M, Savedra R, Perera P-Y, Vogel S, Golenbock D. J Biol Chem. 1994;269:22252–22260. [PubMed] [Google Scholar]
- 40.Poltorak A, He X, Smirnova I, Liu M-Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 41.Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D E, Anderson P. Infect Immun. 2001;69:4870–4873. doi: 10.1128/IAI.69.8.4870-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinfort C, Wilson R, Mitchell T, Feldman C, Rutman A, Todd H, Sykes D, Walker J, Saunders K, Andrew P W, et al. Infect Immun. 1989;57:2006–2013. doi: 10.1128/iai.57.7.2006-2013.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heine H, Kirschning C J, Lien E, Monks B G, Rothe M, Golenbock D T. J Immunol. 1999;162:6971–6975. [PubMed] [Google Scholar]
- 44.Gray B M, Converse G M, III, Dillon H C., Jr J Infect Dis. 1980;142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 45.Austrian R. J Antimicrob Chemother. 1986;18, Suppl. A:35–45. doi: 10.1093/jac/18.supplement_a.35. [DOI] [PubMed] [Google Scholar]
- 46.Bennett N M, Buffington J, LaForce F M. Am J Public Health. 1992;82:1513–1516. doi: 10.2105/ajph.82.11.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breiman R F, Spika J S, Navarro V J, Darden P M, Darby C P. Arch Intern Med (Moscow) 1990;150:1401–1405. [PubMed] [Google Scholar]
- 48.Black S, Shinefield H, Fireman B, Lewis E, Ray P, Hansen J R, Elvin L, Ensor K M, Hackell J, Siber G, et al. Pediatr Infect Dis J. 2000;19:187–195. doi: 10.1097/00006454-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 49.Tweten R K, Parker M W, Johnson A E. Curr Top Microbiol Immunol. 2001;257:15–33. doi: 10.1007/978-3-642-56508-3_2. [DOI] [PubMed] [Google Scholar]
- 50.Palmer M. Toxicon. 2001;39:1681–1689. doi: 10.1016/s0041-0101(01)00155-6. [DOI] [PubMed] [Google Scholar]
- 51.Benton K A, Paton J C, Briles D E. Microb Pathog. 1997;23:201–209. doi: 10.1006/mpat.1997.0150. [DOI] [PubMed] [Google Scholar]
- 52.Berry A M, Ogunniyi A D, Miller D C, Paton J C. Infect Immun. 1999;67:981–985. doi: 10.1128/iai.67.2.981-985.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Means T K, Wang S, Lien E, Yoshimura A, Golenbock D T, Fenton M J. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 54.Echchannaoui H, Frei K, Schnell C, Leib S L, Zimmerli W, Landmann R. J Infect Dis. 2002;186:798–806. doi: 10.1086/342845. [DOI] [PubMed] [Google Scholar]