Abstract
Synthesis of the relaxed-circular (RC) DNA genome of hepadnaviruses requires two template switches during plus-strand DNA synthesis: primer translocation and circularization. Although primer translocation and circularization use different donor and acceptor sequences, and are distinct temporally, they share the common theme of switching from one end of the minus-strand template to the other end. Studies of duck hepatitis B virus have indicated that, in addition to the donor and acceptor sequences, three other cis-acting sequences, named 3E, M, and 5E, are required for the synthesis of RC DNA by contributing to primer translocation and circularization. The mechanism by which 3E, M, and 5E act was not known. We present evidence that these sequences function by base pairing with each other within the minus-strand template. 3E base-pairs with one portion of M (M3) and 5E base-pairs with an adjacent portion of M (M5). We found that disrupting base pairing between 3E and M3 and between 5E and M5 inhibited primer translocation and circularization. More importantly, restoring base pairing with mutant sequences restored the production of RC DNA. These results are consistent with the model that, within duck hepatitis B virus capsids, the ends of the minus-strand template are juxtaposed via base pairing to facilitate the two template switches during plus-strand DNA synthesis.
Hepadnaviruses are a family of DNA viruses that replicate via reverse transcription of an RNA intermediate (1). Like other reverse transcribing elements, hepadnaviruses use template switches or template exchanges for the synthesis of their genomes. Template switching is the process in which the strand of DNA being synthesized switches from its current template to a new template. For retroviruses, two template switches are required for the synthesis of retroviral DNA: the first and second strong stop template switches (2). These are referred to as replicative template switches. Retroviruses also perform a second type of template switching, called recombinogenic, between the two RNA templates or within a single RNA template during the synthesis of the minus-strand DNA (3, 4). However, these recombinogenic template switches, unlike the replicative template switches, are not obligatory steps in each cycle of reverse transcription. Similar to the replicative template switches in retroviruses, three template switches are needed for the synthesis of the relaxed-circular (RC) genome of hepadnaviruses: one during the synthesis of minus-strand DNA and two for plus-strand RC DNA synthesis. In general, replicative template switches involve translocation of the nascent DNA strand from one end to the other end of the template. Sequence identity at the donor and acceptor sites ensures that the nascent DNA strand elongates from the correct position on the new template.
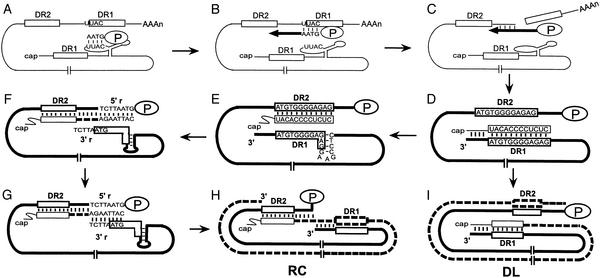
Reverse transcription of hepadnaviruses takes place within the cytoplasmic capsids in hepatocytes (Fig. 1; for a review, see ref. 5). The first template switch occurs shortly after the initiation of minus-strand DNA synthesis. The P protein, acting as reverse transcriptase and primer, initiates minus-strand DNA synthesis within the encapsidation signal, epsilon, which is near the 5′ end of pregenomic RNA (pgRNA; Fig. 1A; refs. 6–8). After four nucleotides are synthesized, the nascent minus-strand DNA switches template to a position overlapping the direct repeat 1 (DR1) near the 3′ end of pgRNA (Fig. 1B; refs. 6 and 8). Resumption of minus-strand DNA synthesis from this position ultimately yields a genome-length minus-strand DNA, with the RNase H activity of the P protein degrading the RNA template (Fig. 1 C and D; ref. 1). The final RNase H cleavage generates the RNA primer for plus-strand DNA replication (Fig. 1D). The primer is 18 or 19 nt and has DR1 sequence at its 3′end (9, 10). In the majority of WT capsids, the RNA primer translocates from DR1, which is at the 3′ end of minus-strand DNA, to base-pair with direct repeat 2 (DR2), which is near the 5′ end of the minus-strand, to prime plus-strand DNA synthesis (Fig. 1E). This template switch is called primer translocation (9, 11). Plus-strand synthesis initiates from DR2 and proceeds until it reaches the 5′ end of the minus-strand DNA (Fig. 1F). At this point, the third template switch, termed circularization, occurs, in which the 3′ end of minus-strand becomes the template for plus-strand synthesis (Fig. 1G; refs. 9 and 12). A short terminal redundancy (5′r and 3′r) facilitates circularization. Further elongation of the plus-strand results in a RC DNA genome (Fig. 1H). For a small fraction of the capsids, the RNA primer is elongated from DR1, in a process called in situ priming, which results in a duplex-linear (DL) form of the genome (11). A small DNA hairpin overlapping the 5′ end of DR1 in the minus strand suppresses in situ priming for avian hepadnaviruses (Fig. 1E; ref. 13).
Figure 1.
Model for DHBV reverse transcription. (A) The pgRNA (thin line) is the template for minus-strand DNA synthesis. The 12-nt direct repeats, DR1 and DR2, are shown as open boxes, with two copies of DR1 on the pgRNA. Four nucleotides are synthesized by P protein (oval labeled P) within the bulge of the stem-loop (epsilon) near the 5′ end of the pgRNA. (B) Minus-strand template switch. The nascent minus-strand DNA switches template to an acceptor sequence that overlaps the 3′ copy DR1. (C and D) Minus-strand DNA synthesis resumes after template switch, resulting in a genome-length minus-strand (thick line). Degradation of pgRNA by RNase H activity of P protein generates the 18-nt RNA primer for plus-strand DNA synthesis. The 3′ terminus of the primer is complementary to DR1. (E) Primer translocation. The plus-strand primer switches template from DR1 to DR2, which is near the 5′ end of the minus-strand DNA. A small hairpin overlapping DR1 suppresses priming from DR1. (F) Plus-strand DNA (thick dashed line) is initiated from DR2 and elongated until the 5′end of the template. The minus-strand template is terminally redundant for 7 or 8 nt (5′r and 3′r). (G) Circularization. The nascent plus-strand moves from 5′r to base pair with the 3′r. This template switch circularizes the genome. (H) Elongation of the plus-strand after circularization results in a RC DNA genome. (I) In situ priming. A small fraction of the plus-strand primer can overcome the inhibitory role of the small DNA hairpin and initiate from DR1, which results in a DL form of the genome.
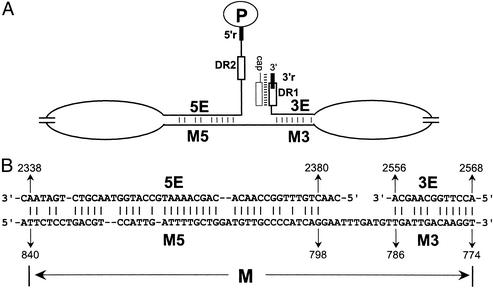
Sequence identity of the donor and acceptor sites contributes to successful template switching for hepadnavirus. The donor and acceptor sites (DR1 and DR2, 5′r and 3′r) for primer translocation and circularization are all located at or near both ends of the minus-strand DNA template (Fig. 2A). However, other cis-acting sequences make crucial contributions to the plus-strand template switches. Studies of plus-strand DNA synthesis of duck hepatitis B virus (DHBV) and heron hepatitis B virus (HHBV) reveal additional cis-acting requirements: 3E, M, and 5E on the minus-strand template for efficient synthesis of RC DNA (Fig. 2A; refs. 14–17). The mechanism by which these cis-acting sequences contribute to plus-strand DNA synthesis was not known. 3E, M, and 5E were shown to contribute to both primer translocation and circularization during plus-strand DNA synthesis, indicating that these template switches share a mechanism. Studies of HHBV/DHBV chimeric viruses demonstrated that these sequences need to be derived from the same virus to function properly, indicating 3E, M, and 5E interact, either directly or indirectly, to contribute to plus-strand DNA synthesis (17). Analysis of the nucleotide sequences revealed the potential for imperfect duplexes between 5E and one portion of M (M5) and between 3E and the other portion of M (M3) (Fig. 2B). We show that disrupting base pairing between 3E and M3 and between 5E and M5 inhibits primer translocation and circularization. Restoring base pairing, albeit with mutant sequences, restores the production of RC DNA. These results show that base pairing between these cis-acting sequences is necessary for RC DNA synthesis. Our results are consistent with the model that these base pairs function by juxtaposing the ends of the minus-strand DNA to facilitate primer translocation and circularization during plus-strand DNA synthesis.
Figure 2.
Locations of 3E, M, and 5E on the minus-strand DNA and the potential base-pairing patterns. (A) Minus-strand DNA (nucleotides 5′-2537–1/3021–2529-3′) is represented by the thin black line. The relative locations of 3E, M3, M5, and 5E are indicated. DR1 (nucleotides 2546–2535) and DR2 (nucleotides 2488–2477) are represented by the open boxes. 3′r (nucleotides 2537–2529) and 5′r (nucleotides 2537–2529) are represented by the thick black lines. The minus strand is not drawn to scale. (B) Putative base-pairing patterns between 3E and M3 and between 5E and M5. The minus-sense nucleotide sequences are shown. The numbers represent the nucleotide coordinates of each element relative to minus-strand.
Methods
Molecular Clones.
All molecular clones were derived from DHBV strain 3 (18). The plasmids contain 1.5 copies of the genome, and support the synthesis of pgRNA. The molecular clone used to express the WT reference virus (p503-3) is null for P protein due to a frameshift mutation in the P gene (19). The molecular clone used to express WT P protein has been described previously (14). All cis-acting mutations were constructed in the p503-3 background. Mutations were created by an oligonucleotide-directed mutagenesis procedure and verified by DNA sequencing (20). Details describing their constructions will be provided on request.
Cell Culture, Isolation, and Analysis of Viral DNA.
Chicken hepatoma cell line LMH was used in all experiments. Culturing LMH cells and transfections were performed as described (21). Viral DNA was isolated from cytoplasmic capsids 3 days posttransfection either by DNaseI-RNaseA-pronase treatment (22) or by micrococcal nuclease-pronase treatment (13).
Southern blotting analysis and primer extension analysis were performed as described (13, 21).
Statistical Analysis.
In both Southern blot and primer extension analyses, we asked three questions: (i) whether the phenotypes of the single mutants are different from those of the WT standard; (ii) whether the phenotypes of the single mutants are different from those of the double mutants; (iii) whether the phenotypes of the double mutants are different from those of the WT standard. To answer these questions, we used the hypothesis testing theory. In particular, we set up three similar hypothesis testing problems, each designed to answer one of the questions. Consider the first aforementioned question as an example. We tested the hypothesis H0 against the alternative H1, where H0 and H1 are defined as follows: H0, phenotypes of the single mutants are the same as those of the WT standard; H1, phenotypes of the single mutants are different from those of the WT standard.
Once the hypothesis testing problem was formulated, we adopted a two-sided Wilcoxon signed rank test using mstat 3.0 software (kindly provided by N. Drinkwater, McArdle Laboratory for Cancer Research) to decide whether to accept H0 or H1. Moreover, the statistical significance of this decision (on H0 or H1) is denoted by the P value and calculated by mstat 3.0. Note that, in general, if P < 0.05, the decision is considered significant. Because there are multiple single and double mutants, multiple hypothesis tests are involved for each of the three questions we asked. We present only the largest P value among the multiple hypothesis tests for each question. For measurements of primer extension analysis, values before normalization to the WT standard were used in statistical analysis, although the values were presented as normalized numbers (Table 1).
Table 1.
Efficiency of template switches during plus-strand DNA synthesis determined by primer extension
| WT | M3A | 3EA | M3A/3EA | M5A | 5EA | M5A/5EA | M5B | 5EB | M5B/5EB | |
|---|---|---|---|---|---|---|---|---|---|---|
| Primer translocation | 1.00 | 0.26 (0.08) | 0.23 (0.05) | 0.97 (0.38) | 0.68 (0.15) | 0.74 (0.16) | 1.17 (0.16) | 0.77 (0.06) | 0.70 (0.09) | 1.06 (0.13) |
| Circularization | 1.00 | 0.11 (0.06) | 0.12 (0.09) | 1.03 (0.46) | 0.47 (0.08) | 0.45 (0.09) | 0.84 (0.09) | 0.53 (0.07) | 0.47 (0.05) | 0.91 (0.17) |
| Primer utilization | 1.00 | 0.34 (0.11) | 0.36 (0.04) | 1.13 (0.46) | 0.77 (0.16) | 0.80 (0.16) | 1.16 (0.14) | 0.85 (0.05) | 0.78 (0.08) | 1.04 (0.11) |
In each experiment, values for WT standard are set to 1. Values for mutants are normalized to WT standard. Shown in the table are the average values and standard deviation (in parentheses) of multiple experiments from independent transfections (M3A and 3EA series, n = 7; M5A and 5EA series, n = 8; M5B and 5EB series, n = 5).
Results
Experimental Design.
Previous analyses identified three cis-acting sequences on the minus-strand template (3E, M, and 5E) necessary for efficient synthesis of RC DNA (11). Additional studies indicated that 5E is located between nucleotides 2342 and 2374, which is near the 5′ end of the minus-strand DNA (15). 3E was discovered in the analysis of a single variant that deleted the 11 nucleotides between DR1 and epsilon, which is near the 3′ end of the minus-strand template (14). The boundaries of 3E were not determined precisely due to its close proximity to other cis-acting sequences: DR1, the small hairpin overlapping DR1, and epsilon. M was located near the middle of the minus-strand template. Previously, M was mapped between nucleotides 723 and 832 (14). We have mapped it more precisely to the sequence between nucleotides 774 and 840 (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org).
Visual inspection of the minus-sense nucleotide sequences of 3E and M identified potential base pairing between 3E (nucleotides 2556–2568) and nucleotides 774–786 of M on the minus-strand DNA. We named this segment of M, M3 (Fig. 2B). Previously, the potential for imperfect base pairing between 5E and an adjacent sequence within M (now called M5) was identified (Fig. 2B; ref. 15). The potential for base pairing between 3E and M3 and between 5E and M5 suggests that these sequences function by forming two imperfect duplexes on the minus-strand template. Because of the distinct locations of 3E, M, and 5E on the minus-strand DNA, formation of these base pairs could juxtapose the ends of minus-strand DNA, thus facilitating primer translocation and circularization, which have their respective donor and acceptor sites near or at the ends of the minus strand.
To determine whether 3E, M, and 5E contribute to plus-strand synthesis by base pairing, we asked whether disrupting and then restoring the potential for base pairing would affect plus-strand DNA synthesis. If these sequences function by base pairing, then disrupting base pairing between 3E and M3 or between 5E and M5 would lead to less efficient primer translocation and circularization. Restoring base pairing, although with mutant sequences, would restore plus-strand template switches. The sequences of M and 5E lie within the P gene. To eliminate the possibility of disrupting P function, we made all of the mutations in a genetic background that was null for P protein production. The WT reference virus and the 3E variants also were placed into the same genetic background. To initiate reverse transcription, plasmids expressing pgRNA of each variant were cotransfected with an expression plasmid for P protein into the chicken hepatoma cell line LMH. Three days postransfection, viral replicative intermediates were isolated from cytoplasmic capsids. Plus-strand DNA synthesis was detected and quantified by Southern blotting and primer extension.
Base Pairing Between 3E and M3 Contributes to both Primer Translocation and Circularization During Plus-Strand DNA Synthesis.
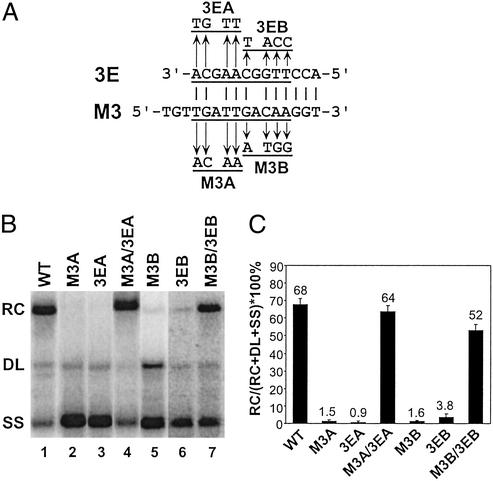
To determine the contribution of the predicted base pairing between 3E and M3 to the synthesis of RC DNA, two sets (A and B) of variants were constructed. Each set contained three variants; four nucleotides were substituted into M3 or 3E to disrupt the predicted base pairing. The third variant combined the M3 and 3E mutations to restore the potential for base pairing (Fig. 3A). The ability of these variants to support plus-strand DNA synthesis was analyzed by Southern blotting. Southern blotting of WT DHBV replicative intermediates reveals three DNA forms at characteristic proportions: RC DNA, DL DNA, and a full-length minus-strand species (SS) of DNA (Fig. 3B, lane 1). When either or both of the plus-strand template switches are inhibited, the proportion of RC DNA on Southern blot will decrease, and the proportion of DL or SS DNA or both will rise, depending on the mutation (11). As shown in Fig. 3 B and C, each of the single mutants, which disrupted base pairing, accumulated much less RC DNA than the WT reference (the largest P value was <0.012; see Methods). Both of the double mutants M3A/3EA and M3B/3EB that restored the potential for base pairing synthesized significantly higher levels of RC DNA than their corresponding single variants (P < 0.012), indicating restoration of function. In addition, the magnitudes of the defect for each of the single mutants and the extent of restoration for the double mutants indicate that base pairing between 3E and M3 makes a large contribution to the synthesis of RC DNA. For each of the single mutants, the decrease in RC DNA was accompanied with an increased accumulation of SS DNA. The proportions of DL DNA in these mutants were not changed compared with the WT reference, indicating that in situ priming was not affected by disruption of the base pairing. One exception was the M3B mutant, in which a 4-fold increase in the proportion of DL DNA was observed, a phenotype not observed in 3EB or M3B/3EB mutants. Overall, analysis of these six variants indicated that base pairing between 3E and M3 was necessary for the synthesis of normal levels of RC DNA.
Figure 3.
Base pairing between 3E and M3 contributes to the synthesis of RC DNA. (A) Putative base-pairing pattern between 3E and M3 elements on the minus-strand DNA. Substitutions designed to disrupt the putative base pairing are shown. Variants to restore the base-pairing potential were constructed by combining the appropriate 3E and M3 substitutions. (B) Southern blotting of WT reference and substitution variants. The positions of RC, DL, and SS DNA are indicated. The blot was hybridized with a genome-length, minus-strand-specific probe. (C) The percentage of RC DNA on the Southern blot was calculated by dividing the level of RC DNA by the total amount of RC, DL, and SS DNA. The mean value is presented for each virus, with the error bars indicating one standard deviation. Each virus was analyzed multiple times from independent transfections (WT, n = 16; M3A, 3EA, and M3A/3EA, n = 8; M3B, 3EB, and M3B/3EB, n = 8).
Although Southern blot analysis indicated that 3E and M3 are required to base-pair for the synthesis of RC DNA, it did not indicate which step or steps in plus-strand DNA synthesis were inhibited by disrupting base pairing. The accumulation of SS DNA seen in the Southern blotting could be due to an inhibition of primer translocation or an inhibition of circularization or both. Therefore a second approach, primer extension, was used to determine the extent that the two template switches were affected (for a diagram of this strategy, see Fig. 6, which is published as supporting information on the PNAS web site). By using a primer that anneals to a position before circularization on the plus strand and a second primer shortly after circularization, the two template switches, primer translocation and circularization, are measured. Here, primer translocation is defined and measured as the amount of plus-strand DNA that initiates from DR2 and elongates to the 5′ end of minus-strand DNA (the precircularization point). Circularization is defined and measured as the fraction of plus-strand DNA initiating from DR2 that elongated beyond the circularization point (see Fig. 6). In situ priming is also measured in the assay, as the amount of plus-strand DNA that initiates from DR1. Total primer utilization is defined and measured as the sum of the amount of priming from DR2 and DR1. A third primer extension reaction measures the level of minus-strand DNA synthesized. All measurements of plus-strand DNA are normalized to the level to minus-strand DNA.
The primer extension analysis was performed on the M3A/3EA series of variants. The results (Table 1) indicated that both single mutants were defective for the two template switches. Specifically, the efficiency of primer translocation for the M3A and 3EA mutants was 26% and 23% of the level of the WT standard, respectively (P < 0.02). A dramatic reduction in circularization was also observed in these mutants (P < 0.02). In addition, disrupting the base pairing decreased the total primer utilization to approximately 1/3 of the WT level (P < 0.02). More importantly, the double mutant M3A/3EA supported primer translocation, circularization, and primer utilization significantly better than each of the single mutations and at a level similar to that of the WT standard. This analysis corroborates and extends the Southern blotting analysis to indicate that base pairing between M3 and 3E contributes to both primer translocation and circularization during plus-strand DNA synthesis.
Base Pairing Between M5 and 5E Contributes to Primer Translocation and Circularization During Plus-Strand DNA Synthesis.
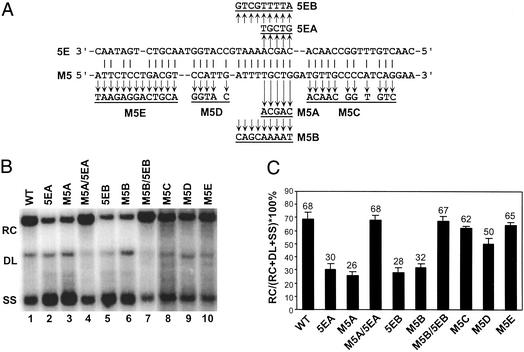
5E and M5 have the potential to form as many as 34 Watson–Crick base pairs over a 43-nt stretch (Fig. 4A). To determine whether base pairing within these sequences contributes to the synthesis of RC DNA, we made substitutions in the M5 and 5E sequences to disrupt and restore the potential for base pairing. We first targeted the region that contains nine consecutive predicted base pairs and analyzed two sets of variants (set A and B). Southern blotting revealed that each of the four single mutants that disrupted the potential for base pairing was moderately defective for synthesizing RC DNA compared with the WT standard (P < 0.008). More importantly, each of the double mutants, M5A/5EA and M5B/5EB, which restored the base-pairing potential, synthesized RC DNA at levels comparable to the WT standard (Fig. 4 B and C). Although the magnitude of the defects for the M5 and 5E single mutants was not as great as the M3 and 3E single variants, they were statistically significant. These results indicate that 5E and M5 base-pair to contribute to the synthesis of RC DNA.
Figure 4.
Base pairing between 5E and M5 contributes to the synthesis of RC DNA. (A) Putative base-pairing pattern between 5E and M5 on the minus-strand DNA is shown. Nucleotide substitutions that disrupt base pairing are shown. Variants that restore the base-pairing potential were constructed by combining appropriate 5E and M5 substitutions. M5E mutation changed both putative base-paired and non-base-paired nucleotides. (B) Southern blotting of WT reference and substitution variants. The positions of RC, DL, and SS DNA are indicated. The blot was hybridized with a genome-length, minus-strand-specific probe. (C) The percentage of RC DNA on the Southern blot was calculated by dividing the level of RC DNA by the total amount of RC, DL, and SS DNA. The mean value is presented for each virus, with the error bars indicating one standard deviation. Each virus was analyzed multiple times from independent transfections (WT, n = 27; M5A, 5EA, and M5A/5EA, n = 10; M5B, 5EB, and M5B/5EB, n = 9; M5C, n = 4; M5D, n = 8; M5E, n = 4).
We then performed primer extension analysis on the M5/5E series of variants. These results were consistent with, and extended, our findings from Southern blotting (Table 1). Similar to the M3/3E analysis, disrupting base pairing between 5E and M5 affected primer translocation and circularization, whereas restoring base-pairing potential restored the efficiency of these template switches during plus-strand synthesis (Table 1). The magnitude of the primer translocation and circularization defects for the 5E and M5 single mutants was moderate compared with the 3E and M3 single mutants, but statistically significant when compared with WT reference virus (P < 0.012 for the M5A and 5EA series, P < 0.044 for the M5B and 5EB series) and with the double mutants (P < 0.012 for the M5A and 5EA series, P < 0.044 for the M5B and 5EB series). In addition, mutants that disrupted base pairing also had defects in total primer utilization (P < 0.057), which was restored with the double mutants. These results demonstrate that base pairing between M5 and 5E contributes to primer translocation and circularization during plus-strand DNA synthesis.
To define better the extent of base pairing between 5E and M5 that contributes to the synthesis of RC DNA, the M5C, M5D, and M5E mutants were analyzed (Fig. 4A). Variants M5C and M5D changed nucleotides of M5 that are putatively base-paired with 5E, whereas M5E changed both base-paired and non-base-paired nucleotides. Southern blotting indicated that M5D had a slight defect in synthesizing RC DNA (73% of the WT RC; P < 0.021), whereas M5C and M5E replicated RC DNA close to WT levels (Fig. 4 B and C). This measurement indicated that these nucleotides had marginal contributions to the synthesis of RC DNA. On consideration of all of the above results, it is possible that only a limited number of base pairs between 5E and M5, as few as five within the central region, are required for function.
Discussion
As part of their replication strategies, reverse-transcribing viruses use template switches to carry out DNA synthesis. Sequence identity of the donor and acceptor sites plays an important role in template switching by ensuring that the nascent DNA anneals to the correct positions on the acceptor template. However, it has been shown in both hepadnaviruses and retroviruses that the process of template switching requires other contributions or mechanisms (14, 23). In DHBV, three cis-acting sequences (3E, M, and 5E) are required for the two plus-strand template switches. The mechanism by which these sequences contribute to template switches was unknown. In this work, we have shown that these sequences function, at least in part, by base pairing with each other. Our analyses suggest that base pairing between 3E and M3 and between 5E and M5 contributes to primer translocation and circularization during plus-strand DNA synthesis. Exactly how these base pairs facilitate template switches remains to be learned, but a straightforward idea can be suggested. The donor and acceptor sites for both plus-strand template switches are located at, or near, both ends of the minus-strand template. Base pairing among 3E, M, and 5E could place the minus-strand into a conformation such that both of its ends are juxtaposed. Juxtaposition of the ends of the minus-strand template inside the capsid could facilitate the annealing of the nascent plus-strand to the acceptor site on minus-strand DNA. Although the data are consistent with this possibility, other ideas can be considered. There are at least two reactions that are required for a successful template switch: annealing of the nascent strand to the acceptor template, followed by initiation and elongation of DNA synthesis. Our results are not inconsistent with the possibility that base pairing among 3E, M, and 5E is important for the initiation and elongation of plus-strand DNA from DR2 or for reinitiation and elongation of plus-strand DNA from 3′r. Base pairing among 3E, M, and 5E could influence initiation and elongation by making the minus-strand a better template for DNA synthesis.
In our analyses, the accumulation of intracellular replicative intermediates was measured. The net accumulation of any replicative intermediate is determined by the balance of its synthesis and loss. There are two potential pathways for the loss of RC DNA-containing capsids: exiting from the cell into the medium or intracellular degradation. We believe these two pathways did not make a significant contribution to the reduction of RC DNA in our analyses. First, most mutants accumulated levels of intracellular DNA intermediates that were similar to the WT reference. In general, to the extent that a variant accumulated less RC DNA, it accumulated more SS and DL DNA. Second, for the A and B series of M5/5E variants, the reduction in the level of RC DNA in the cytoplasm was not due to increased accumulation of RC-containing capsids or virions into the medium (data not shown). Based on these observations, we think that the cis-acting sequences 3E, M, and 5E affect the synthesis of RC DNA.
The phenotypes of 3E and M3 mutations are generally more drastic than 5E and M5 mutations, which suggests that the two duplexes are not equivalent in their contribution to function. This situation would not necessarily be predicted by the juxtaposition model. We cannot offer a coherent explanation for this observation, but it suggests that base pairing is providing additional or other roles.
In general, mutations within 3E, M, and 5E reduced the level of RC DNA and increased the level of SS DNA, with modest or no increase in the level of DL DNA. Two possibilities can be considered to explain the lack of increase in in situ priming when priming from DR2 is inhibited. The small DNA hairpin overlapping DR1 that normally suppresses in situ priming (13) would be intact in 3E, M, and 5E mutants and thus suppress in situ priming. Alternatively, disruption of base pairing between 3E, M, and 5E could lead to an overall reduction in primer utilization at both DR1 and DR2, leading to an apparent lack of increase in in situ priming.
In general, cis-acting sequences are located at the locus at which they act. For example, a protein binds to its cognate cis-acting sequence to manifest a reaction at, or near, the site of binding, such as restriction endonuclease. Or, in the case of template switching, the donor and acceptor sites, which are the cis-acting sequences, are at the site of action. The cis-acting sequences 3E, M, and 5E can be thought as being different because they are away from the site of action, at least on the primary sequence. These sequences could represent a new type of cis-acting element, whose function is to place the nucleic acid into the proper conformation such that other cis-acting sequences can function.
Our data indicate that base pairing among 3E, M, and 5E is important for the synthesis of RC DNA and suggest that this base pairing could be placing the minus-strand DNA into a conformation that juxtaposes the donor and acceptor sites for primer translocation and circularization to facilitate these template switches. This possibility invokes a more general idea: during the process of reverse transcription, which occurs within the capsid, the conformation of the RNA template and then the DNA template, which would likely be dynamic, is important for the execution of the individual steps of DNA synthesis.
Our studies were performed with DHBV, which begs the question whether base pairing among 3E, M, and 5E is a common mechanism in the hepadnavirus family. Because of the nucleotide sequence conservation, we found via sequence inspection the potential for similar but not identical base-pairing patterns in other avian members, such as heron hepatitis B virus, Ross Goose hepatitis B virus, and white stork hepatitis B virus. Whether these other avian hepadnaviruses use a similar mechanism requires testing through experimentation. Obviously, it is a germane question whether the mammalian hepadnaviruses, including HBV, use base pairing between cis-acting sequences to affect plus-strand DNA synthesis. Because of the lack of nucleotide sequence conservation between avian and mammalian hepadnaviruses, base-pairing patterns in the minus-strand of mammalian viruses are much more difficult to predict with confidence. Enumeration of the HBV cis-acting sequences required for plus-strand DNA synthesis has yet to be reported. Finally, based on the use of template switching in their replication strategies, it is not unreasonable to propose that retroviruses and caulimoviruses might also use base pairing to affect template switching.
Supplementary Material
Acknowledgments
We thank Paul Ahlquist, Jeff Habig, Amanda Mack, Kristin Ostrow, Bill Sugden, and Jesse Summers for many helpful discussions and critical review of this manuscript. We thank Norman Drinkwater and Jianzhong Zhang for help with statistical analyses. This work was supported by National Institutes of Health Grants R29 GM50263, P01 CA22443, P30 CA07175, and P30 CA14520 and American Cancer Society Grant JFRA-651.
Abbreviations
- DHBV
duck hepatitis B virus
- pgRNA
pregenomic RNA
- RC
relaxed-circular
- DL
duplex-linear
- SS
single-stranded
- DR1
direct repeat 1
- DR2
direct repeat 2
- r
redundancy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Summers J, Mason W S. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 2.Gilboa E, Mitra S W, Goff S, Baltimore D. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 3.Pfeiffer J K, Telesnitsky A. J Virol. 2001;75:11263–11274. doi: 10.1128/JVI.75.23.11263-11274.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Temin H M. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganem D, Schneider R. In: Fields Virology. Knipe D, Howley P, Griffin D, Lamb R, Martin M, Roizman B, Strauss S, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2923–2969. [Google Scholar]
- 6.Tavis J E, Perri S, Ganem D. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G H, Seeger C. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 8.Wang G H, Seeger C. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lien J M, Aldrich C E, Mason W S. J Virol. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb D D, Hirsch R C, Ganem D. EMBO J. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staprans S, Loeb D D, Ganem D. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loeb D D, Gulya K J, Tian R. J Virol. 1997;71:152–160. doi: 10.1128/jvi.71.1.152-160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habig J W, Loeb D D. J Virol. 2002;76:980–989. doi: 10.1128/JVI.76.3.980-989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havert M B, Loeb D D. J Virol. 1997;71:5336–5344. doi: 10.1128/jvi.71.7.5336-5344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havert M B, Ji L, Loeb D D. J Virol. 2002;76:2763–2769. doi: 10.1128/JVI.76.6.2763-2769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller-Hill K, Loeb D D. J Virol. 1996;70:8310–8317. doi: 10.1128/jvi.70.12.8310-8317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller-Hill K, Loeb D D. J Virol. 2002;76:4260–4266. doi: 10.1128/JVI.76.9.4260-4266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprengel R, Kuhn C, Will H, Schaller H. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 20.Ke S H, Madison E L. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu N, Ostrow K M, Loeb D D. Virology. 2002;295:348–359. doi: 10.1006/viro.2002.1425. [DOI] [PubMed] [Google Scholar]
- 22.Calvert J, Summers J. J Virol. 1994;68:2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topping R, Demoitie M A, Shin N H, Telesnitsky A. J Mol Biol. 1998;281:1–15. doi: 10.1006/jmbi.1998.1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.