Abstract
Group A Streptococcus (GAS) evades polymorphonuclear leukocyte (PMN) phagocytosis and killing to cause human disease, including pharyngitis and necrotizing fasciitis (flesh-eating syndrome). We show that GAS genes differentially regulated during phagocytic interaction with human PMNs comprise a global pathogen-protective response to innate immunity. GAS prophage genes and genes involved in virulence, oxidative stress, cell wall biosynthesis, and gene regulation were up-regulated during PMN phagocytosis. Genes encoding novel secreted proteins were up-regulated, and the proteins were produced during human GAS infections. We discovered an essential role for the Ihk-Irr two-component regulatory system in evading PMN-mediated killing and promoting host–cell lysis, processes that would facilitate GAS pathogenesis. Importantly, the irr gene was highly expressed during human GAS pharyngitis. We conclude that a complex pathogen genetic program circumvents human innate immunity to promote disease. The gene regulatory program revealed by our studies identifies previously undescribed potential vaccine antigens and targets for therapeutic interventions designed to control GAS infections.
Polymorphonuclear leukocytes (PMNs) are critical effectors of the human innate immune system and provide an essential first line of host defense against invading microorganisms. PMNs eliminate invading bacterial pathogens by a process known as phagocytosis (1). Ingested microorganisms are subsequently destroyed by reactive oxygen species (ROS) and microbicidal products contained within PMN granules (2). Although many human pathogens are killed readily by PMNs, some have evolved mechanisms to inhibit phagocytosis and death resulting from exposure to ROS and microbicidal products. The global gene regulatory networks used by pathogens to escape destruction by PMNs have not been studied.
Group A Streptococcus (GAS) successfully evades PMN phagocytosis and killing to cause human infections such as pharyngitis, cellulitis, and necrotizing fasciitis (flesh-eating syndrome). These infections and postinfection sequelae (acute rheumatic fever and glomerulonephritis) are responsible for high morbidity and mortality globally (3). Several extracellular molecules are known to contribute to the ability of GAS to resist phagocytosis (3–6); however, a comprehensive analysis of changes in GAS expression that occur during interaction with the human innate immune system has not been conducted. In addition, although significant progress has been made toward understanding host responses to invading microorganisms, there is limited understanding of the gene regulatory pathways used by pathogens to circumvent the innate immune response, and thereby survive and cause disease. To address this deficiency in knowledge, we studied GAS gene expression during phagocytic interaction with human PMNs.
Materials and Methods
Detailed protocols can be found in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Bacterial Strains.
Serotype M1 GAS strain MGAS5005 was used because it is genetically representative of serotype M1 strains causing abundant disease in North America and Western Europe (7). Serotype M6 GAS strains JRS4 and isogenic irr− mutant, JRS550, have been described (8).
Isolation of Human PMNs.
PMNs were isolated from venous blood of healthy individuals (9) in accordance with a protocol approved by the Institutional Review Board for Human Subjects, National Institute of Allergy and Infectious Diseases.
Phagocytosis Experiments and Electron Microscopy.
Phagocytosis and killing of GAS by human PMNs and production of PMN ROS were determined as described (6, 10). For transmission electron microscopy (TEM), phagocytosis assays were performed in wells of a 24-well culture plate containing serum-coated Thermanox coverslips (Nunc). Samples were examined with a model CM10 transmission electron microscope (Philips, Eindhoven, The Netherlands).
GAS Gene Expression Microarray Experiments.
Phagocytosis of GAS by human PMNs is described in Supporting Methods. Total RNA from each sample was isolated with RNeasy kits (Qiagen, Valencia, CA), and contaminating PMN mRNA was removed with an Oligotex mRNA kit (Qiagen). cDNA was generated from GAS RNA and used to probe a DNA microarray containing 1,705 (of 1,752) M1 GAS ORFs based on strain SF370 (11), and unique M18 and M3 ORFs (12, 13, 15). All genes identified as differentially expressed passed a filter based on signal-minus-background, spot area, and signal-to-noise ratio, and were at least 1 SD above controls. Fold changes for each gene were determined from the median fluorescence ratio of spots containing GAS alone to those of GAS during phagocytosis. The experiment was performed in triplicate (on separate days with different blood donors) with each slide containing 4–12 spots per gene (n ≥ 12 spots).
TaqMan Real-Time RT-PCR and Analysis of irr Gene Expression in Human Pharyngitis.
TaqMan analysis of selected genes (n = 26) was performed in duplicate with RNA from three phagocytosis experiments using an ABI 7700 thermocycler (Applied Biosystems) as described (14, 15).
For analysis of irr expression in human clinical material, throat swabs were taken from the posterior pharynx of 16 pediatric patients presenting with acute GAS pharyngitis in accordance with a protocol approved by the Institutional Review Board for Human Subjects at Baylor College of Medicine (Houston). RNA was isolated from each swab and TaqMan real-time RT PCR was performed as described in Supporting Methods.
Cloning and Expression of Novel GAS Secreted Proteins.
The region of the genes encoding mature (lacking secretion signal sequence) Spy0136 (H28 to E221) and Spy2191 (D36 to 204Y) were cloned from serotype M1 GAS strain MGAS5005 and overexpressed in recombinant Escherichia coli BL21.
Results and Discussion
Phagocytosis and Killing of GAS by Human PMNs.
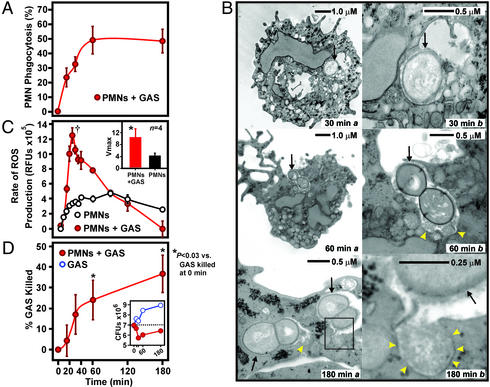
We investigated the ability of human PMNs to ingest and kill GAS, a bacterial pathogen that causes widespread disease in humans (Fig. 1). PMNs ingested opsonized GAS within 30 min of incubation (phagocytosis increased from 0.0% at 0 min to 32.7 ± 5.1% by 30 min) (Fig. 1A). In contrast, there was no increase in phagocytosis of GAS from 60 to 180 min (49.3 ± 9.5% at 60 min vs. 48.7 ± 8.2% at 180 min) (Fig. 1A). We confirmed that GAS were ingested by PMNs and subsequently exposed to PMN microbicidal granule contents and ROS (Fig. 1 B and C). Inasmuch as PMN granule contents and ROS kill most microorganisms effectively, we next determined the extent of GAS killing by PMNs (Fig. 1D). By 180 min of incubation, PMNs had killed only 36.7 ± 9.0% of the GAS (Fig. 1D). Moreover, the rate that GAS were killed by PMNs decreased dramatically after the first 30 min (Fig. 1D). These data indicate that as a population GAS became more resistant to phagocytosis and killing by PMNs and/or produced factors that altered normal PMN function after the initial stages (0–30 min) of GAS–PMN interaction.
Figure 1.
GAS–PMN interaction. (A) Phagocytosis of GAS. Phagocytosis (%) is the percent of PMNs that contain ingested GAS at each time. Results are the mean ± SD of three experiments. (B) Ultrastructural analyses of PMNs containing ingested GAS. Black arrows indicate ingested GAS. (Right) Higher magnification. Yellow arrowheads indicate fusion of PMN granules with GAS phagosomes. (C) ROS production during phagocytosis of GAS. Data shown are from a representative experiment performed four times. Inset illustrates the rate of ROS production at Vmax (†) (mean ± SD of four experiments). *, P = 0.006 vs. unstimulated PMNs. (D) Killing of GAS by PMNs. At each time, PMNs were lysed and GAS were plated on Todd–Hewitt agar containing yeast extract. Colonies were enumerated the following day, and percent GAS killed was calculated by using the equation (1 − (CFUPMN+/CFUPMN−)) × 100 as described in Supporting Methods. Results are the mean ± SD of three experiments. *, P < 0.03 vs. PMNs + GAS at 0 min, one-way ANOVA with Tukey posttest. Inset illustrates representative growth curves of GAS in the presence (red) and absence (blue) of PMNs.
Global Changes in Gene Expression by GAS During PMN Phagocytosis.
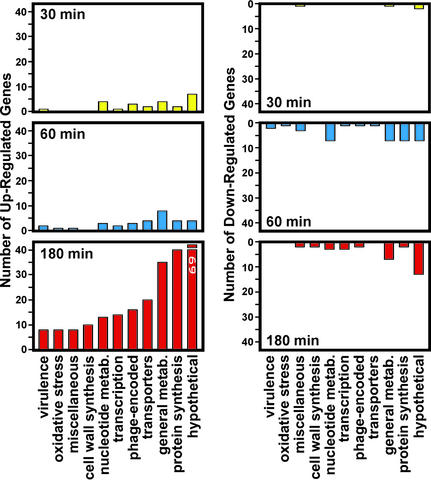
To test the hypothesis that GAS evades the human innate immune response by altering its pattern of gene transcription, we measured global changes in GAS gene expression during GAS–PMN interaction with a DNA microarray comprising 97.3% of the serotype M1 GAS genome. Relatively few GAS genes were differentially regulated at 30 and 60 min after initial GAS–PMN interaction (28 genes at 30 min and 69 genes at 60 min, 1.6% and 4.0% of the genome, respectively) (Fig. 2 and Table 1, which is published as supporting information on the PNAS web site). The greatest changes in GAS gene expression were observed at 180 min, a time when the rate of PMN phagocytosis and GAS killing were significantly reduced compared with earlier time points (Fig. 2). Two hundred seventy-six genes (≈16.0% of the genes comprising the M1 GAS genome) were differentially transcribed at this time point (Fig. 2). Importantly, 69 genes encoding proteins with undefined function were up-regulated, representing ≈20% of all differentially regulated genes (Table 1).
Figure 2.
Differential GAS gene expression during PMN phagocytosis. PMNs were incubated with GAS for the indicated times, and gene expression was measured with a DNA microarray containing 1,705 (of 1,752) serotype M1 GAS ORFs, 97.3% of the genome. Data were analyzed with genespring software, version 4.2 (Silicon Genetics).
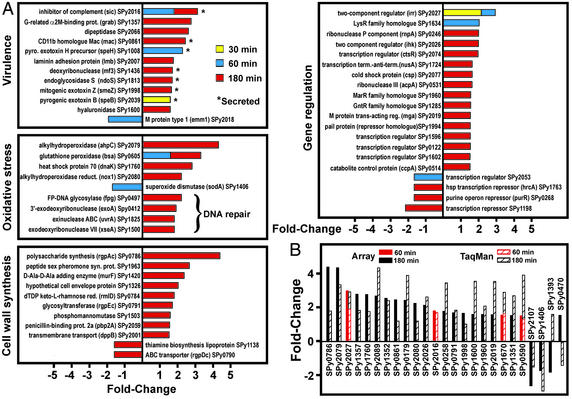
Up-Regulation of GAS Genes Encoding Proteins That Mediate Virulence and Evasion of Host Defense During PMN Phagocytosis.
The GAS genes differentially expressed during PMN phagocytosis were divided into categories based on function or annotation (Fig. 3A, and Fig. 7, which is published as supporting information on the PNAS web site). At least 11 genes associated with GAS virulence, including sic (streptococcal inhibitor of complement), mac (human Mac-1 homologue), speH (streptococcal pyrogenic exotoxin H), ndoS (endoglycosidase S), smeZ (streptococcal mitogenic exotoxin Z), and speB (streptococcal pyrogenic exotoxin B) were up-regulated (Fig. 3A). This finding is important because seven of these genes encode secreted proteins known to inhibit PMN phagocytosis or otherwise modulate the innate immune response of the host (5, 6, 16). For example, SpeB, Sic, and Mac each contribute directly to GAS pathogenesis by blocking PMN phagocytosis and promoting pathogen survival (5, 6, 16). The gene encoding sortase A (srtA) was up-regulated (Fig. 7A). Sortase A is a protein required for surface localization of virulence GAS factors such as GRAB and M protein (17). Consistent with our finding that GAS was exposed to PMN-derived ROS (Fig. 1B), bsa (glutathione peroxidase), ahpC (alkylhydroperoxidase), dnaK (hsp70), and nox1 (alkylhydroperoxidase reductase) were up-regulated (Fig. 3A). These genes encode proteins that detoxify cell-damaging ROS or other free radicals. Four genes encoding proteins involved in DNA repair were up-regulated, including fpg (formamidopyrimidine-DNA glycosylase), an enzyme that removes oxidized purines from oxidatively damaged DNA (Fig. 3A). Taken together, these data demonstrate that evasion of PMN phagocytosis and killing by GAS is regulated at the level of gene transcription.
Figure 3.
GAS genome-wide protective response to PMN phagocytosis. (A) GAS genes differentially expressed during PMN phagocytosis. Results are presented as the mean fold-induction or repression of genes from three experiments (three blood donors with phagocytosis assays done on separate days) and three accompanying microarray experiments (done on separate days). (B) TaqMan confirmation of microarray results. Genes (n = 26) identified as differentially transcribed by GAS microarrays were analyzed by TaqMan real-time PCR. There was a strong positive correlation (r = 0.76) between TaqMan and microarray results, consistent with previous comparisons (15, 25).
Gene-Regulated Cell Wall Biosynthesis and Metabolic Pathways in GAS During PMN Phagocytosis.
During phagocytic interaction with PMNs, GAS up-regulated nine genes encoding proteins that participate in cell wall biosynthesis (Fig. 3A). These findings suggest that there was rapid cell wall turnover and/or repair, presumably in response to PMN-mediated cell wall damage. Consistent with a need for increased energy and biosynthetic activity for production of GAS cell wall components and virulence factors, genes encoding enzymes that represent distinct metabolic pathways were up-regulated (Figs. 3A and 7). For example, seven genes encoding enzymes involved in glycolysis and 17 genes encoding proteins critical to nucleotide and nucleic acid biosynthesis and metabolism, including parE (DNA topoisomerase IV) and holB (DNA polymerase III), were up-regulated (Fig. 7B). These findings provide strong evidence that up-regulation of energy metabolism and multiple biosynthetic processes are critical for immune evasion by GAS.
Phagocytosis Induces Differential Expression of GAS Genes Involved in Transcription and Transcriptional Regulation.
Our finding that GAS differentially regulated expression of genes important for pathogen survival during phagocytic interaction with human PMNs implies a role for transcriptional regulatory proteins. Consistent with this hypothesis, mga, a gene encoding a transcriptional regulator of virulence factors (18), was up-regulated during phagocytosis (Fig. 3A).
Importantly, genes encoding both proteins of a two-component gene regulatory system, ihk and irr (SPy2026 and SPy2027, respectively) (8) were up-regulated (Fig. 3A). These genes have homology with the ArlS-ArlR two-component gene regulatory system of Staphylococcus aureus (19), which regulates production of virulence factors (20). In GAS, the function of Ihk-Irr is not known nor is it known which genes it controls; however, Federle et al. (8) reported that insertional inactivation of irr did not alter in vitro growth. Up-regulation of genes encoding Ihk and Irr during phagocytosis by human PMNs suggests that this gene regulatory system promotes evasion from host innate immunity, a hypothesis tested below.
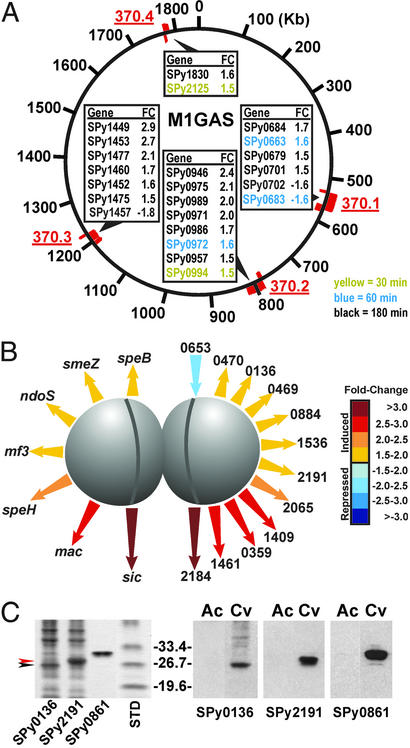
Differential Expression of GAS Genes Encoded by Prophages During GAS–PMN Interaction.
Twenty-three genes encoded by prophages or prophage-like elements in the M1 GAS strain SF370 (11), were differentially regulated during phagocytosis (Fig. 4A). These findings are important because many proven and putative GAS virulence factors are encoded by prophages, including streptococcal pyrogenic toxin superantigens, DNases, and a recently identified phospholipase A2 (13, 21). Our discovery that 20 of the 23 differentially regulated phage-encoded GAS genes are up-regulated during PMN phagocytosis suggests that these genes play a role in host–pathogen interaction.
Figure 4.
Genes encoding GAS prophage and secreted proteins. (A) Prophage-encoded genes differentially regulated during phagocytosis. Genes were assigned to prophage (red) based on homology with serotype M1 GAS SF370 (11). (B) Differential expression of genes encoding secreted proteins of known (left) and unknown (right) function. Length of arrows is relative for gene expression. (C) Production of novel secreted proteins in humans with GAS disease. SPy0136 and SPy2191 were overexpressed in E. coli and separated by SDS-PAGE (Gel-Code protein stain, Left). Comparison of acute (Ac) and convalescent (Cv) paired sera from an individual with GAS invasive disease (Right). Data are representative of paired human sera from three individuals with GAS invasive disease.
Identification of Novel GAS Secreted Proteins Up-Regulated During Phagocytosis.
Given the critical contribution of GAS exoproteins to virulence, we note that 18 genes encoding proteins with predicted secretion signal motifs (http://psort.nibb.ac.jp/form.html) were up-regulated (Fig. 4B). Seven of these genes encode proteins implicated in virulence (3, 5, 6, 22) (Fig. 4B Left). The other 11 genes encode previously uncharacterized secreted proteins (Fig. 4B Right). To determine whether these genes encode proteins that are produced in infected humans, we overexpressed two representative proteins of unknown function (SPy0136 and SPy2191) in E. coli and tested immunoreactivity of the lysates with sera from patients with GAS invasive disease (Fig. 4C). Antibodies to SPy0136, SPy2191, and Spy0861 (Mac, a control protein known to be produced during GAS infections in humans) (5), were absent in sera obtained from the acute stage of infection, but were present during the convalescent stage (Fig. 4C Right), indicating that these proteins are made during human infections. Based on the observation that genes encoding Spy0136 and SPy2191 are up-regulated during PMN phagocytosis, and that each protein contains a secretion signal motif, we speculate that they assist evasion from innate immune surveillance, thereby promoting GAS survival in vivo.
Confirmation of Microarray Data by TaqMan Real-Time RT-PCR.
We next used TaqMan RT-PCR to verify changes in expression detected by microarray analysis (Fig. 3B). We selected 26 genes representative of the entire microarray data set for confirmation (Fig. 3B). These genes were assigned to six different functional categories, and included genes that encode proteins predicted to participate in GAS virulence and/or survival within the host. There was strong correlation between the TaqMan and microarray gene expression data, consistent with previous studies (Fig. 3B) (15). Importantly, we confirmed that ihk and irr (SPy2026 and SPy2027, respectively) were up-regulated (Fig. 3B).
Ihk-Irr Two-Component Gene Regulatory System Contributes to Evasion of Human Innate Immunity.
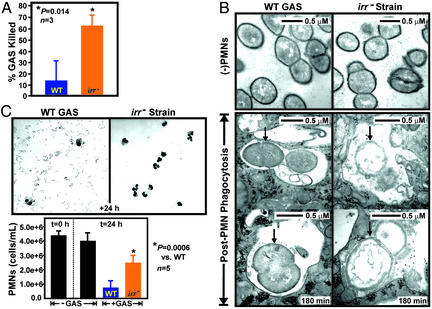
The mga regulon, and fasBCA and covR/covS two-component gene regulatory systems are the only global transcriptional regulatory systems linked to virulence factor production in GAS (23–25). Our finding that genes encoding Ihk and Irr were up-regulated during PMN phagocytosis suggest that this transcriptional regulatory system controls (directly or indirectly) genes that assist GAS immune evasion (Fig. 3). To directly test this hypothesis, we compared the ability of human PMNs to kill an isogenic irr− mutant strain with the parental wild-type strain (Fig. 5A). Significantly more of the mutant strain was killed than the wild-type strain at 180 min (62.7 ± 9.6% GAS mutant killed versus 13.6 ± 17.8% for the wild-type strain, n = 3, P = 0.01) (Fig. 5A), indicating that Irr-controlled genes are necessary for evasion of PMN-mediated killing. Increased killing of the isogenic GAS mutant by human PMNs was due neither to enhanced phagocytosis nor exposure to more ROS, because both of these GAS–PMN interaction sequelae were essentially identical for the wild-type and irr− isogenic mutant strains (43.9 ± 3.4% and 41.1 ± 3.6% phagocytosis at 180 min, respectively, n = 4; PMN ROS production data not shown). These findings suggested that Ihk-Irr enhances GAS survival at a step subsequent to PMN phagocytosis. To test this hypothesis, we analyzed wild-type and mutant GAS strains that had been ingested by human PMNs at 180 min by TEM (Fig. 5B). Significantly more of the mutant strain was destroyed after phagocytosis (at 180 min) compared with the parental wild-type strain (68.0 ± 14.6% of the mutant GAS strain destroyed vs. 38.3 ± 12.4% of the wild type GAS strain destroyed, P < 0.0001, n > 200 GAS each), demonstrating that Ihk-Irr controls expression of genes critical for GAS survival after phagocytosis (Fig. 5B). Importantly, we discovered that the mutant strain was reduced significantly in its ability to cause PMN lysis compared with the wild-type strain, a finding that has important implications for our understanding of streptococcal disease (Fig. 5C).
Figure 5.
Ihk-Irr two-component gene regulatory system is essential for GAS survival and host-cell lysis. (A) Killing of mutant (irr−) strain by PMNs at 180 min. Results are the mean ± SD of three experiments. (B) Ultrastructural analyses of PMNs containing ingested wild-type (WT) or mutant (irr−) strains at 180 min. Black arrows indicate ingested GAS. (C) Isogenic GAS mutant (irr−) strain has reduced ability to cause PMN lysis. Results are the mean ± SD of three to five experiments.
irr Is Expressed in Infected Humans.
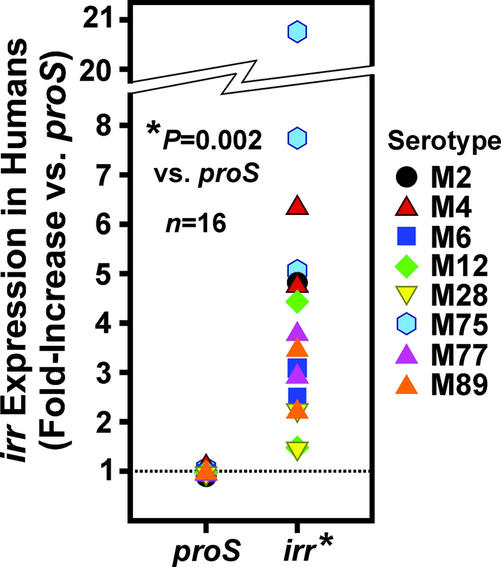
To directly determine whether irr is important for GAS pathogenesis in humans, we measured irr expression in 16 patients with GAS pharyngitis using TaqMan real-time PCR (Fig. 6). Consistent with the finding that Ihk-Irr facilitates escape from innate immunity, irr was highly expressed in GAS isolated from these 16 patients (Fig. 6). Further, expression of irr was increased significantly compared with control gene proS in strains of many distinct M serotypes (P = 0.002) (Fig. 6). Taken together, these findings provide strong evidence that Ihk-Irr is critical for GAS survival and pathogenesis in humans.
Figure 6.
The irr gene is highly expressed in vivo in human infections. Transcript levels for irr were determined by TaqMan real-time PCR in 16 patients with GAS pharyngitis. Results are expressed as fold-increase compared with the GAS proS gene.
Conclusions.
Several lines of evidence indicate that global changes in GAS gene expression are essential for survival and pathogenesis in the human host. First, rates of PMN phagocytosis and killing of GAS decrease after initial host cell–pathogen interaction, presumably because of induction of genes critical for survival. Consistent with this hypothesis, the greatest numbers of differentially expressed genes were detected 180 min after GAS–PMN interaction. We discovered that in addition to virulence factors, multiple gene-regulated pathways and prophage-encoded proteins likely contribute to the ability of GAS to evade innate immunity. Importantly, we also identified an array of novel secreted proteins that were made during human GAS infections and likely promote immune evasion.
Another critical discovery in our study was that the Ihk-Irr two component gene-regulatory system is essential for GAS survival after PMN phagocytosis. This gene-regulatory system was not previously recognized to have a fundamental role in GAS pathogenesis, and it will be important to determine the role of this gene regulatory system in disease progression in vivo. Microarray analysis was key to generating a global understanding of how GAS responds to the human innate immune system, providing new insight into bacterial pathogenesis. The GAS regulatory program revealed by our studies identified many new targets for therapeutic interventions and other maneuvers designed to control disease caused by this human bacterial pathogen.
Supplementary Material
Acknowledgments
We thank M. Graham, S. Beres, and D. Banks for helpful discussion, J. Scott for generously providing GAS strains JRS4 and JRS550, and physicians and patients at Pediatric Medical Group (an affiliate of Texas Children's Pediatric Associates).
Abbreviations
- GAS
Group A Streptococcus
- PMN
polymorphonuclear leukocyte
- ROS
reactive oxygen species
- TEM
transmission electron microscopy
References
- 1.Jones S L, Lindberg F P, Brown E J. In: Fundamental Immunology. Paul W E, editor. New York: Lippincott-Raven; 1999. pp. 997–1021. [Google Scholar]
- 2.Nauseef W M, Clark R A. In: Basic Principles in the Diagnosis and Management of Infectious Diseases. Mandell G L, Bennett J E, Dolin R, editors. New York: Churchill Livingstone; 2000. pp. 89–112. [Google Scholar]
- 3.Cunningham M W. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank M M. Nat Med. 2001;7:1285–1286. doi: 10.1038/nm1201-1285. [DOI] [PubMed] [Google Scholar]
- 5.Lei B, DeLeo F R, Hoe N P, Graham M R, Mackie S M, Cole R L, Liu M, Hill H R, Low D E, Federle M J, et al. Nat Med. 2001;7:1298–1305. doi: 10.1038/nm1201-1298. [DOI] [PubMed] [Google Scholar]
- 6.Hoe N P, Ireland R M, DeLeo F R, Gowen B B, Dorward D W, Voyich J M, Liu M, Burns E H, Jr, Culnan D M, Bretscher A, Musser J M. Proc Natl Acad Sci USA. 2002;99:7646–7651. doi: 10.1073/pnas.112039899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoe N P, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, Dou S J, Pan X, Vuopio-Varkila J, Salmelinna S, et al. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 8.Federle M J, McIver K S, Scott J R. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyum A. Scand J Clin Lab Invest Suppl. 1968;97:77–89. [PubMed] [Google Scholar]
- 10.DeLeo F R, Allen L A, Apicella M, Nauseef W M. J Immunol. 1999;163:6732–6740. [PubMed] [Google Scholar]
- 11.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, et al. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smoot J C, Barbian K D, Van Gompel J J, Smoot L M, Chaussee M S, Sylva G L, Sturdevant D E, Ricklefs S M, Porcella S F, Parkins L D, et al. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beres S B, Sylva G L, Barbian K D, Lei B, Hoff J S, Mammarella N D, Liu M Y, Smoot J C, Porcella S F, Parkins L D, et al. Proc Natl Acad Sci USA. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaussee M S, Watson R O, Smoot J C, Musser J M. Infect Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smoot L M, Smoot J C, Graham M R, Somerville G A, Sturdevant D E, Migliaccio C A, Sylva G L, Musser J M. Proc Natl Acad Sci USA. 2001;98:10416–10421. doi: 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukomski S, Burns E H, Wyde P R, Podbeilski A, Rurangirwa J, Morre-Poveda D K, Musser J M. Infect Immun. 1998;66:771–776. doi: 10.1128/iai.66.2.771-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnett T C, Scott J R. J Bacteriol. 2002;184:2181–2191. doi: 10.1128/JB.184.8.2181-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kihlberg B M, Cooney J, Caparon M G, Olsen A, Bjork L. Microb Pathog. 1995;19:229–3125. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 19.Fournier B, Hooper D C. J Bacteriol. 2000;182:3955–3964. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fournier B, Klier A, Rapoport G. Mol Microbiol. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 21.Banks D J, Beres S B, Musser J M. Trends Microbiol. 2002;10:515–521. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 22.Collin M, Olsen A. EMBO J. 2001;20:3046–3055. doi: 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin J C, Wessels M R. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 24.Kreikemeyer B, Boyle M D, Buttaro B A, Heinemann M, Podbielski A. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 25.Graham M R, Smoot L M, Migliaccio C A, Virtaneva K, Sturdevant D E, Porcella S F, Federle M J, Adams G J, Scott J R, Musser J M. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.