Abstract
Localized mRNAs are thought to be transported in defined particles to their final destination. These particles represent large protein complexes that may be involved in recognizing, transporting, and anchoring localized messages. Few components of these ribonucleoparticles, however, have been identified yet. We chose the strategy to biochemically enrich native RNA–protein complexes involved in RNA transport to identify the associated RNAs and proteins. Because Staufen proteins were implicated in intracellular RNA transport, we chose mammalian Staufen proteins as markers for the purification of RNA transport particles. Here, we present evidence that Staufen proteins exist in two different complexes: (i) distinct large, ribosome- and endoplasmic reticulum-containing granules preferentially found in the membrane pellets during differential centrifugation and (ii) smaller particles in the S100 from rat brain homogenates. On gel filtration of the S100, we identified soluble 670-kDa Staufen1-containing and 440-kDa Staufen2-containing particles. They do not cofractionate with ribosomes and endoplasmic reticulum but rather coenrich with kinesin heavy chain. Furthermore, the fractions containing the Staufen1 particles show a 15-fold enrichment of mRNAs compared with control fractions. Most importantly, these fractions are highly enriched in BC1, and, to a lesser extent, in the α-subunit of the Ca2+/calmodulin-dependent kinase II, two dendritically localized RNAs. Finally, both RNAs colocalize with Staufen1–hemagglutinin in particles in dendrites of transfected hippocampal neurons. We therefore propose that these Staufen1-containing particles may represent RNA transport intermediates that are in transit to their final destination within neurons.
Keywords: Ca2+, calmodulin-dependent kinase II‖double-stranded RNA-binding protein‖dendritic mRNA transport‖BC1
Localizing a certain transcript can restrict the distribution of a protein to a particular cytoplasmic domain of a polarized cell (1–6). In polarized neurons, however, very little is known about the molecular mechanism, especially how dendritically localized transcripts are targeted to distal parts of dendrites. The first hint that RNA may be transported via ribonucleoproteins (RNPs) in neurons came from work on the noncoding BC1 transcript (7) because BC1 RNA was found to be part of a 10S RNP complex (8). Then, it was shown that myelin basic protein mRNA, when microinjected into cultured oligodendrocytes, formed granules, which were transported to the cell's periphery (9). Based on these data, a model was proposed that mRNA localization in a mammalian cell might be achieved through the assembly and translocation of RNA transport particles (10). In the meantime, localized mRNA-containing particles, termed mRNA granules, have been observed in many cell types (3–6). This finding led to the suggestion that such large protein complexes may be involved in recognizing, transporting, and anchoring localized messages. The search for such mRNA granules has been significantly facilitated by the identification of several conserved families of transacting factors (reviewed in refs. 5 and 11) that bind to cis-acting mRNA localization elements residing within the 3′ UTR (2, 12).
We decided to focus on Staufen-containing transport particles for several reasons. First, a previous study succeeded in visualizing the microtubule-dependent transport of Staufen–GFP particles into dendrites of living hippocampal neurons (13). Second, an immuno-electron microscopy study found Staufen1 (Stau1) to be present in RNA granules in neurons (14). This finding was extended by another recent study that identified Staufen to be part of large mRNA–protein complexes associated with rough endoplastic reticulum (ER) (15). Third, a second mammalian homolog of Staufen (Stau2) has been identified (16, 17) that might also play an important role in the delivery of (yet unidentified) RNAs into dendrites of hippocampal neurons (16). These observations along with the evolutionary conservation of Staufen protein structure strongly support the involvement of Staufen proteins in RNA transport via RNPs in mammalian cells.
Methods
Generation of Stau1 Antibodies.
Soluble his6-Stau1 (amino acid 179 to the end in pRSET vector, a gift from J. Ortín, Consejo Superior de Investigaciones Cientificas, Madrid), expressed in Escherichia coli was purified under denaturing conditions on Ni-NTA Sepharose (Qiagen, Chatsworth, CA). The antigens were purified from SDS/PAGE as described (18). Mice were injected i.p. with 50 μg of protein per injection. The immune serum was prepared as described (18, 19). The resulting polyclonal antibodies were tested for their specificity for Stau1 and no cross-reactivity with Stau2 could be observed. For Western blot assay, a dilution of 1:1,000 was used.
Preparation of Rat Brain Homogenates and Differential Centrifugation.
Sprague–Dawley rats (6–8 weeks old, <250 g) were anesthetized with ether and decapitated. All steps were performed under strict RNase-free conditions at 4°C or on ice. Two forebrains were homogenized on ice in 4 ml of lysis buffer (150 mM KCl/50 mM Hepes/1 mM DTT/80 units/ml RNasin and proteinase inhibitors) and centrifuged for 10 min at 16,000 × g (P16 vs. S16). The supernatant was separated and centrifuged for 20 min at 100,000 × g (P100 and S100). To analyze the distribution of the different proteins in the resulting fractions, samples were processed for SDS/PAGE including chloroform/methanol extraction as described (19). Fifty micrograms of total protein per lane was separated on 10% SDS/PAGE gels.
Gel Filtration (GF) Analysis.
Throughout the study, a Sephacryl S300-HR GF column from Amersham Pharmacia was used and calibrated with the following size markers (Sigma): blue dextran (2 MDa), thyroglobulin (669 kDa), apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), and BSA (66 kDa). Two milliliters of S100 (15–19 mg total protein) was loaded onto the GF column; proteins were eluted with GF buffer (150 mM KCl/50 mM Hepes, pH 7.4; 1.3 ml/min). Three-milliliter fractions starting from 90 to 210 ml were collected representing a size range from >2 MDa to <50 kDa. For Fig. 3D, a Sephacryl S500-HR GF column from Amersham Pharmacia was used instead. Proteins were eluted with GF buffer (2.5 ml/min), and 9-ml fractions from 140 to 254 ml were collected.
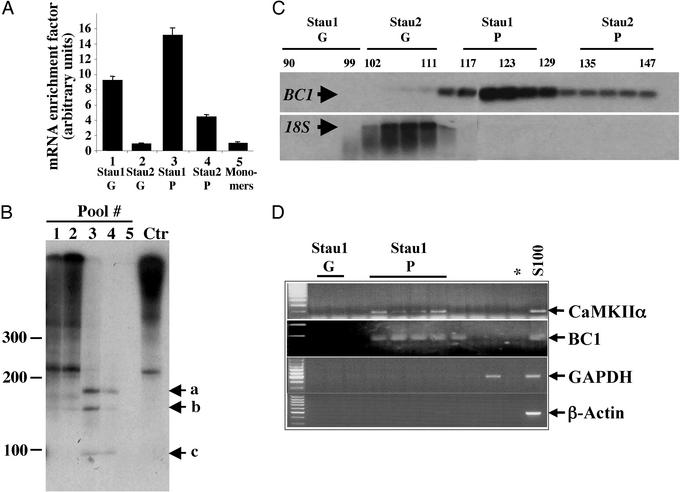
Figure 3.
Staufen particles are enriched in specific mRNAs. (A) Selective enrichment of mRNAs within Stau1 particles. Several GF were performed and the different Staufen complexes were pooled. The associated RNA was detected by radioactive reverse transcription with oligo(dT) primers. Whereas Staufen particles contain only a small amount of total RNA (data not shown), there is a 15.1- and 4.4-fold enrichment for mRNA in both Stau1 and Stau2 particles compared with the free cytosolic mRNA (monomers, pool 5). (B) The same RNAs from A were visualized by autoradiography. Staufen particles contain a specific subset of small RNA fragments (lanes 3 and 4) compared to the Staufen granules (lanes 1 and 2) and the control (Ctr). (C and D) The dendritically localized RNA BC1 is highly enriched in Stau1 particles. RNA from single GF fractions were analyzed by Northern blot (C) or RT-PCR (D). Whereas 18S rRNA is predominantly found in the Stau2 granule pool overlapping with the position of ribosomes, the BC1 transcript is highly enriched in the Stau1 particle pool. In addition, the dendritically transported CaMKIIα, but not cell body-restricted RNAs, e.g., GAPDH and β-actin could be detected in the Stau1 particle pool. Stau1 G, Stau1 granules; Stau1 P, Stau1 particles; *, negative control for PCR.
Proteins were precipitated with trichloroacetic acid (12% final concentration), washed with acetone, and redissolved in SDS sample buffer. A fifth of every fraction was separated on 10% SDS/PAGE gels and blotted onto nitrocellulose (0.2 μm, Schleicher & Schuell). The membrane was blocked with 5% skimmed milk powder in TBS (20 mM Tris⋅Cl, pH 7.5/200 mM NaCl) and immunodecorated with the following monospecific antibodies diluted in blocking buffer: mouse polyclonal Stau1 antibodies (1:1,000, this study); rabbit anti-Stau2 antibodies (1:1,000, a gift from Luc DesGroseillers, University of Montreal, Montreal) (17); rabbit anti-L7a serum (20) (dilution 1:1,000; a gift from A. Ziemiecki, University of Bern, Bern, Switzerland); rabbit anti-calnexin serum (21) (dilution 1:1,000; Stressgene, Victoria, Canada); immunopurified rabbit anti-kinesin serum (dilution 1:25,000; a gift from Marc McNiven, Mayo Cancer Center, Rochester, NY); monoclonal anti-β-tubulin (dilution 1:2,000; Amersham Pharmacia), and antisynaptophysin (dilution 1:500; Roche Diagnostics). Horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG or HRP-conjugated sheep anti-mouse IgG (both from Amersham Pharmacia) diluted either 1:4,000 (ECL, Amersham Pharmacia) or 1:10,000 (SuperSignal West Femto Maximum, Pierce) served as secondary antibodies. Chemiluminescence was detected with either ECL or SuperSignal West Femto Maximum.
RNase and DNase Treatment.
The S16 was prepared under strict RNase-free conditions as described above (see Fig. 2A). Samples were either mock-treated or incubated with 40 μg/ml RNase A for 1 h at 30°C in RNase buffer (10 mM Tris⋅Cl, pH 7.5/15 mM NaCl) and processed for differential centrifugation and Western blotting as described above. For Fig. 2B, the S100 was treated with RNase A (see above) and loaded on the GF column. For DNase (MBI Fermentas, St. Leon-Rot, Germany) treatment, samples were incubated with 2 units of DNase for 30 min at 37°C; an additional 2 units was supplied after 15 min. Samples were then processed as described above.
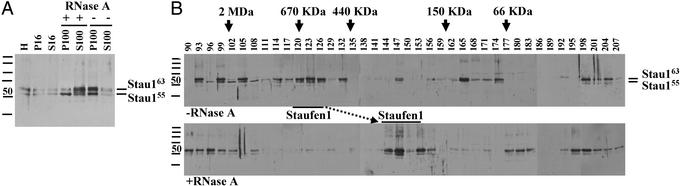
Figure 2.
Stau1 particles but not Stau1 granules are sensitive to RNase treatment. (A) RNase pretreatment of S16 supernatants released Stau1 from the pellet into the S100. Homogenates (H) were prepared and the resulting S16 was treated with RNase A or mock-treated. Samples were processed as described in Fig. 1. (B) RNase A pretreatment partially disassembled Stau1 particles. RNase A pretreated or mock-treated S100 was analyzed by GF. RNase pretreatment selectively disassembled Stau1 particles (marked by bars) but did not affect the large Staufen granules that contain ribosomes and ER.
Northern Blotting and Radioactive Reverse Transcription.
For Northern blot analysis, GF fractions were digested with 200 μg/ml proteinase K (Roche Diagnostics) in 0.5% SDS at 55°C for 30 min. On digestion, total RNA was extracted, precipitated, and resuspended in 20 μl of loading buffer (50% formamide/17.5% formaldehyde/1× Mops), denatured for 15 min at 65°C and loaded onto a 1.5% denaturing agarose gel (5% formaldehyde, 1× Mops). RNA was blotted on nylon membrane (Hybond-N, Amersham Pharmacia), and filters were hybridized with 32P-radiolabeled probes (1 × 106 cpm/ml) specific for BC1 and 18S ribosomal RNA in hybridization buffer (5× Denhart's/6× SSC/0.5% SDS/100 ng/ml herring sperm double-stranded DNA) overnight at 60°C. Filters were washed twice for 30 min in 2× SSC/1% SDS at 60°C before autoradiography. For radiolabeled reverse transcription, fractions from several GFs were collected and combined in five pools: pool 1 (fractions 90–96); pool 2 (99–105); pool 3 (117–123); pool 4 (135–141); and pool 5 (186–192). Total RNA was extracted as described above. One microgram per pool of RNA was retrotranscribed with oligo(dT)20 primer by using the Superscript Kit (Invitrogen) according to the manufacturer's instructions with the following exceptions: [dCTP] in the dNTP mix was 0.5 mM and 1 μl of [32P-dCTP] (3,000 mCi/mM, Amersham Pharmacia) was added to each sample just before incubation. Samples were then purified from the unincorporated radionucleotides by using the High Pure PCR Purification Kit (Roche, Diagnostics). Seventeen percent of the purified samples were subjected to trichloroacetic acid precipitation on 3MM paper, and the relative radioactivity was measured by liquid scintillation (TriCarb 1600, Packard). Twenty microliters was denatured in 2× denaturing buffer (80% formaldehyde/10 mM EDTA, pH 8/1 mg/ml xylene cyanol FF/1 mg/ml bromophenol blue) at 95°C for 5 min and loaded onto a 6% acrylamide/bisacrylamide gel (37:1 vol/vol), 7 M urea, 0.5× TBE (0.089 M Tris-borate/0.089 M boric acid/2 mM EDTA, pH 8). DNA was resolved by running at 350 mV for 2 h. The gel was dried and exposed by autoradiography.
In Situ Hybridization and Immunofluorescence of Cultured Neurons.
All experiments were carried out on Stau1–hemagglutinin (HA)-tagged transfected neurons. The following DNA fragments were cloned into Bluescript KS+ (Stratagene): BC1 (nucleotides 1–80), Ca2+/calmodulin-dependent kinase II α (CaMKIIα) (nucleotides 2401–2571), and GAPDH A (nucleotides 804-1004). Antisense and sense RNA probes labeled with digoxigenin were in vitro-synthesized according to the manufacturer's protocol (Roche Diagnostics). Fluorescent in situ hybridization was performed as described (22) with the following modifications. Briefly, after fixation with 4% paraformaldehyde in Hanks' balanced salt solution, cells on coverslips were washed with PBS containing 5 mM MgCl2 (PBSM) for 5 min at room temperature and then permeabilized for 2 min in PBSM containing 0.1% (vol/vol) Triton X-100. Coverslips were then washed three times with PBSM for 5 min at room temperature and, subsequently, prehybridized for 2 h at 48°C in 30% (vol/vol) deionized formamide (20% for BC1), 2× SSC, 10% dextran sulfate, 10 mM NaH2PO4, 250 μg/ml E. coli tRNA (Roche, Diagnostics), 100 μg/ml denatured herring sperm DNA. The coverslips were then incubated overnight at 48°C with fresh hybridization solution containing 500 ng/ml in vitro-transcribed digoxigenin-labeled probes. Cells were washed twice at 48°C in 2× SSC, 30% deionized formamide (20% for BC1) for 20 min and twice in 2× SSC (15 min each). The probes were detected by using antidigoxigenin rhodamine-conjugated Fab fragments (Roche Diagnostics) according to the manufacturer's description. Stau1-HA was detected by using monoclonal anti-HA (Babco, Richmond, CA; 1:1,000 dilution) and anti-mouse FITC (Dianova, Hamburg, Germany; 1:1,000 dilution)-conjugated antibodies. Coverslips were mounted with a ProLong antifade kit (Molecular Probes). Image analysis was performed as described (22).
Results
We recently reported that in both untransfected and Staufen–GFP-transfected hippocampal neurons, at least two principally different types of Staufen-containing complexes exist (13, 19): large protein complexes associated with the membrane system of the nucleus and the ER and smaller RNA-containing particles more in the periphery of neurons. Whereas the larger Staufen complexes were mostly immobile, the Staufen particles in the periphery showed either Brownian movement or alternatively bidirectional movement in dendrites of hippocampal neurons (13). To study these RNA-containing particles in detail, we set out to biochemically isolate these complexes on the basis of two criteria. First, we chose Stau1 (13, 23, 24) and Stau2 (16, 17) proteins, as biochemical markers to follow their isolation. Second, we assumed based on an attractive model (10) that the observed Staufen-containing moving particles represent soluble RNPs not associated with membranes. To enrich for Staufen-containing RNPs, rat forebrain homogenates were differentially centrifuged and the resulting pellets and supernatants were analyzed for the presence of Stau1 and Stau2 by protein-monospecific antibodies (ref. 17 and this study). Fig. 1A shows that both Staufen proteins are preferentially found in the P100 pellet because they associate with intracellular membranes (19) and ribosomes (14, 23–26). In addition, a significant fraction of Stau1 (19, 23, 24) (labeled Stau163) and all three known differentially spliced isoforms of Stau2 (17) (labeled Stau262, Stau259, and Stau252) are present in the S100 (Fig. 1A).
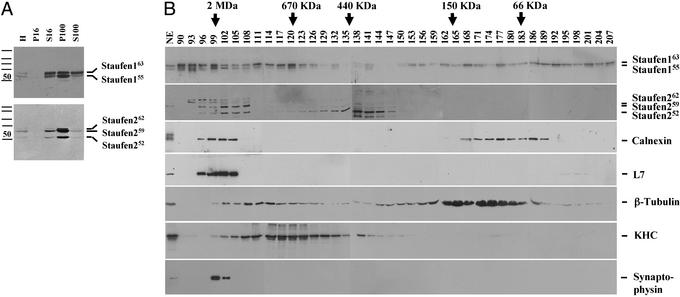
Figure 1.
Identification of soluble Staufen complexes. (A) Fractionation behavior of Staufen complexes on centrifugation. Rat forebrain homogenates (H) were differentially centrifuged. Equal protein amounts of both pellets and supernatants were resolved by SDS/PAGE and blotted against Staufen-specific antibodies. The majority of both Stau1 and Stau2 proteins were detected in the P100. However, a small fraction of both Staufen proteins was routinely found in the S100. Please note: S100 contains 3-fold more total protein than P100. (B) Five different Staufen complexes of various sizes were detected by GF. S100 was applied to a commercial GF column, 3-ml fractions were collected, and aliquots were processed for Western blotting. Blots were immunodecorated for Stau1, Stau2, calnexin, L7a, β-tubulin, KHC, and synaptophysin. Five different Staufen complexes were detected: (i) high-molecular weight granules for Stau1 (fractions 90–96) and (ii) Stau2 (fractions 99–105), that cofractionate with ER and ribosomal markers, respectively; (iii) particles for Stau1 (peak in fraction 120), which cofractionate with KHC, and (iv) Stau2 (peak in fraction 138), which do not contain both ER and ribosomal markers; (v) free Stau1 (peaks in fractions 168 and 189/192).
In a second step, S100 was separated by size on a standard GF column. Interestingly, different Stau1 pools can be detected (Fig. 1B): very large protein complexes with a size of >2 MDa (fractions 90–99); protein particles in the size range of ≈650 kDa (peak in fraction 120); and both Stau1 dimers and monomers with a size of 130 and 60 kDa, respectively. For Stau2, similar results were obtained (Fig. 1B). Large protein complexes (fractions 99–108) in a size range of <2 MDa and protein particles (peak in fraction 138) ≈440 kDa are detected with somewhat different size characteristics than those of Stau1 particles. This finding is in good agreement with immunofluorescent microscopical data that show that the majority of Stau1 and Stau2 proteins are not present in the same particles (17). Although the majority of the ER and ribosomes are found in the P100 (data not shown), we could not exclude that a fraction of these organelles is also present in the S100, e.g., in the observed very large Stau1 and Stau2 complexes (fractions 90–96 and 99–108, respectively). To test this hypothesis, we performed Western blot analysis of the same fractions for both L7a and calnexin established biochemical markers for ribosomes (20) and ER (21). As suspected, both the ER and the ribosomal markers cofractionated only with the large Staufen protein complexes, but not with the Staufen particles (Fig. 1B). Such large Stau1-containing protein complexes have been isolated from cortical neurons and characterized on the ultrastructural level (14); they appear to contain densely packed clusters of ribosomes. We therefore refer to these RNPs as granules.
The existence of two distinct Staufen complexes differing in size verifies previous observations that there might be different Staufen pools inside neurons (13). Time-lapse video microscopy of transiently transfected hippocampal neurons expressing Stau1–GFP suggested that the smaller particles, mostly in the periphery, reflect the mobile Staufen pool along microtubules in dendrites. We went on to test whether microtubules and even more importantly, kinesin heavy chain (KHC), cofractionated with one of the observed Staufen-containing particles. This assumption is based on recent work that implicated the plus end directed motor protein, KHC, in the transport of oskar mRNA in the Drosophila oocyte and of mRNAs in hippocampal neurons (27, 28). The majority of β-tubulin is detected in a broad peak (fractions 159–186) that represents partially depolymerized microtubules (Fig. 1B). A smaller pool is detected in fractions that peak around 111–117, in close proximity to the Stau1 particle peak. Most interestingly, however, was the existence of a single, broad peak for KHC around fraction 120 (Fig. 1B) that appears to cofractionate with the observed Stau1 particle peak. To test whether this KHC might be associated with synaptic vesicles, we immunodecorated another membrane with antisynaptophysin antibodies. Whereas no signal for synaptic vesicles was observed in the fractions representing Stau1 or Stau2 particles, synaptophysin was predominantly found in the fractions (99–102) that also contain Stau2 granules (Fig. 1B). This finding suggests that Staufen particles (and the associated RNAs) appear not to be associated with synaptic vesicles. Taken together, these results gave us a strong hint that the Staufen particles may represent Staufen-containing RNPs in transit on their way from the cell body to the distal tips of dendrites of hippocampal neurons.
To further investigate this hypothesis, we tested whether the association of Stau1 and Stau2 to the ER and/or ribosomes depended on the presence of mRNA and whether the observed Staufen complexes in the S100 were sensitive to RNase treatment. In a first set of experiments, the S16 from brain extracts was treated with RNase A or mock-treated before ultracentrifugation. On RNase treatment, most of the Stau1 was released from the P100 into the S100 (Fig. 2A). This shift of Stau1 proteins, however, did not occur, when samples were treated with DNase instead of RNase (data not shown). This finding indicated that the association of Stau1 with organelles such as ER or ribosomes could be RNA mediated.
In a second step, RNase A- or mock-treated S100 samples were then subjected to GF. Strikingly, the RNase A treatment (Fig. 2B) caused a significant change in the fractionation behavior of the Stau1 particles (fractions 117–123), but not of the Stau1 granules (fractions 90–96). Whereas untreated Stau1 complexes have an estimated size of ≈650 kDa, the RNase-treated particles are now shifted toward 250–300 kDa (fractions 144–146), which is another good indication that the observed Stau1 particles are indeed RNPs. The fact that these Stau1 RNPs might contain KHC (see Fig. 1B) allows us to speculate that Stau1 particles represent the delivery units for RNA from the cell body to dendrites.
Not a single cargo RNA, however, has been identified to date that is specifically recognized and transported by mammalian Staufen proteins (reviewed in refs. 4 and 29). In vitro binding assays are not suitable for identifying specific target mRNAs for mammalian Staufen proteins because Staufen binds all double-stranded RNAs in vitro (23, 24, 30). Therefore, we decided to isolate RNAs from the native Staufen-containing transport particles. We isolated total RNA from each fraction of the GF and measured its optical density to analyze its distribution over the entire gradient (data not shown). Although two major RNA peaks were observed over the entire gradient, we found very little RNA in the Stau1 and Stau2 particle fractions. To test whether there may be a specific enrichment of mRNA in these fractions, we performed several GFs and collected five different pools (Fig. 3 A and B): Stau1 granules (pool 1, fractions 90–96), Stau2 granules (pool 2, fractions 99–105), Stau1 particles (pool 3, fractions 117–123), Stau2 particles (pool 4, fractions 135–141), and Stau1 monomers (pool 5, fractions 186–192). The total RNA isolated from these five pools was reverse-transcribed into radiolabeled cDNA and trichloroacetic acid-precipitated, and radioactivity was measured. Despite the low amount of total RNA in the Stau1 and Stau2 particle fractions, a significant enrichment of mRNA was detected. If the resulting values were normalized to the Stau1 monomer pool, a 15.1- and 4.4-fold enrichment of mRNA in the Stau1 and Stau2 particles, respectively, was obtained (Fig. 3A). We then visualized the radiolabeled cDNAs in each Staufen pool (Fig. 3B). The reverse-transcribed mRNAs derived from both Staufen granule pools (Fig. 3B, lanes 1 and 2) thereby strongly resembled total mRNA (Fig. 3B, Ctr) extracted from S100. In contrast, only few discrete bands were detected in either of the Staufen particles (Fig. 3B, lanes 3 and 4) with sizes between 100 and 250 nt. No detectable RNA, however, was found in the Staufen monomer fractions (Fig. 3B, lane 5). Taken together, this finding is another good indication that these Staufen particles may represent RNA transport intermediates.
We then identified one of the major bands (Fig. 3B, band b) as BC1, a noncoding RNA transcribed from the RNA polymerase III. Interestingly, BC1 RNA is a component of RNPs in neurons (7, 8) and it is not only localized in dendrites in the rodent nervous system, but becomes actively transported into dendrites on cytoplasmic microinjection (31, 32). We therefore investigated whether BC1 RNA is also present in our Staufen particles. Interestingly, the BC1 transcript (Fig. 3C) was highly enriched in the Stau1 particle pool, whereas 18S rRNA, a marker for ribosomes, was predominantly present in the Stau2 granule pool. This result encouraged us to test whether other known dendritically localized mRNAs (12), e.g., CaMKIIα (33) or microtubule-associated protein-2 (34) were also present in these Staufen particles. RNA was isolated from selected fractions, and RT-PCR was performed for BC1, CaMKIIα, microtubule-associated protein-2, and several nonlocalized RNAs such as GAPDH A, β-actin, histone H3, and CaMKIIβ. Whereas CaMKIIα yielded the same cofractionation like BC1, GAPDH A, β-actin (Fig. 3D), MAP2, histone H3, and CaMKIIβ (data not shown) were all absent from the Stau1 particle pool. This result clearly indicates that only a subset of (dendritically localized) mRNAs is present in the Stau1 RNPs.
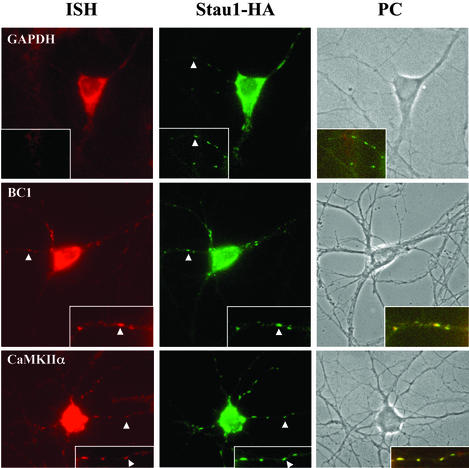
We then investigated whether these potential Stau1 RNA cargos were indeed present in the same particles together with Stau1. In situ hybridization was performed followed by immunostaining on Stau1–HA-transfected hippocampal neurons. As shown in Fig. 4, both the BC1 and CaMKIIα RNA colocalize in the same dendritic Stau1–HA-containing particles, whereas other RNAs, e.g., GAPDH A and CaMKIIβ (data not shown), do not. This experiment strongly suggests that these two RNAs might be transported by Stau1 into dendrites of hippocampal neurons.
Figure 4.
CaMKIIα and BC1 RNA colocalize with Stau1–HA in the same particles within dendrites of transfected hippocampal neurons. In situ hybridization for CaMKIIα, BC1, and GAPDH RNAs was combined with immunocytochemistry for Stau1–HA detected with anti-HA antibodies. Phase contrast (PC) as well as corresponding fluorescent images of the same neurons are shown for each RNA. High magnification images of selected dendrites as well as the merge (within the computer image) are shown (Insets). (Magnifications: ×40.)
Taken together, these results are of particular importance for several reasons. First, the noncoding BC1 transcript localizes to dendrites and is part of brain-derived RNPs (7, 8). Second, inhibition of electrical activity leads to a reversible down-regulation of the somatodendritic BC1 RNA level in neurons. A recent study suggests that BC1 RNA exerts a role in dendritic translational regulation, thereby modulating local protein synthesis in postsynaptic neuronal microdomains (35). This result fits well with the recent observation that Staufen proteins are part of large granules (14, 15) associated with ribosomes/ER (23–26) through a still unknown mechanism. Our study, however, identifies additional, soluble Stau1- and KHC-containing RNPs that cofractionate and colocalize with dendritically transported RNAs, e.g., CaMKIIα and BC1. These results further strengthen our hypothesis that the soluble Stau1- and KHC-containing RNPs indeed represent RNA transport intermediates that are in transit on their way from the cell body to their final destination within dendrites.
Acknowledgments
We are grateful to Barbara Grunewald for excellent technical assistance and Drs. Tilmann Achsel, Martin Bayer, Carlos Dotti, Andreas Mayer, Oliver Müller, and Reinhard Lührmann for advice and comments on earlier drafts of this manuscript. We are also indebted to Dr. Luc DesGroseillers for rabbit anti-Stau2 antibodies, Dr. Marc McNiven for KHC antibodies, Dr. Andrew Ziemiecki for anti-L7a antibodies, and Dr. Juan Ortín for the pRSET-Stau1 construct. This work was supported by a Ph.D. fellowship from Graduiertenkolleg Neurobiologie, University of Tübingen (to A.D.), Sonderforschungsbereich Grant SFB446 (to P.M. and M.A.K.), and a Human Frontier Science Program Organization network grant (to M.A.K.).
Abbreviations
- CaMKII
Ca2+/calmodulin-dependent kinase II
- ER
endoplastic reticulum
- GF
gel filtration
- HA
hemagglutinin
- KHC
kinesin heavy chain
- RNP
ribonucleoprotein
- Stau1
Staufen1
- Stau2
Staufen2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 1465.
References
- 1.St. Johnston D. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 2.Bashirullah A, Cooperstock R L, Lipshitz H D. Annu Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- 3.Bassell G J, Oleynikov Y, Singer R H. FASEB J. 1999;13:447–454. doi: 10.1096/fasebj.13.3.447. [DOI] [PubMed] [Google Scholar]
- 4.Kiebler M A, DesGroseillers L. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 5.Jansen R P. Nat Rev Mol Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 6.Kloc M, Zearfoss N R, Etkin L D. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 7.Tiedge H, Fremeau R T, Jr, Weinstock P H, Arancio O, Brosius J. Proc Natl Acad Sci USA. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi S, Goto S, Anzai K. J Biol Chem. 1991;266:4726–4730. [PubMed] [Google Scholar]
- 9.Ainger K, Avossa D, Morgan F, Hill S J, Barry C, Barbarese E, Carson J H. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm J E, Vale R D. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnapp B J. Curr Biol. 1999;9:R725–R727. doi: 10.1016/s0960-9822(99)80468-0. [DOI] [PubMed] [Google Scholar]
- 12.Kuhl D, Skehel P. Curr Opin Neurobiol. 1998;8:600–606. doi: 10.1016/s0959-4388(98)80087-1. [DOI] [PubMed] [Google Scholar]
- 13.Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti C G, Kiebler M A. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krichevsky A M, Kosik K S. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato T A, Anzai K. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 16.Tang S J, Meulemans D, Vazquez L, Colaco N, Schuman E. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 17.Duchaîne T F, Hemraj I, Furic L, Deitinghoff A, Kiebler M A, DesGroseillers L. J Cell Sci. 2002;115:3285–3295. doi: 10.1242/jcs.115.16.3285. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D, editors. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 19.Kiebler M A, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión R M, Ortín J, Dotti C G. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziemiecki A, Muller R G, Fu X C, Hynes N E, Kozma S. EMBO J. 1990;9:191–196. doi: 10.1002/j.1460-2075.1990.tb08095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou W J, Cameron P H, Thomas D Y, Bergeron J J. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 22. Macchi, P., Hemraj, I., Goetze, B., Grunewald, B., Mallardo, M. & Kiebler, M. A. (2003). Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 23.Marión R M, Fortes P, Beloso A, Dotti C, Ortín J. Mol Cell Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickham L, Duchaîne T, Luo M, Nabi I R, DesGroseillers L. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo M, Duchaîne T F, DesGroseillers L. Biochem J. 2002;365:817–824. doi: 10.1042/bj20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le S, Sternglanz R, Greider C W. Mol Biol Cell. 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brendza R P, Serbus L R, Duffy J B, Saxton W M. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severt W L, Biber T U, Wu X, Hecht N B, DeLorenzo R J, Jakoi E R. J Cell Sci. 1999;112:3691–3702. doi: 10.1242/jcs.112.21.3691. [DOI] [PubMed] [Google Scholar]
- 29.Roegiers F, Jan Y N. Trends Cell Biol. 2000;10:220–224. doi: 10.1016/s0962-8924(00)01767-0. [DOI] [PubMed] [Google Scholar]
- 30.Ramos A, Grunert S, Adams J, Micklem D R, Proctor M R, Freund S, Bycroft M, St. Johnston D, Varani G. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muslimov I A, Santi E, Homel P, Perini S, Higgins D, Tiedge H. J Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muslimov I A, Banker G, Brosius J, Tiedge H. J Cell Biol. 1998;141:1601–1611. doi: 10.1083/jcb.141.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayford M, Baranes D, Podsypanina K, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garner C C, Tucker R P, Matus A. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Iacoangeli A, Popp S, Muslimov I A, Imataka H, Sonenberg N, Lomakin I B, Tiedge H. J Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]