Abstract
The Drosophila tracheal system and mammalian airways are branching networks of tubular epithelia that deliver oxygen to the organism. In mammals, the epithelial Na+ channel (ENaC) helps clear liquid from airways at the time of birth and removes liquid from the airspaces in adults. We tested the hypothesis that related Drosophila degenerin (DEG)/ENaC family members might play a similar role in the fly. Among 16 Drosophila DEG/ENaC genes, called pickpocket (PPK) genes, we found 9 expressed in the tracheal system. By in situ hybridization, expression appeared in late-stage embryos after tracheal tube formation, with individual PPK genes showing distinct temporal and spatial expression patterns as development progressed. Promoters for several PPK genes drove reporter gene expression in the larval and adult tracheal systems. Adding the DEG/ENaC channel blocker amiloride to the medium inhibited liquid clearance from the trachea of first instar larvae. Moreover, when RNA interference was used to silence PPK4 and PPK11, larvae failed to clear tracheal liquid. These data suggest substantial molecular diversity of DEG/ENaC channel expression in the Drosophila tracheal system where the PPK proteins likely play a role in Na+ absorption. Extensive similarities between Drosophila and mammalian airways offer opportunities for genetic studies that may decipher further the structure and function of DEG/ENaC proteins and development of the airways.
Keywords: trachea‖Na+ channel‖absorption‖epithelia‖amiloride
Complex multicellular organisms require an elaborate oxygen delivery system to meet tissue metabolic demands. In mammalian lung, a network of branching epithelial tubes carries oxygen to the alveoli, where it diffuses into blood to be distributed to the tissues. Drosophila melanogaster also possesses a network of branching epithelial tubes through which oxygen diffuses to reach cells throughout the organism (1). In addition to their functional similarity, the growth factors and transcription factors controlling branching morphogenesis in mammalian airways and the fly tracheal system share numerous features (2). Moreover, at specific points in development, both systems must convert from liquid- to air-filled tubules. In mammals, this conversion occurs at birth, when the epithelium actively absorbs salt and hence liquid from the airways. In Drosophila, this conversion occurs 2 h before larvae emerge, also by active epithelial absorption (1).
In mammalian airways, the epithelial Na+ channel (ENaC), composed of α-, β-, and γ-ENaC subunits, contributes to salt and liquid absorption (3, 4). Striking evidence for this role came from mice bearing targeted disruptions of ENaC genes. Mice with a disrupted α-ENaC gene died shortly after birth, because they failed to clear lung liquid, and mice with disrupted β- or γ-ENaC genes showed delayed lung liquid clearance (5–7). In humans, impaired ENaC activity may contribute to neonatal respiratory distress syndrome (8). ENaC-dependent salt and liquid absorption may also help clear pulmonary edema fluid in adult patients (9).
ENaC subunits are members of the degenerin (DEG)/ENaC gene family (3, 10, 11). Members of this family contain intracellular N and C termini and two membrane-spanning domains (M1 and M2) separated by a large, cysteine-rich extracellular domain. The family shows sequence conservation in multiple regions of the protein, especially in M2 and in a conserved pattern of extracellular cysteines. Individual subunits associate as homomultimers or heteromultimers to form voltage-insensitive cation channels, most of which are inhibited by amiloride. DEG/ENaC family members have been identified in many multicellular organisms, from nematode to human; however, they have not yet been discovered in unicellular organisms such as bacteria or the yeast Saccharomyces cerevisiae. The function of DEG/ENaC family members is also diverse: they can form a receptor for the neuropeptide FMRFamide; they may be mechanosensors; they may contribute to acid-evoked nociception; they may participate in salt and sour taste; they may contribute to synaptic plasticity; and they contribute to salt absorption by kidney, gut, and airway epithelia (3, 4, 10, 12–17). We identified two Drosophila DEG/ENaC family members, pickpocket (PPK) and ripped pocket (RPK) (18, 19). We found PPK expressed during the last stage of embryogenesis in three multidendritic sensory neurons per hemi-segment; this location suggested a role in sensory function. We found RPK expressed in the adult ovary; its function there remains unknown.
In this study we tested the hypothesis that DEG/ENaC family members may play a role in the Drosophila tracheal system. We chose Drosophila because it offers several advantages as a model system, and, as described previously, the tracheal system has been well characterized. In addition, the recent completion of the Drosophila genome sequence (20) allowed us to identify additional DEG/ENaC genes.
Materials and Methods
In Situ Hybridization.
We cloned 14 D. melanogaster genes: PPK4, -6, -7, -10, -11, -12, -13, -14, -16, -19, -20, -21, -23, and -28. The sequences have been deposited in GenBank. D. melanogaster embryos (7–24 h) were collected and fixed as described (21). Embryonic stage was determined by the structural pattern in the head, body, and trachea (22). Partial PPK cDNA sequences (0.7–1.2 kb) were used for in situ hybridization. Digoxigenin-labeled sense and antisense probes were synthesized with a digoxigenin-labeling kit (Roche Molecular Biochemicals). Whenever comparing expression patterns for an individual PPK gene, embryos from different stages were labeled at the same time. The expression patterns we show by in situ hybridization were reproducible with multiple embryos from different collections. Photomicrographs of in situ specimens were taken with a Spot (Bio-Rad) high-resolution digital camera.
Construction of PPK Promoter-Driven Gal4 Transgenes.
To assess the pattern of PPK expression we used PCR of genomic sequence to amplify ≈2 kb in the 5′ direction from the translational start site of several PPK genes. The DNA was first ligated into the PCRII vector (Invitrogen) and then into the P element Gal4 vector (a gift from D. F. Eberl, Univ. of Iowa). Transgenic lines were generated by injecting the construct into oocytes of the yw67c23 fly line. After the transgenic lines were made homozygous, the PPK promoter-Gal4 lines (PPK-Gal4) were crossed to a UAS-enhanced GFP (UAS-eGFP) line. Because Gal4 protein production and accumulation requires a few hours, eGFP expression could be delayed relative to the time at which the promoter was first expressed. For each PPK gene studied, three to nine transgenic lines PPK-Gal4 were generated, and the line that showed the strongest expression when crossed to the UAS-eGFP line was chosen for further study.
RNA Interference (RNAi).
Single-stranded RNA was synthesized by using the T7 and SP6 mMACHINE kit (Ambion, Austin, TX). Double-stranded RNA (dsRNA) was generated by mixing the two complimentary single-stranded RNAs together, heating at 80°C for 5 min, and then cooling at room temperature overnight (23). The length of PPK cDNA sequences used to generate dsRNA varied from 0.5 to 1.5 kb (23–25). dsRNA (0.3 nl at 1 μg/μl) was injected into each early-stage embryo. Buffer alone or irrelevant dsRNA were used as controls. We found a consistent mutant phenotype in ≈30–50% of surviving larvae. This proportion is consistent with that obtained with RNAi used to silence other genes (23, 25).
Treatment with Amiloride and Benzamil.
Drosophila medium (Carolina Biological Supply Company, Burlington, NC) was mixed with either amiloride or benzamil (Sigma). In preliminary studies using different concentrations of these drugs, we found that 30 mM amiloride or 10 mM benzamil caused larval lethality during the early larval stages. However, when adult Drosophila were fed medium containing amiloride (30 mM) or benzamil (10 mM), it had no effect on survival or egg laying.
Evaluation of the Tracheal System.
To determine whether the tracheal system was filled with air or liquid, we used phase-contrast microscopy as described (26).
Results
Temporal Expression of PPK Genes During Embryonic Development.
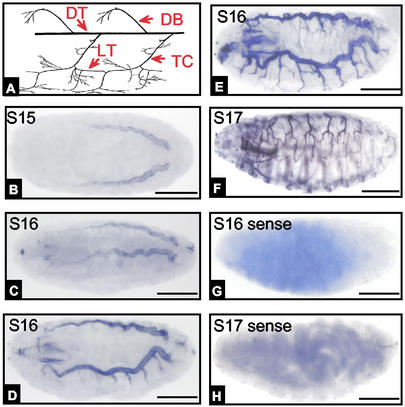
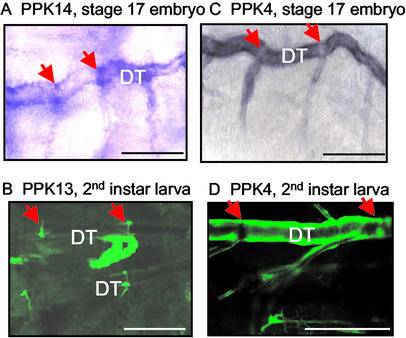
To determine the embryonic expression pattern of PPK genes, we used two methods, in situ hybridization and promoter-driven eGFP expression. By using in situ hybridization, we detected eight PPK genes expressed in the tracheal system: PPK4, -7, -10, -11, -12, -14, -19, and -28. As a negative control for each gene, we used sense probes. Because the antisense probes for PPK6 and PPK13 stained the surface cuticle, their expression in trachea could not be determined by in situ hybridization. The expression pattern for PPK16, -21, -22, and -23 could not be confirmed, because antisense and sense probes both showed labeling. Although in this work we focused on the tracheal expression pattern, each of these genes also showed expression at sites other than the trachea, most notably in neurons. Fig. 1A shows a schematic of the Drosophila tracheal system and the nomenclature in accord with Manning and Krasnow (1).
Figure 1.
In situ hybridization of PPK11 expression at different embryonic stages. (A) Diagram of tracheal system [adopted from Manning and Krasnow (1)]. Anterior is to the left and dorsal is to the top. DT, dorsal trunk; DB, dorsal branch; LT, lateral trunk. (B–F) Antisense probe staining from stages 15–17 (S15–S17). (G and H) Negative control of sense probe staining. Scale bar indicates 100 μM for all images in Figs. 1–6.
The pattern of PPK gene expression varied during embryo development (as an example, Fig. 1 shows results for PPK11). The earliest detectable expression was during late embryogenesis, at stage 15 in the dorsal trunk (Fig. 1B). Previous work has shown that the tracheal system begins to develop from the tracheal precursor cells starting at stage 12 (1, 27). Thus, PPK11 expression lagged behind tracheal tube formation. Although the specific pattern of expression varied from one gene to another (see below), expression of all of the tracheal PPK genes seemed to occur after tracheal system tubule formation.
As development progressed, some PPK genes expanded their expression to other portions of the tracheal tree. For example, PPK11 expression was detected in posterior regions of the dorsal trunk earlier than in anterior regions during developmental stages 15 and 16 (Fig. 1 B and C). PPK11 was not detected in the transverse connective (TC) until stage 16 (Fig. 1D). Expression extended into the primary branches during stage 16 (Fig. 1E). By stage 17, expression was more extensive in the primary and some secondary branches (Fig. 1F). Thus, PPK11 gene expression appeared to occur shortly after tracheal tube formation.
PPK Gene Promoters Drive Reporter Expression in the Tracheal System.
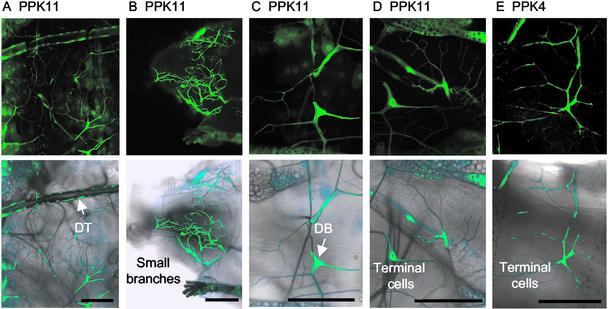
We generated transgenic lines with the PPK promoters linked to Gal4 for six PPK genes, including four that were expressed in the trachea by in situ hybridization (PPK4, -10, -11, and -12) and two that showed a dark appearance caused by cuticular staining (PPK6 and PPK13). When these six lines were crossed to UAS-eGFP, five (PPK4, -10, -11, -12, and -13) showed expression in the larval tracheal system (Figs. 2–5). Fig. 2 shows examples for PPK11 and PPK4. Because of the delay in accumulation of Gal4 and then eGFP, we were not able to study promoter-driven GFP expression in embryos. However, expression was appreciable in first instar larvae and was readily apparent in second and third instar larvae. Consistent with the in situ hybridization, the PPK11 promoter was expressed in the dorsal trunk, TC, and several other tracheal branches of second and third instar larvae (Fig. 2A). PPK11 expression was observed at multiple levels of the tracheal system, including some very small branches (Fig. 2B), dorsal branches (Fig. 2C), and terminal cells (Fig. 2D). The PPK4 promoter showed a very similar pattern of expression (Fig. 2E shows an example of PPK4 expression in terminal cells).
Figure 2.
PPK4 and PPK11 promoter-driven eGFP expression in the tracheal system. Shown are eGFP florescent images (Upper) and eGFP fluorescence laid over light micrographs that show tracheal branches in the same field (Lower). (A–D) PPK11-Gal4;UAS-eGFP. (E) PPK4-Gal4;UAS-eGFP. In A, the dorsal trunk (DT) and smaller branches are shown. In B, the anterior part of the larval tracheal network is shown, including the head, anterior spiracle, and multiple small branches. In C, dorsal branch (DB) epithelial cell is shown. In D and E, terminal cells are shown.
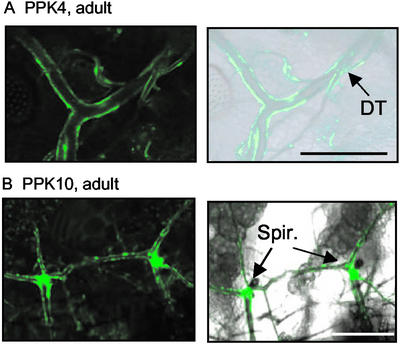
Figure 5.
PPK gene expression at the spiracles and in adult Drosophila. (A) PPK4 promoter-driven expression in the adult tracheal system. Dorsal trunk (DT) is indicated. (B) PPK10 promoter-driven eGFP expression in the adult spiracles.
PPK Genes Show Distinct Spatial Expression Patterns in the Larval Tracheal System.
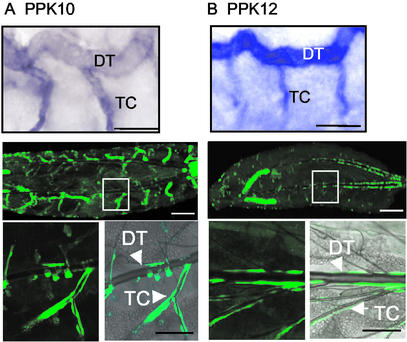
In contrast to the widespread tracheal expression of PPK4 and PPK11, PPK10 and PPK12 had a more restricted expression pattern. PPK10 and PPK12 were expressed predominantly in dorsal trunk and TC as assessed by in situ hybridization and promoter-driven eGFP expression (Fig. 3). However, PPK10 expression was more prominent in cells of the TC than in the dorsal trunk.
Figure 3.
In situ hybridization and promoter-mediated eGFP expression for PPK10 and PPK12. (A and B) PPK10 and PPK12. (Top) In situ hybridization staining pattern in stage-17 embryos. Dorsal trunk (DT) and TC are indicated. (Middle) PPK10 and PPK12 promoters driving eGFP expression (PPK10-Gal4;UAS-eGFP and PPK12-Gal4;UAS-eGFP) in first instar larvae. (Bottom) Enlarged views from Middle.
During formation of the tracheal system, epithelial tubes originating from tracheal pits in each hemi-segment grow toward each other, meet, and form a continuous open tube, i.e., the dorsal trunk. Fusion cells lie at the point where the two epithelial tubes join (28–30). By in situ hybridization, PPK14 was expressed at the fusion point in the dorsal trunk (Fig. 4A). Although PPK13 could not be assessed by in situ hybridization, because cuticle expression obscured the tracheal system (not shown), the PPK13 promoter-Gal4 transgenic drove UAS-eGFP expression at the site of fusion in the tracheal dorsal trunk (Fig. 4B). In contrast, the dorsal trunk expression patterns of PPK4 were nearly the opposite (Fig. 4 C and D): PPK4 was expressed strongly in most regions of the dorsal trunk with the exception of the junction between the dorsal trunk and TC. PPK11 showed a similar pattern (not shown).
Figure 4.
PPK genes showed distinct expression patterns in the dorsal trunk (DT). (A) PPK14 in situ hybridization in stage-17 embryos. Arrows indicate the fusion point between two segmental dorsal trunks. (B) PPK13 promoter driving eGFP expression (PPK13-Gal4;UAS-eGFP). (C and D) PPK4 expression shown by in situ hybridization and promoter-driven eGFP expression (PPK4-Gal4;UAS-eGFP).
The two techniques we used to localize PPK gene expression each have limitations. For the in situ hybridization, the reaction product can leak into the tracheal lumen. For promoter-driven eGFP expression, we used only ≈2 kb of sequence upstream of the translation start site. This amount of sequence likely will not encode the entire region that normally drives expression, and variation could occur depending on positional effects at the site of chromosomal insertion. To minimize such effects, we examined at least three lines for all transgenic PPK promoters, and although we chose the lines showing the strongest expression, the patterns were similar in individual lines. As an additional control, we found that a 2-kb PPK6 promoter was not expressed in trachea but was expressed in cuticle epithelial cells, neurons, and muscles (not shown). Moreover, agreement between results obtained by in situ hybridization in embryos and promoter-driven reporter expression in larvae validate these conclusions. Thus, the data indicate that multiple PPK genes were expressed in the tracheal system of late-stage embryos and in larvae. Moreover, different genes showed very different patterns of expression (Table 1).
Table 1.
Summary of PPK gene expression pattern in the tracheal system
| Gene | DT | Fusion point in DT | TC | Dorsal branch | Terminal cells |
|---|---|---|---|---|---|
| (ISH:Prom) | (ISH:Prom) | (ISH:Prom) | (ISH:Prom) | (ISH:Prom) | |
| PPK4 | +:+ | −:− | +:+ | +:+ | +:+ |
| PPK7 | +:NA | −:NA | +:NA | +:NA | +:NA |
| PPK10 | +:+ | −:− | +:+ | −:− | −:− |
| PPK11 | +:+ | −:− | +:+ | +:+ | +:+ |
| PPK12 | +:+ | −:− | +:+ | −:− | −:− |
| PPK13 | ?:− | ?:− | ?:− | ?:− | ?:− |
| PPK14 | +:NA | +:NA | +:NA | −:NA | −:NA |
| PPK19 | +:NA | −:NA | +:NA | +:NA | +:NA |
| PPK28 | +:NA | −:NA | +:NA | +:NA | +:NA |
ISH, In situ hybridization; Prom, promoter-driven eGFP expression; DT, dorsal trunk. +, expression; −, no observable expression; NA, not available; ?, not clear.
Distinct PPK Gene Expression Patterns in the Adult Tracheal System.
We also examined eGFP expression from the PPK promoters in adults for PPK4, -6, -10, -11, -12, and -13. All six PPK promoters drove expression, and the patterns differed with expression in neurons, intestine, epidermis, and fat body (not shown). However, only two, PPK4 and PPK10, were detected in the adult tracheal system, and they showed different expression patterns. PPK4 was expressed in much of the dorsal trunk but not at the spiracles (Fig. 5A). PPK10 was also expressed in some epithelial cells of the dorsal trunk but additionally, it was expressed strongly at sites surrounding the spiracles (Fig. 5B).
Amiloride and Benzamil Inhibit Tracheal Liquid Clearance.
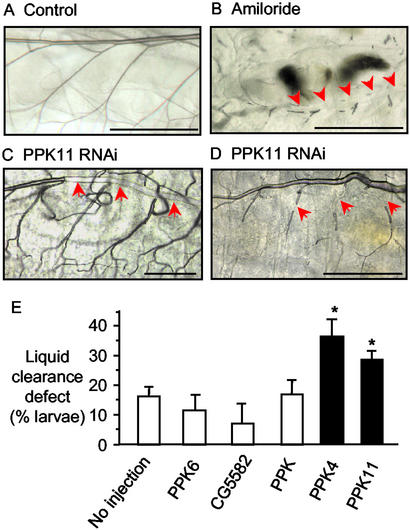
By analogy to the function of ENaC in mammalian lung, we hypothesized that PPK genes might be involved in liquid clearance from fly trachea. To test this hypothesis, we examined the effect of the diuretic amiloride and its analog benzamil. These agents inhibit most known DEG/ENaC channels (3, 31). Moreover, their application into the liquid-filled lung of embryonic guinea pigs delayed lung liquid clearance at birth, causing hypoxemia and respiratory distress (8). We included either drug in the Drosophila medium and evaluated larvae grown under low population density. We examined living larvae with phase-contrast microscopy, which allows air-filled tubes to be identified by their refractive properties (26). The tracheal tubules of untreated larvae were predominantly air-filled (Fig. 6A). In contrast, all first instar larvae treated with amiloride (30 mM) failed to clear liquid from portions of the trachea. (Fig. 6B shows an example of liquid filling the dorsal trunk and most of its branches; however, some areas of air-filled tubules were observed.) We obtained similar results with benzamil (10 mM; data not shown). These data suggest that amiloride-sensitive channels may be important for tracheal liquid clearance. However, this suggestion is limited, because amiloride and benzamil at these high concentrations can block other cellular functions including Na+/H+ antiport and Na+/Ca2+ exchange (32, 33).
Figure 6.
Liquid clearance in the first instar larva. (A) Wild-type larvae with air-filled trachea. (B) Amiloride-treated larvae showing liquid-filled trachea; arrows point to regions of liquid-filled trachea. (C and D) PPK4 and PPK11 RNAi-injected larvae. (E) Percentage of larvae with defective liquid clearance. n = 223–459 for each condition; *, P < 0.01.
Inhibition of PPK Gene Function Can Impair Tracheal Liquid Clearance.
To test further the role of PPK gene function in liquid clearance, we used RNAi, which can be used to silence specific genes, including in D. melanogaster (23–25). With RNAi, injection of gene-specific dsRNA leads to an ATP-dependent cleavage of the target mRNA (34). To target some of the PPK gene transcripts, we injected dsRNA into early-stage wild-type (yw67c23) embryos.
Injection of PPK4 or PPK11 dsRNA produced liquid clearance defects in the tracheal system (examples are shown in Fig. 6 C and D). In contrast, the frequency of liquid clearance defects was significantly lower in noninjected embryos and in embryos injected with PPK, PPK6, or an irrelevant (CG5582) dsRNA or in noninjected embryos (Fig. 6E). PPK, PPK6, and CG5582 were used as negative controls because they were not expressed in the tracheal system by in situ hybridization or promoter-driven GFP detection; however, each was expressed at other sites (ref. 18 and data not shown).
Discussion
Our data indicate that D. melanogaster express several DEG/ENaC genes in the tracheal system. There they may function to clear liquid and generate an air-filled tubule system. Thus, the related Drosophila PPK proteins and mammalian ENaC proteins may share a physiologic function in clearing liquid from the airways.
Potential Contribution of PPK Gene Products to Tracheal Liquid Clearance.
The entire Drosophila tracheal system of epithelial tubes is filled with liquid until 2 h before the larvae emerge. Then gas begins to fill one of the dorsal trunks, expands within a few minutes to fill both trunks, and then spreads to the smaller branches in the next 10 min (1). This switch from a liquid- to a gas-filled lumen has been attributed to active salt and liquid absorption by the tracheal epithelium (1). Expression of the nine (eight detected by in situ hybridization and one by promoter-driven expression) PPK genes in tracheal tubules places them at an appropriate site to mediate salt absorption. Their similarity to ENaC subunits also suggests they are involved in transepithelial cation transport and function as heteromultimers. This hypothesis was supported by two findings. First, amiloride, which blocks DEG/ENaC Na+ channels, inhibited tracheal liquid clearance. Second, when we injected embryos with dsRNA directed toward PPK4 and PPK11 transcripts, portions of the trachea remained liquid-filled. Thus, the PPK genes seem to play a key role in tracheal liquid clearance, and disruption of one subunit could affect the whole channel complex. In rats, the onset of liquid clearance may involve increased epinephrine levels (35). Thus, it would be interesting to know what cell signaling processes modify PPK or associated proteins to initiate liquid clearance 2 h before larvae emerge.
Heterologous expression of α-, β-, and γ-ENaC generates constitutively active channels (36, 37). In contrast, expression of β- and/or γ-ENaC produces no current, and expression of α-ENaC alone generates only very small currents. We expressed several of the PPK genes, including PPK7 and PPK10, alone and in combination in Xenopus oocytes, but have not observed currents (Candice Askwith, L.L. and M.J.W., unpublished observation). Perhaps analogous to the mammalian β- and γ-ENaC subunits (36, 37) and Caenorhabditis elegans DEG/ENaC channels (38), heterologous expression failed to generate current because a key subunit or associated protein was missing (39).
Expression Patterns of PPK Genes in the Drosophila Trachea.
The PPK genes showed a surprising degree of complexity in their tracheal expression. Of the 16 genes cloned to date, 8 were expressed in the Drosophila larval tracheal system as revealed by in situ hybridization. We confirmed the tracheal expression pattern by further studying four of the eight, and found that all four of the PPK promoters drove tracheal eGFP expression. In addition, promoter-driven reporter expression showed PPK13 expression in trachea. However, only two of five PPK genes expressed in the larval trachea were detected by eGFP expression in the adult tracheal system.
Why might the embryonic and larval trachea require expression of a greater number of DEG/ENaC proteins than the adult trachea? We speculate that their requirements for liquid clearance exceed those in the adult. The larval tracheal system comprises a very long series of branching tubes composed of at least 20 different tracheal cell types (1), with air entering only through two anterior and two posterior spiracles. Moreover, larvae live in fruit that has a high moisture content, and often the two anterior spiracles are buried in the fruit. In contrast, the adult trachea has 18 spiracles opening to the air, regions of the adult tracheal system are expanded into air sacs (1), and adults usually live in a drier environment. We speculate that the demands of forming and then maintaining a liquid-free tubular system that delivers oxygen to the larva require a greater diversity of function than needed for the adult. The large number of tracheal PPK subunits could also allow construction of heteromultimeric cation channels exhibiting a wide variety of biophysical properties to match functional needs at various times and locations.
Parallels Between PPK Expression in Drosophila Trachea and ENaC Expression in Mammalian Lung.
Earlier work has drawn several parallels between the fly tracheal system and the mammalian lung (2). Our data extend the parallels in terms of DEG/ENaC gene expression. First, the temporal pattern of PPK gene expression varies. During mammalian lung development, ENaC genes also show distinct temporal expression patterns. For example, in the developing human lung α-ENaC was detected at 4–5 weeks gestation, whereas β- and γ-ENaC were expressed after 17 weeks gestation (40, 41). Second, both PPK and ENaC genes showed striking variation in their spatial expression patterns. For example, during late gestation, mouse and rat α- and γ-ENaC are expressed in all lung regions, whereas β-ENaC is expressed most intensely in small airways (42). Third, Drosophila PPK and mammalian ENaC proteins may share similar functional roles in clearing liquid and maintaining gas-filled airways. Fourth, several PPK genes showed expression not only in the trachea but also in neurons. Likewise, ENaC genes are expressed not only in epithelia but also in specialized cutaneous nerve terminals (43, 44) and in baroreceptor nerve endings (45) where they may be involved in mechanosensation. A dual role in epithelial salt transport and in sensory processes suggests that the function of an individual subunit may depend on the cell type in which it is expressed, its location in the cell, the other DEG/ENaC subunits with which it associates, and the intra- and extracellular proteins to which it binds.
Identification of these new DEG/ENaC genes in the Drosophila tracheal system extends the similarities between fly and mammalian airways. Moreover, because Drosophila offers several experimental advantages, their use as a model system may provide new insight into the biology of both these complex airways and the DEG/ENaC protein family.
Acknowledgments
We thank Lori L. Wallrath, Daniel E. Eberl, Christopher M. Adams, Greg J. Beitel, and Teresa Grunst for many helpful discussions. We thank Yuhong Li for excellent technical assistance. We thank the University of Iowa DNA Core Facility Diabetes and Endocrine Research Center (DK-25295) for assistance with sequencing and oligonucleotide synthesis and the Central Microscopy Research Facility (NHLBI-51670, DK-54759, and the Cystic Fibrosis Foundation) for assistance with microscopy. This work was supported by the Howard Hughes Medical Institute (HHMI). M.J.W. is an Investigator of the HHMI.
Abbreviations
- DEG/ENaC
degenerin/epithelial Na+ channel gene family
- dsRNA
double-stranded RNA
- RNAi
RNA interference
- PPK
pickpocket gene family members
- eGFP
enhanced GFP
- TC
transverse connective
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF024691 U53479 (PPK4S, also called NaCh), AY226538 (PPK4L), AY226539 (PPK6), AY226540 (PPK7), AY226541 (PPK10), AY226542 (PPK11), AY226543 (PPK12), AY226544 (PPK13), AY226545 (PPK14), AY226546 (PPK16), AY226547 (PPK19), AY226548 (PPK20), AY226549 (PPK21), AY226550 (PPK23L), AY226551 (PPK23S), AY226552 (PPK28L), and AY226553 (PPK28S)].
References
- 1.Manning G, Krasnow M A. In: The Development of Drosophila melanogaster. Bate M, editor. Vol. 1. New York: Cold Spring Harbor Lab. Press; 1993. pp. 609–685. [Google Scholar]
- 2.Metzger R J, Krasnow M A. Science. 1999;284:1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- 3.Garty H, Palmer L G. Physiol Rev. 1997;77:359–396. doi: 10.1152/physrev.1997.77.2.359. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez de la Rosa D, Canessa C M, Fyfe G K, Zhang P. Annu Rev Physiol. 2000;62:573–594. doi: 10.1146/annurev.physiol.62.1.573. [DOI] [PubMed] [Google Scholar]
- 5.Hummler E, Barker P, Gatzy J T, Beermann F, Verdumo C, Schmidt A, Boucher R C J, Rossier B C. Nat Genet. 1996;12:325–328. doi: 10.1038/ng0396-325. [DOI] [PubMed] [Google Scholar]
- 6.McDonald F M, Yang B, Hrstka R F, Drummond H A, Tarr D E, McCray P B J, Stokes J B, Welsh M J, Williamson R A. Proc Natl Acad Sci USA. 1999;96:1727–1731. doi: 10.1073/pnas.96.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker P M, Nguyen M S, Gatzy T, Grubb B, Norman H, Hummier E, Rossier B, Boucher R C J, Koller B. J Clin Invest. 1998;102:1634–1640. doi: 10.1172/JCI3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brodovich H M. Proc Assoc Am Phys. 1996;108:345–355. [PubMed] [Google Scholar]
- 9.Pittet J F, Wiener-Kronish J P, McElroy M C, Folkesson H G, Matthay M A. J Clin Invest. 1994;94:663–671. doi: 10.1172/JCI117383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mano I, Driscoll M. BioEssays. 1999;21:568–578. doi: 10.1002/(SICI)1521-1878(199907)21:7<568::AID-BIES5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Benos D J, Stanton B A. J Physiol (London) 1999;520:631–644. doi: 10.1111/j.1469-7793.1999.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Añoveros J, Corey D P. Annu Rev Neurosci. 1997;20:567–594. doi: 10.1146/annurev.neuro.20.1.567. [DOI] [PubMed] [Google Scholar]
- 13.Waldmann R, Lazdunski M. Curr Opin Neurobiol. 1998;8:418–424. doi: 10.1016/s0959-4388(98)80070-6. [DOI] [PubMed] [Google Scholar]
- 14.Boughter J D J, Gilbertson T A. Neuron. 1999;22:213–215. doi: 10.1016/s0896-6273(00)81082-x. [DOI] [PubMed] [Google Scholar]
- 15.Price M P, Lewin G B, McIlwrath S L, Cheng C, Xie J, Heppenstall P A, Stucky C L, Mannsfeldt A G, Brennan T J, Drummond H A, et al. Nature. 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- 16.Price M P, McIllwrath S L, Xie J, Cheng C, Qiao J, Tarr D E, Sluka K A, Brennan T J, Lewin G R, Welsh M J. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 17.Wemmie J A, Chen J, Askwith C C, Hruska-Hageman A M, Price M P, Nolan B C, Yoder P G, Lamani E, Hoshi T, Freeman J H J, Welsh M J. Neuron. 2002;34:463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 18.Adams C M, Anderson M G, Motto D G, Price M P, Johnson W A, Welsh M J. J Cell Biol. 1998;140:143–152. doi: 10.1083/jcb.140.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darboux I, Lingueglia E, Pauron D, Barbry P, Lazdunski M. Biochem Biophys Res Commun. 1998;246:210–216. doi: 10.1006/bbrc.1998.8183. [DOI] [PubMed] [Google Scholar]
- 20.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann R, Tautz D. Methods Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- 22.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin: Springer; 1997. [Google Scholar]
- 23.Misquitta L, Paterson B M. Proc Natl Acad Sci USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 25.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 26.Hartenstein K, Sinha P, Mishra A, Schenkel H, Török I, Mechler B M. Genetics. 1997;147:1755–1768. doi: 10.1093/genetics/147.4.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samakovlis C, Hacohen N, Manning G, Sutherland D C, Guillemin K, Krasnow M A. Development (Cambridge, UK) 1996;122:1395–407. doi: 10.1242/dev.122.5.1395. [DOI] [PubMed] [Google Scholar]
- 28.Nardi J B. Development (Cambridge, UK) 1990;110:681–688. doi: 10.1242/dev.110.3.681. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Development (Cambridge, UK) 1996;122:3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Kolodziej P A. Development (Cambridge, UK) 2002;129:1509–1520. doi: 10.1242/dev.129.6.1509. [DOI] [PubMed] [Google Scholar]
- 31.Benos D J, Awayda M S, Berdiev B K, Bradford A L, Fuller C M, Senyk O, Ismailov I I. Kidney Int. 1996;49:1632–1637. doi: 10.1038/ki.1996.237. [DOI] [PubMed] [Google Scholar]
- 32.Garty H, Benos D J. Physiol Rev. 1988;68:309–373. doi: 10.1152/physrev.1988.68.2.309. [DOI] [PubMed] [Google Scholar]
- 33.Luciani S, Bova S, Cargnelli G, Debetto P. Pharmacol Res. 1992;25:303–310. doi: 10.1016/1043-6618(92)90666-y. [DOI] [PubMed] [Google Scholar]
- 34.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters D V, Olver R E. Pediatr Res. 1978;12:239–242. doi: 10.1203/00006450-197803000-00017. [DOI] [PubMed] [Google Scholar]
- 36.Canessa C M, Schild L, Buell G, Thorens B, Gautschi I, Horisberger J D, Rossier B C. Nature. 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 37.McDonald F J, Price M P, Snyder P M, Welsh M J. Am J Physiol. 1995;268:C1157–C1163. doi: 10.1152/ajpcell.1995.268.5.C1157. [DOI] [PubMed] [Google Scholar]
- 38.Tavernarakis N, Driscoll M. Annu Rev Physiol. 1997;59:659–689. doi: 10.1146/annurev.physiol.59.1.659. [DOI] [PubMed] [Google Scholar]
- 39.Goodman M B, Ernstrom G G, Chelur D S, O'Hagan R, Yao C A, Chalfie M. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 40.Smith D E, Otulakowski G, Yeger H, Post M, Cutz E, O'Brodovich H M. Am J Respir Crit Care Med. 2000;161:1322–1331. doi: 10.1164/ajrccm.161.4.9905064. [DOI] [PubMed] [Google Scholar]
- 41.Gaillard D, Hinnrasky J, Coscoy S, Hofman P, Matthay M A, Puchelle E, Barbry P. Am J Physiol. 2000;278:L177–L184. doi: 10.1152/ajplung.2000.278.1.L177. [DOI] [PubMed] [Google Scholar]
- 42.Talbot C L, Bosworth D G, Briley E L, Fenstermacher D A, Boucher R C, Gabriel S E, Barker P M. Am J Respir Cell Mol Biol. 1999;20:398–406. doi: 10.1165/ajrcmb.20.3.3283. [DOI] [PubMed] [Google Scholar]
- 43.Drummond H A, Abboud F M, Welsh M J. Brain Res. 2000;884:1–12. doi: 10.1016/s0006-8993(00)02831-6. [DOI] [PubMed] [Google Scholar]
- 44.Fricke B, Lints R, Stewart G, Drummond H, Dodt G, Driscoll M, von During M. Cell Tissue Res. 2000;299:327–334. doi: 10.1007/s004419900153. [DOI] [PubMed] [Google Scholar]
- 45.Drummond H A, Price M P, Welsh M J, Abboud F M. Neuron. 1998;21:1435–1441. doi: 10.1016/s0896-6273(00)80661-3. [DOI] [PubMed] [Google Scholar]