Abstract
The majority of mitochondrial phosphatidylethanolamine (PtdEtn), a phospholipid essential for aerobic growth of yeast cells, is synthesized by phosphatidylserine decarboxylase 1 (Psd1p) in the inner mitochondrial membrane (IMM). To identify components that become essential when the level of mitochondrial PtdEtn is decreased, we screened for mutants that are synthetically lethal with a temperature-sensitive (ts) allele of PSD1. This screen unveiled mutations in PHB1 and PHB2 encoding the two subunits of the prohibitin complex, which is located to the IMM and required for the stability of mitochondrially encoded proteins. Deletion of PHB1 and PHB2 resulted in an increase of mitochondrial PtdEtn at 30°C. On glucose media, phb1Δ psd1Δ and phb2Δ psd1Δ double mutants were rescued only for a limited number of generations by exogenous ethanolamine, indicating that a decrease of the PtdEtn level is detrimental for prohibitin mutants. Similar to phb mutants, deletion of PSD1 destabilizes polypeptides encoded by the mitochondrial genome. In a phb1Δ phb2Δ psd1ts strain the destabilizing effect is dramatically enhanced. In addition, the mitochondrial genome is lost in this triple mutant, and nuclear-encoded proteins of the IMM are assembled at a very low rate. At the nonpermissive temperature mitochondria of phb1Δ phb2Δ psd1ts were fragmented and aggregated. In conclusion, destabilizing effects triggered by low levels of mitochondrial PtdEtn seem to account for synthetic lethality of psd1Δ with phb mutants.
INTRODUCTION
Phospholipids are important structural components of cellular membranes and provide a permeability barrier to cells and organelles, but they also affect properties of membrane-associated proteins. Because activities of mitochondrial enzymes of the facultative anaerobic microorganism Saccharomyces cerevisiae are strongly dependent on culture conditions, this experimental system has a high potential to study molecular functions of certain phospholipids linked to respiration and other mitochondrial processes. Mitochondria harbor biosynthetic pathways for some of their phospholipids, namely, phosphatidylglycerol (PtdGro), cardiolipin (CL) (reviewed by Schlame et al., 2000), and phosphatidylethanolamine (PtdEtn) (reviewed by Daum et al., 1998). Although CL is specifically localized to mitochondrial membranes, PtdEtn is found in all subcellular membranes of eukaryotes. Both phospholipids, however, are considered to be important mitochondrial components (Birner et al., 2001; Ostrander et al., 2001).
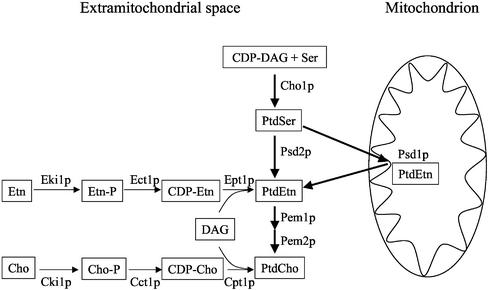
Biosynthesis of PtdEtn in S. cerevisiae can be accomplished by formation and decarboxylation of phosphatidylserine (PtdSer) or by the cytidinediphosphate (CDP)-ethanolamine (Etn) branch of the Kennedy pathway (Figure 1) (reviewed by Daum et al., 1998). PtdSer is synthesized from CDP-DAG (diacylglycerol) and serine (Ser) by PtdSer synthase, Cho1p, which is localized to the endoplasmic reticulum (ER). Mutants deleted of CHO1 do not contain detectable amounts of PtdSer and are auxotrophic for ethanolamine or choline (Cho), indicating that PtdSer is not essential and Cho1p is the only PtdSer synthase in yeast (Atkinson et al., 1980). Decarboxylation of PtdSer by Psd1p occurs in the IMM (Zinser et al., 1991), whereas Psd2p was localized to a Golgi/vacuolar compartment (Trotter and Voelker, 1995). Methylation of PtdEtn by PtdEtn methylases Pem1p and Pem2p in the ER yields phosphatidylcholine (PtdCho), the final product of the de novo route of aminoglycerophospholipid synthesis. Mutants defective in either of the PtdSer decarboxylases, Psd1p or Psd2p, grow like wild type on glucose medium. psd1Δ psd2Δ double mutants are auxotrophic for Etn or Cho on glucose media (Trotter et al., 1995) and strictly auxotrophic for Etn on lactate (Birner et al., 2001; Storey et al., 2001). Etn or Cho exogenously added to a yeast culture are used for PtdEtn or PtdCho synthesis via the CDP-Etn and CDP-Cho branches of the Kennedy pathway (Figure 1). This pathway comprises phosphorylation of Etn or Cho by the kinases Eki1p or Cki1p, activation with CTP to CDP-Etn and CDP-Cho by the cytidylyltransferases Ect1p and Cct1p, and reaction with DAG catalyzed by the phosphotransferases Ept1p and Cpt1p, yielding the final products PtdEtn and PtdCho. Different auxotrophies for Etn or Cho of psd1Δ psd2Δ double mutants on different media are due to the enhanced proliferation of mitochondria on nonfermentable carbon sources, which results in an increased specific requirement for PtdEtn (Birner et al., 2001). Because PtdEtn is imported into mitochondria only with moderate efficiency, the major route of PtdEtn formation is synthesis of PtdSer by Cho1p in the ER, transport of PtdSer from the ER to mitochondria, and decarboxylation to PtdEtn by Psd1p in the IMM. A psd1Δ mutant strain does not grow on nonfermentable carbon sources without supplementation of Etn, Cho, or Ser; contains only small amounts of mitochondrial PtdEtn compared with wild-type; and has a high tendency to form respiration deficient cells (petites) on glucose. These data support the idea that PtdEtn is essential for mitochondrial function, and mitochondrial Psd1p is of major importance for the supply of PtdEtn to mitochondria.
Figure 1.
Biosynthesis of phosphatidylethanolamine in yeast. Biosynthesis of PtdEtn in yeast is accomplished by decarboxylation of PtdSer by either Psd1p in the IMM or by Psd2p in the Golgi/vacuole. Alternatively, exogenous Etn can be incorporated into PtdEtn via the CDP-Etn branch of the Kennedy pathway. PtdCho is formed by either methylation of PtdEtn or from exogenous Cho through the CDP-Cho branch of the Kennedy pathway.
The high level of PtdEtn in mitochondria (Tuller et al., 1999) and the important role of Psd1p led us to identify components sensitive to the level of mitochondrial PtdEtn. Thus, we screened for mutants that are synthetically lethal with a temperature-sensitive (ts) allele of PSD1, psd1ts, in a psd2Δ background. As is shown herein, this screen uncovered mutations in PHB1 and PHB2, which encode the two subunits of the high-molecular-weight prohibitin complex (Steglich et al., 1999). This complex is localized to the inner mitochondrial membrane (IMM) and functions as a chaperone for mitochondrially encoded proteins (Nijtmans et al., 2000). We demonstrate that phb1Δ/phb2Δ mutants can only survive with a high level of mitochondrial PtdEtn, which seems to compensate for the lack of prohibitin. Because this requirement is not fulfilled in the psd1Δ background the double/triple mutation is lethal. Mitochondrial PtdEtn and the prohibitin complex show some functional overlap regarding the stability of mitochondrially encoded proteins and of mitochondrial DNA, suggesting that a combination of destabilizing effects is the reason for synthetic lethality of psd1Δ with phb mutants.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Culture Conditions
Strains and plasmids used in this study are listed in Table 1. Yeast strains were grown under aerobic conditions at 30°C on YP medium (1% yeast extract, 2% bactopeptone) containing 2% glucose (YPD), lactate (YPLac), or galactose (YPGal), respectively, as the carbon source. Precultures grown to the stationary phase were diluted 1:500 (vol/vol) in fresh medium. Optical density at 600 nm was measured at the time points indicated. For selective growth, yeast strains were cultivated on solid synthetic medium (Sherman et al., 1986) containing 2% glucose or lactate, respectively, and 2% bactoagar (Difco, Detroit, MI). Supplementation with Etn, Cho, or Ser was 5 mM. Viability of the phb1Δ phb2Δ psd1ts strain was tested by staining with acridine orange (Molecular Probes, Leiden, The Netherlands) at 30°C for fluorescence microscopic inspection and by dilution tests after 0-, 2-, 4-, 6-, 8-, 10-, 12-, 14-, and 15-h shift to 37°C on YPD plates at 30°C. Loss of plasmids containing the URA3 gene was performed on solid synthetic medium containing 1 mg/ml 5′-fluoroorotic acid (FOA) (BioTech Trade & Service GmbH., St. Leon-Rot, Germany).
Table 1.
Yeast strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Y00000 | BY4741 MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Euroscarf collection (Frankfurt, Germany) |
| FY1679 | MATa his3Δ200 trp1Δ63 ura3-52 | Winston et al., 1995 |
| CH1462 | MATα ade2 ade3 his3 leu2 ura3 | Kranz and Holm, 1990 |
| YRB5 | FY1679 MATa his3Δ200 leu2Δ1 ura3-52 psd1Δ∷KanMX4 psd2Δ∷KanMX4 | Birner et al., 2001 |
| YRB27 | ade2 ade3 his3 leu2 ura3 psd1Δ∷KanMX4 psd2Δ∷KanMX4 | This study |
| YRB28 | BY4741 MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 psd1Δ∷His3Mx | This study |
| YRB31 | FY1679 MATa his3Δ200 trp1Δ63 ura3-52 phb1Δ∷His3Mx phb2Δ∷TRP1 | This study |
| YRB32 | FY1679 MATa his3Δ200 leu2Δ1 ura3-52 PSD1-GFP-KanMx6 | This study |
| YRB33 | FY1679 PSD1-GFP-KanMx6 psd2Δ∷KanMX4 | This study |
| YRB35 | FY1679 MATα his3Δ200 trp1Δ63 ura3-52 PHB1-GFP-KanMx6 | This study |
| YRB36 | his3 trp1Δ63 ura3 PHB1-GFP-KanMx6 psd1Δ∷His3Mx | This study |
| YRB37 | FY1679 MATa/MATα his3Δ200/his3Δ200 leu2Δ1/LEU2 trp1Δ63/trp1Δ63 ura3-52/ura3-52 phb1Δ∷His3Mx/PHB1 phb2Δ∷TRP1/PHB2 psd1Δ∷KanMX4/PSD1 | This study |
| YRB38 | FY1679 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 phb1Δ∷His3Mx psd1Δ∷KanMX4 + pRB1 | This study |
| YRB39 | FY1679 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 phb2Δ∷Trp1Mx psd1Δ∷KanMX4 + pRB1 | This study |
| YRB40 | FY1679 his3Δ200 trp1Δ63 ura3-52 phb1Δ∷His3Mx phb2Δ∷Trp1Mx psd1Δ∷KanMX4 + pRB1 | This study |
| YRB41 | FY1679 MATα his3Δ200 leu2Δ1 trp1Δ63 ura3-52 phb1Δ∷His3Mx psd1Δ∷KanMX4 + pRS315-psd1ts#2 | This study |
| YRB42 | FY1679 MATa his3Δ200 trp1Δ63 ura3-52 phb1Δ∷His3Mx phb2Δ∷TRP1 psd1Δ∷KanMX4 + pRS316-psd1ts#2 | This study |
| YRB43 | FY1679 his3Δ200 trp1Δ63 ura3-52 phb1Δ∷His3Mx phb2Δ∷TRP1 psd2Δ∷KanMX4 | This study |
| YRB48 | BY4742 MATα his3Δ1 lys2Δ0 leu2Δ0 ura3Δ0 psd1Δ∷His3Mx yta10Δ∷KanMX4 | This study |
| YRB49 | BY4741/2 his3Δ1 leu2Δ0 ura3Δ0 psd1Δ∷His3Mx yta12Δ∷KanMX4 | This study |
| YRB50 | BY4742 MATα his3Δ1 lys2Δ0 leu2Δ0 ura3Δ0 psd1Δ∷His3Mx yme1Δ∷KanMX4 | This study |
| Y10148 | BY4742 MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 yta10Δ∷KanMX4 | Euroscarf collection (Frankfurt, Germany) |
| Y16224 | BY4742 MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 yta12Δ∷KanMX4 | Euroscarf collection (Frankfurt, Germany) |
| Y17144 | BY4742 MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 yme1Δ∷KanMX4 | Euroscarf collection (Frankfurt, Germany) |
| YRB55 | ade2 ade3 his3 leu2 ura3 psd1Δ∷KanMX4 psd2Δ∷KanMX4 + pRS313-psd1ts#2 + pRB5 | This study |
| YRB56 | FY1679 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 psd1Δ∷KanMX4 + pRS313-psd1ts#2 | This study |
| rho0 a | W303-1A MATa can1-100 ade2-1 his3-11,15 leu2-3,122 trp1-1 ura3-1 rho0 | T. Langer (University of Köln, Germany) |
| rho0 α | W303-1B MATα can1-100 ade2-1 his3-11,15 leu2-3,122 trp1-1 ura3-1 rho0 | T. Langer (University of Köln) |
| rho− | MATα ade2 oxi2 | T. Langer (University of Köln) |
| pRB1 | YCp50-PSD1 | Birner et al., 2001 |
| pRB4 | 3.9 kb EagI/SalI fragment with PSD1 from pRB1 cloned into pRS313 cut with EagI/SalI | This study |
| pRB5 | 3.9 kb EagI/SalI fragment with PSD1 from pRB1 cloned blunt into pCH1122 cut with SmaI | This study |
| pCH1122 | 3.7kb NheI/BamHI fragment with ADE3 cloned into YCp50 cut with SmaI and SpeI | Kranz and Holm, 1990 |
Standard techniques of Escherichia coli molecular biology were used throughout this study (Ausubel et al., 1996). Plasmids were introduced into yeast cells by lithium acetate transformation (Gietz et al., 1992).
Construction of psd1ts Alleles and Synthetic Lethal Screen with psd1ts
A 3.9-kb EagI/SalI fragment with PSD1 of pRB1 (YCp50-PSD1) was cloned into the centromeric vector pRS313 cut with EagI/SalI (Table 1). The pRS313-PSD1 (pRB4) plasmid was functional to restore Etn prototrophy of a psd1Δ psd2Δ strain. Temperature-sensitive PSD1 alleles were generated by error-prone polymerase chain reaction (PCR) (Stack et al., 1995). Primers Psd1-M1 and Psd1-M2 (Table 2) were chosen to span the 1.2-kb C-terminal region of PSD1, leaving the N-terminal mitochondrial-targeting sequence and the potential transmembrane domain of the protein intact. Infidelity of Taq polymerase was increased by addition of 10 mM MgCl2 and increasing the amount of dGTP (1250 μM) in the standard assay mixture fivefold. The PCR products were cotransformed with a 7-kb NcoI/BsrGI fragment of pRS313-PSD1 into a psd1Δ psd2Δ strain and the recombinant plasmid pool was tested for conferring Etn prototrophy at 30 and 37°C; 50% of 12,000 generated PSD1 alleles were functional at 30°C. We isolated 20 temperature-sensitive PSD1 alleles and 11 of these alleles were sequenced. pRS313-PSD1ts#2 (Lys356→Arg, Phe397→Leu, Glu429→Gly, Met448→Thr) was cut with EagI/SalI, and the 3.9-kb insert containing the PSD1ts#2 allele was cloned into vectors pRS315 and pRS316 cut with EagI/SalI. pRS315-PSD1ts#2 was used for construction of the phb1Δ psd1ts mutant (YRB41) and pRS316-PSD1ts#2 for construction of the phb1Δ phb2Δ psd1ts mutant (YRB42).
Table 2.
Primers
| Primer | Primer sequence from 5′ to 3′ |
|---|---|
| Psd1-M1 | AAAAGGGAGAAGGACAAGAAAAATC |
| Psd1-M2 | ACTGGGGTAAGGTGTAATCTTTGT |
| Psd1-G1 | GGTTAAGATGGGACAGAAATTAGGCATAATTGGAAAGAATGATTTAAAAGGAGCAGGTGCTGGTGCTGGTGCTGGAGCA |
| Psd1-G2 | CAGATAATACTATATACAGCAAAATAAATGCTAACTTTACATATGATTGCTTTCAATCGATGAATTCGAGCTCGTTTAAAC |
| Phb1-G1 | GTAACAGCGAGTCTTCGGGATCACCAAATTCCTTGCTTTTGAACATTGGCCGTGGAGCAGGTGCTGGTGCTGGTGCTGGAGCA |
| Phb1-G2 | GATTAAAAAAAAATTTTCTCCCCTAGTTTATTGTGTTCATAGCTTTTCCAGACTTAATCGATGAATTCGAGCTCGTTTAAAC |
| Psd1-F1 | GCCAGTTAAGAACGCCTTGGCGCAAGGGAGGACGCTCCTCCGGATCCCCGGGTTAATTAA |
| Psd1-R1 | CAGGTATGTGGTTCCAAGTGTTTGTCGCTCTTTGAATTTGGAATTCGAGCTCGTTTAAAC |
| Phb1-F1 | GAAACTTACATTCAAAATCAATAATTTACTTTAGAAAAGACGGATCCCCGGGTTAATTAA |
| Phb1-R1 | TTTTCTCCCCTAGTTTATTGTGTTCATAGCTTTTCCAGACGAATTTGAGCTCGTTTAAAC |
| Phb2-F1 | AAAGCAAGCGGCTGCTAGAAAAGAATATAATTTAGTGCTGCGGATCCCCGTGTTAATTAA |
| Phb2-R1 | TGGCATCTTAACATTGTTACTCAATTCTTAAAGATAATATGAATTCGAGCTCGTTTAAAC |
A psd1Δ psd2Δ ade2 ade3 strain (YRB27) was constructed by mating of YRB5 with CH1462, sporulation of the zygotes, and tetrad dissection. Identity of the strain was confirmed by kanamycin resistance, Etn auxotrophy, no growth on ade−-selective media, and a red-white sectoring phenotype when transformed with an ADE3 plasmid (pCH1122). For construction of a plasmid carrying the wild-type PSD1 gene and ADE3 (pRB5), the sticky ends of a EagI/SalI fragment of pRB1 with PSD1 were filled in with Klenow polymerase and the fragment cloned blunt-end into plasmid pCH1122 cut with SmaI. The plasmids pRB5 and pRS313-psd1ts#2 were introduced into strain YRB27 by transformation. The resulting psd2Δ psd1ts ade2 ade3 + pRB5 (YRB55) strain was transformed with a transposon (mTn-lacZ/LEU2) mutagenized yeast genomic library (Burns et al., 1994) to introduce random knockouts into the genome. Then 24,400 mutagenized colonies were screened for their inability to lose the plasmid pRB5 on YPD by a red-white sectoring screen. Red colonies (unable to lose pRB5) were tested for their inability to grow on selective media containing FOA and Etn. After transformation with plasmid pRB4 growth of the mutants on selective media with the same additives was restored. Isolated genomic DNA of the remaining two mutants was used as template for vectorette PCR (Riley et al., 1990; Burns et al., 1994), and the obtained PCR products were sequenced for identification of the mutations.
Construction of Deletion Strains and Green Fluorescent Protein (GFP)-Fusions
Primers used in this study are listed in Table 2. psd1Δ::His3Mx, and the phb1Δ::His3Mx phb2Δ::TRP1 double deletion strain were constructed as described by Longtine et al. (1998). Primers Psd1-F1 and Psd1-R1 were used to amplify the His3-Mx disruption cassette, which was introduced into the wild-type BY4741 by transformation. Primers Phb1-F1 and Phb1-R1 and Phb2-F1 and Phb2-R1 were used to amplify the His3-Mx and TRP1 disruption cassettes for transformation of the wild-type FY1679. Insertion of the His3-Mx and TRP1 disruption cassettes was tested by growth of the respective strains on selective media without the respective amino acid and by colony PCR with the appropriate primers.
Genomic C-terminal GFP-fusions of PSD1 and PHB1 were constructed by insertion of a GFP-KanMx6 cassette just upstream of the STOP codon of the PSD1 or PHB1 ORF, respectively, in the wild-type FY1679. Primers Psd1-G1 and Psd1-G2 were used to amplify the PSD1-GFP-KanMx6 cassette, and primers Phb1-G1 and Phb1-G2 were used to amplify the PHB1-GFP-KanMx6 cassette from plasmid pFA6a-GA5-GFP(S65T)-KanMx6 (kindly provided by R. E. Jensen, John Hopkins University of Medicine, Baltimore, MD) by PCR. The haploid wild-type strain FY1679 was transformed with these GFP-cassettes. Correct insertion of GFP was tested by resistance of the strain to kanamycin and by colony PCR. Functionality of the generated GFP-fusion proteins was confirmed by Etn prototrophy of a psd2Δ Psd1-GFP strain and by the viability of a psd1Δ Phb1-GFP strain on YPD (Table 1). Double and triple deletion mutants were constructed by mating of the corresponding single deletion mutants, sporulation of the zygotes, and tetrad dissection. Identity of the strain was confirmed by marker-dependent growth and colony PCR.
Psd1p Activity Assay and In Vivo PtdSer Import into Mitochondria
PtdSer decarboxylase activity was measured in homogenates of cells grown in YPD to the logarithmic growth phase as reported by Kuchler et al. (1986) with minor modifications: 100 nmol of [3H]PtdSer with a specific radioactivity of 1.8 μCi/nmol was used as a substrate, and the assay was performed in 0.1 M Tris-HCl, pH 7.2, containing 10 mM EDTA.
For measurement of PtdSer import, yeast strains were radiolabeled with [3H]serine. For each time point, an equivalent of 10 OD of an overnight culture was harvested, washed once, resuspended in 1 ml of YPD, and incubated for 30 min at 30°C or for 1 h at 37°C when testing psd1ts strains. Cells were labeled with 10 μCi of [3H]serine per time point for 0, 0.5, and 1 h; put on ice; and harvested by centrifugation. Three milliliters of chloroform/methanol (2:1, vol/vol) and 3 ml of glass beads were added to cell pellets. After shock freezing in liquid nitrogen samples were shaken vigorously for 10 min at 4°C, and lipids were extracted at room temperature by the method of Folch et al. (1957). Phospholipids were separated by thin layer chromatography (TLC) on Silica gel 60 plates (Merck, Darmstadt, Germany) with chloroform/methanol/25% ammonia (50:25:6, per volume) as a developing solvent. Individual spots were stained with iodine vapor, scraped off the TLC plate, and suspended in 8 ml of scintillation cocktail (J. T. Baker, Deventer, The Netherlands) containing 5% water. Radioactivity was determined by liquid scintillation counting.
Preparation of Mitochondria and Submitochondrial Membranes
Mitochondria and submitochondrial membranes were prepared from spheroplasts by published procedures (Daum et al., 1982; Zinser et al., 1991). Relative enrichment of markers and cross-contamination of subcellular fractions were assessed as described by Zinser and Daum (1995). Protein was quantified by the method of Lowry et al. (1951) by using bovine serum albumin as a standard.
Phospholipid Analysis
Lipids were extracted by the procedure of Folch et al. (1957). Individual phospholipids were separated by two-dimensional TLC by using chloroform/methanol/25% NH3 (65:35:5, per volume) as first and chloroform/acetone/methanol/acetic acid/water (50:20:10:10:5, per volume) as second developing solvent. Phospholipids were visualized on TLC plates by staining with iodine vapor, scraped off the plate, and quantified by the method of Broekhuyse (1968).
Western Blot Analysis
For localization and protein expression studies, Western blot analysis of cellular fractions prepared as described above, or of homogenates prepared by alkaline lysis (Volland et al., 1994), were performed as described by Haid and Suissa (1983) by using primary mouse antibodies against GFP, or rabbit antibodies against porin, Aac1p, Cox4p, and Phb2p. Immunoreactive bands were visualized by enzyme-linked immunosorbent assay by using a peroxidase-linked secondary antibody (Sigma-Aldrich, St. Louis, MO) following the instructions of the manufacturer.
Morphology of Mitochondria
Mitochondrial morphology was examined by indirect immunofluorescence microscopy. Cells were grown to an optical density at 600 nm of 1 at 30 or 37°C, fixed with 4% formaldehyde (Sigma-Aldrich), washed three times with 0.1 M sodium phosphate buffer pH 6.5, and once with solution A (1 M sorbitol, 0.1 M sodium phosphate buffer, pH 6.5). Then cells were resuspended in solution A to an optical density at 600 nm of 10. After adding 2 μl/ml β-mercaptoethanol (Bio-Rad, Hercules, CA) and 40 μl/ml of a 10 mg/ml zymolyase 20T (Seikagaku, Tokyo, Japan) solution, cells were incubated at 37°C for 30 min. After two washes with solution A, spheroplasts were resuspended in solution A. Coverslips were coated by adding 20 μl of 1 mg/ml polylysine onto wells, drying, washing three times with deionized water, and drying again. Then 20 μl of the cell suspension was added onto wells of coated coverslips and incubated for 20 min at 4°C in a humid box. Next, coverslips were incubated in methanol for 6 min at −20°C and in acetone for 30 s at −20°C. After drying on air, coverslips were stored at −70°C. Wells were blocked with 150 mM NaCl, 50 mM potassium phosphate buffer, pH 7, containing 0.1% NaN3, 0.1% Tween 20, and 2% milk-powder for 30 min at room temperature. After washing once with solution B (0.1 M sodium phosphate buffer, pH 6.5, containing 2% bovine serum albumin), fixed cells were incubated with rabbit anti-porin antiserum diluted 1:250 with solution B in a humid box at 4°C overnight. After three washes with solution B, cells were incubated with an anti-rabbit secondary antibody conjugated to fluorescein (Sigma-Aldrich) in a dark humid box for 1 h at 4°C. After four washes with solution B and drying, DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) (Molecular Probes). Fluorescence was observed with an Axiovert 35 fluorescence microscope (Carl Zeiss, Jena, Germany).
Analysis of the Mitochondrial Genome
Respiration-deficient psd1Δ single colonies of each mating type, and phb1Δ psd1ts and phb1Δ phb2Δ psd1ts strains were examined for the presence of the mitochondrial genome by mating with rho− (deletion in the mitochondrial genome) and rho0 tester strains (no mitochondrial genome) (Table 1). Zygotes were analyzed for growth on YPLac. In addition, strains were labeled with the DNA stain DAPI (Molecular Probes), and 100 cells were inspected for presence of mitochondrial DNA with an Axiovert 35 fluorescence microscope (Carl Zeiss).
Stability of Mitochondrially Encoded Proteins
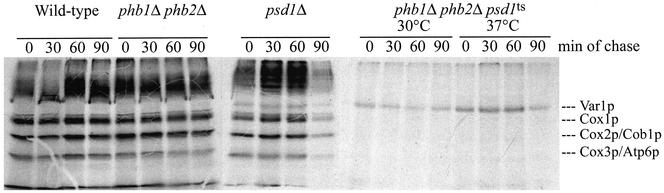
Twenty milliliters of cultures of phb1Δ phb2Δ, psd1Δ and phb1Δ phb2Δ psd1ts strains grown to the mid-logarithmic growth phase on YP medium with 2% galactose as carbon source under aerobic conditions at 30°C were harvested, washed twice with 10 mM Tris-HCl pH 7.5, and suspended in 4 ml of methionine-free minimal medium (Sherman et al., 1986) with 0.3% glucose as the carbon source. Then cells were incubated for 15 min at 30°C, or in the case of the phb1Δ phb2Δ psd1ts strain, for 1 h at 37°C. After further incubation for 10 min at permissive or restrictive temperature with 40 μl of a cycloheximide stock solution (60 mg/ml in ethanol), 100 μCi/ml of EXPRE35S35S protein-labeling mix (PerkinElmer Life Sciences, Boston, MA) was added. Labeling was carried out for 30 min, and after addition of 400 μl of chase solution (0.14 M methionine and 0.06 M cysteine), 1-ml samples were withdrawn at 0, 30, 60, and 90 min and put on ice. Samples were washed twice with ice cold chase solution, and cell pellets were suspended in 200 μl of MTE buffer (0.65 M mannitol, 20 mM Tris, 1 mM EDTA) containing 1 mM phenylmethylsulfonyl fluoride (Calbiochem, La Jolla, CA) and disintegrated by shaking with glass beads for 30 min at 4°C. The supernatant was centrifuged for 20 min at 13,000 rpm at 4°C to sediment a crude mitochondrial fraction. After a wash with MTE, the membrane pellet was solubilized in reducing sample buffer by heating to 95°C for 3 min. Labeled proteins were separated by 12.5% SDS-PAGE, stained with Coomassie Blue, and visualized by autoradiography of the dried gel.
RESULTS
A Strategy to Screen for Synthetic Lethality with psd1ts
To gain more insight into the function of PtdEtn in mitochondria, we screened for mutations that depend on high levels of mitochondrial PtdEtn. For this purpose, a psd1Δ psd2Δ double deletion strain in an ade2 ade3 background carrying two centromeric plasmids, one with a ts allele of PSD1 (PSD1ts) and the other with a wild-type allele of PSD1 linked to the ADE3 gene, was transformed with a transposon-mutagenized yeast genomic library for introduction of random deletions in the genome (see MATERIALS AND METHODS).
The strategy behind this approach was that colonies losing the plasmid with the wild-type allele of PSD1 linked to ADE3 would show a red-white sectoring phenotype, whereas colonies that depend on a fully functional Psd1p and thus are unable to lose the plasmid with PSD1 linked to ADE3 would be red. Such mutants are hence synthetically lethal with psd2Δ psd1ts. Mutants exhibiting this phenotype were isolated on glucose media (YPD). The screen was performed at 30°C, when the in vitro activity of Psd1tsp encoded by pRS313-PSD1ts#2 was 70% compared with wild-type Psd1p. These conditions allowed mitochondria to produce their own PtdEtn, although with lower efficiency than in wild type. PSD2 was deleted to enforce the requirement for mitochondrial PtdEtn biosynthesis by Psd1p.
Mutations in Mitochondrial Prohibitin Are Synthetically Lethal with psd1ts psd2Δ
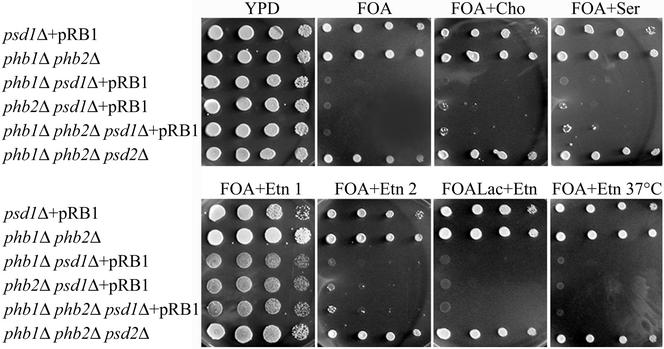
Using this approach, we isolated two mutants synthetically lethal with psd1ts psd2Δ, one with a deletion in PHB1 and the other in PHB2. Because Phb1p and Phb2p are interdependent components (Berger and Yaffe, 1998) mutants deleted of either PHB1 or PHB2 do not contain detectable amounts of the other protein. To confirm the synthetic lethality of phb1/phb2 with psd1 and to analyze the specificity of the genetic interaction, we constructed a series of double and triple deletion mutants for phenotypic analysis (Table 1). Strains bearing psd1Δ phb1Δ, psd1Δ phb2Δ and psd1Δ phb1Δ phb2Δ deletions could not be obtained by tetrad dissection of a psd1Δ/PSD1 phb1Δ/PHB1 phb2Δ/PHB2 diploid strain YRB37 on YPD. Tetrad dissection of YRB37 after transformation with the pRB1 plasmid encoding PSD1 led to the isolation of psd1Δ phb1Δ, psd1Δ phb2Δ and psd1Δ phb1Δ phb2Δ strains bearing pRB1. Viability of these strains (YRB38, 39, and 40) strictly depended on the presence of an intact Psd1p. When these mutants were forced to lose the plasmid on FOA plates (Figure 2), they could only be rescued for a limited number of generations by supplementation with Etn, but not with Cho or Ser. Transfer of these strains from a FOA plate supplemented with Etn to a fresh plate with the same additives resulted in lethality. Thus, depletion of PtdEtn seemed to be the primary reason for the synthetic lethality of prohibitin mutants with psd1Δ. Moreover, exogenous Etn could not rescue psd1Δ phb1Δ, psd1Δ phb2Δ and psd1Δ phb1Δ phb2Δ strains at elevated temperature or on the nonfermentable carbon source lactate. In contrast, a psd2Δ phb1Δ phb2Δ triple deletion strain (YRB43) was viable on full media and not auxotrophic for Etn irrespective of the carbon source.
Figure 2.
psd1Δ phb1Δ and psd1Δ phb2Δ mutants are rescued by Etn on glucose for a limited number of generations, but not on lactate or at elevated temperature. Dilutions (1, 1/10, 1/100, and 1/1000) of cell suspensions were spotted on rich glucose media (YPD) or forced to lose pRB1 on synthetic media containing 2% glucose and FOA (FOA), supplemented with Cho (FOA + Cho), Ser (FOA + Ser), or Etn (FOA + Etn). Strains were transferred from FOA + Etn to a fresh plate (FOA + Etn 2). Additionally, strains were forced to lose pRB1 on synthetic media containing 2% lactate and FOA supplemented with Etn (FOALac + Etn). Plates were incubated at 30°C unless stated otherwise.
Psd1p and the Prohibitin Complex Are Independently Expressed and Assembled
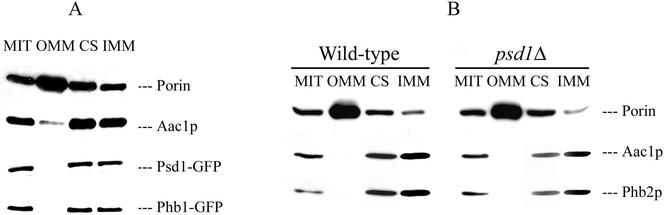
Cell fractionation experiments demonstrated that Psd1p does not affect expression and subcellular localization of the prohibitin complex. Western blot analysis showed that Phb1p-GFP and Psd1p-GFP fusion proteins are components of the IMM in wild-type (Figure 3A), confirming previous results (Zinser et al., 1991; Berger and Yaffe, 1998). In a psd1Δ strain Phb2p was also localized to the IMM (Figure 3B). In vitro Psd1p activity of a phb1Δ phb2Δ strain (0.061 nmol/min × mg protein) was even higher than in wild-type (0.056 nmol/min × mg protein). These results clearly demonstrated that Psd1p and the prohibitin complex are expressed and assembled independently of each other.
Figure 3.
Submitochondrial distribution of Psd1-GFP, Phb1-GFP, and Phb2p. Mitochondria (MIT) and subfractions of mitochondria, OMM, contact sites (CS), and IMM, were subjected to Western blot analysis. Psd1-GFP and Phb1-GFP were detected in wild type (A) bearing the respective hybrid proteins by using anti-GFP antibodies. Phb2p was detected in submitochondrial fractions of wild-type and psd1Δ strains (B) by using anti-Phb2p antiserum. Porin and Aac1p (ATP/ADP carrier) were used as marker proteins for the OMM and IMM, respectively.
Mitochondria of a phb1Δ phb2Δ Mutant Have Increased Levels of PtdEtn
To determine whether prohibitin mutants had a specific requirement for PtdEtn or whether the prohibitin complex was required for mitochondrial lipid homeostasis in yeast, we analyzed phospholipids of a phb1Δ phb2Δ double deletion strain grown to the late-exponential phase on rich lactate media (YPLac). This analysis revealed a significantly elevated amount of PtdEtn at the expense of PtdIns in the homogenate of the phb1Δ phb2Δ strain compared with wild- type (Table 3). This effect was even more pronounced with isolated mitochondria of the prohibitin deletion mutant. Levels of other phospholipids, including PtdSer were not altered in the double mutant. In contrast, a psd1Δ mutant accumulated a significant amount of PtdSer in mitochondria and had a dramatically reduced PtdEtn level compared with wild-type. Thus, phospholipid compositions are altered in opposite directions in the psd1Δ and prohibitin mutants compared with wild-type. Deletion of the prohibitin complex seemed to be compensated by a high level of mitochondrial PtdEtn, which could not be maintained in a psd1Δ deletion strain.
Table 3.
Phospholipid composition of mitochondria and homogenate of a phb1Δ phb2Δ mutant grown on YPLac
| Strain | CF | Phospholipids (mol %)
|
||||||
|---|---|---|---|---|---|---|---|---|
| PA | PtdSer | PtdIns | PtdCho | PtdEtn | CL | LPL | ||
| FY1679 | HOM | 2 | 6 | 17 | 53 | 18 | 3 | 1 |
| phb1Δ phb2Δ | HOM | 2 | 6 | 12 | 54 | 23 | 3 | 1 |
| psd1Δ | HOM | 2 | 7 | 21 | 58 | 8 | 3 | 1 |
| FY1679 | MIT | 1 | 1 | 13 | 51 | 25 | 7 | 2 |
| phb1Δ phb2Δ | MIT | 0 | 2 | 6 | 52 | 33 | 6 | 1 |
| psd1Δ | MIT | 1 | 6 | 13 | 67 | 6 | 5 | 2 |
CF, cellular fraction; HOM, homogenate; LPL, lysophospholipid; MIT, mitochondria.
Mean values of two independent measurements are shown. Mean deviations are <5%.
Cell Biological Effects Leading to Synthetic Lethality of phb1Δ phb2Δ with psd1Δ
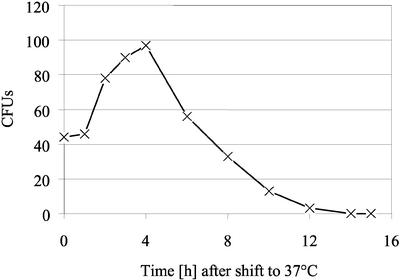
What is the reason for synthetic lethality of prohibitin mutations with psd1Δ? To address this question, we used a temperature-sensitive phb1Δ phb2Δ psd1ts triple mutant (Table 1). Although phb1/phb2 psd2Δ psd1ts strains isolated in the synthetic lethal screen described above were not viable at 30°C on YPD, the phb1Δ phb2Δ psd1ts strain grew at 30°C, but started to lose viability after a 4-h temperature shift to 37°C on YPD (Figure 4). Viability of the strain at the restrictive temperature strictly depended on the presence of Psd1tsp, because loss of the plasmid pRS316-psd1ts on FOA plates was lethal. Temperature sensitivity of the mutant was not rescued by supplementation with Etn, Cho, or Ser, confirming the results obtained with the plasmid rescue experiment (Figure 2).
Figure 4.
Viability of a phb1Δ phb2Δ psd1ts mutant on YPD after a temperature shift to 37°C. A phb1Δ phb2Δ psd1ts mutant grown on YPD was temperature shifted to 37°C. At time points indicated the number of viable cells was determined by dilution plating on YPD plates and counting of the colony-forming units after incubation at 30°C.
The lethality of the phb1Δ phb2Δ psd1ts strain is linked to a decrease of the mitochondrial PtdEtn level (Table 4), which is the result of a decreased Psd1tsp activity to 10% of wild type at the nonpermissive temperature. Mitochondria of the phb1Δ phb2Δ strain exhibited an increased level of PtdEtn compared with wild-type at 30°C similar to cells grown on YPLac (Table 3). At 37°C, however, the mitochondrial phospholipid composition of wild-type and phb1Δ phb2Δ was almost identical. Growth of phb1Δ phb2Δ psd1ts at 30°C, or after a shift to 37°C for only 1 h, resulted in a mitochondrial PtdEtn level that roughly corresponds to the wild-type control. The PtdEtn level of the triple mutant at 30°C was slightly lower than that of phb1Δ phb2Δ at this temperature. Prolonged growth of the triple mutant at the restrictive temperature, however, caused a dramatic decrease of the mitochondrial PtdEtn level. This level seems to be below the critical concentration of PtdEtn in mitochondria and explains the loss of viability of the triple mutant after several hours at 37°C (Figure 4).
Table 4.
Phospholipid composition of mitochondria of a phb1Δ phb2Δ psd1ts mutant grown on YPD at 30°C and after a 1- and 6-h shift to 37°C
| Strain | Temp. (°C) | Phospholipids (mol %)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PA | PtdSer | PtdIns | PtdCho | PtdEtn | CL | DMPE | LPL | ||
| FY1679 | 30 | 2 | 3 | 22 | 35 | 24 | 3 | 6 | 3 |
| 37 | 3 | 7 | 15 | 48 | 23 | 4 | 1 | 0 | |
| phb1Δ phb2Δ | 30 | 3 | 4 | 16 | 34 | 31 | 6 | 5 | 2 |
| 37 | 3 | 9 | 14 | 46 | 23 | 4 | 2 | 0 | |
| phb1Δ phb2Δ psd1ts | 30 | 3 | 4 | 23 | 38 | 26 | 2 | 1 | 3 |
| 37 (1 h) | 3 | 8 | 22 | 38 | 23 | 2 | 2 | 2 | |
| 37 (6 h) | 4 | 11 | 18 | 46 | 14 | 2 | 1 | 4 | |
DMPE, dimethyl-PtdEtn; LPL, lysophospholipid.
Mean values of two independent measurements are shown. Mean deviations are <5%.
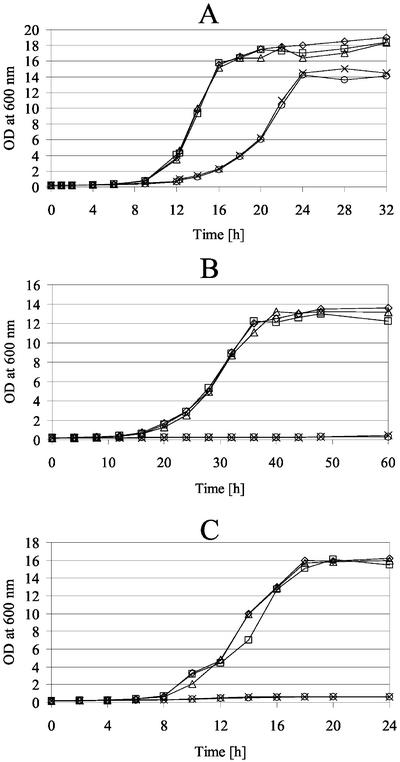
At 30°C, the psd1ts and phb1Δ phb2Δ strains grew like wild- type, whereas the combination of prohibitin mutations with psd1ts resulted in a decreased growth rate on YPD (Figure 5). At the nonpermissive temperature, the phb1Δ psd1ts and phb1Δ phb2Δ psd1ts strains did not grow. On rich lactate media (YPLac), growth of psd1ts and phb1Δ phb2Δ strains was not impaired, indicating that these strains did not accumulate petites (respiratory defects) at the permissive temperature, as was shown in previous work from our laboratory with psd1Δ mutants (Birner et al., 2001). In contrast, the phb1Δ psd1ts and phb1Δ phb2Δ psd1ts strains did not grow on YPLac at 30°C, suggesting a petite phenotype.
Figure 5.
The phb1Δ phb2Δ psd1ts strain does not grow at elevated temperature or on nonfermentable carbon sources. Growth of phb1Δ psd1ts (○), phb1Δ phb2Δ psd1ts (×), psd1ts (▵), phb1Δ phb2Δ (□), and wild-type (⋄) strains on YPD at 30°C (A) or 37°C (C) and on YPLac at 30°C (B) was monitored by measurement of the optical density at 600 nm at time points indicated.
The Prohibitin Complex Is Not Involved in Transport of PtdSer into Mitochondria
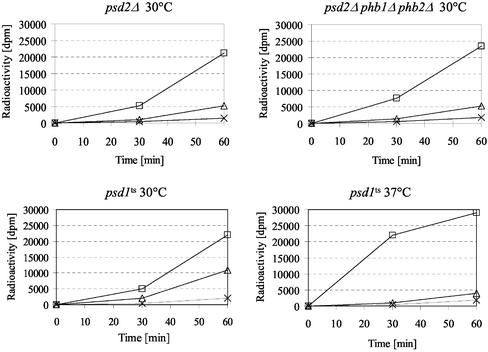
One reason for the synthetic lethality of psd1Δ and the prohibitin complex could be the involvement of the prohibitin complex in import and/or assembly of phospholipids into mitochondria. The elevated level of mitochondrial PtdEtn in the phb mutant, however, suggested that the prohibitin complex was not involved in import of PtdSer or PtdEtn. Moreover, the Etn prototrophy of the psd2Δ phb1Δ phb2Δ strain (Figure 2) indicated that import of PtdSer, the substrate for Psd1p, into mitochondria, and PtdEtn export out of mitochondria were not significantly impaired in this strain. This notion was confirmed by in vivo labeling of the psd2Δ phb1Δ phb2Δ strain with [3H]serine (Figure 6). The triple deletion and the psd2Δ mutant exhibited a similar rate of PtdEtn appearance, indicating that the prohibitin complex was not involved in the supply of PtdSer to mitochondria. As a control, the psd1ts strain was shown to decarboxylate PtdSer to PtdEtn efficiently at 30°C, but to accumulate PtdSer and convert it slowly to PtdEtn due to residual Psd2p activity after a 1-h shift to 37°C. Similar results would be expected with a mutant defective in transport of PtdSer into mitochondria.
Figure 6.
The prohibitin complex is not involved in import of PtdSer into mitochondria. Strains were labeled for 0, 30, and 60 min with [3H]serine at the indicated temperature. Incorporation of label into PtdSer (□), PtdEtn (▵), and PtdCho (×) was determined by liquid scintillation counting of individual phospholipid spots scraped off a TLC plate (see MATERIALS AND METHODS).
Mitochondrial Morphology and Stability of Mitochondrial DNA in phb and psd1 Mutants
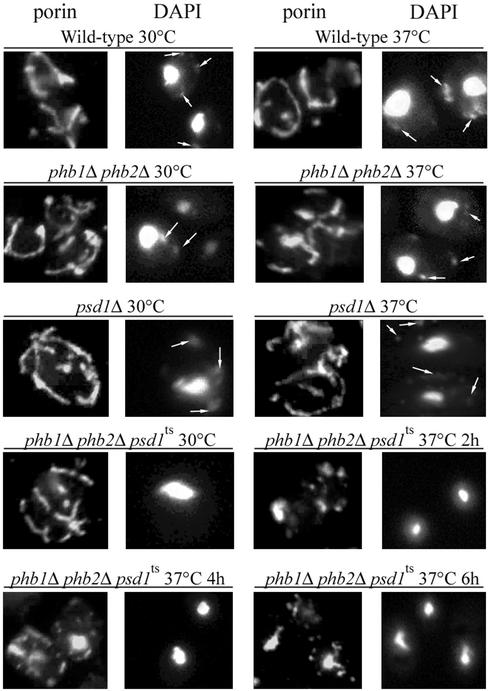
Mitochondrial morphology of strains bearing defects in prohibitin and/or PSD1 was examined by indirect immunofluorescence of the outer mitochondrial membrane (OMM) protein porin (see MATERIALS AND METHODS). Although phb1Δ phb2Δ and psd1Δ cells showed a wild-type mitochondrial morphology at 30 and 37°C (Figure 7), the phb1Δ phb2Δ psd1ts triple mutant lost the wild-type mitochondrial reticulum after a shift to the nonpermissive temperature. Instead, mitochondria were fragmented and partly collapsed. Mitochondria of phb1Δ psd1ts and phb1Δ phb2Δ psd1ts strains had a very low membrane potential already at 30°C and could not be stained with 4-(4-dimethylamino) styryl-N-methylpyridinium iodide (DASPMI) or MitoTracker (Molecular Probes), two membrane potential-dependent dyes (our unpublished data). In contrast, mitochondria of psd1Δ and phb1Δ phb2Δ mutants showed normal membrane potential and morphology, confirming previous results (Berger and Yaffe, 1998; Birner et al., 2001).
Figure 7.
Mitochondrial morphology of phb and psd1 mutants. The OMM protein porin was detected in fixed cells by indirect immunofluorescence (see MATERIALS AND METHODS). Nuclear and mtDNA was stained with DAPI. Arrows indicate mtDNA.
In contrast to the phb1Δ phb2Δ and psd1Δ cells, the phb1Δ phb2Δ psd1ts mutant apparently lacked mitochondrial (mt) DNA (Figure 7). To shed more light on the reason for the respiratory deficiency of phb1Δ phb2Δ psd1ts (Figure 5B), we extended our analysis of the strain to examine the presence of mtDNA. In contrast to wild type and a rho− tester strain, DAPI-stained mtDNA could not be detected in >98% of single cells of the phb1Δ phb2Δ psd1ts and phb1Δ psd1ts strains, and in the rho0 tester strain (Table 5). To study whether stability of the mitochondrial genome was affected in psd1Δ petites that accumulated on glucose, we subjected single respiratory-deficient colonies to microscopic inspection. mtDNA could not be detected in 98% of inspected cells. Moreover, diploid strains produced by mating of eight petite single colonies of psd1Δ strains of each mating type and of the phb1Δ phb2Δ psd1ts and the phb1Δ psd1ts strains with rho0 and rho− tester strains were unable to grow on nonfermentable carbon sources. These results indicated that loss of the majority of the mitochondrial genomes in psd1Δ, phb1Δ phb2Δ psd1ts and phb1Δ psd1ts accounts for the observed growth phenotype of these strains.
Table 5.
Analysis of mitochondrial DNA
| Strain | % Cells containing mtDNA (n = 100)a | Respiratory growth of diploids produced by mating withb
|
|
|---|---|---|---|
| rho0 | rho− | ||
| Wild-type | 95 | + | + |
| rho− | 89 | − | ND |
| rho0 | 0 | − | − |
| Petite psd1Δ MATa | 2 | − | − |
| Petite psd1Δ MATα | 0 | − | ND |
| phb1Δ psd1ts | 1 | − | ND |
| phb1Δ phb2Δ psd1ts | 0 | − | − |
ND, not determined.
Presence of mitochondrial DNA was tested by staining strains with DAPI and microscopic inspection.
+, growth; −, no growth.
Defective Expression and Stability of Mitochondrial Proteins in phb1Δ phb2Δ psd1ts
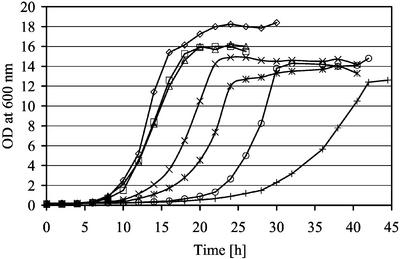
Prohibitin is involved in stability of mitochondrially encoded proteins by directly inhibiting the activity of the matrix-AAA protease Yta10p/Yta12p, but not of the intermembrane space-AAA protease Yme1p (Steglich et al., 1999; Nijtmans et al., 2000). Thus, it was tempting to speculate that Psd1p or mitochondrial PtdEtn may also have an effect on mitochondrial protein stability. phb1Δ and phb2Δ mutants were reported to be synthetic slow with yta10Δ and yta12Δ deletions but not with yme1Δ (Steglich et al., 1999). We observed similar, although less severe growth defects, when PSD1 was deleted in these protease mutants. The psd1Δ mutant was synthetic slow with yta10Δ and yta12Δ but not with yme1Δ deletions on YPD (Figure 8).
Figure 8.
Synthetic slow growth phenotype of psd1Δ and matrix-AAA protease mutants. Growth of yta10Δ (×), yta10Δ psd1Δ (*), yta12Δ (○), yta12Δ psd1Δ (+), yme1Δ (□), yme1Δ psd1Δ (▵), and wild-type (⋄) strains on YPD at 30°C was monitored by measurement of the optical density at 600 nm at time points indicated.
Stability of mitochondrially encoded proteins was slightly affected in the phb1Δ phb2Δ deletion mutant (Figure 9), confirming previous results by Nijtmans et al. (2000). The stability defect was more dramatic in the psd1Δ strain. In the phb1Δ phb2Δ psd1ts strain the only labeled peptide was the 47-kDa ribosomal Var1p, confirming that the strain had lost the majority of its mtDNA. This defect was observed both at the permissive and the nonpermissive temperature.
Figure 9.
Instability of mitochondrial encoded proteins of phb and psd1 mutant strains. Mitochondrial encoded proteins were pulse labeled and analyzed by autoradiography after 0, 30, 60 and 90 min of chase (see MATERIALS AND METHODS).
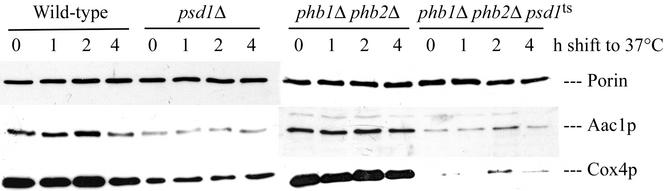
To investigate whether the expression, assembly and/or stability of nuclear encoded mitochondrial proteins was also affected by the prohibitin and/or psd1 mutations, we performed a Western blot analysis of homogenates from phb1Δ phb2Δ psd1ts, phb1Δ phb2Δ, psd1Δ, and wild-type grown on YPD at 30°C and after a 0-, 1-, 2-, and 4-h temperature shift to 37°C (Figure 10). Although assembly of porin into the OMM was not affected in any of these strains, nuclear-encoded proteins of the IMM, such as Aac1p (ATP/ADP carrier) and Cox4p (cytochrome c oxidase subunit IV), were not efficiently formed and/or assembled in psd1Δ and to a more dramatic extent in the phb1Δ phb2Δ psd1ts strain. These defects were already visible at 30°C, and protein levels were not further decreased by the temperature shift to 37°C. The cytochrome c oxidase complex consists of nuclear and mitochondrially encoded proteins. Because the mitochondrially encoded subunits Cox1–3p were not expressed (Figure 9) the nuclear encoded subunits, such as Cox4p, may be degraded in phb1Δ phb2Δ psd1ts and in petite psd1Δ cells. Because the ATP/ADP carrier Aac1p is nuclear encoded transport of proteins into/across the IMM or assembly into the IMM may also be impaired in the psd1Δ and phb1Δ phb2Δ psd1ts mutants. This phenotype correlates with the accumulation of petites during growth of the psd1Δ strain on glucose and the petite phenotype of the phb1Δ phb2Δ psd1ts mutant. In contrast, mitochondria isolated from a psd1Δ strain grown on nonfermentable carbon sources had a wild-type protein composition (Figure 3B), because only 10% of the cells are petite under those conditions (Birner et al., 2001).
Figure 10.
Proteins of the IMM are not efficiently assembled in psd1Δ and phb1Δ phb2Δ psd1ts strains. Homogenates of strains grown at 30°C on YPD were analyzed after a 0-, 1-, 2-, and 4-h temperature shift to 37°C by Western blotting with primary antibodies against the OMM protein porin and the IMM proteins Aac1p and Cox4p.
DISCUSSION
In this study, we provide new evidence for the function of mitochondrial PtdEtn through characterization of synthetic lethality of psd1Δ with mutations in the mitochondrial prohibitin complex Phb1p/Phb2p. We show that phb mutants are sensitive to the level of mitochondrial PtdEtn in a very specific way. Loss of viability of a phb psd1ts mutant after shift to the nonpermissive temperature clearly correlates with a decrease in mitochondrial PtdEtn. The apparent lag time of this effect suggests that upon inactivation of Psd1tsp mitochondrial PtdEtn is diluted out by ongoing cell division. Moreover, phbΔ psd1Δ mutants are rescued only for a limited number of generations by supplementation with Etn. Because extramitochondrial PtdEtn synthesized by the Kennedy pathway is not efficiently imported into mitochondria (Birner et al., 2001), mitochondrial PtdEtn is also steadily decreased by ongoing cell division in these mutants.
Strains deleted of both subunits of the prohibitin complex exhibit significantly higher concentrations of total and mitochondrial PtdEtn at 30°C. It seems that the enhanced level of mitochondrial PtdEtn compensates for the prohibitin defect. This compensation is specific for PtdEtn insofar as mutations affecting the biosynthetic pathway of the other mitochondrially formed phospholipids, CL and PtdGro, are not synthetically lethal with phb1/phb2 (our unpublished data). The observation that the mitochondrial phospholipid composition of the phb mutant is similar to wild-type at 37°C suggests that other factors may compensate for the loss of prohibitin at elevated temperature.
How does prohibitin interact with the mitochondrial PtdEtn-synthesizing machinery? It is not simply inhibition of expression of Psd1p, because enzymatic activity of this enzyme is not affected in the phb mutant. It is also unlikely that prohibitin affects the import of the substrate of Psd1p, PtdSer, from the ER to the IMM, because phb mutations rather cause an increase than a decrease of the mitochondrial PtdEtn level. Similarly, export of PtdEtn from mitochondria does not seem to be affected by the prohibitin mutation, because the effects expected from such a defect, namely, a decrease in total cellular PtdCho and a Cho auxotrophic phenotype of combined psd2Δ phb mutants, were not observed. It is possible, however, that import of extramitochondrially formed PtdEtn into mitochondria is decreased in phb mutants. Such a defect would not affect mitochondrial function in wild-type, because supply of PtdEtn to mitochondrial membranes is much more efficient through Psd1p than through extramitochondrial Psd2p and the CDP-Etn pathway (Bürgermeister, Birner, Nebduer, and Dawn, unpublished data). In phb psd1 double mutants, however, such a defect may become harmful to the cell. As shown in a previous study (Birner et al., 2001) the level of PtdEtn in psd1 becomes critical upon an increased requirement, e.g., when cells are grown on nonfermentable carbon sources. A similar effect may be caused by the combination of psd1 with prohibitin mutations, although direct experimental evidence supporting this hypothesis is missing.
The increased level of PtdEtn in phb and the synthetic lethality of probibitin mutants with psd1 clearly point to antagonistic effects of the two mutations and specific function(s) of mitochondrial PtdEtn in processes governed by prohibitin. It has been shown previously (Nijtmans et al., 2000) that defects in the prohibitin complex negatively affect the stability of proteins encoded by the mitochondrial protein-synthesizing machinery. In our laboratory, we demonstrated that defects in Psd1p result in formation of petites (respiratory-deficient cells) (Birner et al., 2001). In the present study, we extend this finding insofar as we demonstrate that deletion of PSD1 causes loss of mtDNA and also instability of mitochondrially synthesized proteins. We conclude that a certain level of PtdEtn in the IMM may be required for the attachment of mtDNA nucleoids, which is thought to be important for replication, recombination, transcription and segregation (Shadel, 1999). Defects of the mitochondrial translation/transcription machinery, however, were dramatically enhanced in phb1/phb2 psd1ts when the requirement for PtdEtn was increased by the prohibitin mutation on one hand, and the level of PtdEtn was decreased by psd1ts on the other hand. In the triple mutant, the mitochondrial genome seems to be largely lost despite a wild-type level of mitochondrial PtdEtn at 30°C. Thus, the prohibitin complex may also be involved in mtDNA stability. We hypothesize that a high local concentration of PtdEtn may determine the attachment site of mtDNA nucleoids. Prohibitin may be involved in the assembly of this domain, and its deletion may be overcome by elevated levels of PtdEtn in the IMM.
The phb1/phb2 psd1ts triple mutation also affects import and/or assembly of mitochondrial proteins that are formed on cytosolic ribosomes. Transport of polypeptides across/into the IMM, in contrast to protein import into the OMM, requires a membrane potential across the IMM (Martin et al., 1991; Kübrich et al., 1998). Loss of the mitochondrial potential in phb1/phb2 psd1ts is already observed at the permissive temperature and may become fatal when cells are shifted to 37°C. The triple mutation, however, does not only affect the formation and/or assembly of mitochondrial complexes of the respiratory machinery but also the incorporation of the ADP/ATP carrier Aac1p into the IMM. This additional defect on ATP translocation may significantly contribute to the dysfunction of mitochondria. It has been postulated that upon loss of the electron transport chain and the F0-ATPase, e.g., in rho0 mutants, generation of a membrane potential across the IMM can only occur by continuous exchange of cytoplasmic ATP and mitochondrial ADP through ADP/ATP-carriers and be driven by an active F1-ATPase in the mitochondrial matrix (Chen and Clark-Walker, 1999). Ostrander et al. (2001) suggested that the petite lethality, i.e., dependence on intact mtDNA, of strains lacking the PtdGro synthase may be explained by similar effects.
In contrast to psd1Δ and phb1Δ phb2Δ mutants, mitochondria of the phb1Δ phb2Δ psd1ts strain become fragmented and collapse at the nonpermissive temperature. Reasons for the mitochondrial morphology defect may be ATP deficiency and loss of membrane potential. The observation that prohibitin mutants are synthetically lethal with the mitochondrial morphology mutants mmm1Δ, mdm10Δ and mdm12Δ (Berger and Yaffe, 1998) led us to speculate that these strains may also be affected by a psd1Δ mutation. This hypothesis turned out to be wrong, because the respective double mutants were viable (our unpublished results). Thus, prohibitin seems indeed to interact with mitochondrial PtdEtn in a specific way. Mmm1p, Mdm10p, and Mdm12p are required for normal mitochondrial morphology, and deletion results in the formation of large, spherical mitochondria. Moreover, mmm1 mutants have a dramatically disorganized IMM, a collapsed mitochondrial nucleoid structure, and are defective in transmission of mtDNA to daughter cells (Hobbs et al., 2001). Preliminary data from our laboratory indicate that in an mmm1Δ strain, in contrast to prohibitin mutants, the amount of mitochondrial PtdEtn decreases to a level similar to psd1Δ. This observation may explain the synthetic lethality of mmm1Δ with phb1/phb2. Thus, Mmm1p/Mdm10p/Mdm12p in the OMM, the prohibitin complex in the IMM, and mitochondrial PtdEtn may form a functional unit that provides stability of mtDNA and integrity of mitochondrial membranes.
ACKNOWLEDGMENTS
The technical assistance of C. Hrastnik is appreciated. We are grateful to T. Langer (University of Köln, Germany) for providing strains and antibodies, and especially for fruitful discussions by e-mail. This work was financially supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich projects 14468 (to G.D.) and 15210 (to R.S.).
Abbreviations used:
- CDP
cytidinediphosphate
- Cho
choline
- CL
cardiolipin
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- Etn
ethanolamine
- FOA
5′-fluoroorotic acid
- GFP
green fluorescent protein
- IMM
inner mitochondrial membrane
- mtDNA
mitochondrial DNA
- OMM
outer mitochondrial membrane
- PA
phosphatidic acid
- PtdCho
phosphatidylcholine
- PtdEtn
phosphatidylethanolamine
- PtdIns
phosphatidylinositol
- PtdSer
phosphatidylserine
- TLC
thin layer chromatography
- ts
temperature sensitive
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0263. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0263.
REFERENCES
- Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S. Yeast mutants auxotrophic for choline or ethanolamine. J Bacteriol. 1980;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1996. [Google Scholar]
- Berger KH, Yaffe MP. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4043–4052. doi: 10.1128/mcb.18.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R, Bürgermeister M, Schneiter R, Daum G. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:997–1007. doi: 10.1091/mbc.12.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekhuyse RM. Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim Biophys Acta. 1968;152:307–215. doi: 10.1016/0005-2760(68)90038-6. [DOI] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. Alpha and beta subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1999;262:898–908. doi: 10.1007/s004380051156. [DOI] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz JE, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübrich M, Rassow J, Voss W, Pfanner N, Hönlinger A. The import of ATP/ADP carrier into mitochondria separates from the general pathway of cleavable preproteins at the trans side of the outer membrane. J Biol Chem. 1998;273:16374–16381. doi: 10.1074/jbc.273.26.16374. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J Bacteriol. 1986;165:901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import: Δϕ drives the movement of presequences. J Biol Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander DB, Zhang M, Mileykovskaya E, Rho M, Dowhan W. Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J Biol Chem. 2001;276:25262–25272. doi: 10.1074/jbc.M103689200. [DOI] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Prog Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- Shadel GS. Yeast as a model for human mtDNA replication. Am J Hum Genet. 1999;65:1230–1237. doi: 10.1086/302630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol. 1999;19:3435–42. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey MK, Clay KL, Kutateladze T, Murphy RC, Overduin M, Voelker DR. Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J Biol Chem. 2001;276:48539–48548. doi: 10.1074/jbc.M109043200. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Yates R, Voelker DR. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J Biol Chem. 1995;270:6071–6080. doi: 10.1074/jbc.270.11.6071. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Tuller G, Nemec T, Hrastnik C, Daum G. Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild-type strain grown on different carbon sources. Yeast. 1999;15:1555–1564. doi: 10.1002/(SICI)1097-0061(199910)15:14<1555::AID-YEA479>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]