Abstract
We previously identified Arabidopsis genes homologous with the yeast ADA2 and GCN5 genes that encode components of the ADA and SAGA histone acetyltransferase complexes. In this report, we explore the biological roles of the Arabidopsis ADA2b and GCN5 genes. T-DNA insertion mutations in ADA2b and GCN5 were found to have pleiotropic effects on plant growth and development, including dwarf size, aberrant root development, and short petals and stamens in flowers. Approximately 5% of the 8200 genes assayed by DNA microarray analysis showed changes of expression in the mutants, three-fourths of which were upregulated and only half of which were altered similarly in the two mutant strains. In cold acclimation experiments, C-repeat binding factors (CBFs) were induced in the mutants as in wild-type plants, but subsequent transcription of cold-regulated (COR) genes was reduced in both mutants. Remarkably, nonacclimated ada2b-1 (but not gcn5-1) mutant plants were more freezing tolerant than nonacclimated wild-type plants, suggesting that ADA2b may directly or indirectly repress a freezing tolerance mechanism that does not require the expression of CBF or COR genes. We conclude that the Arabidopsis ADA2b and GCN5 proteins have both similar and distinct functions in plant growth, development, and gene expression and may be components of both a common coactivator complex and separate complexes with distinct biological activities.
INTRODUCTION
The regulation of gene expression at the level of transcription underlies many biological processes, such as growth and development, metabolic and physiological balances, and responses to the environment. In eukaryotes, chromatin plays a pivotal role in regulating gene expression (Narlikar et al., 2002). To activate gene expression, transcriptional activator proteins must overcome repressive chromatin structures. The changes in chromatin are accomplished by two broad classes of protein complexes. One class comprises the ATP-dependent chromatin-remodeling machines, which can alter nucleosome positions or conformations relative to the DNA (Peterson and Workman, 2000; Narlikar et al., 2002). The second class comprises complexes that covalently modify the histone proteins in the nucleosome by acetylation, phosphorylation, methylation, ADP-ribosylation, or ubiquitination (Strahl and Allis, 2000; Jenuwein and Allis, 2001; Berger, 2002).
The acetylation of specific Lys residues within the N-terminal tails of core histones, mediated by histone acetyltransferases (HATs), has been implicated in the regulation of transcription and other nuclear processes (Sterner and Berger, 2000; Roth et al., 2001). The yeast Gcn5 protein is the HAT component of at least two distinct transcriptional adaptor complexes, termed ADA and SAGA, that are capable of acetylating histones H3 and H2B in nucleosomes (Grant et al., 1997; Eberharter et al., 1999). The closely related mammalian GCN5 and PCAF proteins are components of similar complexes (Ogryzko et al., 1998). The yeast complexes contain the transcriptional adaptor proteins Ada2 and Ada3 as well as other complex-specific proteins (Grant et al., 1998b; Brown et al., 2000). Transcriptional activation by Gcn4 or Gal4-VP16 is reduced significantly in yeast mutants that do not produce Ada2, Ada3, or Gcn5 (Berger et al., 1992; Georgakopoulos and Thireos, 1992; Marcus et al., 1994). Gcn5 can be targeted to specific promoter regions by transcriptional activators (Kuo et al., 2000), and transcription of the corresponding genes in vivo is dependent on the HAT activity of Gcn5 (Kuo et al., 1998; Wang et al., 1998). Microarray analyses showed that the expression of ∼5% of yeast genes depends on Gcn5 during growth in rich media (Holstege et al., 1998; Lee et al., 2000), although additional Gcn5-dependent genes might be expressed only under specific growth or stress conditions. Several components of the SAGA complex, including Gcn5 and Ada2, are not essential for yeast viability (Roberts and Winston, 1997). However, yeast ada2 and gcn5 null mutants grow slowly in minimal medium and are cold and heat sensitive (Berger et al., 1992; Marcus et al., 1994). In mice, homozygous gcn5 null mutations result in an embryonic-lethal phenotype, whereas the knockout mutation of PCAF has no discernible phenotype (Xu et al., 2000; Yamauchi et al., 2000).
Our interest in the chromatin-remodeling factors in plants stems from our studies of the phenomenon of cold acclimation, the process whereby certain plants, including Arabidopsis, increase in freezing tolerance in response to low, nonfreezing temperatures (Thomashow, 1999). This response involves the action of the C-repeat binding factor (CBF) cold response pathway (Thomashow, 2001). Arabidopsis encodes a family of transcriptional activator proteins, known as CBF/DREB1, which contains AP2/EREBP DNA binding domains (Riechmann and Meyerowitz, 1998). These proteins recognize the CRT (C-repeat)/ DRE (dehydration-responsive) cold- and dehydration-responsive elements (Yamaguchi-Shinozaki and Shinozaki, 1994) present in the promoters of COR (cold-regulated) and dehydration-inducible genes (Thomashow, 1999). These CBF/DREB1 genes are induced rapidly (within 15 min) upon exposure of plants to low temperature followed at ∼2 h by induction of the CBF regulon of genes (i.e., CBF/DREB1-targeted genes). Genes in the CBF regulon contribute to freezing tolerance in multiple ways. At least one of the COR genes, COR15a, encodes a cryoprotective polypeptide that is proposed to help stabilize cellular membranes against freezing injury (Artus et al., 1996; Steponkus et al., 1998). Other COR genes encode proteins involved in amino acid and sugar metabolism that bring about increased levels of compatible solutes with cryoprotective activities, including sucrose, raffinose, and proline (Gilmour et al., 2000; Fowler and Thomashow, 2002; Taji et al., 2002). Constitutive overexpression of CBF1/DREB1b (Jaglo-Ottosen et al., 1998) or CBF3/DREB1a (Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000) in transgenic Arabidopsis plants induced the expression of COR and other cold-responsive genes (Fowler and Thomashow, 2002) without a low-temperature signal and thereby enhanced freezing tolerance. These results indicate that CBF proteins and their targets play important roles in cold acclimation.
Previously, we reported results suggesting that transcriptional activation by Arabidopsis CBF1 might be mediated, at least in part, by homologs of the yeast Ada2 and Gcn5 proteins (Stockinger et al., 2001). The transcriptional activity of CBF1 in yeast was impaired greatly in yeast strains carrying the ada2, ada3, or gcn5 mutations. The Arabidopsis genome encodes two ADA2 proteins (designated ADA2a and ADA2b) and one GCN5 protein, the latter exhibiting HAT activity in vitro. Both AtADA2 proteins can interact with the AtGCN5 protein in vitro or in two-hybrid assays. Moreover, the AtADA2 proteins and GCN5 were found to interact in vitro with CBF1, suggesting that CBF1 might stimulate transcription through the recruitment of the ADA/SAGA-like complexes to the promoters of its target genes (Stockinger et al., 2001).
In this report, we identify Arabidopsis mutants with T-DNA inserts in either the ADA2b or GCN5 gene and determine the effects of the mutations on plant growth, development, and gene expression. Our results indicate that ADA2b and GCN5 are important for normal plant growth and development and influence the low-temperature induction of the COR genes. Differences in the phenotypes of ada2b-1 and gcn5-1 mutant plants suggest that the wild-type proteins have distinct as well as common functions, a conclusion reinforced by gene expression profile analysis. Remarkably, nonacclimated ada2b-1 mutant plants were more freezing tolerant than nonacclimated wild-type plants, suggesting that ADA2b is involved in repressing a freezing tolerance pathway at warm temperature that is independent of the CBF cold response pathway.
RESULTS
Isolation and Molecular Characterization of the ada2b-1 Mutant
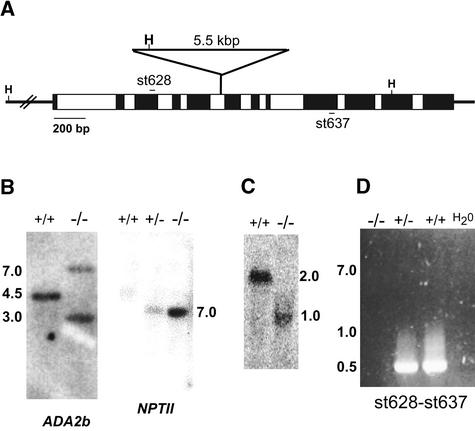
Plants bearing a disruption mutation of the Arabidopsis ADA2b gene were identified by screening a population of T-DNA insertional mutants from the University of Wisconsin Arabidopsis Knockout Facility (Krysan et al., 1996, 1999). The initial allele that we acquired, which we designated ada2b-1, contains a T-DNA insertion in the fifth intron, 1243 bp downstream of the translational initiation codon (Figure 1A). DNA gel blot analysis revealed a single T-DNA insertion in ada2b-1 genomic DNA (Figure 1B). The progeny of heterozygous plants showed a segregation of the ada2b-1 phenotype (described below) of ∼1:3 (χ2 = 1.27, P > 0.05), indicating that the ada2b-1 mutation is a recessive loss-of-function mutation that segregates as a single nuclear locus. RNA gel blot analysis using a probe corresponding to the 5′ half of the ADA2b gene revealed a truncated mRNA of 1 kb rather than the full-length ADA2b mRNA (Figure 1C), indicating that the transcript is terminated in the T-DNA in the ada2b-1 mutant. Reverse transcriptase–mediated (RT) PCR analysis using primers that span the T-DNA insertion confirmed the absence of full-length ADA2b mRNA (Figure 1D). However, transcripts corresponding to the 3′ half of the ADA2b gene in the ada2b-1 mutants were detected by RNA gel blot analysis and RT-PCR (data not shown); these transcripts presumably initiated inside the T-DNA. We conclude that no full-length ADA2b transcripts are expressed in the ada2b-1 mutant and that this allele probably represents a null mutation.
Figure 1.
Molecular Characterization of the ada2b-1 Mutant.
(A) Scheme of the T-DNA insertion in the Arabidopsis ADA2b gene, indicating the positions of exons (black boxes), introns (white boxes), HindIII sites (H), PCR primers (st628 and st637), and the T-DNA insertion.
(B) DNA gel blot analysis of genomic DNA from wild-type (+/+), heterozygous (+/−), and homozygous ada2b-1 (−/−) plants digested with HindIII and probed with radiolabeled fragments of the ADA2b gene or the neomycin phosphotransferase gene (NPTII) from the T-DNA.
(C) RNA gel blot analysis of total cellular RNA from wild-type (+/+) and homozygous ada2b-1 (−/−) plants probed with radiolabeled fragments of the 5′ half of the ADA2b cDNA.
(D) RT-PCR analysis of total cellular RNA from wild-type (+/+), heterozygous (+/−), and homozygous ada2b-1 (−/−) plants using primers that flank the T-DNA insertion site.
Phenotypic Characterization and Complementation of the ada2b-1 Mutant
Homozygous ada2b-1 plants showed pleiotropic effects at several stages of plant development. The ada2b-1 phenotype is characterized by small, dark green, twisted (curled) leaves, dwarf stature, and infertility (Figure 2). The cotyledons of ada2b-1 seedlings were smaller, dark green, and epinastic (i.e., curled down) (Figures 2A and 2B). The hypocotyls of the homozygous mutant seedlings, with an average length of 5 mm at 9 days, were longer than the heterozygote or wild-type hypocotyls, which averaged 3 mm (Figure 2B). Histological analysis indicated that cells in the hypocotyl of ada2b-1 plants were longer than those of wild-type plants (data not shown). In contrast to the extended hypocotyls, the rosette leaves of the homozygous mutant plants were much smaller than the heterozygous or wild-type leaves (Figures 2A and 2D). Laser scanning confocal microscopy revealed that the palisade mesophyll leaf cells from ada2b-1 plants were smaller (approximately one-third by cross-sectional area) than corresponding cells of wild-type plants (Figures 2G and 2H), suggesting that the ada2b-1 dwarfism arises from decreased cell size rather than from decreased cell division. The root system in ada2b-1 plants was shorter than that in wild-type plants, with an increased number of secondary roots (Figure 2C) and with cells in the elongation zone being markedly smaller (data not shown). The inflorescence stems of mutant plants were much smaller than those of wild-type plants (Figures 2E and 2F). The floral organs, particularly petals and stamens, also were affected in ada2b-1 plants. The flowers opened prematurely, with small petals that curled outward and stamens that were markedly shorter than the carpels (Figures 2I and 2J). Cells in the stamen filament of ada2b-1 were shorter than wild-type filament cells (Figures 2K to 2N), although pollen morphology and production appeared normal (data not shown). Manual fertilization resulted in small siliques with few seeds. Thus, the infertility of homozygous mutant plants may be a consequence of the failure of the pollen to reach the stigma as a result of the short stamens.
Figure 2.
Phenotypes of Arabidopsis ada2b-1 Mutant Plants.
(A) Heterozygous (left) and homozygous (right) ada2b-1 plants at 14 days of growth on agar plates under continuous light.
(B) Hypocotyl elongation of heterozygous (left) and homozygous (right) ada2b-1 seedlings grown on plates for 9 days. Representative seedlings were placed vertically before photographing.
(C) Roots of wild-type (wt) and ada2b-1 mutant seedlings grown on a vertical plate for 14 days. Bar = 1 cm.
(D) Heterozygous and homozygous ada2b-1 plants at 21 days of growth in soil. Bar = 1 cm.
(E) Adult plants at 60 days of growth in soil, including mutant, wild-type, and complemented (ada2b-1;35S-ADA2b) plants. Bar = 1 cm.
(F) Higher magnification of the ada2b-1 homozygous plant shown in (E). Bar = 1 cm.
(G) and (H) Scanning laser confocal micrographs of palisade mesophyll cells from leaves of wild-type (G) and ada2b-1 mutant (H) plants. Bars = 25 μm.
(I) and (J) Scanning electron micrographs of mature wild-type (I) and ada2b-1 (J) flowers. Bars = 500 μm.
(K) and (L) Scanning electron micrographs of stamens from wild-type (K) and ada2b-1 (L) flowers. Bars = 100 μm.
(M) and (N) Scanning electron micrographs of stamen filaments from mature wild-type (M) and ada2b-1 (N) flowers. Bars = 50 μm.
To determine whether the mutant phenotypes were caused by the T-DNA insertion in the ADA2b gene, genetic complementation was performed by transforming ada2b-1 heterozygotes with a DNA fragment expressing both the wild-type ADA2b cDNA and an herbicide resistance marker gene from constitutive 35S promoters of Cauliflower mosaic virus. Transformed plants were screened by PCR to identify plants homozygous for the ada2b-1 allele and bearing the transgene. Introduction of the ADA2b cDNA in ada2b-1 plants rescued all of the ada2b-1 phenotypes described above, confirming that the T-DNA insertion was responsible for the observed phenotypes (Figure 2E). By contrast, constitutive expression of the ADA2a cDNA in homozygous ada2b-1 plants did not rescue any of the mutant phenotypes (data not shown).
Isolation and Molecular Characterization of the gcn5-1 Mutant
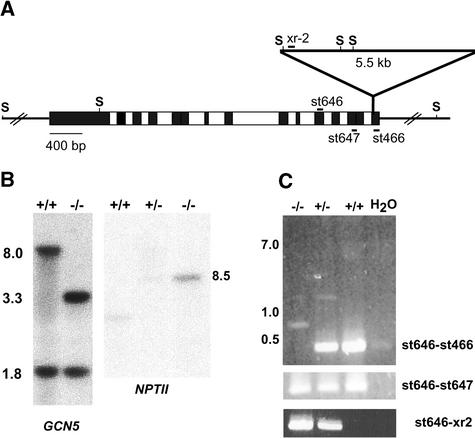
A T-DNA disruption mutant of the Arabidopsis GCN5 gene also was identified from the University of Wisconsin Arabidopsis Knockout Facility collection. This allele, designated gcn5-1, contains a T-DNA insertion in the last exon, 3730 bp from the translational start codon (Figure 3A). The T-DNA disrupts the last 17 amino acids of GCN5, including the C terminus of the bromodomain. DNA gel blot analysis revealed a single T-DNA insertion in gcn5-1 genomic DNA (Figure 3B). RT-PCR analysis using primers that span the insertion site did not detect full-length GCN5 mRNA in homozygous mutant plants (Figure 3C). However, transcripts corresponding to the region upstream of the insertion site were detected in the gcn5-1 mutants by both RT-PCR and RNA gel blot analysis. This transcript is terminated inside the T-DNA (Figure 3C) and represents a nearly full-length mRNA. Therefore, the gcn5-1 allele is likely to be hypomorphic rather than null.
Figure 3.
Molecular Characterization of the gcn5-1 Mutant.
(A) Scheme of the T-DNA insertion in the Arabidopsis GCN5 gene, indicating the relative positions of exons (black boxes), introns (white boxes), SacI sites (S), PCR primers (xr-2, st646, st647, and st466), and the T-DNA insertion.
(B) DNA gel blot analysis of genomic DNA from heterozygous (+/−) and homozygous (−/−) gcn5-1 plants digested with SacI and probed with radiolabeled fragments of the GCN5 gene or the neomycin phosphotransferase gene (NPTII) from the T-DNA. The plants used here as wild-type controls (+/+) have a single T-DNA insertion at an undefined locus.
(C) Expression analysis of the GCN5 gene in wild-type, heterozygous, and homozygous gcn5-1 plants using RT-PCR with several primer pairs. No template was added to control reactions shown in the lanes at right (H2O).
Phenotypic Characterization and Complementation of the gcn5-1 Mutant
Homozygous gcn5-1 plants were identified and confirmed by PCR. The phenotype of gcn5-1 homozygotes resembled the ada2b-1 phenotype in some respects, most notably in root growth, the dwarf size of rosette and adult plants, and tissue-specific deformities in flowers. By contrast, other phenotypic features were distinctly different, including hypocotyl elongation, leaf morphology, and the number of floral shoots. Specific features of the gcn5-1 phenotype, compared with those of wild-type and ada2b-1 plants, are detailed below.
In seedlings and until the formation of the second pair of leaves, no morphological difference was apparent between gcn5-1 mutants and wild-type plants. In plate-grown gcn5-1 plants, the first pair of leaves had slightly longer petioles (data not shown). The second pair of leaves in the gcn5-1 plants initially was folded upward, suggesting that the abaxial cells had divided or expanded faster than the adaxial cells. When the leaves unfolded, they developed a serrated morphology (Figures 4A and 4B). In contrast to ada2b-1 plants, gcn5-1 leaves were not dark green but instead were chlorotic (especially under increased light intensity). The size of the gcn5-1 palisade mesophyll cells was smaller than those in the wild type but larger than those in the ada2b-1 mutants (data not shown). The root development of gcn5-1 seedlings was similar to that of ada2b-1 plants (Figure 4C). The timing of the initiation of reproductive development was unaffected in the gcn5-1 mutants, but the growth of the inflorescence was strongly affected, resulting in dwarf plants (Figures 4D and 4E). Moreover, the gcn5-1 mutants displayed a loss of apical dominance. The number of secondary inflorescences increased from a typical wild-type number of two or three to an average in gcn5-1 plants of approximately six (Figures 4D and 4E). The flower morphology of the gcn5-1 mutants also was similar to that of the ada2b-1 mutants, with short stamens and petals (Figures 4F to 4H) and reduced fertility. As in ada2b-1 plants, cells in the stamen filament of gcn5-1 were shorter than corresponding cells in wild-type plants (Figures 4I to 4L). The gcn5-1 flowers showed a phenotypic gradient: early-arising flowers had shorter petals and stamens than late flowers (Figures 4G and 4H). The latest flowers produced small siliques that contained only a few seeds.
Figure 4.
Phenotypes of the gcn5-1 Mutant.
(A) and (B) Wild-type (wt) Arabidopsis Wassilewskija-2 (A) and gcn5-1 mutant (B) plants at 14 days of growth in soil under continuous light. Bars = 1 cm.
(C) Roots of wild-type, ada2b-1, and gcn5-1 homozygous mutant plants grown on a vertical plate for 14 days. Bar = 1 cm.
(D) Complementation of the gcn5-1 mutation. Wild-type and gcn5-1 homozygous plants, and gcn5-1;35S-MYCGCN5 at 60 days of growth in soil. Bar = 1 cm.
(E) Higher magnification of the gcn5-1 homozygous plant shown in (D). Bar = 1 cm.
(F) to (H) Scanning electron micrographs of mature wild-type (F), early-formed mature gcn5-1 (G), and late-formed mature gcn5-1 (H) flowers. Bars = 500 μm.
(I) and (K) Scanning electron micrographs of stamens from wild-type (I) and gcn5-1 (K) plants. Bars = 100 μm.
(J) and (L) Scanning electron micrographs of stamen filaments of mature wild-type (J) and early-formed mature gcn5-1 (L) flowers. Bars = 50 μm.
To determine whether the mutant phenotypes were caused solely by the T-DNA insertion in the GCN5 gene, genetic complementation was performed by transforming gcn5-1 heterozygous plants with the intact GCN5 cDNA linked to six repeats of the Myc epitope tag driven by a constitutive 35S promoter of Cauliflower mosaic virus. The expression of the Myc-tagged GCN5 transgene in plants homozygous for the gcn5-1 disruption allele rescued all gcn5-1 phenotypes, indicating that no other mutations were responsible for the phenotypes (Figure 4D).
Low-Temperature Induction of COR Genes Is Delayed in the ada2b-1 and gcn5-1 Mutants
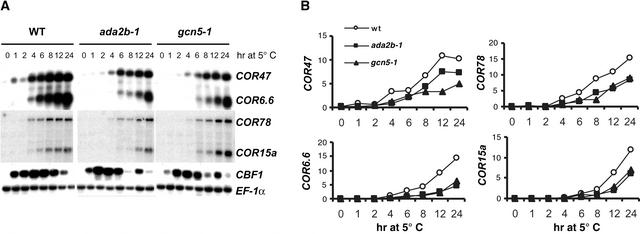
Our previous studies demonstrated that transcriptional activation by the Arabidopsis CBF1 protein, when expressed in yeast, requires the activity of three components of the yeast ADA and SAGA complexes (Stockinger et al., 2001). These results suggested that CBF activators might function through the action of similar complexes in Arabidopsis. To test that hypothesis, we compared the induction of COR gene expression upon low-temperature treatment of ada2b-1, gcn5-1, and wild-type plants. Seedlings were grown on agar plates and transferred to 5°C for various periods of time before total cellular RNA was harvested for examination by RNA gel blot analysis. As expected, in wild-type plants, COR78, COR47, COR15a, and COR6.6 transcript levels increased within 4 h and continued to increase until 24 h (Figure 5). In ada2-1 and gcn5-1 plants, COR gene transcripts also accumulated in response to low temperature, but the rate of induction was slower and the final levels of transcripts attained were less than in wild-type plants. COR6.6 transcript levels were reduced approximately threefold, whereas transcript levels for COR15a, COR47, and COR78 were reduced approximately twofold. These results indicate that ADA2b and GCN5 are not required for COR gene induction during cold acclimation but appear to play a contributory role that enhances the levels of COR expression.
Figure 5.
Transcript Levels of COR and CBF Genes in ada2b-1 and gcn5-1 Mutants.
(A) RNA gel blot analysis of total RNA prepared from whole seedlings of control Arabidopsis Wassilewskija-2 wild-type (WT) and ada2b-1 and gcn5-1 mutant plants. Plants were grown for 14 days at 20°C and then cold treated at 5°C for the times indicated. The RNA gel blots were hybridized with a probe corresponding to CBF1 and then stripped and hybridized with probes corresponding to COR47, COR6.6, COR78, or COR15a. Hybridization with a probe representing EF-1α, a constitutively expressed gene, was used to normalize signals from the COR and CBF genes.
(B) Quantitative representation of the relative expression of COR genes (COR/EF-1α expression) during a low-temperature time course. wt, wild type.
The observed reduction in COR gene expression in the ada2b-1 and gcn5-1 mutants might have been attributable to either defects in transcriptional activation by the CBF proteins or reduced expression of the CBF genes themselves. To distinguish between these possibilities, we assessed the expression of three CBF genes (CBF1, CBF2, and CBF3) collectively by RNA gel blot analysis using as a probe a cDNA for the entire CBF1 sequence, which hybridizes with all three CBF transcripts (Gilmour et al., 1998). The results shown in Figure 5A indicate that CBF transcripts were induced by 1 h of cold treatment in both wild-type and mutant plants. In wild-type plants, CBF transcript levels remained high until gradually declining at 12 and 24 h. By contrast, CBF expression in ada2b-1 and gcn5-1 mutant plants showed a more rapid reduction, reaching a nadir at 8 h, before a second wave of expression evident in 12-h samples. This biphasic pattern of CBF expression in the mutant plants was reproduced in three separate experiments. We infer that the signal transduction pathway leading to initial CBF transcription is unimpaired in the ada2b-1 and gcn5-1 mutants but that some aspect of sustained CBF expression is altered.
Nonacclimated ada2b-1 Plants Display Freezing Tolerance
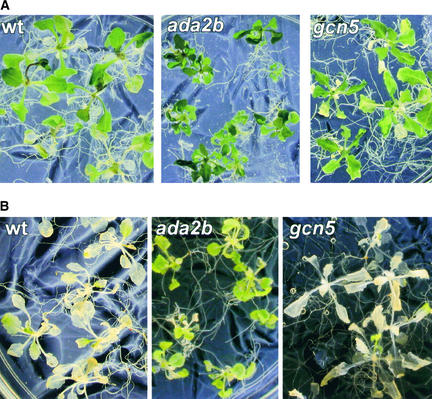
Because both the ada2b-1 and gcn5-1 mutations modestly affect the low-temperature induction of COR gene expression, we investigated the freezing tolerance of acclimated and nonacclimated plants. After 1 week of acclimation to low but nonfreezing temperatures, ada2b-1 and gcn5-1 mutants survived a freezing challenge (−5°C for 2 days), as did wild-type plants (Figure 6A). This result indicates that the effect of the ada2b-1 and gcn5-1 mutations on COR gene expression was not sufficient to appreciably impair the ability of the mutants to cold acclimate.
Figure 6.
Effect of ada2b-1 and gcn5-1 Mutations on Plant Freezing Tolerance.
Seedlings of control Arabidopsis Wassilewskija-2 wild-type (wt) and ada2b-1 and gcn5-1 mutant plants were grown at 20°C on plates. To test freezing tolerance, plants were transferred to −2°C for 24 h followed by 24 h at −5°C and then warmed to 4°C for 24 h followed by 24 h at 20°C.
(A) Plants were cold acclimated at 5°C for 7 days before freezing challenge.
(B) Nonacclimated plants were transferred directly from 20°C to freezing challenge.
As expected, nonacclimated wild-type and gcn5-1 plants were killed by freezing temperatures. Remarkably, however, the nonacclimated ada2b-1 plants survived the freezing challenge without significant damage (Figure 6B). This experiment was repeated six times with similar results. Altogether, 86% of the nonacclimated ada2b-1 plants (n = 70) survived the freezing temperatures, in contrast to only 18% of the nonacclimated wild-type plants (n = 70). Given that neither the CBF nor the COR gene is expressed constitutively in the ada2b-1 plants (Figure 5), these results indicate that the freezing tolerance of the ada2b-1 mutants is achieved by a novel, undefined pathway.
Gene Expression Profiles in the ada2b-1 and gcn5-1 Mutants
To identify genes whose expression is affected by the ada2b-1 or gcn5-1 mutation, we used high-density oligonucleotide arrays (Affymetrix) representing ∼8200 different Arabidopsis genes (Zhu and Wang, 2000), or approximately one-third of the genome (Arabidopsis Genome Initiative, 2000). Fluorescent cDNA probes were synthesized from total RNA harvested from leaves of wild-type and mutant plants in two separate biological experiments, permitting four pair-wise comparisons of mutant and wild-type leaves for both ada2-1 and gcn5-1. Genes were considered to be induced or repressed if differences in hybridization signals were threefold or more in at least three of the four comparisons. Only genes with discernible signals (i.e., those designated “present”) in either wild-type or mutant plants, with an average expression level of ≥100, were considered in the comparisons; on average, ∼5400 genes met this criterion in each pair-wise comparison. Based on these criteria, 401 transcripts (or 5% of the genes represented on the chip) were identified as being affected by ada2b-1 and gcn5-1. The affected genes are listed in the supplemental data online. Complete data files for all single chips and all pair-wise combinations are available at www.arabidopsis.org/info/expression/ada_gcn.html.
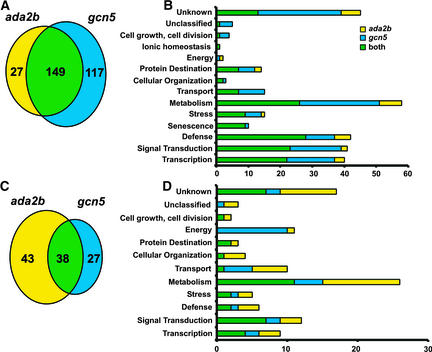
Of the affected genes, 293 (or 73%) were induced and 108 (or 27%) were repressed at least threefold in the mutant plants (Figures 7A and 7C). Of the induced genes, 149 (51%) were increased in both mutants, whereas 117 (40%) were induced only in gcn5-1 and 27 (9%) were upregulated solely in ada2b-1. Of the repressed genes, 38 (35%) were repressed in both mutants, whereas 43 (40%) were decreased only in ada2b-1 and 27 (25%) were downregulated in gcn5-1.
Figure 7.
Gene Expression Profiles of ada2b-1 and gcn5-1 Mutations.
(A) and (B) Venn diagram (A) and functional categories (B) of genes with increased expression in ada2b-1 and gcn5-1 mutants.
(C) and (D) Venn diagram (C) and functional categories (D) of transcripts with decreased expression in ada2b-1 and gcn5-1 mutants.
Approximately 85% of the genes exhibiting significantly higher or lower expression levels in ada2b-1 or gcn5-1 could be assigned to groups according to known or putative functions (Figures 7B and 7D; see also supplemental data online), whereas the remaining genes were characterized as encoding unknown or unclassified proteins. Genes whose expression was increased in ada2b-1 or gcn5-1 mutant plants (Figure 7B; see also supplemental data online) represent a wide range of biological functions, including cellular metabolism (20%), signal transduction (14%), transcription regulation (14%), defense against pathogens (14%), various abiotic stresses (5%), and senescence (4%). The remaining 15% include genes putatively involved in transport, protein destination, cellular organization, energy, or cell growth and development. Genes exhibiting significantly lower expression levels in ada2b-1 and gcn5-1 than in wild-type plants also represent a range of biological functions (Figure 7D; see also supplemental data online). Interestingly, transcripts of genes involved in energy and particularly in photosynthesis accumulated to lower levels in gcn5-1 than in ada2b-1 or wild-type plants.
DISCUSSION
In yeast, the results of both biochemical and genetic analyses indicate that ADA2 and GCN5 have a common set of functions: the yeast Ada2 and Gcn5 proteins are components of the same multiple-protein complexes, ADA and SAGA, and yeast strains carrying ada2 and gcn5 null mutations have very similar phenotypes (Berger et al., 1992; Marcus et al., 1994; Grant et al., 1997, 1998a). Here, we describe the effects of mutations in ADA2b and GCN5 on gene expression and on growth and development in Arabidopsis. Our results indicate that the Arabidopsis ADA2b and GCN5 genes have both common and distinct functions, suggesting that the plant ADA2b and GCN5 proteins are likely to be components of a common coactivator complex, as in yeast, as well as being components of separate complexes with distinct biological activities.
Arabidopsis Ada2b and Gcn5 Proteins Have Common and Distinct Functions
The similar phenotypes observed in ada2b-1 and gcn5-1 mutant plants suggest that the corresponding wild-type proteins work in a common pathway to regulate a wide range of growth and developmental processes ranging from cell elongation and expansion in leaves and roots to stamen and petal development in flowers. Interestingly, disruption of ADA2b or GCN5 did not result in homeotic transformations, indicating that they are not involved in regulating genes that govern cell, tissue, or organ identity.
In other respects, however, the phenotypes of the ada2b-1 and gcn5-1 mutants are quite distinct, suggesting that the corresponding proteins also have separate biological functions. For instance, some features of the ada2b-1 phenotype are similar to those of auxin-overproducing mutants, including elongated hypocotyls, small and epinastic cotyledons, reduced leaf expansion rates, increased number of lateral roots, and dwarf plants (Boerjan et al., 1995; Ljung et al., 2001; Zhao et al., 2001). By contrast, some features of the gcn5-1 phenotype resemble those observed in mutants with reduced auxin responses, including short hypocotyls, upcurled leaves, and reduced apical dominance (Liscum and Reed, 2002; Swarup et al., 2002).
Gene expression patterns in the ada2b-1 and gcn5-1 mutants also support the conclusion that ADA2b and GCN5 might function in both the same and distinct coactivator complexes. We analyzed the presence of some 8200 transcripts in leaf tissue from wild-type and mutant plants and observed changes in relative expression levels for some 400 genes in the ada2b-1 and gcn5-1 mutants using rather stringent criteria. Of these genes, nearly half (187 of 401) were affected similarly in both mutants. Thus, these genes might depend on coactivator complexes that contain both ADA2b and GCN5. However, a significant number of genes were affected more significantly in one mutant than in the other (Figure 7). For instance, 27 transcripts that were more abundant in ada2b-1 than in wild-type plants did not show a similar increase in gcn5-1 plants. Conversely, 117 transcripts that were more abundant in gcn5-1 than in wild-type plants did not show a similar increase in ada2b-1 plants. Although in a few cases these distinctions may be rather subtle (if transcript levels barely miss the cutoff criterion), many genes are affected markedly by the ada2b-1 mutation but not by the gcn5-1 mutation (or vice versa), indicating that the ADA2b and GCN5 proteins have distinct biological activities.
As with any such microarray analysis, our results cannot distinguish between direct and indirect contributions of ADA2b and GCN5 to the expression levels of specific genes. Moreover, although some of the changes in gene expression in ada2b-1 and gcn5-1 mutant plants are likely to be responsible for the dramatic phenotypes observed, the converse also may be true—that is, some gene expression changes may be the consequence rather than the cause of the pronounced phenotypes. More direct biochemical assays, such as chromatin immunoprecipitation or promoter-based microarray analysis, will be necessary to distinguish between these possibilities. The analysis reported here was limited to a single time point under a single set of growth conditions. Other differences in gene expression are likely to be revealed by examining these mutants at earlier or later times in development, by analyzing additional tissues, and by testing the effects of various growth and stress conditions.
The Arabidopsis genome encodes two homologs of the yeast ADA2 gene, both of which are expressed at the mRNA level in plant roots, leaves, and flowers (Stockinger et al., 2001). The pronounced phenotype of the ada2b-1 mutant suggests that the plant ADA2b and ADA2a proteins are not functionally redundant. It is conceivable that the two genes may be expressed in different cells or may be expressed differentially in response to developmental or environmental cues. However, ectopic overexpression of ADA2a did not complement the ada2b-1 mutation (our unpublished results), suggesting that the two ADA2 proteins may be components of distinct complexes that mediate different regulatory signals. At present, the only T-DNA disruption mutation in AtADA2a that we have characterized has no apparent phenotype (our unpublished results). Additional experimentation will be required to define the distinct biological and biochemical roles for the two Arabidopsis ADA2 homologs.
Disruption or knockout mutations of GCN5 have been described previously only in budding yeast (Berger et al., 1992; Georgakopoulos and Thireos, 1992; Marcus et al., 1994; Stockinger et al., 2001) and mice (Xu et al., 2000; Yamauchi et al., 2000). Mammalian genomes encode two GCN5 homologs (known as GCN5 and PCAF), disruptions of which create strikingly different phenotypes. The mouse gcn5 knockout mutants are embryonic lethal, with defects in the formation of dorsal mesoderm cell lineages at 8.5 days after conception. By contrast, the mouse pcaf disruption results in no apparent phenotype. The disruption of the single Arabidopsis GCN5 gene described here presents a markedly different effect: the mutation is not lethal, but it does cause dramatic phenotypes at a wide range of developmental stages and sites. These differences in the types of effects of GCN5 mutations may reflect differences between plants and animals in the biological pathways in which these homologous proteins function as transcriptional coactivators.
Ada2b and Gcn5 Contribute to Cold-Regulated Gene Expression
Our hypothesis that Arabidopsis coactivator proteins are involved in the mechanism by which the CBF proteins activate the expression of the COR genes was prompted by two results. First, when CBF1 was expressed in yeast, its transcriptional activity required intact yeast ADA2, ADA3, and GCN5 genes (Stockinger et al., 2001). Second, we observed direct in vitro interactions between CBF1 and the ADA2 proteins of Arabidopsis (Stockinger et al., 2001), consistent with previous reports of yeast Ada2b interactions with transcriptional activators (Barlev et al., 1995). A recent report proposed that another subunit of the yeast SAGA complex, Tra1, is more likely than Ada2 to be the direct contact for acidic activation domains (Brown et al., 2001). The Arabidopsis genome encodes two apparent homologs of Tra1, but the association of these proteins with plant coactivator complexes remains to be established.
The induction of COR gene expression during low-temperature treatment was modestly diminished or delayed in ada2b-1 and gcn5-1 plants compared with wild-type plants. The reduced COR gene expression does not result from an inability to induce CBF gene expression (Figure 5), although the curious decrease in apparent CBF expression at 8 h of cold treatment might reflect a transient difficulty in maintaining CBF expression. If COR gene expression depends exclusively on the CBF regulatory proteins, then our observations suggest that transcriptional activation by CBF proteins is only partially dependent on the ADA2b and GCN5 coactivators. This partial dependence might reflect the existence of other mechanisms for transcriptional activation, perhaps by direct recruitment of basal transcription factors or the RNA polymerase II holoenzyme. A yeast gcn5 mutant strain also showed a delay in induced gene expression, associated with slow remodeling of chromatin at the PHO5 promoter (Barbaric et al., 2001). Together, these results are consistent with a model in which gene induction can be affected kinetically by HATs, which accelerate the remodeling of chromatin and thus transcriptional activation but do not determine the eventual mRNA expression level. An alternative explanation is that activation by the CBF proteins per se might be fully ablated in the ada2b and gcn5 mutants, but the residual COR gene expression might be supported by other transcription factors that bind COR gene promoters (Kim et al., 2001) that use other activation mechanisms.
In another Arabidopsis mutant, sfr6, the expression of CBF genes (but not COR genes) was induced by low temperature (Knight et al., 1999), suggesting that the SFR6 protein might mediate transcriptional activation by CBF. However, the physiological consequences of the sfr6 mutation are quite different from those of ada2b-1 or gcn5-1 in that cold-acclimated sfr6 plants were freezing sensitive but cold-acclimated ada2b-1 and gcn5-1 plants were freezing tolerant. Thus, the SFR6 protein likely plays some role distinct from that of ADA2b or GCN5 in transcriptional activation.
Enhanced Freezing Tolerance in ada2b-1 Plants
Quite surprisingly, ada2b-1 plants were more freezing tolerant than wild-type plants without previous acclimation and without overexpression of CBF or COR genes. This result suggests the possibility that a novel pathway can induce freezing tolerance without cold acclimation and without previous expression of COR genes.
Freezing tolerance in warm-grown Arabidopsis plants has been observed previously in at least two circumstances. First, constitutive expression of CBF proteins themselves can result in COR gene expression and freezing tolerance in warm-grown plants (Jaglo-Ottosen et al., 1998; Liu et al., 1998). Soluble sugars, including sucrose, galactinol, and raffinose, accumulate during low-temperature stress (Gilmour et al., 2000), and the biosynthetic enzymes galactinol synthase and raffinose synthase are cold inducible and expressed in CBF/DREB1 overexpressor lines (Fowler and Thomashow, 2002; Taji et al., 2002). By contrast, it seems likely that the freezing tolerance of ada2b-1 plants arises from some other mechanism, because these sugar biosynthetic genes were not upregulated in our microarray analysis. Moreover, CBF and COR gene expression was not increased constitutively in the ada2b-1 mutant (Figure 5). Second, the esk1 mutant of Arabidopsis displays similar characteristics (Xin and Browse, 1998). Nonacclimated esk1 plants overexpress Δ1-pyrolline-5-carboxylase synthetase (P5CS), the first enzyme in Pro synthesis, and have 30-fold higher levels of Pro as well as 2-fold higher levels of total soluble sugars than do wild-type plants. Again, we surmise that the freezing tolerance of ada2b-1 likely arises from a different mechanism, because our microarray data indicate that expression of the P5CS gene is not increased in ada2b-1 plants. We conclude that the loss of ADA2b may result in the activation of a freezing tolerance mechanism that differs markedly from the precedents established for the CBF and ESK1 signal transduction pathways.
What, then, makes ada2b-1 plants freezing tolerant? Some of the transcription factors upregulated in nonacclimated ada2b-1 plants, including ERF5 and ERF6, WRKY33 and WRKY53, ZAT10 and ZAT12, and other putative zinc-finger proteins, also are upregulated in cold-acclimated wild-type plants (Fowler and Thomashow, 2002). Overexpression of these regulatory proteins may activate downstream genes, resulting in freezing tolerance. Surprisingly, gcn5-1 plants are not freezing tolerant, despite the considerable overlap among genes whose expression levels are affected in ada2b-1 and gcn5-1. The difference may be caused by the expression of specific transcription factors (and their target genes) in ada2b-1 but not in gcn5-1. Alternatively, gcn5-1 plants may have the potential to withstand freezing stress, but defects in the expression of the photosynthetic machinery (evident in the DNA microarray analysis) might render these plants unable to tolerate frost damage, because photosynthesis is required for enhanced freezing tolerance (Wanner and Junttila, 1999).
Positive and Negative Regulation by Arabidopsis ADA2b and GCN5
In yeast cells grown in rich medium, the expression of ∼5% of the genome is dependent on Gcn5 activity (Holstege et al., 1998; Lee et al., 2000). Similarly, the Arabidopsis gcn5-1 and ada2b-1 mutants have altered expression of ∼5% of the genes represented in the 8200-element Affymetrix microarray. However, the gene expression profiles of Arabidopsis mutants are different from the yeast gcn5 expression profile. Whereas in gcn5 yeast cells, most of the affected genes show a decrease relative to wild-type cells, as expected for a coactivator protein, the opposite is true in the Arabidopsis mutants, in which most of the affected genes show increased transcript levels.
These findings suggest that ADA2b and GCN5 can function as both positive and negative regulators of transcription. This hypothesis is consistent with a recent report that the yeast SAGA complex plays dual roles, acting to repress and activate specific target genes in different growth conditions (Ricci et al., 2002). Our present experiments, however, cannot distinguish whether those positive and negative effects represent direct activity of ADA2 and GCN5 at the corresponding promoters or if the effects are secondary or indirect. Direct assays of the presence of ADA2b and GCN5 at these putative target genes, such as with chromatin immunoprecipitation experiments, might address this important issue.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Wassilewskija-2 was grown in controlled-environment chambers at 22°C under constant illumination with cool-white fluorescent lights (100 to 150 μmol·m−2·s−1) in Bacto planting mix (Michigan Peat, Houston, TX). Plants used for low-temperature experiments were grown on Gamborg's B5 medium (Gibco BRL) for 14 days at 22°C and then placed at 5°C under continuous light (20 to 60 μmol·m−2·s−1).
Identification of T-DNA Mutants
The ada2b-1 and gcn5-1 plants were isolated from a T-DNA–mutagenized population of Arabidopsis using a PCR-based reverse genetic screen (Krysan et al., 1996, 1999). The primers used to detect T-DNA insertions into the ADA2b and GCN5 genes are listed in Table 1. The PCR products were separated by agarose gel electrophoresis, blotted onto a nylon membrane, and probed with the gene of interest to determine if the amplified fragments contained gene-specific sequences. DNA sequencing of PCR-amplified fragments that spanned the insertion site determined the position of the T-DNA insertion in each gene. The genotypes of isolated mutants were confirmed by PCR with two gene-specific primers or with one gene-specific primer and the T-DNA border–specific primer. In the case of homozygous plants, only one band representing T-DNA insertion was detectable, whereas in the case of heterozygous plants, two bands were detected: the T-DNA insertion and the band amplified with gene-specific primers. The T-DNA insertions also were confirmed by DNA gel blot analysis. Two micrograms of genomic DNA isolated from wild-type and mutant plants were digested with HindIII and SacI, respectively, separated by electrophoresis on 1% agarose gels, and transferred to a nylon membrane. The blots were hybridized with radiolabeled cDNA probes for ADA2b or GCN5 or with a T-DNA–specific probe. Hybridization was performed with PerfectHyb Plus (Sigma) at 68°C, and the blots were washed in 0.5× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.1% SDS, and 0.01% sodium pyrophosphate at 65°C for 30 min and then with 0.2× SSC, 0.1% SDS, and 0.01% sodium pyrophosphate at 65°C for 30 min.
Table 1.
Oligonucleotides Used as PCR Primers for the Identification and Characterization of T-DNA Disruption Mutants of ADA2b and GCN5 Genes
| Primer Name | Sequence | Location | Stranda |
|---|---|---|---|
| JL202 | 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ | T-DNA left border | NA |
| XR-2 | 5′-TGGGAAAACCTGGCGTTACCCAACTTAAT-3′ | T-DNA right border | NA |
| ST461 | 5′-AGGGTTCTTCTTCTCTGTGGTTTCGCATA-3′ | 5′ end of ADA2b | NT |
| ST468 | 5′-GGTTTAGATTGTGTGGCTCTGATTCACCT-3′ | 3′ end of ADA2b | T |
| ST628 | 5′-ACTCCTCACAAATGTGATCACCCATACCG-3′ | Exon 3 of ADA2b | NT |
| ST637 | 5′-CTCCATCTCCGCCAAGAGTTGCTCAG-3′ | Exon 9 of ADA2b | T |
| ST496 | 5′-GAATATTCCGTTCGTCGGCGTAGTCTTTC-3′ | 5′ end of GCN5 | NT |
| ST497 | 5′-AAATGACCCATGACGCTCATTCTCTTCAC-3′ | 3′ end of GCN5 | T |
| ST646 | 5′-AGTGGGGGCACACTCGTTTCAAATTATTC-3′ | Exon 10 of GCN5 | NT |
| ST647 | 5′-TCCGCAACAAACATATCCAATGTCACGTA-3′ | Exon 12 of GCN5 | T |
| ST466 | 5′-TTGAGATTTAGCACCAGATTGGAGACCTG-3′ | Exon 13 of GCN5 | T |
Primers used to prime DNA synthesis on the template (T) or nontemplate (NT) strand. NA, not applicable.
Complementation of the T-DNA Mutants
The full-length ADA2b cDNA was subcloned into a binary vector, pKVG1 (35S promoter, bar1 selection), which was derived from pCAMBIA 3301 (CAMBIA, Canberra, Australia), using the restriction sites BamHI and SacI to create pKVA31. The 6Myc epitope-tagged full-length GCN5 cDNA (pKVB32) was subcloned into a binary vector, pKVB24 (35S promoter, bar1 selection), using the restriction sites ClaI and SacI to create pKVB34. The binary vectors pKVA31 and pKVB34 were introduced into Agrobacterium tumefaciens strain GV3101 and used to transform mutant plants heterozygous at the ada2b-1 locus and the gcn5-1 locus, respectively, by the floral-dip method (Clough and Bent, 1998). T1 plants were selected in saturated soil with 200 μg/L BASTA. PCR methods were used to distinguish mutant plants homozygous at the ada2b-1 or gcn5-1 locus from wild-type plants in T1-transformed populations.
RNA Analysis
Total RNA from Arabidopsis tissues was isolated with the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Reverse transcriptase–mediated PCR was performed using 2 μg of DNase-treated RNA from the appropriate plants and gene-specific primers for ADA2b or GCN5 with the Access RT-PCR System (Promega, Madison, WI). RNA gel blot analysis was performed with 5 or 10 μg (per lane) of total RNA. After electrophoresis and blotting to a Hybond N+ membrane (Amersham Pharmacia) using standard procedures (Sambrook et al., 1989), RNAs were hybridized with radiolabeled cDNA probes that encode COR6.6, COR15a, COR47, COR78, or CBF1. Subsequently, the membranes were stripped and rehybridized with probes for EF-1α. Hybridization was performed in PerfectHyb Plus (Sigma) at 65°C overnight, and the blots were washed in 0.5× SSC, 0.1% SDS, and 0.01% sodium pyrophosphate at 65°C for 30 min and then with 0.2× SSC, 0.1% SDS, and 0.01% sodium pyrophosphate at 65°C for 30 min.
Confocal Microscopy and Scanning Electron Microscopy
Plants were grown for 14 days in soil. Leaves were dissected with a razor blade, and optical sections of the fluorescing chloroplasts of mesophyll palisade cells were collected with a Zeiss 210 laser scanning confocal microscope (Thornwood, NY) using light of wavelength 488 nm from a dual-line argon ion laser.
Flowers from the mutant and wild-type plants were fixed at 4°C for 2 h in 4% glutaraldehyde buffered with 0.1 M sodium phosphate, pH 7.4. After a brief rinse in that buffer, samples were dehydrated in an ethanol series (25, 50, and 75%) for 10 to 15 min and with three 10-min changes in 100% ethanol. Samples were dried in a Balzers critical point dryer (Balzers, Liechtenstein) using liquid carbon dioxide as the transitional fluid and then mounted on aluminum stubs using adhesive tabs. Samples were coated with gold (20 nm thickness) for 9 min in an Emscope Sputter Coater (model SC 500; Ashford, Kent, UK) purged with argon before being examined with a JEOL-35S scanning electron microscope.
Whole Plant Freezing Test
Wild-type, ada2b-1, and gcn5-1 plants were grown for 14 days on Gamborg's B5 basal medium with minimal organics (Sigma) under sterile conditions in Petri dishes. The plants were tested for freezing tolerance by first placing the plates at −2°C in the dark for 3 h followed by ice nucleation with ice chips. The plants then were incubated for 21 h at −2°C, transferred to −5°C for 2 days, and placed at 4°C in the dark for 18 h, followed by 2 days at 22°C under constant illumination from cool-white fluorescent light (100 to 150 μmol·m−2·s−1). The plates then were scored for freezing damage.
Microarray Experiments
Sterilized seeds from mutant and wild-type plants were germinated on plates. After 6 days, the homozygous mutant and wild-type seedlings were transplanted to soil (nine seedlings per pot). Before bolting occurred (i.e., approximately 21 to 25 days after germination), the aerial tissues were collected and frozen in liquid N2. We collected 9 wild-type, 18 gcn5-1, and 27 ada2b-1 plants for RNA isolation. cDNA synthesis was accomplished with the Superscript Choice system (Gibco BRL Life Technologies) using 24 μg of total RNA and 100 pmol of an oligo-dT(24) primer containing a 5′ T7 RNA polymerase promoter sequence (Genset Oligos, La Jolla, CA). Approximately 6 μg of these cDNAs was used as a template to produce biotinylated complementary RNA probes by in vitro transcription using T7 RNA polymerase (BioArray High-Yield RNA Transcript Labeling Kit, Enzo Biochem, Farmingdale, NY). Labeled complementary RNAs were purified using Qiagen RNeasy spin columns, and 15 μg was fragmented randomly to produce molecules of approximately 35 to 200 bases.
Hybridization to the GeneChip Arabidopsis Genome Array (Affymetrix, Santa Clara, CA), scanning, and data analysis were performed at the Michigan State University Genomic Technology Support Facility according to standard Affymetrix protocols. In all GeneChip experiments, hybridization signals are scaled such that the average intensity from all genes is set equal to an arbitrary intensity (which in our case was 1500). By scaling all experiments to the same target intensity, comparisons can be made between the various GeneChips. In our experiments, the scaling factors required to achieve the target intensity ranged from 7 to 28. Two biological replicates of each type of plant (wild type, ada2b-1, or gcn5-1) were obtained, permitting four pair-wise comparisons of wild-type and mutant expression patterns.
Several criteria were applied to identify genes whose expression in mutant plants was different from that in wild-type plants. The gene must have been expressed in one or both of the samples (P or M in the Affymetrix nomenclature; approximately 5400 genes met this criterion). The differences in corrected hybridization signals must have been threefold or greater in at least three of the four pair-wise comparisons. The differences in signal strength must have been designated as either increased (I or MI) or decreased (D or MD) by the Affymetrix software. Within any replicate experimental pair (i.e., wild type versus wild type, ada2b-1 versus ada2b-1, or gcn5-1 versus gcn5-1), only 2% of the genes met these criteria. These differences presumably represent random experimental noise, because few of genes that were identified as different in one replicate pair were determined to be different in the other replicate pairs. Thus, by requiring that differences between wild-type and mutant signals be discerned in three of four pair-wise comparisons, the likelihood of a given gene appearing in our lists by random chance is remote. (Applying the binomial theorem, only ∼0.0032% of the 5400 expressed genes, or 0.17 genes total, would be expected to be included in the lists.) Complete data sets for each of the Affymetrix GeneChip hybridizations and comparisons are available at the TAIR World Wide Web site (under Functional Genomics) or at www.arabidopsis.org/info/expression/ada_gcn.html.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Annette Thelen of the Michigan State University Genomic Technology Support Facility and Sarah Fowler for advice and assistance with Affymetrix microarrays, Joanne Whallon and Shirley Owens for assistance with confocal and electron microscopy, Megan Rich for assistance with plant husbandry, and Kurt Stepnitz for photographic help. We also thank Min-Hao Kuo, Amy Hark, and Yaopan Mao for comments on the manuscript. This work was supported by a grant from the National Science Foundation (MCB 9728462) to M.F.T. and S.J.T., by funds from the Michigan Agricultural Experiment Station and Department of Energy (DEFG 0291ER20021) to M.F.T., and by funds from the Michigan State University Department of Biochemistry and Molecular Biology to S.J.T.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007922.
Footnotes
Online version contains Web-only data.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Artus, N.N., Uemura, M., Steponkus, P.L., Gilmour, S.J., Lin, C., and Thomashow, M.F. (1996). Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 93, 13404–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric, S., Walker, J., Schmid, A., Svejstrup, J.Q., and Horz, W. (2001). Increasing the rate of chromatin remodeling and gene activation: A novel role for the histone acetyltransferase Gen5. EMBO J. 20, 4944–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev, N.A., Candau, R., Wang, L., Darpino, P., Silverman, N., and Berger, S.L. (1995). Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270, 19337–19344. [DOI] [PubMed] [Google Scholar]
- Berger, S.L. (2002). Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148. [DOI] [PubMed] [Google Scholar]
- Berger, S.L., Pina, B., Silverman, N., Marcus, G.A., Agapite, J., Regier, J.L., Triezenberg, S.J., and Guarente, L. (1992). Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70, 251–265. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inze, D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.E., Howe, L., Sousa, K., Alley, S.C., Carrozza, M.J., Tan, S., and Workman, J.L. (2001). Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Brown, C.E., Lechner, T., Howe, L., and Workman, J.L. (2000). The many HATs of transcription coactivators. Trends Biochem. Sci. 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Eberharter, A., Sterner, D.E., Schieltz, D., Hassan, A., Yates, J.R., III, Berger, S.L., and Workman, J.L. (1999). The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6621–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos, T., and Thireos, G. (1992). Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11, 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Sebolt, A.M., Salazar, M.P., Everard, J.D., and Thomashow, M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Duggan, L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., Berger, S.L., and Workman, J.L. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Schieltz, D., Pray-Grant, M.G., Steger, D.J., Reese, J.C., Yates, J.R., III, and Workman, J.L. (1998. a). A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Sterner, D.E., Duggan, L.J., Workman, J.L., and Berger, S.L. (1998. b). The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8, 193–197. [DOI] [PubMed] [Google Scholar]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. (1998). Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kim, J.C., Lee, S.H., Cheong, Y.H., Yoo, C.M., Lee, S.I., Chun, H.J., Yun, D.J., Hong, J.C., Lee, S.Y., Lim, C.O., and Cho, M.J. (2001). A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 25, 247–259. [DOI] [PubMed] [Google Scholar]
- Knight, H., Veale, E.L., Warren, G.J., and Knight, M.R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M.H., vom Baur, E., Struhl, K., and Allis, C.D. (2000). Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Kuo, M.H., Zhou, J., Jambeck, P., Churchill, M.E., and Allis, C.D. (1998). Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I., Causton, H.C., Holstege, F.C., Shen, W.C., Hannett, N., Jennings, E.G., Winston, F., Green, M.R., and Young, R.A. (2000). Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405, 701–704. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung, K., Bhalerao, R.P., and Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474. [DOI] [PubMed] [Google Scholar]
- Marcus, G.A., Silverman, N., Berger, S.L., Horiuchi, J., and Guarente, L. (1994). Functional similarity and physical association between GCN5 and ADA2: Putative transcriptional adaptors. EMBO J. 13, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar, G.J., Fan, H.Y., and Kingston, R.E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Ogryzko, V.V., Kotani, T., Zhang, X., Schiltz, R.L., Howard, T., Yang, X.J., Howard, B.H., Qin, J., and Nakatani, Y. (1998). Histone-like TAFs within the PCAF histone acetylase complex. Cell 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Peterson, C.L., and Workman, J.L. (2000). Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10, 187–192. [DOI] [PubMed] [Google Scholar]
- Ricci, A.R., Genereaux, J., and Brandl, C.J. (2002). Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 22, 4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Roberts, S.M., and Winston, F. (1997). Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, S.Y., Denu, J.M., and Allis, C.D. (2001). Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Steponkus, P.L., Uemura, M., Joseph, R.A., Gilmour, S.J., and Thomashow, M.F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95, 14570–14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D.E., and Berger, S.L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Mao, Y., Regier, M.K., Triezenberg, S.J., and Thomashow, M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Parry, G., Graham, N., Allen, T., and Bennett, M. (2002). Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol. Biol. 49, 411–426. [DOI] [PubMed] [Google Scholar]
- Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29, 417–426. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 570–599. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (2001). So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 125, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Liu, L., and Berger, S.L. (1998). Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, L.A., and Junttila, O. (1999). Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 120, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z., and Browse, J. (1998). Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95, 7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Edmondson, D.G., Evrard, Y.A., Wakamiya, M., Behringer, R.R., and Roth, S.Y. (2000). Loss of Gcn512 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26, 229–232. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, T., Yamauchi, J., Kuwata, T., Tamura, T., Yamashita, T., Bae, N., Westphal, H., Ozato, K., and Nakatani, Y. (2000). Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97, 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. [DOI] [PubMed] [Google Scholar]
- Zhu, T., and Wang, X. (2000). Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol. 124, 1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.