Abstract
We have identified a ku80 mutant of Arabidopsis and show that telomerase is needed to generate the longer telomeres observed in this mutant. Telomeres are specialized nucleoprotein structures at the ends of chromosomes that permit cells to distinguish chromosome ends from double-strand breaks, thus preventing chromosome fusion events. Ku80 deficiency results in the lengthening of telomeres, a phenotype also seen in an Arabidopsis ku70 mutant. Furthermore, homogeneous populations of ku80 mutant cells show a steady increase in the length of telomere tracts, which reach an equilibrium length and then stabilize. In contrast to that in mammals, Ku80 deficiency in Arabidopsis cells does not cause end-to-end fusion of chromosomes. This telomere lengthening is dependent on the presence of telomerase, although it is not attributable to a significant increase in telomerase activity per se. These results demonstrate the essential role of the Ku80 protein as a negative regulator of telomerase function in plant cells.
INTRODUCTION
Telomeres are specialized nucleoprotein structures at the ends of chromosomes that protect them from fusion and degradation. They consist of a repeated DNA sequence conforming to the consensus Tx(A)Gy and an unknown number of proteins. Telomeric repeats are extended by telomerase, a reverse transcriptase with an RNA component that serves as a template for the synthesis of de novo telomeric repeats. Telomere length is stable and species specific, suggesting mechanisms to regulate telomerase and limit the addition of repeats. In mammals, telomeres have been shown to end in a large duplex loop, termed the telomeric T-loop (Griffith et al., 1999). Two proteins, TRF1 and TRF2, bind directly to the double-stranded telomeric DNA (Bilaud et al., 1997; van Steensel and de Lange, 1997; van Steensel et al., 1998; Smogorzewska et al., 2000). Two other proteins, TIN2 and hRAP1, specifically localize to mammalian telomeres by binding to TRF1 and TRF2, respectively (Kim et al., 1999; Li et al., 2000). Telomere structure and maintenance, in general, have been the subject of a number of recent reviews (Evans and Lundblad, 2000; Blackburn, 2001; Shore, 2001; Chan and Blackburn, 2002; de Lange, 2002; McKnight et al., 2002).
A fundamental question concerning telomere regulation is the means by which cells distinguish between DNA double-strand breaks (DSBs) and chromosome ends. DSBs are caused by cellular processes such as replication as well as by DNA-damaging agents such as ionizing radiation, and tight regulatory and repair systems have evolved to respond to these breaks. DSB repair is performed by genetic recombination, and two general mechanisms have been identified: homologous recombination and nonhomologous end joining (NHEJ). These mechanisms are present in species from yeast to mammals and plants. However, yeast cells use mainly homologous recombination, whereas higher eukaryotes favor NHEJ (Paques and Haber, 1999; Khanna and Jackson, 2001; van Gent et al., 2001).
Several proteins have been identified in yeast and mammals as essential for NHEJ. In yeast cells, the Ku heterodimer Ku70/Ku80 and the Rad50-Mre11-Xrs2 complex play key roles in NHEJ. These two complexes respectively recognize the DNA DSB and prepare it to be ligated by the Ligase IV-Xrcc4 complex. In mammalian cells, the DNA-PK complex, which consists of the Ku heterodimer associated with the DNA-PK catalytic subunit, senses DNA damage and is an integral component of the NHEJ system (Karran, 2000; Khanna and Jackson, 2001; Pierce et al., 2001). Mice mutated in any of these genes are radiation sensitive, present growth retardation, and are deficient in V(D)J recombination (Blunt et al., 1996; Nussenzweig et al., 1996; Zhu et al., 1996; Gu et al., 1997; Ouyang et al., 1997; Gao et al., 1998).
In yeast cells, the Rad50-Mre11-Xrs2 complex and the Ku heterodimer also are involved in telomere metabolism. Yeast cells deficient in any of these genes present shorter but stable telomeres (Boulton and Jackson, 1996, 1998; Porter et al., 1996; Kironmai and Muniyappa, 1997; Nugent et al., 1998). The role of the Ku80 protein in telomere regulation has been shown recently in mammals. Telomere length is deregulated in Ku80 mutant mouse cells; in fact, contradictory results showing both shortening and lengthening of telomeres have been reported. However, in both studies, a high frequency of end-to-end chromosome fusions was found, suggesting a role for the Ku80 protein in protecting telomeres from fusion (Bailey et al., 1999; Samper et al., 2000; d'Adda di Fagagna et al., 2001; Espejel et al., 2002).
Plant homologs of proteins involved in NHEJ have been characterized. Arabidopsis Ku70 and Ku80 proteins form a heterodimer with DNA binding activity (Riha et al., 2002; Tamura et al., 2002; West et al., 2002). Plants homozygous for T-DNA insertions in these genes are hypersensitive to DNA-damaging agents (Bundock et al., 2002; Riha et al., 2002; West et al., 2002). Arabidopsis homologs of Ligase IV and its interacting protein Xrcc4 also have been characterized (West et al., 2000), but no mutants have been analyzed to date. Arabidopsis mutants harboring a T-DNA insertion in the RAD50 gene are sterile and present a hyperrecombination phenotype (Gherbi et al., 2001). Furthermore, cultured cells derived from rad50 mutant plants are hypersensitive to methyl methanesulfonate (Gallego et al., 2001).
Telomere structure and regulation in plants has been reviewed recently (McKnight et al., 2002). An Arabidopsis telomerase mutant presents progressive shortening of telomeres, and late-generation mutants are defective in vegetative growth, with chromosome fusions occurring in 40% of the cells (Fitzgerald et al., 1999; Riha et al., 2001). We have shown previously that the Arabidopsis Rad50 protein is involved in plant telomere metabolism (Gallego and White, 2001). Mutant cells present progressive shortening of telomeres associated with cell senescence. However, surviving mutant cells have longer telomeres than those of nonmutant RAD50/rad50 heterozygote control cells. During the preparation of this article, Riha et al. (2002) and Bundock et al. (2002) reported the presence of long telomeres in Arabidopsis ku70 mutants, suggesting a role for Ku70 in telomere metabolism. A telomere binding protein homologous with the mammalian TRF1 protein has been identified in Arabidopsis, but no mutant has been studied (Chen et al., 2001).
Here, we present an analysis of the possible role of the Ku80 protein in telomere metabolism in plant cells. Arabidopsis plants homozygous for a T-DNA insertion in the KU80 gene develop and grow normally. However, they present progressive lengthening of telomeric repeats both in planta and in callus cultures in vitro. This telomere extension is dependent on the presence of telomerase, although it is not correlated with a significant increase in telomerase activity per se, as measured by the in vitro telomerase repeat amplification protocol (TRAP) assay. Thus, the Ku protein controls telomere length in Arabidopsis by directly or indirectly inhibiting the action of the telomerase at telomere ends.
RESULTS
The ku80 Mutant Presents Longer Telomeres in Plant Cells
Based on the Arabidopsis genome sequence, we used reverse transcriptase–mediated PCR to clone the cDNA of the Arabidopsis homolog of the human and yeast KU80 genes. The sequence of our AtKU80 clone corresponds exactly to that submitted previously to GenBank by Y. Adachi, K. Oguchi, K. Tamura, and H. Takahashi. This cDNA gives a predicted protein of 680 amino acids encoded by 12 exons.
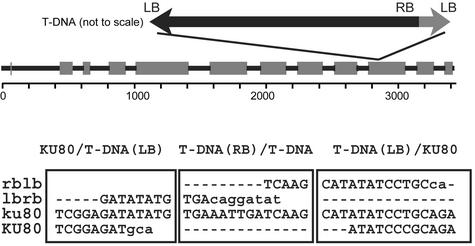
We then screened the Institut National de la Recherche Agronomique Versailles Arabidopsis T-DNA insertion collection using the flanking insertion site FLAG sequence database (Samson et al., 2002) and identified a putative mutant plant with a T-DNA insertion in the KU80 gene. PCR analysis using oligonucleotides specific for the KU80 gene and the T-DNA showed that the T-DNA is inserted in exon 10 of the KU80 gene (Figure 1). Should any protein be produced by this mutant ku80 locus, it would lack the C-terminal 125 amino acids. The inserted T-DNA has a 564-bp inverted duplication of the T-DNA left border sequence downstream of the right border. The beginning of the insertion left border has a three-nucleotide homology with the KU80 sequence. The left border at the end of the T-DNA insertion has a nine-nucleotide homology with the KU80 gene (Figure 1). The kanamycin resistance marker of the inserted T-DNA in progeny of selfed heterozygotes segregates with a 3:1 ratio for resistance to sensitivity, as expected for a single-locus insertion (282 kanamycin-resistant:102 kanamycin-sensitive plants; χ2 [1 df] = 0.5). DNA gel blot analysis confirmed that the T-DNA insertion is present at a single locus (data not shown). Plants homozygous for the T-DNA insertion develop normally and are fertile.
Figure 1.
T-DNA Insertion into the Arabidopsis KU80 Gene.
Boxes represent the exons in the KU80 gene. The position of the T-DNA insertion is indicated. At bottom, the junction sequences of the inserted T-DNAs are shown. The three boxes show the three junctions: KU80–T-DNA, T-DNA–T-DNA, and T-DNA–KU80. The mutant ku80 genomic DNA sequence is aligned with the KU80 genomic sequence and the ends of the inserted T-DNA. The mutant sequence and the sequences homologous with it (from which it derives) are shown in uppercase letters. Orientations of the T-DNA sequence are indicated as right border–left border (rblb) and left border–right border (lbrb). LB, left border; RB, right border.
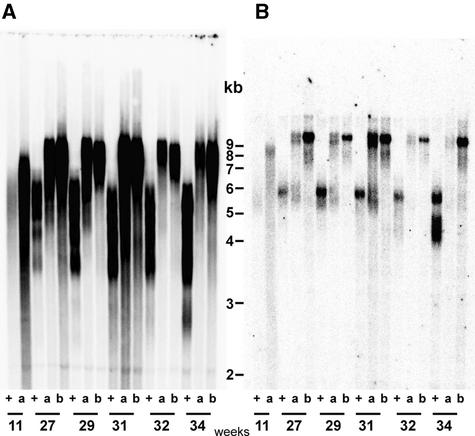
To investigate whether Ku80 plays a functional role in telomere regulation in Arabidopsis, callus suspension cultures were derived from two individual ku80 mutant plants and a wild-type parent as a control. The genotypes of the cultured cells were verified by DNA gel blot analysis using a KU80 DNA probe (data not shown). The telomere length of these cells was measured by DNA gel blot analysis of MboI-digested genomic DNA using the telomere repeat as a probe. Wild-type cells presented the expected telomeric smear, whereas cell lines derived from homozygous ku80 plants showed much longer telomeres (Figure 2A). Longer telomeres were observed after 11 weeks of growth in ku80 cells compared with the wild type. After 27 weeks, telomeres had reached their maximum length, and this length was stable for at least 34 weeks of growth. Figure 2B shows the same DNA gel blot filter reprobed with a subtelomeric probe specific for the telomere of the long arm of chromosome 2 (Gallego and White, 2001). As reported previously for Arabidopsis (Gallego and White, 2001) and tobacco (Fajkus et al., 1998), telomere length was stable in wild-type cells, in contrast to the elongation reported upon culture in Melandrium album (Riha et al., 1998) and barley (Kilian et al., 1995).
Figure 2.
Telomere Dynamics in Homogeneous Populations of ku80 Mutant Dividing Cells.
DNA was prepared from wild-type (lanes +) and two independent ku80 (lanes a and b) cell cultures grown in liquid for the indicated number of weeks. MboI-digested DNA was examined by DNA gel blot analysis using the telomeric repeat probe (A). The DNA gel blot was washed off and reprobed with a subtelomeric region from the long arm of chromosome 2 (B), which specifically detects the telomeric MboI fragment of this chromosome arm.
Thus, the lack of a functional Ku80 protein in Arabidopsis resulted in a telomere elongation phenotype. The fact that ku80 Arabidopsis plants presented a telomere phenotype similar to that of ku70 plants, together with the known physical association of these two proteins, indicates that these two proteins work as a heterodimer for telomere length regulation (Bundock et al., 2002; Riha et al., 2002; Tamura et al., 2002; West et al., 2002).
Extended Telomeres in ku80 Mutant Cells Do Not Result from Chromosome Fusion or Circularization
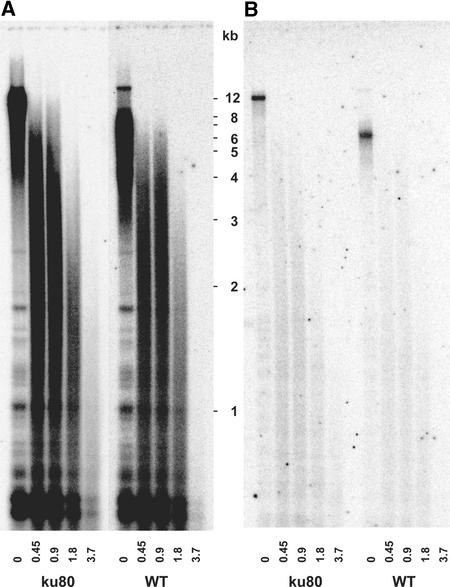
Mammalian ku80 mutant cells present a 24-fold increase in telomeric fusions compared with wild-type cells (Samper et al., 2000). Therefore, we asked whether the elongated telomeric repeats we observed in the Arabidopsis ku80 mutant cells could be the result of chromosome fusions. End-to-end chromosome fusions would result in the fused telomeric repeat sequences being internal to the chromosome. Thus, these fused telomeric repeats would become resistant to Bal31 exonuclease treatment. However, if the extended telomeres were generated by the addition of new repeats, they should be sensitive to Bal31 exonuclease attack. Genomic DNA from wild-type and ku80 mutant cells was treated with different concentrations of Bal31, digested with MboI, and analyzed by DNA gel blot hybridization with the telomere repeat probe. Bal31 digestion of genomic DNA from both ku80 mutant and wild-type cells showed progressive shortening of the telomeric repeats with increasing Bal31 concentration (Figure 3A). Thus, the longer telomeric sequences in the ku80 mutant cells were the result of lengthening of the telomeric repeats. Reprobing this blot with the chromosome II subtelomeric probe (see above) showed sharp, nondegraded bands in the untreated lanes, confirming the integrity of the genomic DNA samples used (Figure 3B). These results are consistent with the absence of observable chromosome fusions in the male meiotic cells that we analyzed (data not shown). As mentioned above, this observation is in contrast to the mammalian ku80 phenotype of chromosome fusion and genome instability (a sixfold increase in broken chromatids and chromosome fragments in ku80 cells compared with the wild type). Furthermore, this phenotype is associated with severe infertility in ku80 mice. Arabidopsis ku80 plants were fertile at least until mutant generation 4, and we observed no cytological aberrations in male meiosis in these plants (data not shown).
Figure 3.
Arabidopsis ku80 Mutant Telomeres Are Sensitive to Bal31 Exonuclease.
Genomic DNA from wild-type (WT) and ku80 mutant cells lines was treated with the indicated amounts (units) of Bal31 endonuclease, digested with MboI, and analyzed by DNA gel blot hybridization with the telomere repeat probe (A). The DNA gel blot was washed off and reprobed with a subtelomeric region from the long arm of chromosome 2 (B), which specifically detects the telomeric MboI fragment of this chromosome arm. ku80 and wild-type DNA were equally sensitive to Bal31 treatment.
The Ku80 Protein Provides a Negative Signal to Telomerase
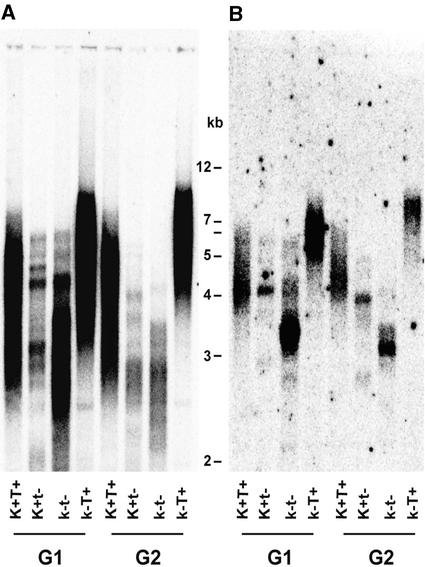
Two mechanisms might explain the new telomeric repeat addition in ku80 mutant cells: direct addition by the telomerase and recombination between telomeric sequences (alternative lengthening of telomeres [ALT] pathway). The Ku80 protein could function as a regulator of the ALT pathway by preventing recombination between telomeres. Alternatively, Ku80 could directly regulate telomerase activity or modulate its access to the telomeres. To distinguish between these two hypotheses, we generated double-mutant ku80 attert plants by crossing the ku80 mutant with the telomerase mutant attert (Fitzgerald et al., 1999) (a kind gift of T. McKnight and D. Shippen, Texas A&M University, College Station, TX). Genotypes of F2 progeny of this cross were verified by DNA gel blot analysis, and wild-type, ku80 mutant, attert mutant, and ku80 attert double-mutant plants were identified. These plants are labeled G1 (for generation 1) in Figure 4. Progeny (selfed) of the plants are labeled G2 (for generation 2) in Figure 4.
Figure 4.
Long Telomeres in ku80 Require Telomerase.
DNA gel blot analysis of telomere length using the telomeric repeat probe (A) and the chromosome II subtelomeric probe (B). MboI-digested DNA from wild-type (K+T+), attert mutant (K+t−), ku80 mutant (k−T+), and the ku80 attert double mutant (k−t−) plants was analyzed. Data from two subsequent generations of plants (G1 and G2) are shown.
We expect the appearance of extended telomeres in the double mutant if a telomerase-independent recombination mechanism is involved in the generation of the ku80 telomeres. Telomere length was measured by DNA gel blot analysis of MboI-digested genomic DNA using the telomere repeat as a probe. As expected, Figure 4A shows longer telomeres for the ku80 single mutant and shorter telomeres for the attert single mutant compared with those for the wild-type plants. Telomeres present in the double mutant and in the single attert mutant were much shorter than those in both the wild type and the ku80 single mutants. Furthermore, both the attert and ku80 telomerase double-mutant telomeres were shorter in the subsequent generation. We also checked the length dynamics of one particular telomere in these mutants. Figure 4B shows the same DNA gel blot filter reprobed with a subtelomeric probe specific for the long arm of chromosome 2. These results confirmed that ku80 attert double-mutant plants presented shorter telomeres for chromosome 2 compared with the ku80 single mutant plant or the wild type. Interestingly, an accelerated rate of telomere shortening was observed in the double mutant compared with the attert single mutant, indicating that the Ku80 protein has another, positive influence on telomere length in the absence of telomerase.
Given the telomerase dependence of the extended telomeres in ku80 cells, we performed an in vitro telomerase assay to check for an increase in telomerase activity in these cells. The modified TRAP assay was used to measure telomerase activity in vitro in total protein extracts of wild-type and ku80 mutant callus culture cells (see Methods). No telomere elongation was detected in the absence of protein extracts (Figure 5, lane 5) or when the extracts were pretreated with RNaseA to inactivate the telomerase enzyme (Figure 5, lanes 1 and 2). Protein extracts from both wild-type and ku80 mutant cells showed similar levels of telomerase activity (Figure 5, lanes 3 and 4). Thus, we were unable to detect major changes in telomerase activity per se in this in vitro test. However, in vivo deregulation of telomerase activity in the absence of the Ku80 protein cannot be excluded.
Figure 5.
Telomerase Activity in Ku−/− Mutant Cells.
Protein extracts were prepared from wild-type (+) and homozygous (k) ku80 mutant cells. The fifth lane (−) contained no protein extract. Telomerase activity was measured by the modified TRAP assay as described in Methods. Extracts were pretreated (+) or not (−) with RNase. Band intensity was quantified by phosphorimager analysis and is shown graphically at right for the wild type (+) and the ku80 mutant (ku80).
DISCUSSION
We have described the role of the Arabidopsis Ku80 protein in telomere homeostasis by studying an Arabidopsis ku80 mutant that carries a T-DNA insertion in the KU80 gene. Mutant plants presented longer telomeres than wild-type plants. This phenotype is similar to that reported recently for an Arabidopsis ku70 mutant (Bundock et al., 2002; Riha et al., 2002). A progressive increase in the number of telomeric repeats was detected in ku80 mutants, which in callus culture reached a maximum length and stabilized. The Bal31 exonuclease sensitivity of these long telomeres indicates that they do not result from end-to-end chromosome fusion or circularization. This finding was confirmed by cytological analysis of male meiotic cells from generation-4 ku80 mutant plants. Additional generations of mutant plants will be monitored for cytological aberrations.
Two mechanisms could be involved in the generation of the extended telomeric repeats: recombination and direct addition of telomeric repeats by the telomerase. Analysis of Arabidopsis ku80 attert double-mutant plants demonstrated that telomere elongation in Ku80-deficient plants was telomerase dependent. Furthermore, Ku80 deficiency did not significantly affect telomerase activity measured in cell extracts, suggesting that Ku80 acts a negative telomere length regulator at individual chromosomes in plants.
The essential role of the Ku70/Ku80 heterodimer in telomere metabolism has been demonstrated in several organisms. These data indicate species-specific mechanisms for Ku protein function in telomeres. Saccharomyces cerevisiae and Schizosaccharomyces pombe cells deficient in Ku70 present shortened but stable telomeres (Boulton and Jackson, 1996; Porter et al., 1996; Baumann and Cech, 2000). In neither case are chromosome fusions observed. Interestingly, in S. cerevisiae, simultaneous deletion of the telomerase catalytic subunit and Ku70 or Ku80 causes lethality (Gravel et al., 1998; Nugent et al., 1998). By contrast, ku70 telomerase double-mutant fission yeast cells senesce, but a large number of survivors are generated. These surviving cells present circularization of all three chromosomes, as is observed for telomerase-deficient survivors in this organism (Nakamura et al., 1998; Baumann and Cech, 2000). Conflicting results concerning telomeres have been obtained in ku80 knockout mice: in one study, extended telomeres were observed, whereas a second study showed a decrease in telomeric repeats (Samper et al., 2000; d'Adda di Fagagna et al., 2001). Nevertheless, a high frequency of end-to-end fusions was found in both studies. These results suggest a role for the Ku80 protein in telomere end capping. ku70 disruption in chicken DT40 cell lines has no effect on telomere length and does not result in chromosome fusions (Wei et al., 2002). Recently, it was shown that Arabidopsis plants carrying a T-DNA insertion in the KU70 gene have extended telomeres, and up to mutant generation 2, chromosome fusions are not observed (Riha et al., 2002). Here, we show that Ku80-deficient Arabidopsis presented an elongation of telomeres, also without chromosome fusions. Thus, it appears that in all organisms studied to date, with the exception of chicken DT40 cells, the Ku80 protein plays a role in telomere length homeostasis, with an additional role in mammalian cells to protect telomeres from fusion.
Telomeres have a species-specific length that is constant over generations, implying tight regulatory mechanisms to measure and modulate the length of the telomeric repeat at individual chromosome ends. In mammalian cells, the TRF1, TRF2, and TIN2 telomere binding proteins have been suggested to function in the negative regulation of telomerase action (Kim et al., 1999; Smogorzewska et al., 2000). Disruption of the function of any of these proteins gives rise to the generation of longer telomeres. However, direct evidence that the extended telomeres are telomerase dependent must await the study of telomerase-deficient cells. TRF1 and TRF2 are double-stranded TTAGGG-repeat binding proteins. It has been proposed that they control telomere length in cis by inhibiting the action of telomerase. In fact, both proteins are needed for the generation and maintenance of the T-loop (Bianchi et al., 1997, 1999; Griffith et al., 1998, 1999; Stansel et al., 2001). In the T-loop, the 3′ end DNA strand overhang required for telomerase action is sequestered into the duplex part of the telomeric repeat tract. TIN2 interacts with TRF1 and has been suggested to stabilize the T-loop (Kim et al., 1999). The Ku80 protein interacts with TRF1 and TRF2 and may contribute to the stabilization of the T-loop structure and control the action of telomerase (Hsu et al., 2000; Song et al., 2000).
Alternatively, the role of the Ku80 protein could be to suppress the accessibility to telomeres of the homologous recombination machinery. S. cerevisiae and S. pombe ku mutants show an increase in subtelomeric and telomeric recombination, notwithstanding the presence of shorter telomeres. S. cerevisiae and mammalian cells lacking telomerase activity present heterogeneous telomere length as a result of a telomerase-independent pathway of telomere maintenance that involves recombination (Lundblad and Blackburn, 1993; Bryan et al., 1995; Teng and Zakian, 1999; Dunham et al., 2000). A recent analysis of yeast ku telomerase double mutants by Tsai et al. (2002) has shown that the Ku complex plays an additional role in protecting telomeres and in generating survivors in telomerase mutants. Thus, Ku80 protein action in telomere homeostasis involves blocking telomerase activity as well as stabilizing the telomeres and facilitating the access of the homologous recombination proteins to telomeres. Both longer and shorter telomeres have been described in mouse ku80 mutants, and telomere lengthening has been shown to be telomerase dependent (Espejel et al., 2002).
The gradual elongation and subsequent stabilization of telomeres we observed in ku80 mutant plant cells suggests a telomerase-mediated, rather than a recombination (ALT), mechanism in Arabidopsis. We have confirmed this by showing that the appearance of long telomeres in Ku80-deficient Arabidopsis depends on the presence of telomerase. An alternative recombination-based (unequal exchange) and telomerase-dependent mechanism also could explain these results; however, such a mechanism would not be expected to lead to the observed stabilization of the elongated telomere length (Figure 2). Interestingly, the ku telomerase double-mutant plants have even shorter telomeres than the single telomerase mutants. This synergism between the ku80 and telomerase mutations indicates a role for the Ku80 protein in telomere stability in addition to its role as a negative regulator of telomerase action. As mentioned above, Tsai et al. (2002) recently reported a role for the Ku complex in telomere protection and survivor generation in yeast. A similar role(s) in Arabidopsis would explain this synergistic effect.
Thus, the Arabidopsis Ku80 protein could directly regulate the action of telomerase at telomeres or possibly regulate other proteins essential for telomere homeostasis as well as play a role in telomere end protection or stability. Full understanding of Ku80's role in telomere metabolism in plant cells will require the identification of the other protein partners present at the telomeres.
METHODS
Plant Growth and Callus Induction
Arabidopsis thaliana seeds were sown directly into damp compost, and plants were grown in a greenhouse under standard conditions. Callus cultures were derived from leaves and maintained as described previously (Gallego and White, 2001).
DNA Isolation and DNA Gel Blot Analysis
DNA was prepared from plants or callus cells as described previously (Gallego and White, 2001). TRF analysis was performed in MboI-digested DNA (0.5 to 1 μg), which was separated by electrophoresis on 0.8% agarose/Tris-borate-EDTA gels and blotted to positively charged nylon membranes (Hybond N+; Amersham). The subtelomeric chromosome II probe was labeled with α-32P-dCTP using the Prime It II kit (Stratagene), and the telomeric repeat probe was 5′ labeled using T4 polynucleotide kinase and γ-32P-ATP. The chromosome 2 long-arm subtelomeric probe specifically detects the telomeric MboI fragment of this chromosome arm. Specific details have been described previously (Gallego and White, 2001).
Telomerase Repeat Amplification Protocol Assay
Whole cell extracts from suspension culture cells were prepared according to Fitzgerald et al. (1996). The telomerase TRAP assay was performed according to the protocol of Szatmari and Aradi (2001) with modifications. The following oligonucleotides were used: forward primer o177 (5′-CACTATCGACTACGCGATCGG-3′), tagging primer o278 (5′-GATCTCGAGCTCGATATCGGATCCCCTAAACCCTAAAGG-3′), and tag primer o274 (5′-GATCTCGAGCTCGATATCGGATC-3′). The assay was performed as follows: 0.5 μg of extract and 1 μL (10 pmol) of primer o177 were added to 40 μL of assay buffer (50 mM Tris acetate, pH 8.3, 5 mM MgCl2, 10 mM EGTA, 1 mM DTT, 50 mM potassium glutamate, 0.1% Triton X-100, 1 mM spermidine, 50 μM each deoxynucleotide triphosphate, and 100 ng/μL BSA). This mixture was incubated at 22°C for 30 min and then heated to 90°C for 3 min. During the 90°C incubation, 6.5 μL of PCR mix (15 pmol of primer o177, 25 pmol of primer o274, 0.5 pmol of primer o278, 5 μCi of α-32P-dCTP, and 0.5 units of Taq DNA polymerase [Qiagen, Valencia, CA]) was added. PCR then was performed as follows: 2 cycles of 94°C for 30 s, 55°C for 60 s, and 72°C for 90 s, followed by 27 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s, incubation at 72°C for 90 s, and cooling to 4°C. Products were separated on 6% denaturing acrylamide sequencing gels, and the gels were dried and autoradiographed with a phosphorimager (Bio-Rad Personal FX).
Bal31 Sensitivity Assay
Total genomic DNA (1 μg) was treated with different concentrations of Bal31 exonuclease (New England Biolabs, Beverly, MA) for 30 min at 37°C in the manufacturer's buffer in a volume of 50 μL. After the addition of EGTA to 20 mM and 10 min of incubation at 65°C, the DNA samples were extracted in phenol/chloroform and precipitated in sodium acetate/ethanol. MboI digestion and DNA gel blot analysis were as described above.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for the AtKU80 clone is AF283758.
Acknowledgments
We thank Dorothy Shippen and Tom McKnight for the attert mutant. This work was financed partly by grants from the Commissariat à l'Energie Atomique France (Laboratoire de Recherche Conventionné Commissariat à l'Energie Atomique No. 19V) and the European Union (Contract QLG2-CT-2001-01397).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008623.
References
- Bailey, S.M., Meyne, J., Chen, D.J., Kurimasa, A., Li, G.C., Lehnert, B.E., and Goodwin, E.H. (1999). DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, P., and Cech, T.R. (2000). Protection of telomeres by the Ku protein of fission yeast. Mol. Biol. Cell 11, 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, A., Smith, S., Chong, L., Elias, P., and de Lange, T. (1997). TRF1 is a dimer and bends telomeric DNA. EMBO J. 16, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, A., Stansel, R.M., Fairall, L., Griffith, J.D., Rhodes, D., and de Lange, T. (1999). TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud, T., Brun, C., Ancelin, K., Koering, C.E., Laroche, T., and Gilson, E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Blunt, T., Gell, D., Fox, M., Taccioli, G.E., Lehmann, A.R., Jackson, S.P., and Jeggo, P.A. (1996). Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. USA 93, 10285–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S.J., and Jackson, S.P. (1996). Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S.J., and Jackson, S.P. (1998). Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, T.M., Englezou, A., Gupta, J., Bacchetti, S., and Reddel, R.R. (1995). Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., van Attikum, H., and Hooykaas, P. (2002). Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30, 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W., and Blackburn, E.H. (2002). New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene 21, 553–563. [DOI] [PubMed] [Google Scholar]
- Chen, C.M., Wang, C.T., and Ho, C.H. (2001). A plant gene encoding a Myb-like protein that binds telomeric GGTTAG repeats in vitro. J. Biol. Chem. 276, 16511–16519. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna, F., Hande, M.P., Tong, W.M., Roth, D., Lansdorp, P.M., Wang, Z.Q., and Jackson, S.P. (2001). Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11, 1192–1196. [DOI] [PubMed] [Google Scholar]
- de Lange, T. (2002). Protection of mammalian telomeres. Oncogene 21, 532–540. [DOI] [PubMed] [Google Scholar]
- Dunham, M.A., Neumann, A.A., Fasching, C.L., and Reddel, R.R. (2000). Telomere maintenance by recombination in human cells. Nat. Genet. 26, 447–450. [DOI] [PubMed] [Google Scholar]
- Espejel, S., Franco, S., Rodriguez-Perales, S., Bouffler, S.D., Cigudosa, J.C., and Blasco, M.A. (2002). Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, S.K., and Lundblad, V. (2000). Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113, 3357.–3364. [DOI] [PubMed] [Google Scholar]
- Fajkus, J., Fulneckova, J., Hulanova, M., Berkova, K., Riha, K., and Matyasek, R. (1998). Plant cells express telomerase activity upon transfer to callus culture, without extensively changing telomere lengths. Mol. Gen. Genet. 260, 470–474. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M.S., McKnight, T.D., and Shippen, D.E. (1996). Characterization and developmental patterns of telomerase expression in plants. Proc. Natl. Acad. Sci. USA 93, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, M.S., Riha, K., Gao, F., Ren, S., McKnight, T.D., and Shippen, D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., Jeanneau, M., Granier, F., Bouchez, D., Bechtold, N., and White, C.I. (2001). Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 25, 1–13. [DOI] [PubMed] [Google Scholar]
- Gallego, M.E., and White, C.I. (2001). RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Chaudhuri, J., Zhu, C., Davidson, L., Weaver, D.T., and Alt, F.W. (1998). A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 9, 367–376. [DOI] [PubMed] [Google Scholar]
- Gherbi, H., Gallego, M.E., Jalut, N., Lucht, J.M., Hohn, B., and White, C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2, 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel, S., Larrivee, M., Labrecque, P., and Wellinger, R.J. (1998). Yeast Ku as a regulator of chromosomal DNA end structure. Science 280, 741–744. [DOI] [PubMed] [Google Scholar]
- Griffith, J., Bianchi, A., and de Lange, T. (1998). TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol. 278, 79–88. [DOI] [PubMed] [Google Scholar]
- Griffith, J., Comeau, J.D., Rosenfeld, S., Stansel, R.M., Bianchi, A., Moss, H., and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Jin, S., Gao, Y., Weaver, D.T., and Alt, F.W. (1997). Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA 94, 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H.L., Gilley, D., Galande, S.A., Hande, M.P., Allen, B., Kim, S.H., Li, G.C., Campisi, J., Kohwi-Shigematsu, T., and Chen, D.J. (2000). Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran, P. (2000). DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10, 144–150. [DOI] [PubMed] [Google Scholar]
- Khanna, K.K., and Jackson, S.P. (2001). DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 27, 247–254. [DOI] [PubMed] [Google Scholar]
- Kilian, A., Stiff, C., and Kleinhofs, A. (1995). Barley telomeres shorten during differentiation but grow in callus culture. Proc. Natl. Acad. Sci. USA 92, 9555–9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Kaminker, P., and Campisi, J. (1999). TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai, K.M., and Muniyappa, K. (1997). Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2, 443–455. [DOI] [PubMed] [Google Scholar]
- Li, B., Oestreich, S., and de Lange, T. (2000). Identification of human Rap1: Implications for telomere evolution. Cell 101, 471–483. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and Blackburn, E.H. (1993). An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73, 347–360. [DOI] [PubMed] [Google Scholar]
- McKnight, T.D., Riha, K., and Shippen, D.E. (2002). Telomeres, telomerase, and stability of the plant genome. Plant Mol. Biol. 48, 331–337. [DOI] [PubMed] [Google Scholar]
- Nakamura, T.M., Cooper, J.P., and Cech, T.R. (1998). Two modes of survival of fission yeast without telomerase. Science 282, 493–496. [DOI] [PubMed] [Google Scholar]
- Nugent, C.I., Bosco, G., Ross, L.O., Evans, S.K., Salinger, A.P., Moore, J.K., Haber, J.E., and Lundblad, V. (1998). Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8, 657–660. [DOI] [PubMed] [Google Scholar]
- Nussenzweig, A., Chen, C., da Costa Soares, V., Sanchez, M., Sokol, K., Nussenzweig, M.C., and Li, G.C. (1996). Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382, 551–555. [DOI] [PubMed] [Google Scholar]
- Ouyang, H., Nussenzweig, A., Kurimasa, A., Soares, V.C., Li, X., Cordon-Cardo, C., Li, W., Cheong, N., Nussenzweig, M., Iliakis, G., Chen, D.J., and Li, G.C. (1997). Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 186, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, A.J., Stark, J.M., Araujo, F.D., Moynahan, M.E., Berwick, M., and Jasin, M. (2001). Double-strand breaks and tumorigenesis. Trends Cell Biol. 11, S52–S59. [DOI] [PubMed] [Google Scholar]
- Porter, S.E., Greenwell, P.W., Ritchie, K.B., and Petes, T.D. (1996). The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., Fajkus, J., Siroky, J., and Vyskot, B. (1998). Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 10, 1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha, K., Watson, J.M., Parkey, J., and Shippen, D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper, E., Goytisolo, F.A., Slijepcevic, P., van Buul, P.P., and Blasco, M.A. (2000). Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, D. (2001). Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11, 189–198. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K., Jung, D., Jung, Y., Lee, S.G., and Lee, I. (2000). Interaction of human Ku70 with TRF2. FEBS Lett. 481, 81–85. [DOI] [PubMed] [Google Scholar]
- Stansel, R.M., de Lange, T., and Griffith, J. (2001). T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20, 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari, I., and Aradi, J. (2001). Telomeric repeat amplification, without shortening or lengthening of the telomerase products: A method to analyze the processivity of telomerase enzyme. Nucleic Acids Res. 29, E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Adachi, Y., Chiba, K., Oguchi, K., and Takahashi, H. (2002). Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: Evidence for a role in the repair of DNA double-strand breaks. Plant J. 29, 771–781. [DOI] [PubMed] [Google Scholar]
- Teng, S.C., and Zakian, V.A. (1999). Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, Y.L., Tseng, S.F., Chang, S.H., Lin, C.C., and Teng, S.C. (2002). Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 22, 5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent, D.C., Hoeijmakers, J.H., and Kanaar, R. (2001). Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2, 196–206. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., and de Lange, T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Smogorzewska, A., and de Lange, T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Wei, C., Skopp, R., Takata, M., Takeda, S., and Price, C.M. (2002). Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 30, 2862–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, C.E., Waterworth, W.M., Jiang, Q., and Bray, C.M. (2000). Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 24, 67–78. [DOI] [PubMed] [Google Scholar]
- West, C.E., Waterworth, W.M., Story, G.W., Sunderland, P.A., Jiang, Q., and Bray, C.M. (2002). Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 31, 517–528. [DOI] [PubMed] [Google Scholar]
- Zhu, C., Bogue, M.A., Lim, D.S., Hasty, P., and Roth, D.B. (1996). Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86, 379–389. [DOI] [PubMed] [Google Scholar]