Abstract
In an attempt to disclose mechanisms of hepatocarcinogenesis and discover novel target molecules for the diagnosis and treatment of hepatocellular carcinomas (HCCs), we previously analyzed expression profiles of HCC tissues by means of human cDNA microarray. Among the genes upregulated in tumor tissues compared with their nontumor counterparts, we focused on a novel gene, termed WDRPUH, and characterized its biologic function. WDRPUH encodes a predicted 620-amino acid protein containing 11 highly conserved WD40-repeat domains. Multiple-tissue Northern blot analysis revealed its specific expression in the testis among 16 normal tissues examined. Transfection of plasmids designed to express WDRPUH-specific siRNA significantly reduced its expression in HCC cells and resulted in growth suppression of transfected cells. Interestingly, we found that WDRPUH associated with HSP70, proteins of the chaperonin-containing TCP-1 (CCT1) complex, as well as BRCA2. These findings have disclosed a novel insight into hepatocarcinogenesis and suggested that WDRPUH may be a molecular target for the development of new strategies to treat HCCs.
Keywords: WDRPUH, hepatocellular carcinoma, HSP70, CCT, BRCA2
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common neoplasm in the world [1], and its incidence is gradually increasing in developed countries. It is the most common primary epithelial malignancy occurring in the liver. Although recent medical advances have made great progress in its diagnosis, many patients with HCC are not diagnosed until the disease has reached an advanced stage. A wide variety of chemotherapeutic agents as well as physical methods that cause obstruction of tumor nutrition vessels or induce necrosis of tumor cells with ethanol have been tried and are in use to treat HCCs, but no regimen has proved sufficiently to be curative and a substantial number of patients suffer from severe adverse effects [2]. Therefore, development of more effective chemotherapeutic drugs or chemoprevention strategies is an urgent issue.
A new class of anticancer drugs that target a specific molecule has proved its effectiveness in the clinical field. For example, imatinib, a small molecule that associates with the kinase domain of bcr-abl fusion protein, is now applied for chronic myelogenous leukemia and gastrointestinal stromal tumor that frequently harbors an activating mutation in the c-kit protein. Because these proteins play an essential role for their development, imatinib showed striking effectiveness against the growth of these neoplasms. Additionally, gefitinib, a drug that was developed to compete with the binding of ATP to the intracellular tyrosine kinase domain of epidermal growth factor receptor (EGFR), was utilized for non-small cell lung carcinoma that expresses abundant EGFR [3]. Because EGFR mediates growth-promoting signals from the extracellular domain by the phosphorylation of downstream mediators, the inhibition of kinase activity results in a drastic growth-suppressive effect on a subset of lung cancers. These evidences strongly suggest that genes that are exclusively expressed in tumors and involved in their proliferation seem to be promising anticancer targets.
Epidemiologic studies have clarified that chronic liver inflammation caused by viral infection and exposures to chemical carcinogens are main risk factors for tumorigenesis. Accumulating evidence from molecular studies suggests that multiple processes involving qualitative and quantitative alterations in several gene products underlie hepatocarcinogenesis. Several growth factors including transforming growth factor-α (TGF-α) [4,5] and insulin-like growth factor-2 (IGF-2) have been implicated in the development of HCC. Altered expression of a number of other genes such as E-cadherin, MMP-1, MMP-7, MT1-MMP, gankyrin, and PEG10 may also be involved in the development and/or progression of HCC [6,7]. In addition, tumor-suppressor genes such as p53, AXIN1, and p16INK4a play a crucial role in hepatocarcinogenesis [8–10]. Regardless of all these findings, the complete understanding of the molecular events that lead to the development and progression of HCC is still far away.
In a previous study of genomewide analysis of gene expression in HCC by cDNA microarray [11], we identified 165 genes upregulated in HCC. Among the 165 transcripts in the list, we focused here on a gene encoding a novel WD-repeat protein. Proteins containing WD repeats have been reported to play crucial roles in a wide range of physiological functions including signal transduction, RNA processing [12], remodeling of cytoskeleton [13], regulation of vesicular trafficking [14], and cell division [15]. For example, RACK1 protein containing seven WD40-repeats functions as an anchor for protein kinase C (PKC) [16] in the PKC signal transduction pathway. Another WD domain-containing protein, TRIP-1, specifically associates with the type II TGF-β receptor in a kinase-dependent manner and mediates TGF-β signaling [17]. The WD-40 domain of Cdc4p binds directly to phospho-Sic1p and controls cell division by the recruitment of Skp1p [15,18]. Other WD-repeat-containing proteins have also been shown to be involved in a variety of functions [e.g., transcriptional regulation (TUPI), RNA processing (PRP4 and PRP17), cell cycle progression (CDC4 and CDC20), and growth regulation (G protein β-subunits and MSII)] [19].
In the present study, we identified a novel gene with growth-promoting activity, WDRPUH, which is overexpressed in a great majority of HCCs. Because suppression of WDRPUH leads to growth retardation of cancer cells, WDRPUH is a novel potential target for the development of therapeutic strategies for HCC.
Materials and Methods
Cell Lines
Human embryonic kidney 293 (HEK293) cells were obtained from IWAKI; human hepatoma cell lines Alexander and HepG2, a testicular cancer cell line Tera-1, and mouse fibroblast cell line NIH3T3 were from the American Type Culture Collection (ATCC, Rockville, MD). Another human hepatoma cell line, Huh7, was obtained from the Japanese Collection of Research Bioresources (JCRB, Tokyo, Japan), whereas SNU449 and SNU475 were obtained from the Korea Cell Line Bank (KCLB Seoul, Korea). All cell lines were grown in monolayers in appropriate media: Dulbecco's modified Eagle's medium for Alexander, Huh7, HepG2, and HEK293; RPMI 1640 for SNU423, SNU449, and SNU475; and McCoy's 5A for Tera-1, supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (Sigma, St. Louis, MO). All cells were maintained at 37°C in humid air with 5% CO2.
RNA Preparation and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted with a Qiagen RNeasy kit (Qiagen, Valencia, CA) or Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturers' protocols. Ten-microgram aliquots of total RNA were reversely transcribed for single-stranded cDNA using poly dT12–18 primer (Amersham Pharmacia Biotechnologies, Little Chalfont, UK) with Superscript II reverse transcriptase (Invitrogen). Each single-stranded cDNA preparation was diluted for subsequent PCR amplification by standard RT-PCR experiments carried out in 12.5-µl volumes of PCR buffer (Takara, Otsu, Japan). Amplification proceeded for 4 minutes at 94°C for denaturing, followed by 35 cycles for WDRPUH or 20 cycles for GAPDH, of 94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 45 seconds, in the GeneAmp PCR system 9700 (Perkin-Elmer, Foster City, CA). Primer sequences were: for GAPDH: forward, 5′-ACAACAGCCTCAAGATCATCAG-3′ and reverse, 5′-GGTCCACCACTGACACGTTG-3′; for WDRPUH: forward, 5′-CAGGTGGAAATGACCATCTGGTCAAAG-3′ and reverse, 5′-CATCAGCTTCAGGAGGTATATGGGTAC-3′.
Northern Blot Analysis
Human multiple-tissue blots (Clontech, Palo Alto, CA) were hybridized with a 32P-labeled WDRPUH cDNA. Prehybridization, hybridization, and washing were performed according to the supplier's recommendations. The blots were autoradiographed with intensifying screens at -80°C for 120 hours.
5′ Rapid Amplification of cDNA Ends (5′ RACE)
5′ RACE experiments were carried out using a Marathon cDNA amplification kit (Clontech) according to the manufacturer's instructions. For the amplification of the 5′ part of WDRPUH cDNA, a gene-specific reverse primer (5′-TTACCGTCGTTCCATGCTGAAATGATGC-3′) and AP-1 primer supplied in the kit were used. The cDNA template was synthesized from human testis mRNA (Clontech). The PCR products were cloned using a TA cloning kit (Invitrogen, Carlsbad, CA) and their sequences were determined with an ABI PRISM 3700 DNA sequencer (Applied Biosystems, Foster City, CA).
Construction of Plasmids Expressing WDRPUH
The entire coding region of WDRPUH was amplified by RT-PCR using a set of gene-specific primers: 5′- GGGGTACCACCATGGATAACAAAATTTCGCCGGAG-3′ and 5′-CGGAATTCTCAGGAGGTATATGGGTACTTCCATCG-3′. The PCR products were cloned into an appropriate cloning site of either pcDNA3.1 (Invitrogen), p3xFLAG-CMV-10 (Sigma), or pcDNA3.1myc/His (Invitrogen) vector.
Immunopreciptation
HEK293 cells that do not express WDRPUH were transfected with p3xFLAG-WDRPUH, mock vector, or p3xFLAG-SMYD3 (as a control). The cells were harvested at 48 hours after transfection and lysed with Nonidet P40 lysis buffer [20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1x complete Protease Inhibitor Cocktail EDTA (-) (Roche, Basel, Switzerland)]. Cell lysates were immunoprecipitated with anti-FLAG M2 antibody conjugated with agarose overnight at 4°C. After being washed five times with lysis buffer, precipitated protein complex was eluted with 25 µg of FLAG peptide for 1 hour at 4°C. The eluted complex was centrifuged and supernatant was resuspended in 5x Laemmli sample buffer and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Immunoblotting
Cells transfected with p3xFLAG-WDRPUH were washed twice with PBS and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 25 M NaCl, 0.1% SDS, 5% NP40, and 1x complete Protease Inhibitor Cocktail (Roche)). After the cells had been homogenized and centrifuged at 10,000g for 15 minutes, each supernatant was standardized for protein concentration by the Bradford assay (Bio-Rad, Hercules, CA). Proteins were separated by 10% SDS-PAGE and immunoblotted with mouse anti-FLAG M2 (Sigma) antibody, anti-HSP70 antibody (Stressgen, Victoria, Canada), anti-chaperonin-containing TCP-1 (CCT1) delta subunity (Santa Cruz Biotechnology, Santa Cruz, CA), anti-CTT1-alpha subunity (Stressgen), or anti-BRCA2 (Stressgen). HRP-conjugated goat anti-mouse IgG (Amersham Pharmacia Biotechnologies) served as the secondary antibody for the ECL Detection System (Amersham Pharmacia Biotechnologies).
Colony Formation Assay
Plasmids expressing WDRPUH (pcDNA-WDRPUH) or the control vector (pcDNA-antisense-WDRPUH) were transfected into NIH3T3 cells in triplicate. After 2 weeks of incubation with an appropriate concentration of geneticin (Invitrogen), cells were fixed with 100% methanol and stained with Giemsa solution. The data were subjected to analysis of variance (ANOVA) and Student's t test.
Gene-Silencing Effect of WDRPUH sIRNA
The genomic fragment of snRNA U6 promoter region was amplified by PCR using a set of primers, 5′-GGGGATCAGCGTTTGAGTAA-3′ and 5′-TAGGCCCCACCTCCTTCTAT-3′. The product was ligated with a fragment of pcDNA3.0 vector containing the neomycin-resistant gene and constructed psiU6BX vector as described elsewhere [20]. Plasmids expressing WDRPUH siRNA were prepared by cloning of double-stranded oligonucleotides into the psiU6BX vector. The oligonucleotides used for WDRPUH siRNA were: 5′-CACCAATGTGATCTTCTCCAGGTGCTTCAAGAGAGCACCTGGAGAAGATCACATT-3′ and 5′-AAAAAATGTGATCTTCTCCAGGTGCTCTCTTGAAGCACCTGGAGAAGATCACATT-3′ for si-01; 5′-CACCAAGGACACCAGTTTCTCGTAGTTCAAGAGACTACGAGAAACTGGTGTCCTT-3′ and 5′-AAAAAAGGACACCAGTTTCTCGTAGTCTCTTGAACTACGAGAAACTGGTGTCCTT-3′ for si -02; 5′-CACCAAAGAGACGCTCATAGCGACTTTCAAGAGAAGTCGCTATGAGCGTCTCTTT-3′ and 5′-AAAAAAAGAGACGCTCATAGCGACTTCTCTTGAAAGTCGCTATGAGCGTCTCTTT-3 for si-03; 5′-CACCAAGGATATCAGGGTGTGGCACTTCAAGAGAGUGCCACACCCTGATATCCTT-3′ and 5′-AAAAAGGATATCAGGGTGTGGCACTCTCTTGAAGTGCCACACCCTGATATCCTTAAAAAGGATATCAGGGTGTGGCACTCTCTTGAAGTGCCACACCCTGATATCCTT-3′ for si-04. The single-stranded oligonucleotides were phosphorylated with T4 polynucleotide kinase, and forward and corresponding reverse oligonucleotides were annealed after denaturation by boiling. Double-stranded oligonucleotides were ligated into psi-U6BX to create an expression vector expressing siRNA. psiU6BX-WDRPUH, psiU6BX-EGFP, and psiU6BX-mock (empty) plasmids were transfected into HepG2 and Alexander cells using a FuGENE6 reagent according to the supplier's recommendations (Roche). Total RNA was extracted from the cells 48 hours after transfection. Silencing of the WDRPUH gene by the specific constructs was confirmed by RT-PCR.
Cell Viability Assays
Cells were plated at a density of 5 x 105 cells/100 mm dish. At 24 hours after seeding, the cells were transfected in triplicate with psiU6BX-WDRPUH, EGFP, or mock plasmids using FuGENE6 (Roche) and subsequently maintained with an appropriate concentration of G418. At 7 days after transfection, the medium was replaced with fresh medium containing 500 µg/ml (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) (Sigma) and the plates were incubated for 4 hours at 37°C. Subsequently, the cells were lysed by the addition of 0.01 N HCl/10% SDS, and absorbance of lysates was measured with an ELISA plate reader at a test wavelength of 570 nm (reference, 630 nm). The cell viability was represented by the absorbance compared to that of control cells. The data were subjected to ANOVA and Student's t test.
Matrix-Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) Mass Spectrometry Analysis
Proteins immunoprecipitated with anti-FLAG antibody from cells transfected with plasmid p3xFLAG-WDRPUH were separated by SDS-PAGE. We also prepared a p3xFLAG-SMYD3 plasmid expressing an unrelated protein and transfected it, or mock vector as controls, and obtained immunoprecipitants with anti-FLAG. Associated proteins were separated by SDS-PAGE, stained with Sypro-Ruby (Bio-Rad), and compared by the bands between WDRPUH and the unrelated protein. Specific bands stained by Sypro-Ruby were excised and digested with trypsin as previously described [21]. In the gel, tryptic digest was analyzed by MALDI-TOF mass spectrometry analysis at Shimadzu Biotechnologies (Tokyo, Japan). The mass spectral data were evaluated using the Mascot search engine (http://www.matrixscience.com) to identify proteins from primary sequence databases.
Results
Identification of a Novel Human Gene Upregulated in HCC
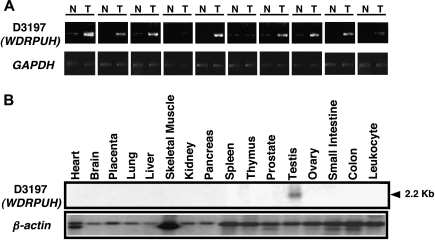
We previously analyzed expression profiles of 20 HCC tissues by the cDNA microarray, and compared them with expression profiles of their corresponding noncancerous liver tissues [11]. Out of 23040 genes screened by the array slides, 165 transcripts were frequently upregulated in the HCCs [11]. Among the 165 genes, we focused on a novel gene with an in-house accession name of D3197 because the gene was overexpressed in 11 of 12 HCCs that passed our cutoff filter (data not shown). The gene corresponded to an EST archived as Hs.232270, and was annotated as LOC146845 in the UniGene database of the National Center for Biotechnology Information. Consistent with the microarray data, semiquantitative RT-PCR corroborated its elevated expression in 8 of 10 additional HCC tissues (Figure 1A). We also analyzed its expression in hepatoma cell lines (Figure W1). Expression of the gene was not frequently elevated in breast, colon, bladder, or prostate cancer in our microarray data (data not shown).
Figure 1.
Expression of D3197 (WDRPUH). (A) Semiquantitative RT-PCR analysis of WDRPUH in 10 HCCs (T) and their corresponding noncancerous liver tissues (N). Expression of GAPDH served as a control. (B) Northern blot analysis of WDRPUH in a panel of 16 adult normal human tissues. β-Actin served as a control.
Expression, Isolation, and Characterization of the Novel Gene
Analysis with multiple-tissue Northern blots using the D3197 cDNA as a probe revealed a 2.2-kb transcript that was abundantly expressed in the testis (Figure 1B). Expression of this transcript was not detected in any other of the 15 organs examined. Because no EST clones containing the 5′ part of this gene were identified in public databases, we searched the genomic sequence encompassing the gene in human genome sequence and obtained a sequence (GenBank accession no. BC025392) that had been assigned to chromosomal band 17p13.1. Using GENSCAN and the Gene Recognition and Assembly Internet Link (GRAIL) program, with the genomic sequences, candidate exon sequences were predicted. Additional exon connection experiments as well as 5′ RACE determined the sequence of the 5′ part of the transcript, and obtained an assembled cDNA sequence of 2152 nucleotides that contained an open reading frame encoding a 620-amino acid protein (GenBank accession no. AB065281). The gene consisted of 14 exons spanning approximately 60 kb on the chromosomal band 17p13.1. The Simple Modular Architecture Research Tool (SMART) program suggested that the predicted protein contained 11 WD40-repeat domains. Thus, we termed the gene as WDRPUH (WD40-repeat protein upregulated in HCC). A homology search with WDRPUH amino acid sequence in public databases identified several homologous proteins, including a mouse protein (accession no. NP_082239), a rat protein (accession no. XP_220582), and a hypothetical Macaca fascicularis protein (accession no. BAB63111), which shared 88%, 89%, and 84% identity with WDRPUH, respectively. These homologous proteins were unnamed, hypothetical, or WD-repeat proteins, and their functions remained to be clarified.
Subcellular Localization of WDRPUH
We prepared plasmids expressing Myc-tagged WDRPUH and transiently transfected them into SNU475 and HepG2 hepatoma cells, and Tera-1 testicular tumor cells. Immunoblot analysis using extracts from HEK293 cells transfected with the plasmid showed a 70-kDa band corresponding to the tagged WDRPUH protein (Figure 2A). Immunocytochemical staining of the SNU475, HepG2, and Tera-1 cells revealed that the tagged WDRPUH protein was present in their cytoplasms by fluorescent microscopy (Figure 2B). The PSORT II program consistently predicted the subcellular localization of WDRPUH protein in the cytoplasm with 69.6% probability, and did not predict any nuclear localization signals (data not shown).
Figure 2.
Subcellular localization of WDRPUH. (A) Western blot analysis of exogenously expressed Myc-tagged WDRPUH protein in HEK293 cells. Empty pcDNA3.1myc/His (mock) vector was used as a negative control. (B) Myc-tagged WDRPUH protein was expressed exogenously into SNU475 and HepG2 hepatoma cells and in Tera-1 testicular cancer cells, and stained with anti-Myc monoclonal antibody. The protein was visualized by FITC-conjugated secondary anti-mouse IgG antibody (left panel). Nuclei were counterstained with DAPI (middle panel). Merged image of FITC and DAPI (right panel).
Growth-Promoting Effect of WDRPUH
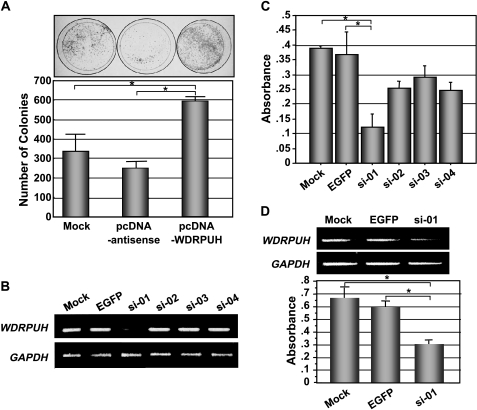
To investigate the effect of WDRPUH on cell growth, we carried out a colony formation assay by transfecting NIH3T3 cells with a plasmid-expressing WDRPUH (pcDNA-WDRPUH). When compared to cells transfected with a mock plasmid (pcDNA) or those with control plasmid expressing the complementary strand of WDRPUH (pcDNA-antisense), transfection with pcDNA-WDRPUH markedly increased the number of colonies in NIH3T3 cells (Figure 3A), suggesting that the elevated expression of WDRPUH may promote the growth of cells. This observation was confirmed by three independent experiments.
Figure 3.
Growth-promoting effect of WDRPUH. (A) Colony formation assay of WDRPUH in NIH3T3 cells. Cells transfected with pcDNA-WDRPUH, pcDNA-WDRPUH-antisense strand, or pcDNA (mock) were cultured with an appropriate concentration of G418 and stained with Giemsa solution. The number of colonies was counted by electric densitometry in a triplicate experiment of colony formation assay. *A significant difference (P < .05) determined by Student's t test. (B) Gene knockdown effect of plasmids expressing four different WDRPUH-siRNA or EGFP-siRNA, and mock plasmid. Semiquantitative RT-PCR was carried out to examine the expression of WDRPUH, 48 hours after the transfection of the plasmids in HepG2 cells. (C) Effect of WDRPUH-siRNA on the growth of HepG2 cells. Viability of HepG2 cells transfected with WDRPUH-siRNA was analyzed in triplicate using MTT assay at day 7 after the transfection and subsequent maintenance of cells with an appropriate concentration of G418. (D) Effect of WDRPUH-siRNA on the growth of Alexander cells was analyzed as described in (C). *A significant difference (P < .05) determined by Student's t test.
Effect of WDRPUH Suppression on the Viability of Cancer Cells
To test whether elevated expression of WDRPUH may play a crucial role in the proliferation of HCC cells, we prepared plasmids expressing four kinds of siRNA to WDRPUH together with a neomycin-resistant gene, and transfected them into HepG2 cells that expressed WDRPUH abundantly (Figure W1). As shown in Figure 3B, plasmid-expressing si-01 most effectively suppressed the expression of WDRPUH compared with that of mock (psiU6BX) and EGFP (psiU6BX-EGFP) controls, whereas other WDRPUH siRNAs showed no or mild gene-silencing effect. Subsequently, to test whether suppression of WDRPUH may result in growth retardation and/or cell death, we transfected HepG2 cells with these plasmids and examined the growth of transfected cells cultured in media containing an appropriate concentration of G418, a derivative of neomycin, by MTT assay. Interestingly, transfection of si-01 into HepG2 cells reduced their viability when compared with cells transfected with mock or EGFP (Figure 3C). However, si-02, si-03, and si-04 showed a mild decrease in the number of surviving cells compared with cells transfected with si-01 (Figure 3C), indicating that suppressed expression of WDRPUH associates with a decrease in cell viability, either by reduction of cell growth or induction of apoptosis. We also investigated the effect of si-01 in Alexander HCC cells, which revealed similar results (Figure 3D). These data suggest that WDRPUH may play an essential role in the growth or survival of HCC cells.
Identification of WDRPUH Interacting Proteins by Immunoprecipitation Mass Spectrometry (IP-MS)
To investigate the biologic function of WDRPUH, we searched its interacting protein(s) by IP-MS. We transfected HEK293 cells, with plasmids expressing FLAG-tagged WDRPUH, empty vector (mock), or plasmid designed to express an unrelated FLAG-tagged protein. After the incubation of cells, we then immunoprecipitated the proteins using an anti-FLAG antibody conjugated with agarose beads. Comparison of the bands on SDS-PAGE among the immunoprecipitants from these cells identified three bands corresponding to proteins of 70, 58, and >250 kDa, which were specifically immunoprecipitated in cells transfected with p3XFLAG-WDRPUH. These bands were excised and subsequently analyzed by MALDI mass spectrometry. The 70-kDa protein was found to be HSP70, the 58-kDa protein was identified as the CCT1-δ, a subunit of the CCT1 protein complex, and the >250-kDa protein was disclosed to be BRCA2 (data not shown). Using anti-FLAG antibody, we immunoprecipitated extracts from cells transfected with p3XFLAG-WDRPUH or mock vector, and confirmed that HSP70 was coimmunoprecipitated from cells with p3XFLAG-WDRPUH, but not from those with mock vector (Figure 4A). Conversely, immunoprecipitaion with anti-HSP70 antibody coimmunoprecipitated FLAG-tagged WDRPUH from cells transfected with p3XFLAG-WDRPUH, but not those with mock (Figure 4B). These results indicated the association between WDRPUH and HSP70. Furthermore, we examined the interaction between exogenous WDRPUH and endogenous CCT1-δ in HEK293 cells. After immunoprecipitation of FLAG-tagged WDRPUH with anti-FLAG antibody, we performed immunoblotting with CCT1-δ antibody and identified a band of 58 kDa confirming their association (Figure 4C). In addition, we detected the association of WDRPUH with CCT1-α subunit by Co-IP, showing that WDRPUH associates with both α and δ subunits of CCT complex proteins (Figure 4D). We also confirmed the association between WDRPUH and BRCA2 in HEK293 cells (Figure 4E), as well as HeLa cells (data not shown). In addition, immunoprecipitation with anti-BRCA2 antibody coimmunoprecipitated FLAG-tagged WDRPUH from cells transfected with p3XFLAG-WDRPUH, but not those with mock vector (Figure 4F). However, because Hek293 cells do not express endogenous WDRPUH, tissue-specific differences may exist in binding partners in normal testes and hepatomas.
Figure 4.
Interaction of WDRPUH with HSP70, CCT1-δ, CCT1-α, or BRCA2. HEK293 cells were transiently transfected with p3xFLAG-WDRPUH or p3xFLAG empty vector (Mock). Cell lysates were immunoprecipitated with anti-FLAG M2 antibody (mouse) conjugated with agarose beads or specific antibodies to interacting protein. (A and B) Coimmunoprecipitation of FLAG-WDRPUH and endogenous HSP70. Whole cell extracts from the cells exogenously expressing FLAG-WDRPUH were used as positive controls. (C) Coimmunoprecipitation of FLAG-WDRPUH and endogenous CCT1-δ. (D) Coimmunoprecipitation of FLAG-WDRPUH and endogenous CCT1-α. (E and F) Coimmunoprecipitation of FLAG-WDRPUH and endogenous BRCA2.
Discussion
In the study presented here, we characterized a novel gene, WDRPUH, which was frequently upregulated in the majority of HCCs, and demonstrated its possible involvement in cancer proliferation. Moreover, we revealed that WDRPUH protein interacts with HSP70 and the CCT chaperonin complex proteins as well as the tumor-suppressor protein BRCA2.
WDRPUH encodes a putative 620-amino acid protein with 11 highly conserved WD40-repeat domains. Structural analysis has clarified that WD-repeat proteins form a propeller-like structure with several blades that is composed of a four-stranded antiparallel β-sheet. This β-propeller-like structure serves as a platform to which proteins can bind either stably or reversibly [22]. The number of blades in the β-propeller-like structure varies depending on the number of WD-repeats that are structural elements of the β-propeller. Therefore, WDRPUH should form a propeller-like structure with 11 blades responsible for its interaction with other proteins. Among other WD-repeat proteins, endonuclein containing five WD-repeat domains was also shown to be upregulated in pancreatic cancer [23]. Although immunohistochemical staining of endonuclein revealed that its subcellular localization was in the nucleus as well as in the endoplasmic reticulum, the functions of endonuclein have not yet been elucidated. Another WD-repeat protein, beta-transducin repeat-containing protein (BTRC), contains an F-box and seven WD-repeats, and functions as a component of SCF (SKIP1, CUL1, and F-protein) complex that degrades IkappaBalpha and β-catenin [24]. A recent report revealed that Btrc null mice showed hypoplasia of mammary glands, and that transgenic mice expressing human BTRC in mammary gland conversely displayed increased ductal branching, leading to the development of mammary tumors [25]. Similarly, accumulated WDRPUH may play a role in development of tumors through unidentified mechanism(s).
In normal tissues examined, WDRPUH is highly expressed in the testis, but not in other normal organs examined, which suggests that WDRPUH may be involved in spermatogenesis. Alternatively, WDRPUH may play a role of maintenance of progenitor properties of spermatocytes because cancers are thought to include some characteristics of progenitor cells [26]. These views may help future studies to reveal the physiological function of WDRPUH in the testis. In addition, because its expression is enhanced in HCC tissues, WDRPUH may be a novel cancer-testis antigen. Therefore, immunotherapies targeting WDRPUH may be a novel therapeutic option.
A number of proteins containing WD40-repeats bind to CCT complex. A proteomic analysis demonstrated that among the 21 CCT-interacting proteins in yeast cells, 16 proteins commonly possessed WD40 motifs [27]. CCT is a molecular chaperone with a double toroidal structure that contains a central cavity in which protein folding and the achievement of the correct conformation of newly translated proteins are assisted [28]. For example, Camasses et al. [29] have elegantly shown that the central cavity of CCT prevents nonnative interactions of the nascent WD-repeats of Cdc20, which in turn facilitates its native association with the WD-repeats. Because WDRPUH also interacts with CCT through its α and δ subunits, the functional structure of WDPURH protein and/or its stability may be achieved and maintained by CCT. It may be possible that CCT also exerts a different role from the maintenance of WDRPUH protein structure because it also regulates the activity of WD-repeat-containing proteins by controlling their liberation into the cytosol or binding to other proteins in assembly, such as with the Von Hippel-Lindal (VHL) protein elongin BC (VBC) tumor-suppressor complex formation [30]. The VHL-VBC complex was also shown to interact with both CCT and HSP70 on expression in mammalian cells [31]. Consistent with reports that CCT interacts with HSP70 either directly [28] or through interaction with the HSP70/HSP90 organizing protein [31], we further identified HSP70 as another WDRPUH-interacting protein but failed to detect association with HSP90 (data not shown). Because a CCT-HSP70 complex was shown to mediate the folding of some newly synthesized proteins [32,33], these chaperones may cooperate to stabilize the conformation of WDRPUH and keep the high level of WDRPUH in HCC cells. Future studies will be required to clarify the role of these chaperones in the function of WDRPUH.
Our experiments revealed an association of WDRPUH with BRCA2. BRCA2 plays an important role in DNA repair by direct binding through its BRC repeats to the DNA repair protein, RAD51 [34,35]. In addition, a protein complex of BRCA2 and BRAF35 was suggested to play a significant role in the regulation of cell cycle progression [36]. Disruption of associations of BRCA2 with BRCA2-binding partners could compromise the DNA repair functions of RADS1, or alter the functions of other binding partners [37]. Combining the facts that WDRPUH was transactivated in HCC cells and interacted with BRCA2, we hypothesize that elevated expression of WDRPUH may abrogate transcriptional modulation, DNA repair, or control of cell cycle progression through the interference of the binding between BRCA2 and its interacting proteins including RAD51, BRAF35, or others. Because WDRPUH is localized in the cytoplasm, its interaction to BRCA2 may impair the translocation of BRCA2 into the nucleus. Therefore, it is interesting to test whether growth promotion by WDRPUH in HCC cells depends on the interaction between WDRPUH and BRCA2.
In this report, we revealed that the expression of WDRPUH was enhanced in the majority of HCC tissues. Because it is very low or absent among all the vital organs and its elevated expression was associated with growth of HCC cells, suppression of WDRPUH may be an optional therapeutic strategy for treating primary HCCs. Because it is evidenced that WDRPUH interacts with HSP70 and CCT as well as BRCA2, the impairment of these interactions implicates a possible strategy to inhibit the function of WDRPUH. Although additional functional analyses of WDRPUH are required, the findings provided here should contribute to a novel insight in hepatocarcinogenesis and to the development of novel therapeutic approaches to HCC.
Acknowledgements
We appreciate the technical assistance of Yuka Yamane and the contributions of Toyomasa Katagiri in the fabrication of cDNA microarray.
Footnotes
This work was supported, in part, by a Research for the Future Program grant (no. 00L01402) from the Japan Society for the Promotion of Science.
This article refers to supplementary material, which is designated by “W” (ie, Table W1, Figure W1) and is available online at www.bcdecker.com.
References
- 1.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Bruix J. Early diagnosis and treatment of hepatocellular carcinoma. Bailliere's Best Pract Res Clin Gastroenterol. 2000;14:991–1008. doi: 10.1053/bega.2000.0143. [DOI] [PubMed] [Google Scholar]
- 3.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 4.Kawakita N, Seki S, Sakaguchi H, Yanai A, Kuroki T, Mizoguchi Y, Kobayashi K, Monna T. Analysis of proliferating hepatocytes using a monoclonal antibody against proliferating cell nuclear antigen/cyclin in embedded tissues from various liver diseases fixed in formaldehyde. Am J Pathol. 1992;140:513–520. [PMC free article] [PubMed] [Google Scholar]
- 5.Grisham JW. The Molecular Basis of Human Cancer. Totowa: Humana Press; 2001. Molecular genetic alterations in primary hepatocellular neoplasms: hepatocellular adenoma, hepatocellular carcinoma, and hepatoblastoma; pp. 269–346. [Google Scholar]
- 6.Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–2604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 7.Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, Yamaoka Y, Nakamura Y. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–3048. [PubMed] [Google Scholar]
- 8.Tanaka S, Toh Y, Adachi E, Matsumata T, Mori R, Sugimachi K. Tumor progression in hepatocellular carcinoma may be mediated by p53 mutation. Cancer Res. 1993;53:2884–2887. [PubMed] [Google Scholar]
- 9.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto A, Demetrick DJ, Spillare EA, Hagiwara K, Hussain SP, Bennett WP, Forrester K, Gerwin B, Serrano M, Beach DH, Harris CC. Mutation and altered expression of pI6INK4 in human cancer. Proc Natl Acad Sci USA. 1994;91:11045–11049. doi: 10.1073/pnas.91.23.11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y, Nakamura Y. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- 12.Bjorn SP, Soltyk A, Beggs JD, Friesen JD. PRP4 (RNA4) from Saccharomyces cerevisiae: its gene product is associated with the U4/U6 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1989;9:3698–3709. doi: 10.1128/mcb.9.9.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaisman N, Tsouladze A, Robzyk K, Ben-Yehuda S, Kupiec M, Kassir Y. The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol Gen Genet. 1995;247:123–136. doi: 10.1007/BF00705642. [DOI] [PubMed] [Google Scholar]
- 14.Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 16.Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen RH, Miettinen PJ, Maruoka EM, Choy L, Derynck R. A WD-domain protein that is associated with and phosphorylated by the type II TGF-beta receptor. Nature. 1995;377:548–552. doi: 10.1038/377548a0. [DOI] [PubMed] [Google Scholar]
- 18.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 19.Duronio RJ, Gordon JI, Boguski MS. Comparative analysis of the beta transducin family with identification of several new members including PWP1, a nonessential gene of Saccharomyces cerevisiae that is divergently transcribed from NMT1. Proteins. 1992;13:41–56. doi: 10.1002/prot.340130105. [DOI] [PubMed] [Google Scholar]
- 20.Shimokawa T, Furukawa Y, Sakai M, Li M, Miwa N, Lin YM, Nakamura Y. Involvement of the FGF18 gene in colorectal carcinogenesis, as a novel downstream target of the beta-catenin/T-cell factor complex. Cancer Res. 2003;63:6116–6120. [PubMed] [Google Scholar]
- 21.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 22.Dell EJ, Connor J, Chen S, Stebbins EG, Skiba NP, Mochly-Rosen D, Hamm HE. The betagamma subunit of heterotrimeric G proteins interacts with RACK1 and two other WD repeat proteins. J Biol Chem. 2002;277:49888–49895. doi: 10.1074/jbc.M202755200. [DOI] [PubMed] [Google Scholar]
- 23.Honore B, Baandrup U, Nielsen S, Vorum H. Endonuclein is a cell cycle regulated WD-repeat protein that is up-regulated in adenocarcinoma of the pancreas. Oncogene. 2002;21:1123–1129. doi: 10.1038/sj.onc.1205186. [DOI] [PubMed] [Google Scholar]
- 24.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, Yamasaki L, Pagano M. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:84–94. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 27.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 28.Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 29.Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 30.Hansen WJ, Ohh M, Moslehi J, Kondo K, Kaelin WG, Welch WJ. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol Cell Biol. 2002;22:1947–1960. doi: 10.1128/MCB.22.6.1947-1960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gebauer M, Melki R, Gehring U. The chaperone cofactor Hop/p60 interacts with the cytosolic chaperonin-containing TCP-1 and affects its nucleotide exchange and protein folding activities. J Biol Chem. 1998;273:2975–2980. doi: 10.1074/jbc.273.45.29475. [DOI] [PubMed] [Google Scholar]
- 32.Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 33.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 34.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 35.Kerr P, Ashworth A. New complexities for BRCA1 and BRCA2. Curr Biol. 2001;11:R668–R676. doi: 10.1016/s0960-9822(01)00389-x. [DOI] [PubMed] [Google Scholar]
- 36.Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA, Yen TJ, Shiekhattar R. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 37.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]