Abstract
Tumor cells and tumor-associated endothelial cells express activated epidermal growth factor receptor (EGFR) due to production of EGF-related ligands in the tumor microenvironment. To investigate the effect of perpetual EGFR activation on endothelial cells, we developed a novel method to generate constitutively active EGFR. We fused the entire intracellular domain of the EGFR to the N-terminus of the CD3ζ component of the T-cell receptor signaling complex. Expression of the chimeric receptor CD3-EGFR in EGFR-deficient human embryonic kidney cells resulted in ligand-independent sustained EGFR phosphorylation and in the induction of Akt, mitogen-activated protein kinase, and signal transducer and activator of transcription 3 (Stat3). Next, CD3-EGFR, was stably expressed in murine brain endothelial cells where it signaled for the initiation of angiogenic programs, Stat3 activation, and continuous proliferation. A comparison between brain endothelial cells encoding CD3ζ and CD3-EGFR revealed that proangiogenic phenotype was modulated by the intracellular effector Stat3 and that suppression of this downstream target with the EGFR tyrosine kinase inhibitor PKI166 could revert this phenotype. Thus, our results validate the use of chimeric constitutively active receptors to replicate critical features observed in pathophysiological processes that can expedite the identification of novel therapeutic agents targeting EGFR activation and function.
Keywords: EGFR, Stat3, angiogenesis, tumor endothelial cell, chimeric receptor
Introduction
Epidermal growth factor receptor (EGFR) is a member of the erbB superfamily, which shares structurally related receptor tyrosine kinases [1]. EGFR comprises a ligand-binding extracellular domain, a transmembrane domain containing a single hydrophobic anchor sequence, and a cytoplasmic region containing the kinase domain and distinctive C-terminal tyrosine residues [2]. The receptor forms homodimers and heterodimers as a result of ligand-induced alterations in conformation and activation. On activation, the dimerized receptor transphosphorylates its C-terminal tyrosine residues, which leads to the recruitment of SH2 domain-containing molecules to the plasma membrane. In turn, these molecules activate several downstream effectors, including mitogen-activated protein (MAP) kinase, phosphatidylinositol 3-kinase/Akt, and signal transducer and activator of transcription (STAT) [3–5]. EGFR is activated in response to various physiological and pathologic stimuli [6], and overexpression or mutation of EGFR is a hallmark of a variety of tumors, in which it transduces signals that promote cell proliferation, invasion, angiogenesis, and, ultimately, metastasis [7,8]. Consequently, much effort has been directed toward developing agents that target EGFR activation and function.
An expanding body of evidence indicates that tumor cell expression of transforming growth factor alpha (TGF-α), a primary ligand for EGFR, correlates with a more aggressive phenotype [9–11]. In addition to enhancing cell growth, TGF-α may also provide a survival advantage for malignant cells by signaling for the expression of antiapoptotic proteins (e.g., Bcl-xL and nuclear factor kappa B) [12,13]. Recent evidence suggests that tumor-secreted TGF-α may act in a paracrine manner to convey information to blood vessels that perfuse malignant tissues. Molecular profiles constructed on tumor endothelial cells indicate that they differ from endothelial cells residing in normal tissues [14], and that one distinguishing feature of tumor endothelial cells is their tendency to express EGFR [15]. In fact, examination of tumor vasculature in xenograft models [16] and of spontaneously arising tumors in man [17] has suggested that EGFR is activated in the tumor endothelium. The importance of EGFR signaling on tumor blood vessels to malignant progression is exemplified by a growing number of preclinical reports demonstrating that pharmacologic suppression of EGFR activation on tumor-associated endothelial cells significantly impedes the growth of primary tumors and, more importantly, reduces the frequency of metastasis [17–20]. Collectively, these data suggest that EGFR expression is restricted to endothelial cells in pathologic tissues and that EGFR activation is critical to the growth and dissemination of tumors. However, a detailed study of the molecular and cellular role of EGFR signaling on tumor endothelial cells has been problematic because no tumor endothelial cell lines are available for study and because replicating the tumor microenvironment in cell culture is not feasible.
Previously, we reported the establishment of a broad panel of organ-specific endothelial cell lines with which to study angiogenesis and metastasis [21]. Each of these cell lines expresses a temperature-sensitive SV40 large T antigen, thereby allowing regulation of the rate of cell division and differentiation by adjusting environmental conditions. More recently, we demonstrated that these lines can be used to define the molecular basis of therapeutic intervention [22]. Because EGFR expression is characteristic of tumor endothelium and because EGFR activation may enhance the growth and spread of neoplasms, we wanted to generate an endothelial cell line that constitutively expresses activated EGFR. In addition to replicating a key component in tumor vasculature, such a line would provide novel information on intracellular signaling pathways that may lead to the identification of new targets for anticancer therapy. Herein, we describe our unique approach to generating cells that express constitutively active EGFR and we report the consequences of persistent EGFR activation in brain endothelial cells. Stable introduction of the ligand-independent CD3-EGFR chimeric receptor into brain endothelial cells initiated the activation of programmed angiogenic responses and signals for continuous proliferation, both of which were modulated by the intracellular effector Stat3. The newly acquired phenotype of brain endothelial cells was susceptible to the small-molecule tyrosine kinase inhibitor PKI166. These results validated our tumor endothelial cell model. Our approach to constructing a constitutively active chimeric receptor might be broadly applied to defining downstream effects of other diseases and developing molecular targeted therapies.
Materials and Methods
Cell Cultures and Antibodies
Human embryonic kidney (HEK) cells (American Type Culture Collection, Manassas, VA) and SB2 cells (human primary melanoma cells; generously provided by Dr. Menashe Bar-Eli, M. D. Anderson Cancer Center, Houston, TX) were grown in Dulbecco's minimum essential medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. Murine brain endothelial cells were isolated from H-2Kb-tsA58 mice (Charles Rivers Laboratories, Wilmington, MA) as previously described [21]. Antibodies directed against the phosphorylated forms of EGFR, MAP kinase, Akt, and Stat3 were purchased from Cell Signaling Technology (Beverly, MA), and those recognizing the intracellular domain of EGFR (sc-03) and CD3ζ (sc-20919) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Reagents
PKI166 (4-phenethylamino-6-[hydroxy]phenyl-7H-pyrrolo[2.3-d]-pyrimidine), an EGFR tyrosine kinase inhibitor, was synthesized and provided by Novartis Pharma (Basel, Switzerland) [23]. Human recombinant epidermal growth factor was obtained from Invitrogen (Carlsbad, CA).
Expression Plasmids and Recombinant Lentiviruses
The full length of the human T-cell receptor signaling complex CD3ζ fragment, which was derived from a human lymph node cDNA library (Edge Biosystems, Gaithersburg, MD), was obtained by high-fidelity polymerase chain reaction using Pfu DNA polymerase (Stratagene, La Jolla, CA). The amplified fragment was constructed in the pLCMV6.1 lentiviral vector in which the human cytomegalovirus (CMV) promoter drives the expression of the CD3ζ gene. pLCMV6.1 is a modified version of the lentivirus vector pLL3.7 [24], and a fragment containing the U6 promoter and enhanced green fluorescence protein expression cassette in the pLL3.7 vector was replaced by the CMV promoter and a multiple cloning site. The CD3-EGFR chimeric receptor was constructed by fusing the fragment containing an N-terminus of CD3ζ (amino acids 1–70) with the entire intracellular domain of human EGFR derived from a pCHC/EGFR expression plasmid. The fusion fragment of CD3-EGFR was then constructed in the pLCMV6.1 vector.
Recombinant lentivirus was generated by transient transfection of the lentiviral vector together with packaging plasmids consisting of expression plasmids for Gag-Pol, Rev, and VSV-G (Invitrogen). Virus-containing supernatants were collected 48 hours after transfection and introduced into target cells according to the manufacturer's instructions.
Transfection and Immunoblot Analysis
HEK cells were plated into individual wells of six-well plates at a density 1 x 106 cells/well. After 24 hours of incubation, the cells were transiently transfected with expression plasmids using PolyFect reagent (Qiagen,Valencia, CA). Twenty-four hours later, the cells were lysed with buffer [20 mM Tris-HCl (pH 8.0), 137 mM NaCl, 10% glycerol, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 mM sodium orthovanadate, 20 µM leupeptin, and 0.15 U/ml aprotinin], and 20 µl of total protein was resolved in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblot analysis as previously described [25].
Cell Proliferation Assay, Soft Agar Assay, and Fluorescence Imaging Analysis
Methods for assessing the in vitro proliferation of cells, for examining the anchorage-independent growth of cells in soft agar, and for visualizing the activation status of intracellular signaling molecules were carried out as described previously [25].
Cell Migration, Invasion, and Production of Matrix Metalloproteinase (MMP) 9
To determine the migration of brain endothelial cells expressing either CD3ζ or CD3-EGFR in response to an angiogenic stimulus, we placed 10 ng/ml basic fibroblast growth factor (bFGF; R&D Systems, Minneapolis, MN) in 500 µl of DMEM containing 2.5% fetal bovine serum into the lower chambers of 24-well plates. Sterile 8-µm migration inserts (BD Biosciences, Bedford, MA) were prewetted with serum-free medium for 30 minutes, and then the medium was aspirated. The inserts were placed onto 24-well plates; 4.0 x 105 brain endothelial cells expressing CD3ζ or CD3-EGFR were resuspended in 1 ml of DMEM containing 2.5% fetal bovine serum; and 100 µl of cell-containing suspension was added to the upper compartment of individual inserts. The assay was terminated after 6 hours, and the inserts were fixed and processed for cell counting. The number of migrating cells in four high-power (x200) fields was recorded. Invasion assays were performed by coating 8-µm inserts with Matrigel matrix (5 ng/filter; BD Biosciences) and placing them into wells containing 500 µl of serum-free medium. Cells expressing CD3ζ or CD3-EGFR were resuspended in serum-free medium at a density of 5.0 x 105 cells/ml, and 100 µl of this suspension was added to the upper compartment. After 24 hours, the inserts were processed, and the numbers of invading cells were recorded as described above. Levels of collagenase activity were determined as previously described [26].
Results
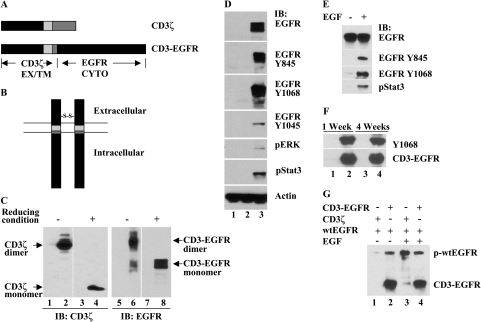
To generate a constitutively active EGFR, we created an inherently dimerized chimeric receptor CD3-EGFR by genetic reconstruction of wild-type EGFR (wtEGFR) (Figure 1A). The cytoplasmic domain of wtEGFR was fused with the extracellular and transmembrane domains derived from CD3ζ by exploiting a unique feature of the CD3ζ chain that allows it to form homodimers through its extracellular disulfide bond [27]. This protein was targeted to the cell membrane, where it spontaneously homodimerized due to inherent dimerization (Figure 1B). To validate this dimerization, we transiently expressed CD3-EGFR in HEK cells, which are deficient in EGFR. As shown in Figure 1C, the full length of CD3ζ was detected as a 16-kDa monomeric protein under reducing electrophoretic conditions and as a 32-kDa dimeric protein under nonreducing conditions. Likewise, the monomeric form of CD3-EGFR was observed as a 75-kDa protein under reducing conditions and as a 150-kDa homodimer in nonreducing conditions. Both the monomeric and dimeric forms of CD3-EGFR were detected with an antibody recognizing the intracellular domain of EGFR. Collectively, these results demonstrate that the CD3-EGFR chimera expressed in mammalian cells forms a homodimer independently of ligand.
Figure 1.
CD3-EGFR is a chimeric receptor containing a constitutively active EGFR tyrosine kinase. (A) Construction of the CD3-EGFR chimeric receptor. A fragment containing the extracellular and transmembrane (EX/TM) domains of CD3ζ was fused with the cytoplasmic (CYTO) domain of EGFR. (B) Schematic depiction of the CD3-EGFR chimeric receptor. The predicted structure of CD3-EGFR is that of a membrane-bound dimer resulting from inherent homodimerization through the extracellular CD3ζ disulfide bond. (C) Evaluation of dimerized CD3-EGFR in transiently transfected HEK cells using immunoblot analysis. Lanes 1, 3, 5, and 7 contain protein lysates from vector-transfected HEK cells, whereas lanes 2 and 4 show dimeric and monomeric CD3ζ proteins, respectively. The symbols (-) and (+) denote nonreducing and reducing electrophoretic conditions, respectively. Lanes 6 and 8 show the dimeric and monomeric forms of CD3-EGFR, respectively, which were detected by an antibody directed against the intracellular domain of EGFR. (D) Examination of the tyrosine kinase activity of CD3-EGFR. Cellular protein extracts from HEK cells transfected with vector (lane 1), CD3ζ (lane 2), or CD3-EGFR (lane 3) were analyzed with the indicated antibodies. Actin was used as a loading control. (E) Activation of wtEGFR by EGF. wtEGFR was transfected into HEK cells, and the basal level of EGFR phosphorylation was compared with that of cells stimulated with 40 ng/ml EGF for 15 min. (F) SB2 cells were stably transduced with lentivirus encoding CD3ζ (lanes 1 and 3) or CD3-EGFR (lanes 2 and 4). The total protein extracts prepared from the cells 1 or 4 weeks after transduction were analyzed for phosphorylated CD3-EGFR (EGFR Y1068) and total CD3-EGFR protein. (G) wtEGFR was cotransfected with CD3ζ (lanes 1 and 3) or CD3-EGFR (lanes 2 and 4) into HEK cells. Cells were harvested for evaluation of phosphorylated wtEGFR and CD3-EGFR 24 h after transfection.
To determine the activity of CD3-EGFR, we mapped phosphorylation patterns of the chimeric receptor on distinct cytoplasmic tyrosine residues in transiently transfected HEK cells. In the absence of EGF, ectopic expression of CD3-EGFR resulted in profound tyrosine phosphorylation of the receptor predominantly at Y845 and Y1068 (Figure 1D). Phosphorylation was also evident on Y1045, albeit to a lesser extent. Y845 is the tyrosine residue phosphorylated by activated c-Src, and Y1068 is a prominent autophosphorylation site. Both tyrosine sites are essential for activating downstream Stat3 in EGFR signaling [28]. The strong tyrosine phosphorylation at these two residues indicated that the expression of CD3-EGFR in cells induced the activation of c-Src and Stat3. By comparison, transient transfection of wtEGFR in HEK cells was associated with a low basal level of tyrosine phosphorylation that was significantly enhanced by stimulating the cells with EGF (Figure 1E).
To determine the integrity of kinase activity in CD3-EGFR-expressing cells, we stably transduced human melanoma SB2 cells (which are endogenously EGFR-deficient cells) by using a recombinant lentivirus encoding CD3ζ or CD3-EGFR and monitored the resulting activity. The kinase activity of CD3-EGFR was sustained for at least 4 weeks without any notable decline (Figure 1F). To test the possibility that introduction of CD3-EGFR stimulates the activation of wtEGFR, we cotransfected the chimera and wtEGFR into HEK cells. In the absence of ligand, CD3-EGFR overexpression induced only a slight increase in wtEGFR phosphorylation; in the presence of EGF, wtEGFR phosphorylation was unchanged (Figure 1G). Collectively, these data suggest that CD3-EGFR exhibits constitutive kinase activity resulting from spontaneous generation of a homodimer and that this activation duplicates wtEGFR activity in the presence of ligand.
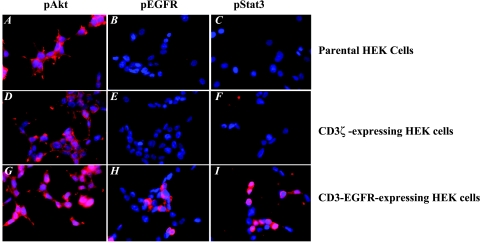
Next, we examined CD3-EGFR-expressing HEK cells to determine the activation status of the downstream effectors MAP kinase, Akt, and Stat3. As we have described, in the absence of EGF, both MAP kinase and Stat3 were constitutively activated in these cells. These activation patterns were consistent with those of EGF-treated HEK cells previously transfected with wtEGFR: in control HEK cells transduced with CD3ζ, the activity of Stat3 was at a basal level, and the MAP kinase level was below detectable limits. These results were supported by fluorescence imaging data, which demonstrated that activated CD3-EGFR anchored to the cell membrane (Figure 2, panel H) and tyrosine-phosphorylated Stat3 localized to the nuclear compartment (Figure 2, panel I). Akt was constitutively tyrosine-phosphorylated in parental HEK cells (Figure 2, panel A), and expression of CD3-EGFR enhanced Akt phosphorylation (Figure 2, panel G).
Figure 2.
Parental HEK cells and cells expressing CD3ζ or CD3-EGFR were plated onto four-chamber slides at a density of 2 x 104 cells per chamber in DMEM containing 10% fetal bovine serum. After a 24-hour incubation period, the cells were fixed in acetone and stained with antibodies recognizing phosphorylated forms of Akt (A,D, and G), EGFR (B,E, and H), and Stat3 (C,F and I).
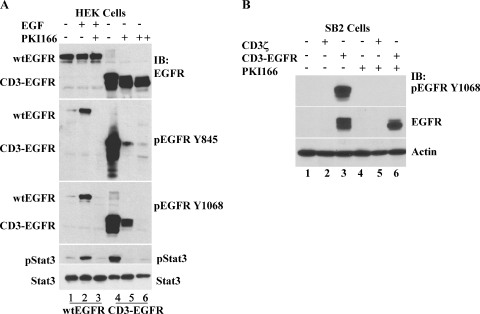
In general, small-molecule inhibitors of EGFR are designed to repress the receptor kinase activity that results from binding of the ligand. To determine whether our CD3-EGFR chimera was sensitive to such inhibition, we transfected HEK cells with wtEGFR and then treated the transfected cells with EGF alone or with PKI166. Stimulation of cells with EGF resulted in a strong tyrosine phosphorylation of both wtEGFR and Stat3 (Figure 3A, lane 2); these activities were completely suppressed by 1 µM PKI166 (Figure 3A, lane 3). Next, we introduced CD3-EGFR into HEK cells and noted that the ligand-independent phosphorylation of the chimeric receptor was also sensitive to treatment with PKI166 (Figure 3A, lane 5). The ectopic expression of CD3-EGFR was at a basal level higher than that of wtEGFR (Figure 3A, lanes 1 and 4), presumably as a result of the enhanced stability of CD3-EGFR. Thus, an increased concentration of PKI166 (5 µM) was required to completely quench the kinase activity of the chimera (Figure 3A, lane 6). We thought it interesting that CD3-EGFR still exhibited detectable activity in the presence 1 µM PKI166 even though the chimera-induced Stat3 tyrosine phosphorylation had declined to the basal level (Figure 3A, lane 5). These data suggest that Stat3 is exquisitely sensitive to minor alterations in EGFR activity. Furthermore, in SB2 cells that stably expressed CD3-EGFR, persistent kinase activity of the chimera was observed and, similar to HEK cells expressing CD3-EGFR, this activity was completely inhibited by 1 µM PKI166 (Figure 3B). These experiments demonstrate that the conformation of the kinase domain of CD3-EGFR approximates the conformation of ligand-activated wtEGFR and that both are sensitive to inhibition by PKI166.
Figure 3.
The CD3-EGFR chimeric receptor is sensitive to the EGFR tyrosine kinase inhibitor PKI166. (A) The kinase activity of HEK cells expressing wtEGFR (lanes 1–3) and CD3-EGFR (lanes 4–6) is inhibited by PKI166. The expression plasmid for wtEGFR or CD3-EGFR (1 µg) was transfected into HEK cells. wtEGFR-transfected cells were treated with EGF (40 ng/ml) alone or in combination with PKI166 (1 µM) for 60 minutes. CD3-EGFR-transfected cells were treated with 1 µM (+) or 5 µM (++) PKI166 for 60 minutes. Total cellular protein extracts were prepared to analyze expression and phosphorylation at specified tyrosine residues of wtEGFR and CD3-EGFR. Tyrosine-phosphorylated and total Stat3 protein levels are also shown. (B) Suppression of the constitutive activity of CD3-EGFR by PKI166 in human melanoma SB2 cells. Parental SB2 cells (lanes 1 and 4) and SB2 cells stably transduced with either CD3ζ (lanes 2 and 5) or CD3-EGFR (lanes 3 and 6) were used. Untreated cells (lanes 1–3) or cells treated with 1 µM PKI166 (lanes 4–6) for 60 minutes were prepared and analyzed for CD3-EGFR phosphorylated at Y1068 and total CD3-EGFR. Actin was used as a loading control.
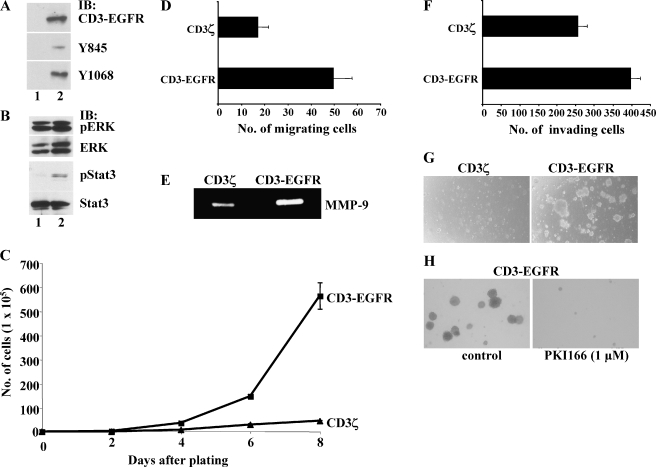
Ectopic expression of CD3-EGFR in murine brain endothelial cells signals for angiogenic program activation and continuous proliferation. Activation of EGFR in tumor-associated blood vessels may be essential for the sustained growth and metastasis of some tumors [17–20]. To determine the effect of CD3-EGFR on endothelial cell processes, we stably expressed the CD3-EGFR transgene in murine microvascular brain endothelial cells. To exclude any potential confounding effects of SV40 large T protein expression on our analyses, we cultured the cells at 37°C at least 72 hours before experimental analysis. At this time, SV40 large T antigen is no longer detectable in the cells (data not shown). As observed in CD3-EGFR-expressing HEK cells, we noted the phosphorylation of the receptor at Y845 and Y1068 in CD3-EGFR-expressing brain endothelial cells (Figure 4A). Introduction of CD3-EGFR, but not of CD3ζ, stimulated the autophosphorylation and activation of Stat3 (Figure 4B). MAP kinase, however, was activated in both CD3ζ- and CD3-EGFR-expressing cells. The persistent activity of MAP kinase did not result from the introduction of transgenes because parental brain endothelial cells also exhibited spontaneous MAP kinase activity (data not shown).
Figure 4.
Ectopic expression of CD3-EGFR in murine brain endothelial cells induces Stat3 activation, angiogenic response, and anchorage-independent growth. (A) The constitutive activity of CD3-EGFR expressed in cells. Cells were stably transduced with CD3ζ (lane 1) or CD3-EGFR (lane 2), and the cells were maintained in culture for at least 4 weeks before analysis, when total protein extracts were prepared. Total CD3-EGFR protein, the form phosphorylated at Y845, and the form phosphorylated at Y1068 are shown. (B) MAP kinase (ERK) and Stat3 expression in cells expressing CD3ζ (lane 1) or CD3-EGFR (lane 2). Protein extracts prepared from stably transduced cells were analyzed for phosphorylated ERK, total ERK, phosphorylated Stat3, and total Stat3. (C) Enhanced proliferation of cells that express CD3-EGFR. Cells were plated at a density of 2 x 105 cells in large dishes and counted at the indicated time points. (D) Enhanced migration of CD3-EGFR-expressing cells to 20 ng/ml bFGF. The number of migrating cells was counted under x200 magnification 6 hours after the addition of cells. (E) Enhanced production of MMP-9 by CD3-EGFR-expressing cells. (F) Cells expressing CD3-EGFR have enhanced invasiveness. This increased invasiveness through 8-µm inserts coated with Matrigel (5 ng/insert) correlated with increased MMP-9 production. The assay was conducted in serum-free medium and ended 24 hours after the plating of 5.0 x 104 cells. Cells were counted under x200 magnification. (G) Anchorage-independent growth of CD3-EGFR-expressing cells in soft agar (0.3%). Cell growth was evaluated 2 weeks after plating. (H) Reversion of the anchorage-independent growth phenotype of CD3-EGFR-expressing cells by 1 µM PKI166.
Because activated EGFR has been identified in the vasculature of tumors and not in the blood vessels of normal adjacent tissue [17,19], we questioned whether this tyrosine kinase might be signaling for processes involved in angiogenesis. Because endothelial cell proliferation is a prerequisite for the growth of malignant tumors, we compared the rates of cell division in brain endothelial cells that are genetically reconstructed to express CD3ζ or CD3-EGFR. Our examination revealed that the cells expressing the chimeric EGFR grew at a rate that was six-fold higher than that of CD3ζ-expressing cells (Figure 4C). Next, we sought to determine whether other endothelial cell processes necessary for the assembly of new vascular structures were also affected by continuous signaling through the EGFR. We found that the migration of CD3-EGFR-expressing cells in response to bFGF was almost three times that of CD3ζ-expressing cells (Figure 4D). The migration of endothelial cells in response to an angiogenic stimulus is known to be coupled to the synthesis of proteolytic enzymes that target the parent vessel and modify the surrounding extracellular matrix [29]. In this study, we found that the brain endothelial cells expressing CD3-EGFR released significantly more MMP-9 than did the CD3ζ-expressing cells (Figure 4E). In vivo, the regional destruction of the surrounding microenvironment is accompanied by endothelial cell invasion. We found that sustained EGFR signaling in brain endothelial cells significantly increased their invasive capacity (Figure 4F). We also noted that the CD3-EGFR-expressing cells were capable of continuous growth on soft agar,whereas those expressing only CD3ζ were not (Figure 4G). Furthermore, the proangiogenic phenotype of CD3-EGFR-expressing cells could be reverted by treating them with 1 µM PKI166 (Figure 4H). Collectively, these results suggested that persistent activation of EGFR on brain endothelial cells signals for processes necessary for neovascularization and leads to excessive proliferation. The latter finding lends credibility to the hypothesis that sustained activation of EGFR on blood vessels supplying malignant tumors may result in the generation of genetically unstable cells [30]. In fact, our phenotypic assessment of these cells is remarkably similar to that from a recent report, which determined that activated Stat3 is localized to tumor endothelial cells in human gliomas and medulloblastomas [31].
Discussion
The primary motivation for the current study was to establish a relevant cell-based system that could be used to identify EGFR-activated intracellular effectors that may serve as potential targets for therapy. To achieve this objective, we first established a new methodology to generate a chimeric constitutively active EGFR. We demonstrated that the conformation of the kinase domain of CD3-EGFR closely approximates that of ligand-activated wtEGFR and that both forms are sensitive to the protein tyrosine kinase inhibitor PKI166. We chose to introduce the chimera into brain endothelial cells to pattern the phenotype of endothelial cells found in malignant tissues. Previous studies have utilized the EGFR mutant EGFRvIII to examine the effects of ligand-independent EGFR activation [32,33]. Our decision to model constitutive wtEGFR activation was based on those studies describing differences between wtEGFR and EGFRvIII intracellular signaling cascades [34] and sensitivity to receptor tyrosine kinase inhibitors [35,36]. Moreover, we are unaware of any reports documenting EGFRvIII expression in tumor-associated endothelial cells; thus, introduction of the mutant receptor into brain endothelial cells would not accurately reproduce a key feature of tumor vasculature. Stable expression of CD3-EGFR in brain endothelial cells signaled for cell division, migration to an angiogenic stimulus, elaboration of proteolytic enzymes, and invasion—each of which is necessary for the generation of a new blood vessel. By comparing the phosphorylation patterns between the CD3ζ- and CD3-EGFR-expressing brain endothelial cells, we determined that the intracellular effector Stat3 is a central mediator that regulates the response of these cells. Our findings (that sustained activation of Stat3 is observed only in CD3-EGFR-expressing cells and that PKI166 suppressed Stat3-induced proliferation) support this determination.
STAT proteins are members of a family of transcription factors that exist in a hypophosphorylated state in the cell cytoplasm. Binding of various ligands (e.g., cytokines, hormones, and growth factors) to their cell surface receptors results in STAT activation and leads to the generation of homodimers and heterodimers that enter the nucleus and bind target gene promoters [37]. STAT proteins play a central role in a number of biologic processes, including cell development, differentiation, proliferation, and immunologic responses [38]. Recently, much attention has focused on the contribution of aberrant STAT signaling to human disease processes. In cancer, constitutive Stat3 activity appears to be prevalent: it has been identified in malignancies originating from the breast [39], lung [40], brain [41], head and neck [42], and other human tissues. Many of these tumors appear dependent on Stat3 signaling for their growth and survival; consequently, Stat3 is rapidly emerging as an important target for drug intervention [43]. However, considerably less is known about the role of Stat3 signaling in vascular endothelial cells. Our data suggest that targeting of Stat3 activation, particularly in tumors expressing TGF-α, could effectively halt neoplastic growth by affecting both tumor cells and tumor vasculature.
The approach we used might be applied more broadly to study receptor function in the context of disease processes. Indeed, the generation of constitutively active receptors in itself may be useful for performing high-throughput analyses in a cell-free system. Because it functions in a specific fashion, the chimeric receptor system avoids much of the interference and bias that may arise from the activation of secondary receptors induced by some ligands. As we have demonstrated, the introduction of continuously active receptors into relevant cells provides important information at the molecular and functional levels. We anticipate that our novel approach will be useful for advancing knowledge about different disease processes and for identifying new targets for intervention.
Acknowledgements
We thank Luk Van Parijs for the gift of the pLL3.7 lentivirus vector, Elizabeth Hess for critical editorial review, and Lola López for expert preparation of the manuscript.
Abbreviations
- EGFR
epidermal growth factor receptor
- TGF-α
transforming growth factor alpha
- HEK
human embryonic kidney
- STAT
signal transducer and activator of transcription
- DMEM
Dulbecco's minimum essential medium
- CMV
cytomegalovirus
- bFGF
basic fibroblast growth factor
- EGF
epidermal growth factor
- wtEGFR
wild-type epidermal growth factor receptor
- MMP
matrix metalloproteinase
Footnotes
This work was supported, in part, by Cancer Center Support Grant CA16672 and SPORE in Prostate Grant CA90270 from the National Cancer Institute, National Institutes of Health.
Hua Cheng and Robert R. Langley contributed equally to this work.
References
- 1.Coussens L, Yang-Feng TL, Liao YC, Chen E, Gray A, McGrath J, Seeburg PH, Libermann TA, Schlessinger J, Francke U, et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science. 1985;230:1132–1139. doi: 10.1126/science.2999974. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter G, Cohen S. Epidermal growth factor. J Biol Chem. 1990;265:7709–7712. [PubMed] [Google Scholar]
- 3.Hu P, Margolis B, Skolnik EY, Lammers R, Ullrich A, Schlessinger J. Interaction of phosphatidylinositol 3-kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Mol Cell Biol. 1992;12:981–990. doi: 10.1128/mcb.12.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wennstrom S, Downward J. Role of phosphoinositide 3-kinase in activation of ras and mitogen-activated protein kinase by epidermal growth factor. Mol Cell Biol. 1999;19:4279–4288. doi: 10.1128/mcb.19.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park OK, Schaefer TS, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci USA. 1996;93:13704–13708. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alroy I, Yarden Y. The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 7.Mendelsohn J. The epidermal growth factor receptor as a target for cancer therapy. Endocr Rev Cancer. 2001;8:3–9. doi: 10.1677/erc.0.0080003. [DOI] [PubMed] [Google Scholar]
- 8.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 9.Umekita Y, Ohi Y, Sagara Y, Yoshida H. Co-expression of epidermal growth factor receptor and transforming growth factor-alpha predicts worse prognosis in breast-cancer patients. Int J Cancer. 2000;89:484–487. doi: 10.1002/1097-0215(20001120)89:6<484::aid-ijc3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 10.De Jong KP, Stellema R, Karrenbeld A, Koudstaal J, Gouw AS, Sluiter WJ, Peeters PM, Sloof MJ, De Vries EG. Clinical relevance of transforming growth factor alpha, epidermal growth factor receptor, p53, and Ki67 in colorectal liver metastases and corresponding primary tumors. Hepatology. 1998;28:971–979. doi: 10.1002/hep.510280411. [DOI] [PubMed] [Google Scholar]
- 11.Uhlman DL, Nguyen P, Manivel JC, Zhang G, Hagen K, Fraley E, Aeppli D, Niehans GA. Epidermal growth factor receptor and transforming growth factor alpha expression in papillary and nonpapillary renal cell carcinoma: correlation with metastatic behavior and prognosis. Clin Cancer Res. 1995;8:913–920. [PubMed] [Google Scholar]
- 12.Ramljak D, Coticchia CM, Nishanian TG, Saji M, Ringel MD, Conzen SD, Dickson RB. Epidermal growth factor inhibition of c-Myc-mediated apoptosis through Akt and Erk involves Bcl-xL upregulation in mammary epithelial cells. Exp Cell Res. 2003;287:397–410. doi: 10.1016/s0014-4827(03)00135-6. [DOI] [PubMed] [Google Scholar]
- 13.Biswas DK, Cruz AP, Gansberger E, Pardee AB. Epidermal growth factor-induced nuclear factor kappa B activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci USA. 2000;97:8542–8547. doi: 10.1073/pnas.97.15.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, et al. Genes expressed in human tumor endothelium. Science. 2000;18:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 15.Hida K, Hida Y, Amin DN, Flint AF, Panigraphy D, Morton CC, Klagsbrun M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004;64:8249–8255. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- 16.Weber KL, Doucet M, Price JE, Baker C, Kim SJ, Fidler IJ. Blockade of epidermal growth factor receptor signaling leads to inhibition of renal cell carcinoma growth in the bone of nude mice. Cancer Res. 2003;63:2940–2947. [PubMed] [Google Scholar]
- 17.Yokoi K, Thaker PH, Yazici S, Rebhun RR, Nam DH, He J, Kim SJ, Abbruzzese JL, Hamilton SR, Fidler IJ. Dual inhibition of epidermal growth factor receptor and vascular endothelial growth factor receptor phosphorylation by AEE788 reduces growth and metastasis of human colon carcinoma in an orthotopic nude mouse model. Cancer Res. 2005;65:3716–3725. doi: 10.1158/0008-5472.CAN-04-3700. [DOI] [PubMed] [Google Scholar]
- 18.Yigitbasi OG, Younes MN, Doan D, Jasser SA, Schiff BA, Bucana CD, Bekele BN, Fidler IJ, Myers JN. Tumor cell and endothelial cell therapy by dual tyrosine kinase receptor blockade. Cancer Res. 2004;64:7977–7984. doi: 10.1158/0008-5472.CAN-04-1477. [DOI] [PubMed] [Google Scholar]
- 19.Kim SJ, Uehara H, Karashima T, Shepherd DL, Killion JJ, Fidler IJ. Blockade of epidermal growth factor receptor signaling in tumor cells and tumor-associated endothelial cells for therapy of androgen-independent human prostate cancer growing in the bone of nude mice. Clin Cancer Res. 2003;9:1200–1210. [PubMed] [Google Scholar]
- 20.Baker CH, Kedar D, McCarty MF, Tsan R, Weber KL, Bucana CD, Fidler IJ. Blockade of epidermal growth factor receptor signaling on tumor cells and tumor-associated endothelial cells for therapy of human carcinomas. Am J Pathol. 2002;161:929–938. doi: 10.1016/S0002-9440(10)64253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langley RR, Ramirez KM, Tsan RZ, Van Arsdall M, Nilsson MB, Fidler IJ. Tissue-specific microvascular endothelial cell lines from H-2Kb-tsA58 mice for studies of angiogenesis and metastasis. Cancer Res. 2003;63:2971–2976. [PubMed] [Google Scholar]
- 22.Langley RR, Fan D, Tsan RZ, Rebhun R, He J, Kim SJ, Fidler IJ. Activation of the platelet-derived growth factor-receptor enhances survival of murine bone endothelial cells. Cancer Res. 2004;64:3727–3730. doi: 10.1158/0008-5472.CAN-03-3863. [DOI] [PubMed] [Google Scholar]
- 23.Traxler P, Buchdunger E, Furet P, Gschwind H-P, Ho P, Mett H, O'Reilly T, Pfaar U, Thomas H. Preclinical profile of PKI166: a novel and potent EGF-R tyrosine kinase inhibitor for clinical development. Clin Cancer Res. 1999;5:3750s. [Google Scholar]
- 24.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, Cenciarelli C, Nelkin G, Tsan R, Fan D, Cheng-Mayer C, Fidler IJ. Molecular mechanism of hTid-1, the human homolog of Drosophila tumor suppressor l(2)Tid, in the regulation of NF-κB activity and suppression of tumor growth. Mol Cell Biol. 2005;25:44–59. doi: 10.1128/MCB.25.1.44-59.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Van Arsdall M, Tedjarati S, McCarty M, Wu W, Langley RR, Fidler IJ. Contributions of stromal metalloproteinase-9 to angiogenesis and growth of human ovarian carcinoma in mice. J Natl Cancer Inst. 2002;94:1134–1142. doi: 10.1093/jnci/94.15.1134. [DOI] [PubMed] [Google Scholar]
- 27.Gold DP, Clevers H, Alarcon B, Dunlap S, Novotny J, Williams AF, Terhorst C. Evolutionary relationship between the T3 chains of the T-cell receptor complex and the immunoglobulin supergene family. Proc Natl Acad Sci USA. 1987;84:7649–7653. doi: 10.1073/pnas.84.21.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbst RS, Fukuoka M, Baselga J. Gefitinib—a novel targeted approach to treating cancer. Nat Rev Cancer. 2004;4:956–965. doi: 10.1038/nrc1506. [DOI] [PubMed] [Google Scholar]
- 29.Stetler-Stevenson WG. Matrix metalloproteinases in angiogenesis: a moving target for therapeutic intervention. J Clin Invest. 1999;103:1237–1241. doi: 10.1172/JCI6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hida K, Klagsbrun M. A new perspective on tumor endothelial endothelial cells: unexpected chromosome and centrosome abnormalities. Cancer Res. 2005;65:2507–2510. doi: 10.1158/0008-5472.CAN-05-0002. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer LK, Ren Z, Fuller GN, Shaefer TS. Constitutive activation of Stat3alpha in brain tumors: localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2) Oncogene. 2002;27:2058–2065. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HG. Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Oncogene. 2004;23:4594–4602. doi: 10.1038/sj.onc.1207602. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Barth RF, Wu G, Ciesielski MJ, Fenstermaker RA, Moffat BA, Ross BD, Wikstrand CJ. Development of a syngeneic rat brain tumor model expressing EGFRvIII and its use for molecular targeting studies with monoclonal antibody L8A4. Clin Cancer Res. 2005;11:341–350. [PubMed] [Google Scholar]
- 34.Moscatello DK, Montgomery RB, Sundareshan P, McDanel H, Wong MY, Wong AJ. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene. 1996;13:85–96. [PubMed] [Google Scholar]
- 35.Heimberger AB, Learn CA, Archer GE, McLendon RE, Chewning TA, Tuck FL, Pracyk JB, Friedman AH, Friedman HS, Bigner DD, et al. Brain tumors in mice are susceptible to blockade of epidermal growth factor receptor (EGFR) with the oral, specific EGFR-tyrosine kinase inhibitor ZD1839 (Iressa) Clin Cancer Res. 2002;8:3496–3502. [PubMed] [Google Scholar]
- 36.Learn CA, Hartzell TL, Wikstrand CJ, Archer GE, Rich JN, Friedman AH, Friedman HS, Bigner DD, Sampson JH. Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Clin Cancer Res. 2004;10:3216–3224. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- 37.Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 38.Leonard WJ, O'Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 39.Watson CJ, Miller WR. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150–4165. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 41.Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 42.Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, Endo S, Johnson DE, Huang L, He Y, Kim JD. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci USA. 2000;97:4227–4232. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turkson J, Jove R. STAT proteins: novel molecular targetfor cancer discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]