Abstract
Genomes of prokaryotes differ significantly in size and DNA composition. Escherichia coli is considered a model organism to analyze the processes involved in bacterial genome evolution, as the species comprises numerous pathogenic and commensal variants. Pathogenic and nonpathogenic E. coli strains differ in the presence and absence of additional DNA elements contributing to specific virulence traits and also in the presence and absence of additional genetic information. To analyze the genetic diversity of pathogenic and commensal E. coli isolates, a whole-genome approach was applied. Using DNA arrays, the presence of all translatable open reading frames (ORFs) of nonpathogenic E. coli K-12 strain MG1655 was investigated in 26 E. coli isolates, including various extraintestinal and intestinal pathogenic E. coli isolates, 3 pathogenicity island deletion mutants, and commensal and laboratory strains. Additionally, the presence of virulence-associated genes of E. coli was determined using a DNA “pathoarray” developed in our laboratory. The frequency and distributional pattern of genomic variations vary widely in different E. coli strains. Up to 10% of the E. coli K-12-specific ORFs were not detectable in the genomes of the different strains. DNA sequences described for extraintestinal or intestinal pathogenic E. coli are more frequently detectable in isolates of the same origin than in other pathotypes. Several genes coding for virulence or fitness factors are also present in commensal E. coli isolates. Based on these results, the conserved E. coli core genome is estimated to consist of at least 3,100 translatable ORFs. The absence of K-12-specific ORFs was detectable in all chromosomal regions. These data demonstrate the great genome heterogeneity and genetic diversity among E. coli strains and underline the fact that both the acquisition and deletion of DNA elements are important processes involved in the evolution of prokaryotes.
Horizontal gene transfer and gene reduction represent two mechanisms contributing to the evolution of prokaryotic genomes “in quantum leaps” (20). Thus, the acquisition of plasmids and phages, as well as large DNA regions called “genomic islands,” plays an important role in the development of new species, subspecies, and pathotypes. Among others, the species Escherichia coli represents an excellent model to study evolution of prokaryotic genomes in detail (36). E. coli is an ideal example for these studies, as numerous ecotypes, adapted to the intestines of humans and various animals, exist. In addition, the species E. coli comprises various pathotypes, which act as causative agents in human as well as veterinary medicine. They can be grouped as extraintestinal pathogenic E. coli (ExPEC), causing urinary tract infections, newborn meningitis, or sepsis, and as intestinal pathogenic E. coli (IPEC), causing enteric and diarrheal diseases. The broad spectrum of pathogenic features and of different clinical symptoms caused by E. coli pathotypes mirrors the presence of different subsets of virulence-associated genes in certain pathotypes which are absent in commensal isolates (42, 45). The localization of many virulence-associated genes on mobile genetic elements, such as bacteriophages, plasmids, and pathogenicity islands (PAIs), indicates that horizontal gene transfer plays a major role in the evolution of different bacterial pathotypes (14). The genome sizes of natural E. coli isolates have been shown to be very heterogenous and may differ by as much as 1 Mb (5). This heterogeneity is thought to be the result of the deletion, as well as the acquisition, of genetic elements. The estimation that ∼18% of all open reading frames (ORFs) of the E. coli strain MG1655 were horizontally acquired and that, for the majority of them, this occurred relatively recently (36) underlines the high variability of the gene content within this species.
To characterize the genetic diversity and genome structures among different pathogenic and commensal variants of E. coli, we applied a whole-genome approach. Using DNA arrays, the genome contents of seven ExPEC (including a fecal O18:K1 strain) and eight IPEC isolates, as well as of eight commensal strains from healthy volunteers (including the laboratory strains MG1655 and B), were determined by investigation of the presence of all 4,290 translatable ORFs of the sequenced nonpathogenic E. coli K-12 strain MG1655 (6). In addition, a newly developed “E. coli pathoarray,” which currently consists of 456 probes specific for typical virulence-associated genes of ExPEC, IPEC, and Shigella, was used to assess the distribution of these genes or their homologues among the pathogenic and commensal strains used in this study. Furthermore, the genome contents of three uropathogenic E. coli (UPEC) isolates were compared with those of their derivatives which had lost large chromosomal regions called PAIs. With these approaches, we address several issues concerning the large genetic diversity and the mechanisms involved in genome optimization in E. coli. Our results underline the fact that E. coli can be used as a paradigm to analyze the evolution of bacteria by whole-genome approaches.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Twenty-six E. coli strains which belong to the Institut für Molekulare Infektionsbiologie (Würzburg, Germany) strain collection were used in this study. These strains exhibit different genome sizes and contain different additional mobile genetic elements (Table 1). The UPEC strains 536, J96, and 764; the asymptomatic bacteriuria strain 83972; the newborn meningitis-causing E. coli (MNEC) strain IHE3034, and the enterohemorrhagic E. coli (EHEC) strains 4797/97, 5714/96, 1639/77, and SF493/89, as well as E2348/69 (enteropathogenic E. coli [EPEC]), have been described elsewhere (3, 7, 25, 31, 32, 35, 37, 46, 52). The deletion mutants 536-21, J96-M1, and 764-2 have been described before (7, 8, 34). The enteroaggregative E. coli (EAEC) strain DPA065 was provided by A. Giammanco (University of Palermo, Palermo, Italy). The enterotoxigenic E. coli (ETEC) strain C9221a and the enteroinvasive E. coli (EIEC) strain EDL1284 belong to the strain collection of the Institut für Molekulare Infektionsbiologie. The UPEC strain P42 and the fecal O18:K1 isolate F54 belong to the strain collection of the Department of Microbiology, Immunology and Glycobiology, Lund University, Lund, Sweden. The E. coli K-12 strain MG1655 and E. coli strain B, as well as the six commensal fecal isolates, have been described before (6, 41, 56). All of the strains were grown in Luria-Bertani medium (51).
TABLE 1.
Strains used in this study
| Pathotype | Straina | Serotype | Chromosomal size (Mb) | ECOR groupb | Known virulence-associated genes on horizontally transferred DNA elementsc | Known horizontally transferred DNA elements |
|---|---|---|---|---|---|---|
| UPEC | 536 | O6:K15:H31 | 4.92 | B2 | sfaI, prf (papG allele III), hly (PAI I536 + PAI II536), kps (group II), fyuA, iro, malX | PAIs |
| UPEC | 536-21∗ | O6:K15:H31 | 4.69 | B2 | sfaI, kps (group II), fyuA, iro, malX | PAIs |
| UPEC | J96 | O4:K:H5 | 5.11 | B2 | foc, pap (papG allele I), prs (papG allele III), hly (PAI IJ96 + PAI IIJ96), kps (group III), fyuA, iro, cnf1, malX | PAIs |
| UPEC | J96-M1∗ | O4:K:H5 | 5.06 | B2 | foc, pap (papG allele I), hly (PAI IJ96), kps (group III), fyuA, iro, malX | PAIs |
| UPEC | 764 | O18:K5:H5/11 | 5.07 | B2 | foc, hly, fyuA, iutA, kps (group II), malX | PAIs |
| UPEC | 764-2∗ | O18:K5:H5/11 | 5.02 | B2 | foc, fyuA, iutA, kps (group II), malX | PAIs |
| UPEC | P42 | O1:K1:H7 | 5.06 | B2 | foc, pap (papG allele II), fyuA, kps (group II), malX | PAIs |
| UPEC (ABU)d | 83972 | OR:K5:H− | 4.88 | B2 | kps (group II), iutA, fyuA, malX | PAIs |
| MNEC | IHE3034 | O18:K1:H7/9 | 4.84 | B2 | sfaII, ibeA, iro, kps (group II), fyuA, malX | PAIs |
| Fecal isolate | F54 | O18:K1 | 5.02 | B1 | traT, fyuA, kps (group II), malX | ND |
| ETEC | C9221a | O6:K15:H16 | 4.79 | A | est, elt, fyuA, kps (group II) | PAIs, plasmid, phage |
| EPEC | E2348/69 | O127:H6 | 4.7 | B2 | eae, esp, astA, bfp, per | PAIs, plasmid |
| EIEC | EDL1284 | O124:H− | 4.68 | A | ipaBCD, mxiAB, icsA, iuc, fyuA | PAIs, plasmid |
| EHEC | 4797/97 | O91:H− | 5.25 | B1 | stx2d | PAI, phage |
| EHEC | 5714/96 | O103:H2 | 4.89 | D | stx1, eae (ɛ), hlyA, etp, sfp | PAI, plasmid, phage |
| EHEC | 1639/77 | O111:H− | 4.85 | B1 | stx1, eae (γ) | PAI, plasmid, phage |
| EHEC | SF493/89 | O157:H− | 4.87 | D | stx2, eae (γ), hlyA, etp, sfp | PAI, plasmid, phage |
| EAEC | DPA065 | O119 | 4.69 | A | astA, fyuA, kps (group II) | PAI, plasmid |
| Commensal isolates | MGS 6 | NDe | 4.71 | A | ND | |
| MGS 32 | ND | 4.95 | B1 | ND | ||
| MGS 73 | ND | 4.99 | A | ND | ||
| MGS 89 | ND | 5.03 | A | ND | ||
| MGS 104 | ND | 5.03 | A | ND | ||
| MGS 124 | ND | 5.07 | A | traT; papEF, fyuA, iutA, kps (group II) | PAIs | |
| Laboratory strains | B | ND | 4.7 | A | Phages | |
| MG1655 | K-12 | 4.64 | A | Phages |
PAI deletion mutants of the UPEC strains 536, J96, and 764 are marked by asterisks.
Strains were grouped based on the triplex PCR as recently described (13).
The presence of virulence-associated genes of ExPEC and of the fecal nonpathogenic E. coli isolates was determined using the multiplex PCR described by Johnson and Stell (28).
ABU, asymptomatic bacteriuria.
ND, not determined.
Genome comparison using E. coli gene arrays.
Total genomic DNAs from the different E. coli strains were used to probe Panorama E. coli gene arrays (Sigma-Genosys, Cambridge, United Kingdom). Two micrograms of total genomic DNA of each of the different strains was used as a template for direct incorporation of [33P]dATP (Amersham Pharmacia, Freiburg, Germany) by a randomly primed polymerization reaction using 0.75 μg of random hexamer primers (New England Biolabs, Frankfurt [Main], Germany) and 10 U of Klenow fragment of DNA polymerase I (New England Biolabs) according to the manufacturers' recommendations. Unincorporated nucleotides were removed with Microspin S 200 HR spin columns (Amersham Pharmacia). Prior to hybridization, the DNA macroarrays were rinsed in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) solution and subsequently prehybridized for 3 h at 65°C in 5 ml of hybridization solution (5× SSPE, 2% sodium dodecyl sulfate, 1× Denhardt's solution, 100 μg of sheared denatured herring sperm DNA/ml). After the addition of the probe denatured in 3 ml of hybridization solution, the arrays were incubated for 12 to 18 h at 65°C. After hybridization, the blots were washed according to the manufacturer's guidelines. The washed filters were air dried and exposed overnight to a PhosphorImager screen (type, super resolution) prior to being scanned on a Typhoon 8600 variable mode imager (Molecular Dynamics). Before rehybridization, the E. coli gene arrays were stripped following the manufacturer's recommendations. Complete removal of radioactivity was confirmed by phosphorimaging after overnight exposure to a PhosphorImager screen.
For every E. coli strain investigated in this study, the E. coli gene arrays were hybridized in four different experiments using independently labeled DNA probes. The scanned E. coli gene arrays were analyzed with ArrayVision software (Imaging Research, St. Catharines, Canada), followed by visual inspection. Calculation of normalized intensity values of the individual spots was performed using the overall spot normalization function of ArrayVision. Background values were measured in the surrounding region of every secondary grid, which contains probes for four ORFs arrayed in duplicate. The mean of the normalized intensity values of the duplicate spots of each gene was used for further analysis. To avoid extreme intensity ratios for genes close to or below the detection limit, signal intensity values corresponding to a signal-to-noise (S/N) ratio of <1.0 were scaled up to a value corresponding to an S/N ratio of 1.0. ORFs were recorded as lacking-undetectable if the S/N ratio was <1.0 in at least three of the four hybridization experiments. In addition, E. coli K-12 strain MG1655-specific ORFs were recorded as lacking-undetectable if the ratio of the individual S/N ratios of the analyzed strain and that of the reference strain MG1655 was <0.3 in at least three of four experiments. The missing-undetectable ORFs were then aligned with their chromosomal locations to determine the number and the sizes of chromosomal regions absent in the different E. coli strains. In addition, the fact that the ORFs are arranged on the DNA macroarrays without regard to their chromosomal localization minimizes the record of false-negative spots, at least with respect to regions consisting of more than one gene, because the probability that two adjacent ORFs would be recorded as absent due to hybridization artifacts is very low. The E. coli gene arrays were hybridized with labeled genomic DNA of the E. coli strain MG1655 as a positive control and that of the Staphylococcus aureus strain Wood 46 as a negative control. Generally, the quotients of the individual S/N ratios obtained from hybridization with genomic DNA of S. aureus and those of E. coli strain MG1655 were <0.3.
Development of the E. coli pathoarray.
To facilitate detection of virulence-associated genes of ExPEC or diarrheagenic E. coli, a DNA array was developed which contains probes (n = 212) specific for the majority of ORFs located on five PAIs of UPEC strain 536, as well as for all other ExPEC-specific virulence genes (n = 100) described so far. The E. coli pathoarray also contains probes specific for several typical virulence-associated genes of IPEC and Shigella (n = 95) (Fig. 1). PCR products of 300 to 500 bp which were specific for the individual genes of interest were generated. The quality and concentration of the PCR products were checked prior to spotting them as double spots onto nylon membranes. These probes enable the detection of genes encoding typical toxins, siderophores, and fimbrial and nonfimbrial adhesins, as well as of other genes which have been described as being involved in the virulence of ExPEC and/or IPEC or which are present on PAIs and other mobile genetic elements frequently present in pathogenic E. coli variants. Primers were generated on the basis of available DNA sequences of PAI I536 to PAI V536 of UPEC strain 536, as well as on the basis of publicly available DNA sequences, including those of PAIs of other ExPEC and ETEC isolates (J96, CFT073, and 10407), complete genomes (O157:H7 strains EDL933 and Sakai), and single virulence genes of interest. Genomic DNAs of the strains from which the DNA sequences of the different virulence-associated genes had been initially determined were used as templates for probe generation. Supplementary information on primer sequences and genes included in the E. coli pathoarray are available (http://www.uni-wuerzburg.de/infektionsbiologie).
FIG. 1.
Analysis of the presence of virulence-associated genes among pathogenic, commensal, and laboratory E. coli strains using the E. coli pathoarray. (A) Signal intensities after DNA-DNA hybridization with labeled genomic DNA of the UPEC strain 536. (B) Signal intensities after DNA-DNA hybridization with labeled genomic DNA of the commensal E. coli isolate MGS 73 isolated from a healthy volunteer.
PCR.
DNA primers were purchased from MWG Biotech (Ebersberg, Germany). The Taq DNA polymerase used for the detection of genes in different E. coli strains was purchased from Qiagen (Hilden, Germany). The grouping into the main phylogenetic lineages of the E. coli Collection of Reference Strains (ECOR) was done by a triplex PCR described previously (13). Virulence assessment of the ExPEC and nonpathogenic E. coli strains included a multiplex PCR specific for a set of typical virulence-associated genes of ExPEC (28).
PFGE.
The chromosomal sizes of the different E. coli isolates were determined by pulsed-field gel electrophoresis (PFGE). Genomic DNA for the analysis by PFGE was prepared in agarose plugs and cleaved with the restriction enzyme I-CeuI. PFGE was performed with the CHEF-Dr III system (Bio-Rad, Munich, Germany) in 0.5× Tris-borate-EDTA buffer at 6.5 kV/cm2 and 12°C. Electrophoresis was carried out with pulse times increasing from 5 to 50 s over a period of 22 h.
DNA sequence analysis.
The genomic region which replaces the Rac prophage, present in E. coli strain MG1655, in UPEC strain 536 was sequenced by primer walking (using an ABI-310 sequencer) starting from the DNA fragment A2 obtained by subtractive hybridization analysis of genomic DNA of strain 536 with that of the nonpathogenic E. coli strain MG1655 (27). Primer walking was continued until at least 2 kb of the E. coli MG1655-specific chromosomal backbone had been sequenced at both ends of this DNA region. In addition, primer walking was also started from the conserved chromosomal backbone of E. coli to confirm that the up- and downstream regions of the ORFs b1367 and b1344, respectively, overlap with the so-far-unknown DNA sequence found on subtractive hybridization analysis fragment A2.
Nucleotide sequence accession number.
The nucleotide sequence of the genomic region replacing the Rac prophage in E. coli strain 536 and the flanking DNA stretches was submitted to the EMBL database (accession number AJ496193).
RESULTS
Overall analysis of genetic diversity in E. coli using the E. coli K-12-specific DNA arrays.
In our study, we analyzed the genome structures of 23 E. coli isolates representing different pathotypes of E. coli, as well as nonpathogenic commensal isolates, including the laboratory strain E. coli B. The group of ExPEC isolates consisted of five UPEC strains and one MNEC strain. In addition to MNEC strain IHE3034, the genome contents of another O18:K1 strain, i.e., fecal isolate F54, was investigated, as well as those of deletion mutants of the UPEC strains 536, J96, and 764, which had lost PAIs. The genetic diversity of four EHEC isolates of serotypes O157:H−, O111, O103, and O91, as well as those of four additional intestinal pathogens representing the ETEC, EPEC, EIEC, and EAEC pathotypes, were also characterized. Six nonpathogenic commensal isolates from feces of healthy volunteers, as well as the laboratory strains E. coli B and MG1655, were included in this study to assess whether genetic diversity in pathogenic isolates differs from that in nonpathogenic strains. According to a rapid method to group E. coli strains into the different main phylogenetic groups of the ECOR (13), most of the pathogenic strains tested in this study belong to the ECOR groups B2 and D, to which virulent E. coli isolates mainly belong, whereas the nonpathogenic strains are members of ECOR groups A and B1. The different isolates differ with respect to their chromosomal sizes (between ∼4.67 and 5.25 Mb, as analyzed by PFGE) and to the presence of already known virulence-associated genes located on different types of mobile genetic elements, such as bacteriophages, plasmids, and PAIs (Table 1).
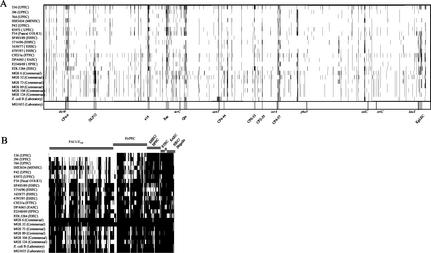
The genome contents of the 22 pathogenic and commensal E. coli isolates were compared to that of the nonpathogenic E. coli K-12 strain MG1655 by DNA-DNA hybridization using an E. coli strain MG1655-specific DNA array which has been designed on the basis of the previously published annotated genome sequence (6). Generally, the results of the DNA-DNA hybridizations of genomic DNA isolated from these E. coli strains with E. coli K-12 gene arrays demonstrated that the genome contents of the different E. coli isolates differ markedly from that of E. coli strain MG1655 (Fig. 2A). On average, 5.8% of the translatable ORFs present in the nonpathogenic reference strain (6) were absent in the individual isolates, in which between 3 and 10% of the translatable ORFs of E. coli strain MG1655 were not detectable (Tables 2 and 3). Based on the functional GenProtEC database classification of the chromosomally encoded genes and proteins of E. coli K-12 (http://genprotec.mbl.edu), the majority of these missing ORFs in every strain can be functionally grouped as coding for hypothetical, unclassified, or unknown gene products. The 22 E. coli isolates also exhibited a great diversity in ORFs which represent mobile genetic elements or which code for structural components of the cell. The alterations were found to be scattered over the entire E. coli MG1655 chromosome. However, prophages of strain MG1655 represent chromosomal variation “hot spots.” Generally, the presence of 10 prophages described in E. coli strain MG1655 is variable in the 22 strains tested (Fig. 2A). In most of the E. coli isolates used in this study, several DNA segments covering the genomic regions described as prophages CP4-6, e14, Qin, and CP4-57 showed a very high density of undetectable clusters of ORFs in comparison to the MG1655 chromosome. The prophages CP4-6 and CP4-57 are located downstream of the tRNA or tRNA-like loci thrW and ssrA, respectively. Whereas the prophage DLP12 was predominantly absent in nonpathogenic isolates, pathogenic strains frequently lacked the Rac prophage. The chromosomal contexts of several other tRNA-encoding genes (e.g., serX, argW, ileY, pheV, and leuX) were also found to contain alterations in some of the pathogenic and commensal isolates in comparison to the corresponding sequences in E. coli MG1655. Several variable chromosomal regions among the studied isolates contain ORFs with homology to ORFs of other accessory genetic elements, e.g., insertion sequence elements.
FIG. 2.
Detection of genomic alterations among pathogenic, commensal, and laboratory E. coli strains using DNA arrays. (A) Comparison of genomes of different pathogenic, commensal, and laboratory E. coli isolates and the E. coli K-12 strain MG1655 using Panorama E. coli gene arrays. The individual chromosomes are displayed linearly and in equal lengths. Missing-undetectable ORFs are marked by vertical black lines in the individual chromosomes. The positions of the undetectable ORFs refer to the E. coli MG1655 chromosome. The positions of tRNA genes frequently used as chromosomal insertion sites of horizontally acquired DNA elements and those of 10 prophages of strain MG1655, as well as the chromosomal origin and terminus of replication, are marked within the map of E. coli strain MG1655. (B) Detection of virulence-associated genes of E. coli and Shigella among pathogenic and commensal E. coli isolates using the E. coli pathoarray. Virulence-associated genes are grouped with regard to typical E. coli pathotypes. Missing-undetectable ORFs are marked by vertical black lines.
TABLE 2.
Number of E. coli K-12-specific ORFs absent in different pathogenic, commensal, and laboratory E. coli strains
| Strain | No. of absent ORFs in functional category:
|
Total | |||
|---|---|---|---|---|---|
| Hypothetical unclassified, unknown | Phage, transposon, or plasmid | Cell structure | Other | ||
| 536 | 206 | 40 | 21 | 116 | 383 |
| 536-21 | 206 | 29 | 21 | 116 | 372 |
| J96 | 132 | 8 | 11 | 46 | 197 |
| J96-M1 | 132 | 8 | 11 | 46 | 197 |
| 764 | 84 | 8 | 11 | 22 | 125 |
| 764-2 | 84 | 8 | 11 | 22 | 125 |
| P42 | 78 | 9 | 2 | 23 | 112 |
| 83972 | 148 | 19 | 15 | 52 | 234 |
| IHE3034 | 114 | 9 | 18 | 61 | 202 |
| F54 | 184 | 9 | 11 | 90 | 294 |
| C9221a | 196 | 14 | 14 | 185 | 409 |
| E2348/69 | 119 | 10 | 15 | 65 | 209 |
| EDL1284 | 184 | 11 | 22 | 129 | 346 |
| 4797/97 | 125 | 9 | 14 | 64 | 212 |
| 5714/96 | 84 | 25 | 10 | 59 | 178 |
| 1639/77 | 105 | 7 | 10 | 37 | 159 |
| SF493/89 | 75 | 9 | 11 | 37 | 132 |
| DPA065 | 145 | 9 | 11 | 107 | 272 |
| MGS 6 | 208 | 23 | 15 | 181 | 427 |
| MGS 32 | 230 | 9 | 16 | 124 | 379 |
| MGS 73 | 202 | 20 | 12 | 116 | 350 |
| MGS 89 | 136 | 12 | 12 | 120 | 280 |
| MGS 104 | 152 | 20 | 13 | 127 | 312 |
| MGS 124 | 142 | 14 | 13 | 107 | 276 |
| E. coli B | 140 | 9 | 30 | 48 | 227 |
| E. coli MG1655 | 0 | 0 | 0 | 0 | 0 |
TABLE 3.
Presence of E. coli MG1655-specific genes and virulence-associated genes of E. coli in different pathogenic, commensal, and laboratory E. coli isolates
| Strains | % of genes detected
|
|||
|---|---|---|---|---|
| E. coli MG1655-specific genes (n = 4,290) | Genes present on PAI I536-PAI V536 (n = 212) | Virulence-associated genes of ExPEC (n = 100) | Virulence-associated genes of IPEC (n = 95) | |
| ExPEC (n = 7) | 94.8 | 61.3 | 42 | 4.8 |
| IPEC (n = 8) | 94.4 | 27.4 | 24 | 24.2 |
| Nonpathogenic E. coli (n = 8)a | 93.4 | 22.6 | 14 | 3.9 |
| All strains (n = 23) | 94.2 | 37.1 | 26.7 | 11.3 |
Including commensal isolates and E. coli laboratory strains B and MG1655.
According to the results of the DNA-DNA hybridization, the presence of 1,165 translatable ORFs (27.2% of all translatable ORFs) of the E. coli K-12 strain was variable among the pathogenic and commensal strains tested. The vast majority of these variable chromosomal regions represent hypothetical and so far uncharacterized ORFs and prophages of E. coli strain MG1655, as well as ORFs involved in lipopolysaccharide biosynthesis, which belong to the functional ORF category “cell structure.” The conserved E. coli-specific genetic backbone, or “core genome,” is therefore estimated to contain at least 3,100 translatable ORFs of strain MG1655. This number of translatable ORFs includes all 232 supposedly essential genes of E. coli as they are compiled in the Profiling of Escherichia coli chromosome database (http://www.shigen.nig.ac.jp/ecoli/pec/About.html).
E. coli K-12 genes absent in pathogenic, commensal, and laboratory E. coli strains.
Certain genes present in E. coli K-12 were not detectable in some of the pathogenic and nonpathogenic strains investigated in our study. The fec operon encoding a ferric citrate uptake system was not detectable in UPEC strain 536, all four EHEC strains tested, strain EDL1284 (EIEC), or fecal isolate MGS 6. Strain EDL1284 also lacks the fim gene cluster, which codes for type 1 fimbriae. The genome of the EAEC strain DPA065 does not contain the hsd-encoded type I restriction modification system. Some of the hsd genes were also absent in other strains tested. Whereas seven of the investigated strains showed no hybridization signal with the hipB-specific probe, the entire hip operon, which is involved in the resistence to the lethal consequences of inhibition of peptidoglycan or DNA synthesis, was not detectable in ETEC strain C9221a. With the exception of UPEC strain P42 and the six fecal isolates from healthy volunteers, the relEB genes coding for a cytotoxin-antitoxin system could not be detected in any strains. Similarly, the complete mcr operon involved in methylation restriction was present only in UPEC strains J96 and 764, in EHEC strain 1639/77, and in the nonpathogenic isolates MGS 32, MGS 104, and MGS 124, as well as in E. coli strain B. Several csp genes whose products are involved in cold shock adaptation were not present in some of the tested pathogenic E. coli strains. The DNA-DNA hybridizations demonstrated that the different E. coli isolates differed markedly in the composition of the waa and wbb operons (formerly known as rfa and rfb, respectively), which are required for lipopolysaccharide biosynthesis. Several of the above-mentioned determinants are supposed to have been acquired via horizontal gene transfer. This is also supported by our results, which indicate that the presence of these genes is variable among the different E. coli strains tested, underlining the importance of horizontal gene transfer for the evolution of different E. coli variants.
Detection of virulence-associated genes in pathogenic, commensal, and laboratory E. coli strains.
The 23 pathogenic and commensal isolates, including the laboratory strains B and MG1655, were screened for the presence of PAI-specific sequences of the UPEC strain 536, as well as for typical virulence-associated genes of ExPEC, IPEC, and Shigella using the E. coli pathoarray. Using sequences of the known PAIs of UPEC strain 536 and of virulence-associated genes of other pathogenic E. coli strains, this array was recently designed in our laboratory. Generally, sequences specific for PAI I536 to PAI V536, as well as typical virulence-associated genes of ExPEC, were more frequently detectable in ExPEC strains (61.3 and 42% of the probes specific for PAI I536 to PAI V536 ORFs and other ExPEC virulence genes spotted on the pathoarray) than in IPEC (27.4 and 24%, respectively). Accordingly, IPEC isolates harbored more genes which are typical for these pathotypes (24.2% of the probes specific for virulence genes of IPEC) than ExPEC (4.8%). However, even in commensal strains, a considerable number of “virulence-associated” genes or their homologues present on PAI I536 to PAI V536 ORFs (22.6%), in ExPEC (14%), and in IPEC (3.9%) were detectable (Fig. 1). The “pathogene” content varies considerably among the different pathogenic and commensal isolates (Fig. 2B and Table 3). The results of the E. coli pathoarray hybridization were partially checked by PCR using the primers designed for probe generation. The vast majority of strong hybridization signals on E. coli pathoarrays could be confirmed or resulted in strong PCR products, suggesting the presence of these genes or their homologues in the different genomes.
Comparison of the genomes of two O18:K1 isolates (E. coli strains IHE3034 and F54) indicated that the overall hybridization patterns of both strains resemble each other and differ from those of the other ExPEC strains tested in the absence of PAI II536-specific sequences (Fig. 2B). Although their virulence gene contents are not identical, they contain typical virulence-associated genes of E. coli O18:K1 strains which frequently cause newborn meningitis: the K1 capsule-encoding determinant, as well as the ibeA gene, which has been shown to be required for the ability to cross the blood-brain barrier (24), were detectable. In addition, both strains contain the type 1 fimbrial gene cluster (fim) and genes whose products are involved in iron acquisition, i.e., genes coding for the hemin receptor ChuA, as well as for the irobactin and yersiniabactin siderophore systems. The MNEC isolate IHE3034 also carries the cdt gene cluster coding for the cytolethal distending toxin. According to these results, the strains of the same serotype exhibit very similar but not identical sets of virulence genes.
Comparison of the virulence gene contents of four EHEC isolates of different serotypes showed that these strains differ with respect to their virulence gene repertoires, but several genes which have been previously described for EHEC O157:H7 strain EDL933 are also present in these strains, i.e., the type 1 fimbrial gene cluster, the stx1 or stx2 determinant coding for shigatoxin, the eae and tir genes involved in the attaching-and-effacing phenotype, and the O-island ORFs Z0250, Z1542, Z4326, and Z4852 (47). Other EHEC PAI genes, e.g., the tellurite resistance gene cluster tlrABCD (54), were detectable only in the O157:H− and the O111 strains. Interestingly, the O157:H− strain also carries the cdtIII gene cluster coding for cytolethal distending toxin III. Probes specific for the P-fimbrial gene cluster usually found in ExPEC gave strong hybridization signals upon hybridization with genomic DNA of the O111 isolate. Virulence genes which are located on pO157, e.g., genes coding for the EHEC hemolysin or the plasmid-encoded catalase, have been detected only upon DNA-DNA hybridization using genomic DNAs of the O157:H− and O103 EHEC strains. These results underline the fact that non-O157:H7 EHEC isolates differ with regard to their virulence gene pools from O157:H7 strains, such as strain EDL933 or Sakai (23, 47).
Comparison of genome contents of the wild-type UPEC strains and their PAI deletion mutants.
It is well known that pathogenic bacteria have a tendency to delete large chromosomal regions (PAIs) comprising virulence-associated genes (7, 8, 17). In order to analyze the mechanisms resulting in the deletion of PAIs, the genome contents of three UPEC isolates and one corresponding PAI deletion mutant were compared. Comparison of the genomes of the uropathogenic wild-type isolates 536, J96, and 764 with those of their corresponding PAI deletion mutants 536-21, J96-M1, and 764-2, respectively, demonstrated that only strain-specific genetic information was deleted but no chromosomal regions which belong to the K-12-specific genetic backbone (Table 4). These results underline the fact that the deletion of PAIs is a specific process which is independent of general genome plasticity in these organisms. The hybridization patterns of the E. coli pathoarray indicate that fewer E. coli pathogenes were detectable in PAI deletion mutants than in the corresponding wild-type strains. The observation that PAIs of different ExPEC isolates are not completely identical with regard to genetic organization and gene content, although they share a great number of identical and homologous genes (15, 29), is also supported by the comparison of the E. coli pathoarray hybridization patterns of the ExPEC strains 536, J96, and 764 with those of their PAI deletion mutants 536-21, J96-M1, and 764-2. Although the deleted chromosomal regions are not identical in these deletion mutants, the proportions of undetectable sequences specific for PAI I536 to PAI V536 compared to other undetectable virulence-associated genes of ExPEC or IPEC are similar in the three UPEC isolates and their PAI deletion mutants (Table 4).
TABLE 4.
Presence of E. coli MG1655-specific genes and virulence-associated genes of E. coli in different UPEC isolates and their PAI deletion mutants
| Strain | % of genes detected
|
|||
|---|---|---|---|---|
| E. coli MG1655-specific genes (n = 4,290) | Genes present on PAII536-PAI V536 (n = 212) | Virulence-associated genes of ExPEC (n = 100) | Virulence-associated genes of IPEC (n = 95) | |
| 536 | 91.1 | 100 | 42 | 2.1 |
| 536-21 | 91.2 | 63.7 | 39 | 2.1 |
| J96 | 95.4 | 71.2 | 40 | 2.1 |
| J96-M1 | 95.4 | 51.9 | 25 | 2.1 |
| 764 | 97.1 | 62.7 | 53 | 6.3 |
| 764-2 | 97.1 | 57.1 | 51 | 6.3 |
DISCUSSION
Variants of the species E.coli are adapted to various host organisms, e.g., humans, monkeys, horses, and birds, in which they belong to the normal intestinal flora. In addition, pathogenic strains have the capacity to cause sepsis or local infections of the intestines, as well as of the kidney, bladder, and brain, in different hosts. The ecological and pathogenic diversity of E. coli strains is a prerequisite for the use of E. coli as a model to study processes of bacterial genome evolution. These mechanisms are reflected in the fact that the genomes of different strains show remarkable variations. The genomes of E. coli K-12 and of strains of the ECOR vary in size from 4.5 to 5.5 Mb (5, 44). The high variability in gene content among different E. coli strains is mainly due to the acquisition of foreign DNA and to deletion of genetic information. As indicated in Table 1, the strains investigated in this study also exhibit marked differences in their chromosomal sizes. The affiliation of the majority of the pathogenic strains with the major phylogenetic ECOR groups B2 and D, as well as that of the nonpathogenic and commensal isolates with ECOR groups A and B1, respectively, was determined by PCR and confirms the general view of the distribution of pathogenic and nonpathogenic isolates in the ECOR groups (10). All ExPEC and IPEC strains, as well as the commensal strain MGS 124, carry genomic islands in their genomes whose products contribute to their individual traits. In addition, EHEC isolates were able to pick up bacteriophages encoding Shiga toxins. Furthermore, the genomes of IPEC strains comprise large plasmids encoding adhesins, toxins, and other virulence factors.
In order to systematically assess the genetic variability of bacteria, several genome comparison techniques, e.g., macrorestriction analysis, PFGE, and genomic subtraction, have been used (9, 39, 49, 58). DNA array technology is a powerful new tool for comparative genomics which has recently been used for the analysis of genome variability among bacterial species or closely related bacteria (2, 4, 18, 19, 43, 50, 53), as well as for detection of specific groups of trait-conferring genes (12, 21). DNA array technology has been utilized in this study to characterize the amount of genome variation among pathogenic and commensal E. coli isolates in general and between ExPEC and IPEC strains in particular. The use of PCR product-based DNA arrays to determine the presence or absence of DNA regions does not allow explicit answers to the question of whether specific genes are present, as hybridization signals can also result from cross hybridization of homologues or of genes with conserved domains. Additionally, no statement can be made as to whether the detected genes are functional, as nonfunctional genes and even gene fragments which are frequently present, especially on PAIs, will result in a hybridization signal. However, we think that, based on the criteria for signal intensities which we described in Materials and Methods, the use of DNA array technology allows us to sufficiently assess the genetic diversity and genome content among different isolates of one species, as discussed below.
The genome contents of different pathogenic, commensal, and laboratory E. coli isolates, as mirrored by the hybridization signals on the E. coli gene arrays, is very heterogenous (Fig. 2A). Depending on the individual isolate, the number of ORFs absent in comparison to strain MG1655 varies between 112 (UPEC strain P42) and 427 (fecal isolate MGS 6). Generally, these alterations show no preferential chromosomal localization and can be scattered all over the chromosome (Fig. 2A). However, we found recently that the E. coli MG1655-specific chromosomal region downstream of gene thrW, which represents the prophage CP4-6, shows a high density of chromosomal variation in all other E. coli strains tested so far (16). Genome comparison shows that this is true for several other prophage sequences present in strain MG1655. Therefore, these regions can be regarded as chromosomal variation hot spots and seem to be E. coli K-12 specific. It has been speculated that the 40-kb chromosomal segment downstream of the tRNA-encoding gene thrW (b0245 to b0286) has been acquired by horizontal gene transfer (55, 57). The fact that the prophages which have been identified in E. coli strain MG1655 (36) are considered to have been recently acquired by E. coli laboratory strains, as they are missing in some natural E. coli isolates, has been mentioned before (43). As a result of our study, a 20-kb DNA stretch between ORFs b1345 and b1375 was characterized by the frequent occurrence of clusters of undetectable ORFs in almost all pathogenic E. coli isolates studied. This chromosomal region is covered in E. coli strain MG1655 by prophage Rac. Sequence analysis of the chromosomal region of the UPEC strain 536 replacing prophage Rac (data not shown; available under accession number AJ496193) indicates that this strain originally possessed Rac-specific sequences which were later deleted. Instead, a 2-kb genomic region is located within the K-12 chromosomal backbone between the putative ORFs b1344 and b1376. This fragment has a G+C content of 35% and exhibits no significant homology on the nucleotide level. A small putative ORF coding for a protein with 72% identity to amino acids 18 to 104 of the SitD protein of Salmonella enterica serovar Typhimurium (AF128999) is located within this region. The fact that a fragment of intR (b1345), which encodes the putative transposase-integrase of prophage Rac, is still present in the chromosome of strain 536 implies that this mobile genetic element was lost upon integration of the DNA region currently present in E. coli strain 536.
Genetic mechanisms of bacterial-genome evolution are often associated with tRNA genes. Several lysogenic bacteriophages preferentially use the 3′ ends of tRNA genes as chromosomal insertion sites, implying that they may have served as vehicles for the integrated foreign DNA (11, 48). Other genes coding for tRNAs or tRNA-like molecules are also associated with bacteriophage integrase genes in many species. Additionally, in many prokaryotes, including E. coli, tRNA genes are frequently associated with genetic elements designated genomic islands. The insertion of genomic islands or PAIs and plasmids into tRNA loci has been described in many pathogenic and nonpathogenic bacteria (22, 26, 30, 33, 40, 44). The sequence contexts of several tRNA-encoding genes of different E. coli isolates (e.g., thrW, ssrA, serX, argW, ileY, pheV, and leuX) differ from that of strain MG1655. Also among these tRNA loci were some of those which are known to be frequently associated with PAIs in several pathogenic bacteria. These results demonstrate that the chromosomal contexts of many tRNA loci represent variable regions of the E. coli chromosome.
Observed sequence alterations in the vicinity of tRNA genes may be due to the integration of foreign DNA fragments. However, they may also be the result of gene reduction. The latter process, from our point of view, is underestimated as a fundamental mechanism of genome evolution, at least in prokaryotes. It is known that the genome of Shigella flexneri, in contrast to the very similar genome of E. coli, lacks the cadA fragment, encoding the enzyme lysine decarboxylase. This “hole” seems to be important for the evolution of Shigella as a pathogen (38). The same is true for the recently evolved human pathogen Yersinia pestis, which has lost many functions compared to its ancestor Yersinia pseudotuberculosis (1). The determination of E. coli K-12-specific genomic regions, which are absent from E. coli pathogens, may lead to the identification of other genes whose loss could be a benefit for E. coli virulence or adaptation under the corresponding growth conditions. A group of genes which was not detectable in many IPEC isolates but which was present in all extraintestinal and commensal strains tested comprises several genes (hofG, hofH, pshM, bfr, and yjhN) which can be functionally grouped as coding for transport and binding proteins. The fec operon, which encodes a siderophore system, was not detectable in any EHEC strains tested and is also absent in the two sequenced O157:H7 strains. Whether the frequent lack of these genes represents a putative characteristic of EHEC or IPEC strains will have to be further analyzed. The eight K-12-specific ORFs missing in all other tested strains seem to represent E. coli strain MG1655-specific genetic information. In contrast, 44 ORFs present in strain MG1655 were absent in fecal commensal isolates of healthy volunteers but present in pathogenic isolates. Genome comparison and determination of the chromosomal sizes of three UPEC strains and their PAI deletion mutants demonstrated that no E. coli MG1655-specific genetic information has been deleted together with PAIs. The chromosomal-size differences of strains 536 and 536-21, together with the results of the different whole-genome approaches, confirm earlier findings that the mutant strain 536-21 lost only PAI I536 and PAI II536, which represent chromosomal regions of ∼75 and 102 kb, respectively (7, 15), and demonstrate the specific character of PAI deletion. This was further substantiated by comparison of two other UPEC isolates and their corresponding PAI deletion mutants. Deletion of PAIs is thought to be mediated by specific enzymes, such as bacteriophage integrases (22). The genetic structures of the deleted PAIs of the UPEC strains J96 and 764 resemble that of strain 536, as the deletion mutants J96-M1 and 764-2 lost more sequences specific for PAI I536 to PAI V536 from their homologues than other ExPEC or IPEC-specific DNA stretches.
Analysis of the presence of virulence-associated genes of E. coli and Shigella among different ExPEC and IPEC pathotypes, as well as among commensal isolates, indicated that sequences specific for PAI I536 to PAI V536 are widespread in pathogenic and many commensal isolates. Our results also demonstrate that the presence of DNA regions described for ExPEC or IPEC is markedly higher in strains belonging to the corresponding pathotype. However, virulence-associated sequences, or at least their homologues, have also been detected to a lesser extent in commensal isolates or strains of other pathotypes. The relatively frequent occurrence of sequences specific for PAI I536 to PAI V536, not only among ExPEC isolates, results from the fact that many putative ORFs present on PAIs of the UPEC strain 536 are similar to fragments of genes of strain MG1655 or belong to different accessory DNA elements, such as transposons, insertion sequence elements, plasmids, and bacteriophages, whose presence is not restricted to PAIs and which can also be found frequently on chromosomal or extrachromosomal DNAs of other bacteria. Therefore, the high frequency with which PAI-specific sequences of strain 536 have been detected in other, even commensal, isolates can be due to similarity or identity to genes or gene fragments which are present on PAIs but which are not important for virulence. This is also true of many ORFs located on the locus of enterocyte effacement-PAI of EHEC and EPEC. The great number of ExPEC-specific DNA sequences even among intestinal pathogenic and nonpathogenic isolates can also be partially explained by the fact that several determinants we included in this group of genes encode gene products which are fitness rather than virulence factors, e.g., aerobactin or type 1 fimbriae, and which are known to be frequently present in pathogenic and commensal E. coli variants. The results obtained with the E. coli pathoarray demonstrate on one hand that the accumulation of virulence-associated sequences determines the pathotypes and pathogenicity of E. coli strains and on the other the great heterogeneity of the gene contents of different E. coli variants, even among members of the same pathotype or serotype.
Taken together, the results of this study clearly indicate that the genetic diversity among pathogenic and commensal E. coli isolates is very high with respect to genomic alterations. This may be indicative of frequent acquisition of foreign DNA by horizontal gene transfer, as well as of a high frequency of deletions during the evolution of bacterial genomes.
Acknowledgments
Our work was supported by the DFG (SFB479; European Graduate College) and by the Fonds der Chemischen Industrie.
We thank B. Plaschke for technical assistance, as well as U. Hentschel for helpful discussions.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akman, L., and S. Aksoy. 2001. A novel application of gene arrays: Escherichia coli array provides insight into the biology of the obligate endosymbiont of tsetse flies. Proc. Natl. Acad. Sci. USA 98:7546-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, P., I. Engberg, G. Lidin-Janson, K. Lincoln, R. Hull, S. Hull, and C. Svanborg. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Bergthorsson, U., and H. Ochman. 1998. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 15:6-16. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Villes, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 7.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum, G., V. Falbo, A. Caprioli, and J. Hacker. 1995. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol. Lett. 126:189-195. [DOI] [PubMed] [Google Scholar]
- 9.Bonacorsi, S. P. P., O. Clermont, C. Tinsley, I. Le Gall, J.-C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheetham, B. F., and M. E. Katz. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201-208. [DOI] [PubMed] [Google Scholar]
- 12.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobrindt, U., and J. Hacker. 1999. Plasmids, phages and pathogenicity islands: lessons on the evolution of bacterial toxins, p. 3-23. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins, 2nd ed. Academic Press, London, United Kingdom.
- 15.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Fünfstück, and J. Hacker. 2001. S-fimbria encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt, U., U. Hentschel, J. B. Kaper, and J. Hacker. 2002. Genome plasticity in pathogenic and nonpathogenic enterobacteria. Curr. Top. Microbiol. Immunol. 264:157-175. [PubMed] [Google Scholar]
- 18.Dong, Y., J. D. Glasner, F. R. Blattner, and E. W. Triplett. 2001. Genomic interspecies microarray hybridization: rapid discovery of three thousand genes in the maize endophyte, Klebsiella pneumoniae 342, by microarray hybridization with Escherichia coli K-12 open reading frames. Appl. Environ. Microbiol. 67:1911-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falkow, S. 1996. The evolution of pathogenicity in Escherichia coli, Shigella and Salmonella, p. 2723-2729. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. Brooks Low, Jr., B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 21.Gill, R. T., S. Wildt, Y. T. Yang, S. Ziesman, and G. Stephanopoulos. 2002. Genome-wide screening for trait conferring genes using DNA microarrays. Proc. Natl. Acad. Sci. USA 99:7033-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 24.Huang, S. H., C. Wass, Q. Fu, N. V. Prasadarao, M. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hull, R. A., E. R. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, R. W., J. W. Mansfield, D. L. Arnold, A. Sesma, C. D. Paynter, J. Murillo, J. D. Taylor, and A. Vivian. 2000. Excision from tRNA genes of a large chromosomal region, carrying avrPphB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola. Mol. Microbiol. 38:186-197. [DOI] [PubMed] [Google Scholar]
- 27.Janke, B., U. Dobrindt, J. Hacker, and G. Blum-Oehler. 2001. A subtractive hybridisation analysis of genomic differences between the uropathogenic E. coli strain 536 and the E. coli K-12 strain MG1655. FEMS Microbiol. Lett. 199:61-66. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, J. R., T. T. O′Bryan, M. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper, J. B., and J. Hacker. 1999. Pathogenicity islands, plasmids and other mobile elements. ASM Press, Washington, D.C.
- 31.Karch, H., H. Böhm, H. Schmidt, F. Gunzer, S. Aleksic, and J. Heesemann. 1993. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J. Clin. Microbiol. 31:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiewitz, C., K. Larbig, J. Klockgether, C. Weinel, and B. Tümmler. 2000. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNA(Lys) gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology 146:2365-2373. [DOI] [PubMed] [Google Scholar]
- 34.Knapp, S., J. Hacker, I. Then, D. Müller, and W. Goebel. 1984. Multiple copies of hemolysin genes and associated sequences in the chromosomes of uropathogenic Escherichia coli strains. J. Bacteriol. 159:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Väisänen-Rhen, J. Finne, I. Ørskov, F. Ørskov, S. B. Svenson, and P. H. Mäkelä. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Sotman, and B. Rowe. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119-1122. [DOI] [PubMed]
- 38.Maurelli, A. T., R. E. Fernández, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melkerson-Watson, L. J., C. K. Rode, L. Zhang, B. Foxman, and C. A. Bloch. 2000. Integrated genomic map from uropathogenic Escherichia coli J96. Infect. Immun. 68:5933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mühldorfer, I., G. Blum, A. Donohue-Rolfe, H. Heier, T. Ölschläger, H. Tschäpe, U. Wallner, and J. Hacker. 1996. Characterization of Escherichia coli strains isolated from environmental water habitats and from stool samples of healthy volunteers. Res. Microbiol. 147:625-635. [DOI] [PubMed] [Google Scholar]
- 42.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 45.Ørskov, F., and I. Ørskov. 1992. Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 38:699-704. [PubMed] [Google Scholar]
- 46.Ott, M., J. Hacker, T. Schmoll, T. Jarchau, T. K. Korhonen, and W. Goebel. 1986. Analysis of the genetic determinants coding for the S-fimbrial adhesin (sfa) in different Escherichia coli strains causing meningitis or urinary tract infections. Infect. Immun. 54:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Reiter, W., D. P. Palm, and S. Yeats. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rode, C. K., L. J. Melkerson-Watson, A. T. Johnson, and C. A. Bloch. 1999. Type-specific contributions to chromosome size differences in Escherichia coli. Infect. Immun. 19:230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Schmidt, H., C. Geitz, P. I. Tarr, M. Frosch, and H Karch. 1999. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J. Infect. Dis. 179:115-123. [DOI] [PubMed] [Google Scholar]
- 53.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor, D. E., M. Rooker, M. Keelan, L. K. Ng, I. Martin, N. T. Perna, N. T. Burland, and F. R. Blattner. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184:4690-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Vliet, F., A. Boyen, and N. Glansdorff. 1988. On interspecies gene transfer: the case of the argF gene of Escherichia coli. Ann. Inst. Pasteur Microbiol. 139:493-496. [DOI] [PubMed] [Google Scholar]
- 56.Wriston, J. C., and T. O. Yellin. 1973. l-Asparaginase: a review. Adv. Enzymol. Relat. Areas Mol. Biol. 39:185-248. [DOI] [PubMed] [Google Scholar]
- 57.York, H. K., and M. Stodolsky. 1981. Characterization of P1argF derivatives from Escherichia coli K12 transduction. I. IS1 elements flank the argF gene segment. Mol. Gen. Genet. 181:230-240. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, L., B. Foxman, S. D. Manning, P. Tallman, and C. F. Marrs. 2000. Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect. Immun. 68:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]