Abstract
SsrA, or tmRNA, is a small RNA found in all bacteria that intervenes in selected translation reactions to target the nascent polypeptide for rapid proteolysis. We have found that the abundance of SsrA RNA in Caulobacter crescentus is regulated with respect to the cell cycle. SsrA RNA abundance increases in late G1 phase, peaks during the G1-S transition, and declines in early S phase, in keeping with the reported role for SsrA in the timing of DNA replication initiation. Cell cycle regulation of SsrA RNA is accomplished by a combination of temporally controlled transcription and regulated RNA degradation. Transcription from the ssrA promoter peaks late in G1, just before the peak in SsrA RNA abundance. SsrA RNA is stable in G1-phase cells and late S-phase cells but is degraded with a half-life of 4 to 5 min at the onset of S phase. This degradation is surprising, since SsrA RNA is both highly structured and highly abundant. This is the first observation of a structural RNA that is cell cycle regulated.
Regulation of mRNA and protein levels is fundamental to control of the cell cycle, both in eukaryotes and in bacteria. We now report that a small structural RNA is cell cycle regulated in the bacterium Caulobacter crescentus. This RNA, known as SsrA (also known as tmRNA and 10Sa RNA), is a small, highly structured RNA that has been found in every species of bacteria as well as in some chloroplasts and mitochondria of eukaryotes (27). SsrA RNA intervenes in selected translation reactions to release ribosomes from the mRNAs and to target the nascent polypeptides for proteolysis. SsrA provides a quality control function for translation by recognizing stalled ribosomes and targeting the incomplete proteins for degradation before they are released into the cell (8, 14, 29). In addition, findings regarding the phenotypes of SsrA mutations in a number of bacteria (3, 4, 10, 20) and the substrates that have been identified for SsrA (1, 19, 22) suggest that SsrA also has a role in regulating gene expression. Recently, we found that SsrA activity is required for correct timing of initiation of DNA replication in Caulobacter (12). The requirement for SsrA activity at a single point in the cell cycle led us to examine the possibility that the SsrA RNA is itself regulated as a function of the cell cycle.
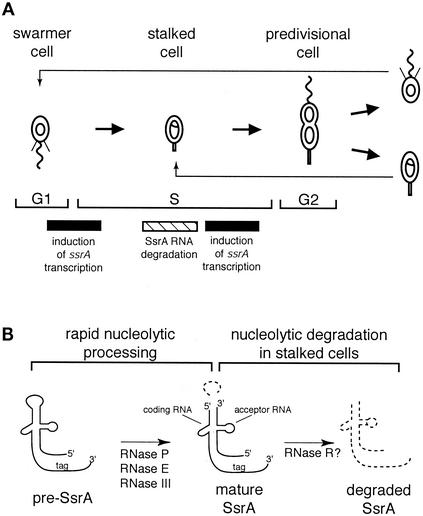
Caulobacter is ideal for cell cycle studies, because it is easy to isolate a population of cells in G1 phase and this population passes synchronously through the cell cycle. In addition, the cell cycle of Caulobacter is coordinated with a program of differentiation which culminates in an asymmetric cell division (see Fig. 4) (24). The progeny swarmer and stalked cells differ with respect to morphology and cell fate. The G1 phase of the Caulobacter cell cycle coincides with the swarmer cell stage, in which cells are motile and have a single polar flagellum. Swarmer cells cannot initiate DNA replication or undergo cell division until they differentiate into stalked cells. This differentiation includes the loss of the polar flagellum, pilus, and chemotaxis apparatus, growth of a new appendage called a stalk at the same pole, and initiation of DNA replication. Thus, the swarmer- to stalked-cell transition is coincident with the G1-S phase transition. After differentiation, the stalked cell continues to replicate the chromosome, elongates, and eventually forms a division plane, becoming a predivisional cell. The predivisional cell divides asymmetrically to produce a swarmer cell and regenerate the stalked cell.
FIG. 4.
Mechanisms of regulation of the abundance of SsrA RNA through the Caulobacter cell cycle. (A) Schematic diagram of the Caulobacter cell cycle showing the time of induction of ssrA transcription and degradation of SsrA RNA. The circles and theta structures within the cells represent nonreplicating and replicating chromosomes, respectively, and the G1, S, and G2 phases of the cell cycle are indicated. The single polar flagellum (squiggly line) and polar pili (thin lines) are lost during the swarmer- to stalked-cell transition and are replaced at the same pole by a stalk (thick line). (B) Schematic diagram of nucleolytic processing and degradation of SsrA RNA. A 30-nt loop is excised from pre-SsrA RNA by specific nucleases to produce the two-piece mature SsrA. The mature SsrA RNA is then completely degraded by one or more nucleases in stalked cells. Degraded RNA is indicated by dashed lines. Candidates for these different nucleases, based on results from E. coli experiments, are indicated.
In Caulobacter and related α-proteobacteria, the ssrA gene is unusual in that it contains a circular permutation that results in a two-piece SsrA RNA (13). The ssrA gene is transcribed as a single transcript with an internal loop connecting the tRNA-like 5′ and 3′ ends. The 30-nucleotide (nt) internal loop of this pre-SsrA RNA is excised by specific ribonucleases, producing the tRNA-like 5′ and 3′ ends and resulting in a mature SsrA composed of two RNA molecules, the coding RNA and the acceptor RNA. Despite this two-piece composition, the Caulobacter SsrA RNA is predicted to have a structure very similar to those of the one-piece SsrA RNAs from other bacteria. As in Escherichia coli, SsrA RNA in Caulobacter tags proteins made from mRNA with no stop codon and the Caulobacter SsrA-encoded peptide targets proteins for rapid degradation in vivo (13). We report here that the Caulobacter SsrA RNA is transcribed and nucleolytically processed to its mature form at the swarmer- to stalked-cell transition and then subjected to nuclease digestion in the stalked cell. Thus, the abundance of this small structural RNA is under strict cell cycle control. There are many examples of mRNAs that peak in expression during specific stages of the cell cycle (16), but Caulobacter SsrA RNA is the first example of a structural RNA that is cell cycle regulated.
MATERIALS AND METHODS
Strains and plasmids.
The wild-type Caulobacter strain used in this work is CB15N (6). Caulobacter strains were grown in M2G or PYE medium at 28°C (5), and growth was monitored by observing the increase in optical density at 660 nm. Using PCR, the ssrA promoter transcriptional reporter was constructed by amplifying the 602-bp segment of DNA 5′ of the SsrA transcriptional start site and cloning the fragment into pRKlac290 5′ of the promoterless lacZ gene (2). The plasmid was verified by DNA sequencing and mobilized into the Caulobacter strains by conjugation from E. coli strain S17 (5). The wild-type E. coli strain used in this work was MG1655, which was grown in Luria-Bertani broth at 30°C, and culture density was monitored by observing optical density at 600 nm.
Cell cycle studies.
Synchronized cultures of Caulobacter were obtained by centrifugation in a Ludox density gradient followed by isolation of swarmer cells (6). Swarmer cells were released into M2G medium, and aliquots were removed from synchronized cultures every 15 min for analysis by Northern blotting.
RNA analysis.
Total RNA was isolated using the hot phenol method (23). Northern blotting was performed after separating equal quantities of total RNA on polyacrylamide-urea gels. 32P-labeled DNA probes were generated from PCR products by using QuickPrime protocol (Amersham Biosciences), visualized using a PhosphorImager (Molecular Dynamics), and quantified with ImageQuant software. RNA decay experiments were performed by inhibiting transcription in a culture by addition of rifampin to achieve a concentration of 300 μg/ml and removing aliquots into an equal volume of stop buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 25 mM sodium azide, 500 μg of chloramphenicol/ml) on ice. RNA was prepared from these samples and analyzed by Northern blotting. The half-life was determined by fitting the data with a single exponential function. To measure the RNA decay in pure populations of swarmer cells, stalked cells, and predivisional cells, synchronous cultures were prepared and analyzed at 15 min for swarmer cells, at 45 min for stalked cells, and at 80 min for predivisional cells.
Promoter activity assays.
Cells bearing lacZ reporter constructs were synchronized, and at 15-min time points, 1-ml aliquots were removed, labeled with 25 μCi of [35S]methionine for 5 min, and added to 50 μl of 100% trichloroacetic acid on ice. Proteins were collected by centrifugation and resuspended in 50 μl of buffer containing 10 mM Tris-HCl (pH 8.0), 1% sodium dodecyl sulfate (SDS), and 1 mM EDTA. After resuspension, 0.75 ml of RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS) was added and the lysates were incubated with agarose conjugated to anti-β-galactosidase antibody (Abcam) for 4 h at 4°C. Immunoprecipitated protein was collected by centrifugation, washed three times with RIPA buffer, and boiled in SDS-polyacrylamide gel electrophoresis sample loading buffer. Proteins were separated on SDS-6% polyacrylamide electrophoresis gels, and the amount of radiolabeled β-galactosidase was quantified using a PhosphorImager. The signal for each synchronous culture was normalized to the level of the 15-min time point, and three normalized experiments were averaged.
RESULTS
Levels of SsrA RNA change through the cell cycle.
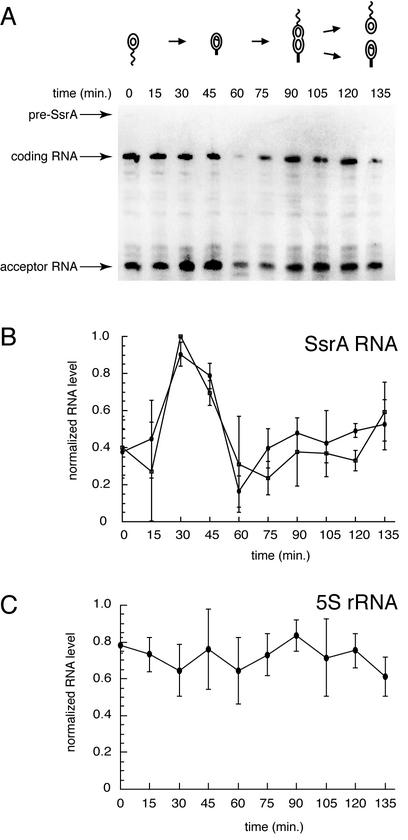
Our observation that SsrA activity is required for proper timing of DNA replication raises the possibility that SsrA RNA is cell cycle regulated. To determine whether the abundance of SsrA RNA changes through the cell cycle, total RNA was isolated from synchronized populations of Caulobacter and the amount of SsrA RNA was measured by Northern blotting (Fig. 1). The amount of the SsrA coding RNA (214 bp) and acceptor RNA (83 bp) increased by 2.5-fold in 15 min during the swarmer- to stalked-cell transition and then decreased to near or below the initial levels over 30 min in stalked cells. The amount of the coding and acceptor RNAs then increased slowly over 60 min in predivisional cells. For comparison, the same Northern blots were probed for 5S rRNA and the amount of 5S rRNA did not change significantly over the cell cycle.
FIG. 1.
Cell cycle regulation of SsrA RNA abundance. (A) A representative Northern blot of total RNA from synchronized cultures probed for SsrA RNA. The schematic diagram indicates the stage of the cell cycle for each time point. The bands corresponding to the pre-SsrA RNA, the 214-nt coding RNA, and the 83-nt acceptor RNA are indicated. (B) The amount of SsrA RNA was quantified and normalized to the peak level of SsrA. The averages of the results of four independent experiments are shown, with error bars indicating the standard deviations. Filled squares correspond to the SsrA coding RNA, and filled circles correspond to the SsrA acceptor RNA. (C) The blots were reprobed for 5S rRNA and quantified in the same manner as described for panel B.
A priori, it is possible that the amount of mature SsrA RNA is regulated at several different steps, including transcription of pre-SsrA RNA, processing of pre-SsrA RNA to the mature form, and degradation of mature SsrA RNA. Because there was little accumulation of pre-SsrA at any point during the cell cycle (Fig. 1A), processing of pre-SsrA RNA to the mature form occurs at the time of its transcription. Thus, transcription of the ssrA gene and degradation of SsrA RNA were examined as possible regulatory pathways for cell cycle control of the abundance of SsrA RNA.
Cell cycle-regulated transcription of ssrA.
The cell cycle dependence of ssrA transcription was determined by assaying the production of β-galactosidase from a lacZ reporter under the control of the ssrA promoter (Fig. 2). This reporter construct contains the PssrA promoter followed by a Shine-Dalgarno sequence and the lacZ gene, such that transcription from PssrA results in production of β-galactosidase. In log-phase cultures, the PssrA promoter generated ∼6,500 Miller units of activity, making it one of the most active promoters reported for Caulobacter. This high level of transcription is consistent with the high abundance of SsrA RNA in the cell. Transcription from PssrA at different points in the cell cycle was compared by pulse-labeling cell samples from a synchronous culture with [35S]methionine and immunoprecipitating the β-galactosidase to determine how much new protein was made from the PssrA reporter construct during the pulse. Transcription increased approximately twofold during the swarmer- to stalked-cell transition and then decreased early in the stalked cell stage (Fig. 2). Transcription from the ssrA promoter peaked immediately before accumulation of SsrA RNA, and the increase in transcription during the swarmer- to stalked-cell transition was similar to the increase in SsrA RNA steady-state levels in magnitude. This correlation suggests that the increase in SsrA RNA levels during the swarmer- to stalked-cell transition is the result of increased transcription. There was a smaller peak of transcriptional activity late in the stalked cell stage, immediately before SsrA RNA steady-state levels began to increase.
FIG. 2.
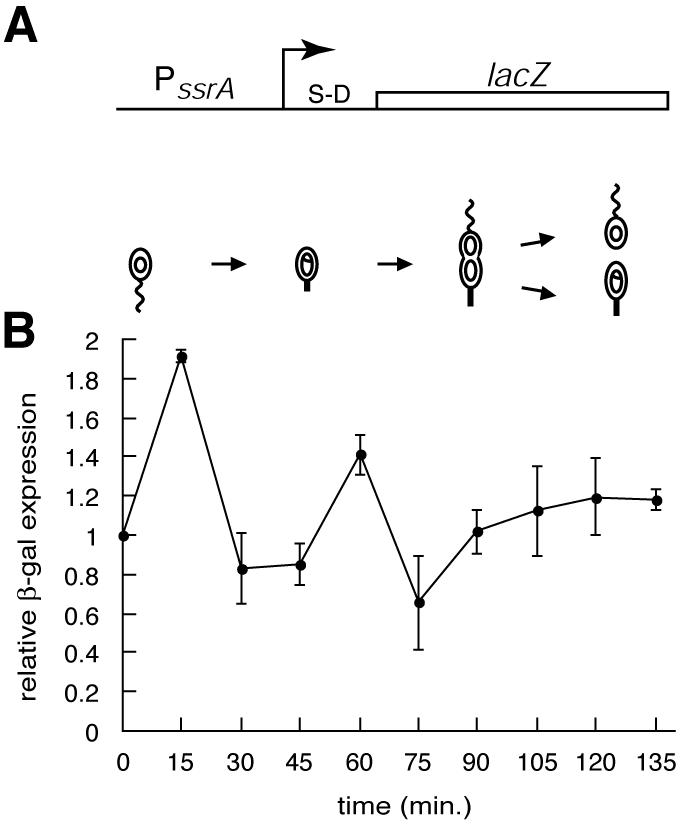
Regulation of transcription from the ssrA promoter. (A) Schematic diagram of the reporter construct used to assay cell cycle regulation of transcription from the ssrA promoter. The arrow indicates the transcriptional start site, and the relative location of the Shine-Dalgarno sequence (S-D) is shown. (B) Promoter activity was measured by immunoprecipitation of β-galactosidase from pulse-labeled samples of a synchronized culture. The amount of β-galactosidase was normalized to the initial time point, and the averages of the results of three independent experiments are shown, with error bars indicating the standard deviations.
Degradation of SsrA RNA.
Cell cycle-regulated transcription of ssrA can account for the increase in SsrA RNA levels during the swarmer- to stalked-cell transition, but the rapid decrease in SsrA RNA levels during the stalked cell stage suggests that SsrA was degraded. To determine whether this was the case, the half-life of SsrA RNA in both log-phase and synchronized cultures was measured by inhibiting transcription with rifampin and monitoring the decay of SsrA RNA by Northern blotting (Fig. 3). In log-phase cultures that contain a mixture of swarmer, stalked, and predivisional cells, both the coding and acceptor RNAs decayed with a half-life of 4 to 5 min (Table 1). The pre-SsrA RNA decayed with a half-life of 2.5 min. However, loss of pre-SsrA RNA is not due solely to RNA degradation; it includes processing into mature SsrA, as well as any complete degradation of the full-length transcript (Fig. 4B). As controls, the Northern blots were stripped and reprobed to measure the decay of two mRNAs, that encoding pilA and that encoding a variant of λ repressor, as well as to measure the decay of 5S rRNA (Table 1). For both mRNAs the half-life was 1 to 2 min, consistent with the rapid degradation expected of mRNAs. The amount of 5S rRNA was constant, indicating that there was no general loss of RNA over the course of the experiment. Thus, mature Caulobacter SsrA RNA is more stable than many mRNAs but is degraded rapidly enough to be completely turned over during a single cell cycle.
FIG. 3.
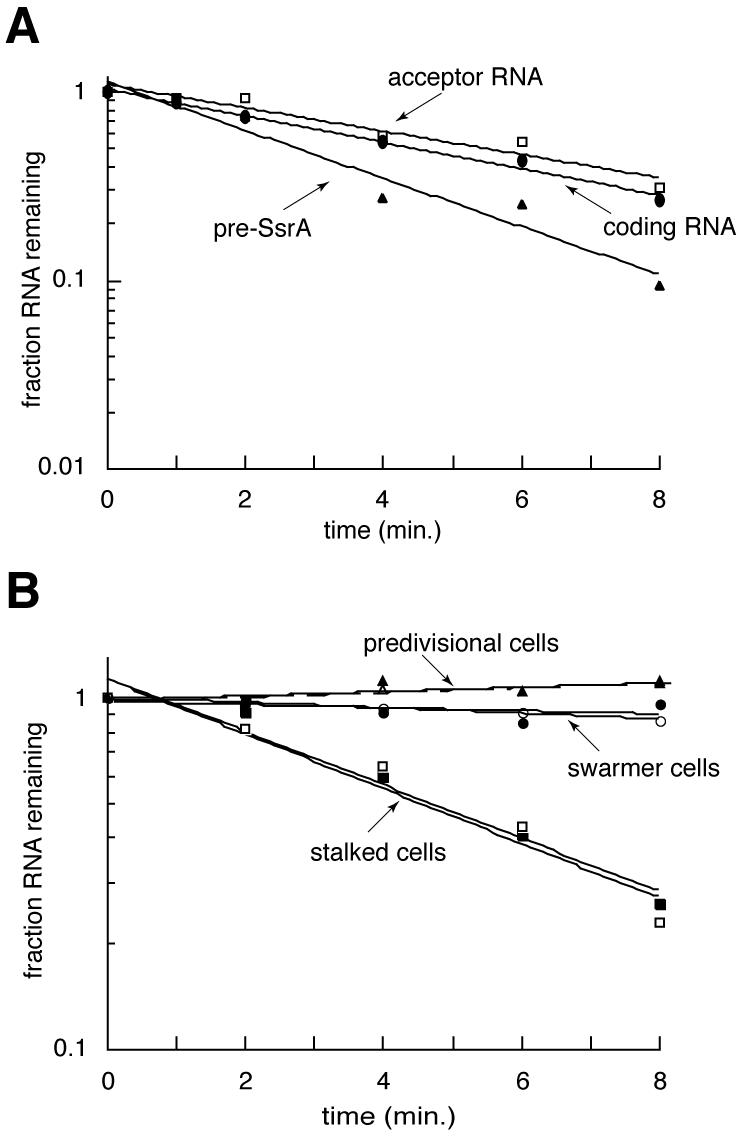
Degradation of SsrA RNA. (A) The decay of SsrA RNAs in a log-phase Caulobacter culture was determined by inhibiting transcription and monitoring the loss of SsrA by Northern blotting. Single-exponential functions fit to each data set are shown. (B) The experiment was repeated on pure populations of swarmer cells (circles), stalked cells (squares), and predivisional cells (triangles). Filled symbols correspond to SsrA coding RNA, and open symbols correspond to SsrA acceptor RNA.
TABLE 1.
Half-lives of RNA species in Caulobacter and E. coli
| Cell type | RNA species | Half-life (min) |
|---|---|---|
| Caulobacter log-phase cultures | SsrA acceptor RNA | 5.2 ± 0.2 |
| SsrA coding RNA | 4.4 ± 0.3 | |
| Pre-SsrA RNA | 2.5 ± 0.2 | |
| 5S rRNA | Stable | |
| pilA mRNA | <2 | |
| λ repressor mRNAa | <2 | |
| Caulobacter swarmer cells | SsrA acceptor RNA | 51 ± 8 |
| SsrA coding RNA | 56 ± 9 | |
| 5S rRNA | Stable | |
| Caulobacter stalked cells | SsrA acceptor RNA | 4.5 ± 1.0 |
| SsrA coding RNA | 4.2 ± 0.4 | |
| 5S rRNA | Stable | |
| Caulobacter predivisional cells | SsrA acceptor RNA | Stable |
| SsrA coding RNA | Stable | |
| 5S rRNA | Stable | |
| E. coli log-phase cultures | SsrA RNA | 89 ± 16 |
| 5S rRNA | Stable |
λ repressor-M2-H6-trpAt variant mRNA measured from Caulobacter constitutively expressing a plasmid-borne copy of this gene (13).
Although it has been assumed that SsrA in E. coli is stable because it is highly abundant, a half-life has not been reported. The decay of SsrA in log-phase cultures of E. coli was measured using the same protocol as for Caulobacter. The half-life of E. coli SsrA under these conditions was 89 min (Table 1), considerably longer than in Caulobacter. The difference in the half-life durations is even more pronounced when the relative length of the cell cycle is considered; in Caulobacter the half-life of SsrA is ∼0.03 cell cycles, whereas in E. coli the half-life of SsrA is ∼3 cell cycles. Thus, with respect to the length of the cell cycle, the degradation of Caulobacter SsrA is 100-fold faster than that of SsrA from E. coli.
To determine whether the degradation of SsrA RNA is cell cycle regulated, the RNA decay experiments were repeated using pure populations of swarmer, stalked, and predivisional cells (Fig. 3B). In swarmer cells, both forms of SsrA RNA decayed with a half-life of more than 50 min. In stalked cells, however, SsrA RNA was degraded with a half-life of 4 to 5 min. This rate of degradation, coupled to the decrease in transcription from the ssrA promoter, is sufficient to account for the fourfold decrease in SsrA RNA levels over 30 min in the stalked-cell stage. In predivisional cells, no degradation of SsrA RNA was detected. These results demonstrate that the amount of SsrA RNA is cell cycle regulated by at least two mechanisms: temporally controlled transcription and temporally controlled degradation (Fig. 4).
DISCUSSION
The data presented here show that SsrA RNA abundance is cell cycle regulated, with peak expression during the G1-S transition, and that at least two mechanisms are employed for this regulation. The regulation of SsrA RNA is not a response to changes in the amount of translation, because the amount of translation in Caulobacter increases at an even rate through most of the cell cycle and does not increase sharply during the G1-S transition (7). If the only function of SsrA RNA is quality control for translation and the total amount of translation does not increase during the G1-S transition, then there must be a dramatic increase in the rate of ribosome stalling during this transition. Alternatively, it is possible that SsrA activity plays a regulatory role in addition to its translational quality control function and that the increase in SsrA RNA levels generates or facilitates a change in gene expression during the G1-S transition. In fact, the increase in the abundance of SsrA RNA during the G1-S transition is correlated with the requirement for SsrA activity for proper timing of DNA replication initiation (12).
The control of both transcription and degradation provides redundant mechanisms to ensure that there is sufficient SsrA RNA during the G1-S transition and that SsrA is cleared from the cell at a critical time after the initiation of DNA replication. Early in the G1-S transition, transcription of ssrA increases and there is no degradation of SsrA RNA; in early S phase, transcription decreases and SsrA RNA degradation is induced (Fig. 4A). Late in S phase, transcription of ssrA increases again and degradation of SsrA RNA is turned off (Fig. 4A). This redundant control by transcription and degradation is analogous to the regulation of CtrA, in which both proteolysis and dephosphorylation are used to ensure that CtrA is not active during the G1-S transition. It is possible that there are also additional mechanisms to regulate SsrA activity in Caulobacter, including control of the abundance and activity of the SsrA RNA-binding protein SmpB, which is required for SsrA activity (12), charging of SsrA RNA with alanine, interaction of SsrA RNA with target ribosomes, and cell cycle regulation of SsrA substrates.
The mechanisms for control of the ssrA promoter are not evident from the DNA sequence. The temporal transcription pattern is similar to that of dnaA, but the sequence elements that have been identified in the dnaA promoter (11, 21, 28) are not found in the ssrA promoter. There are also no consensus CtrA binding sites or binding sites for other characterized transcription factors in Caulobacter. Therefore, understanding the cell cycle regulation of transcription of the ssrA gene requires mutagenesis of the promoter and identification of the transcription factors that regulate its activity.
The regulation of SsrA by ribonucleases includes both the processing of pre-SsrA RNA to the mature form and complete degradation of mature SsrA RNA (Fig. 4B). The processing of pre-SsrA RNA to the mature form is fast enough to prevent accumulation of pre-SsrA RNA throughout the cell cycle. Several ribonucleases, including RNase P, RNase E, and RNase III, have been implicated in the processing of SsrA RNA in E. coli (15, 17, 18, 25, 26). Caulobacter contains homologues of each of these RNases, and it has been proposed that they are responsible for excising the 30-nt loop from Caulobacter pre-SsrA RNA (13). Because each of these nucleases is also involved in the processing of tRNAs and other RNA molecules, it is likely that they are constitutively active in Caulobacter, accounting for the rapid processing of pre-SsrA RNA.
How is degradation of SsrA RNA controlled through the cell cycle? Two possibilities are that SsrA RNA is specifically stabilized in swarmer and predivisional cells and that SsrA RNA is specifically degraded in stalked cells. It is possible that SsrA RNA is stabilized by RNA-binding proteins such as SmpB. Notably, the level of SsrA RNA in log-phase cultures is reduced by 90% in the absence of SmpB (12), although it is not known whether the lower steady-state level is due to decreased transcription or increased degradation. If SmpB is expressed only in swarmer and predivisional cells, then its absence in stalked cells might cause degradation of SsrA RNA. It is also possible that the regulated degradation of SsrA RNA is due to the presence of a specific endo- or exoribonuclease. One-piece SsrAs are likely to be resistant to exoribonucleases, since the 5′ and 3′ ends of SsrA are folded in a tRNA-like conformation, but the two-piece Caulobacter SsrA has additional 5′ and 3′ ends that are possibly available to an exoribonuclease. RNase activity taking place exclusively in the stalked cell would result in the observed regulation of SsrA RNA degradation. One candidate for the nuclease that degrades SsrA RNA in stalked cells is the 3′-5′ exoribonuclease RNase R. RNase R has been found in association with SsrA RNA in E. coli (9), and Caulobacter also has an RNase R homologue. Identification and characterization of the RNase or RNases that degrade SsrA RNA are required to understand this mode of regulation.
Are SsrA RNAs in other organisms also degraded? It has been assumed that SsrA RNA is stable in E. coli and other species because of its abundance and structure, and SsrA RNA in E. coli is in fact very stable (Table 1). However, SsrA RNA in Caulobacter is also highly abundant and structured (13) but is degraded. If the two-piece composition of Caulobacter SsrA is important for its degradation, then two-piece SsrAs in other species, including all α-proteobacteria and some cyanobacteria (13), should be degraded as well. In addition, RNase R is conserved throughout bacteria, so if RNase R is responsible for the degradation of SsrA RNA in Caulobacter strains, it is possible that other bacteria also use RNase R to degrade SsrA RNA. One intriguing possibility is that the regulation of SsrA is correlated with its role in cellular physiology. In Caulobacter strains, SsrA activity is required for correct timing of replication initiation during the G1-S transition (12) and SsrA RNA is degraded once per cell cycle. In E. coli strains, SsrA activity is not required for log-phase growth and SsrA is very stable. It would be instructive to examine the regulation of SsrA RNA in other bacteria that have a requirement for SsrA activity.
Acknowledgments
We thank Quyen-Anh Tran for technical assistance and Sarah Ades for critical reading of the manuscript.
This work was supported by National Institutes of Health Grants GM32506/5120 M2 and GM51426. K.C.K. was supported in part by a DOE-Energy Biosciences Research Fellowship of the Life Sciences Research Foundation.
REFERENCES
- 1.Abo, T., T. Inada, K. Ogawa, and H. Aiba. 2000. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J. 19:3762-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alley, M. R. K., S. L. Gomes, W. Alexander, and L. Shapiro. 1991. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics 129:333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler, A. J., J. G. Kusters, I. Stojiljkovic, and F. Heffron. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 62:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebeling, S., C. Kundig, and H. Hennecke. 1991. Discovery of a rhizobial RNA that is essential for symbiotic root nodule development. J. Bacteriol. 173:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 6.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iba, H., A. Fukuda, and Y. Okada. 1978. Rate of major protein synthesis during the cell cycle of Caulobacter crescentus. J. Bacteriol. 135:647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation, and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 9.Karzai, A. W., and R. T. Sauer. 2001. Protein factors associated with the SsrA.SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. USA 98:3040-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karzai, A. W., M. M. Susskind, and R. T. Sauer. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keiler, K. C., and L. Shapiro. 2001. Conserved promoter motif is required for cell cycle timing of dnaX transcription in Caulobacter. J. Bacteriol. 183:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiler, K. C., and L. Shapiro. 2003. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 185:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keiler, K. C., L. Shapiro, and K. P. Williams. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. USA 97:7778-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 15.Komine, Y., M. Kitabatake, T. Yokogawa, K. Nishikawa, and H. Inokuchi. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:9223-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laub, M. T., H. H. McAdams, T. Feldblyum, C. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 17.Li, Z., S. Pandit, and M. P. Deutscher. 1998. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin-Chao, S., C. L. Wei, and Y. T. Lin. 1999. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 96:12406-12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranquet, C., J. Geiselmann, and A. Toussaint. 2001. The tRNA function of SsrA contributes to controlling repression of bacteriophage Mu prophage. Proc. Natl. Acad. Sci. USA 98:10220-10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Retallack, D. M., L. L. Johnson, and D. I. Friedman. 1994. Role for 10Sa RNA in the growth of λ-P22 hybrid phage. J. Bacteriol. 176:2082-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts, R. C., and L. Shapiro. 1997. Transcription of genes encoding DNA replication proteins is coincident with cell cycle control of DNA replication in Caulobacter crescentus. J. Bacteriol. 179:2319-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roche, E. D., and R. T. Sauer. 2001. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J. Biol. Chem. 276:28509-28515. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89-98. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava, R. A., N. Srivastava, and D. Apirion. 1992. Characterization of the RNA processing enzyme RNase III from wild type and overexpressing Escherichia coli cells in processing natural RNA substrates. Int. J. Biochem. 24:737-749. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava, R. K., A. Miczak, and D. Apirion. 1990. Maturation of precursor 10Sa RNA in Escherichia coli is a two-step process: the first reaction is catalyzed by RNase III in presence of Mn2+. Biochimie 72:791-802. [DOI] [PubMed] [Google Scholar]
- 27.Williams, K. P. 2002. The tmRNA website: invasion by an intron. Nucleic Acids Res. 30:179-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winzeler, E., and L. Shapiro. 1996. A novel promoter motif for Caulobacter cell cycle-controlled DNA replication genes. J. Mol. Biol. 264:412-425. [DOI] [PubMed] [Google Scholar]
- 29.Withey, J., and D. Friedman. 1999. Analysis of the role of trans-translation in the requirement of tmRNA for λimmP22 growth in Escherichia coli. J. Bacteriol. 181:2148-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]