Abstract
Background:
Large gene expression studies, such as those conducted using DNA arrays, often provide millions of different pieces of data. To address the problem of analyzing such data, we describe a statistical method, which we have called 'gene shaving'. The method identifies subsets of genes with coherent expression patterns and large variation across conditions. Gene shaving differs from hierarchical clustering and other widely used methods for analyzing gene expression studies in that genes may belong to more than one cluster, and the clustering may be supervised by an outcome measure. The technique can be 'unsupervised', that is, the genes and samples are treated as unlabeled, or partially or fully supervised by using known properties of the genes or samples to assist in finding meaningful groupings.
Results:
We illustrate the use of the gene shaving method to analyze gene expression measurements made on samples from patients with diffuse large B-cell lymphoma. The method identifies a small cluster of genes whose expression is highly predictive of survival.
Conclusions:
The gene shaving method is a potentially useful tool for exploration of gene expression data and identification of interesting clusters of genes worth further investigation.
Background
Through the use of recently developed DNA arrays, it is now possible to obtain accurate, quantitative (relative) measurements of a large proportion of the mRNA species present in a biological sample. DNA arrays have been used to monitor changes in gene expression during important biological processes (for example, cellular replication and the response to changes in the environment), and to study variation in gene expression across collections of related samples (such as tumor samples from patients with cancer). A major challenge in interpreting these results is to understand the structure of the data produced by such studies, which often consist of millions of measurements. A variety of clustering techniques have been applied to such data, and have proved useful for identifying biologically relevant groupings of genes and samples [1,2,3,4,5,6,7,8,9,10,11,12,13]. Although the underlying principles and computational details of these methods differ, they share the goal of organizing the elements under consideration (such as genes) into groups (clusters) with coherent behavior across relevant measurements (such as samples). Generally absent is any consideration of the nature of the coherent variation. For example, one might want to identify groups of genes that have coherent patterns of expression with large variance across samples, or groups of genes that optimally separate samples into predefined classes (such as different clinical response groups in tumor samples). Here, we introduce a new statistical method, which we call gene shaving, that attempts to identify groups of elements (genes) that have coherent expression and are optimal for various properties of the variation in their expression.
Figure 1 shows the dataset used in our study, which consisted of 4673 gene expression measurements on 48 patients with diffuse large B-cell lymphoma (DLCL). These data have been described in detail previously [14]. The column labels refer to different patients, and the rows correspond to genes. The order of rows and columns is arbitrary.
Figure 1.
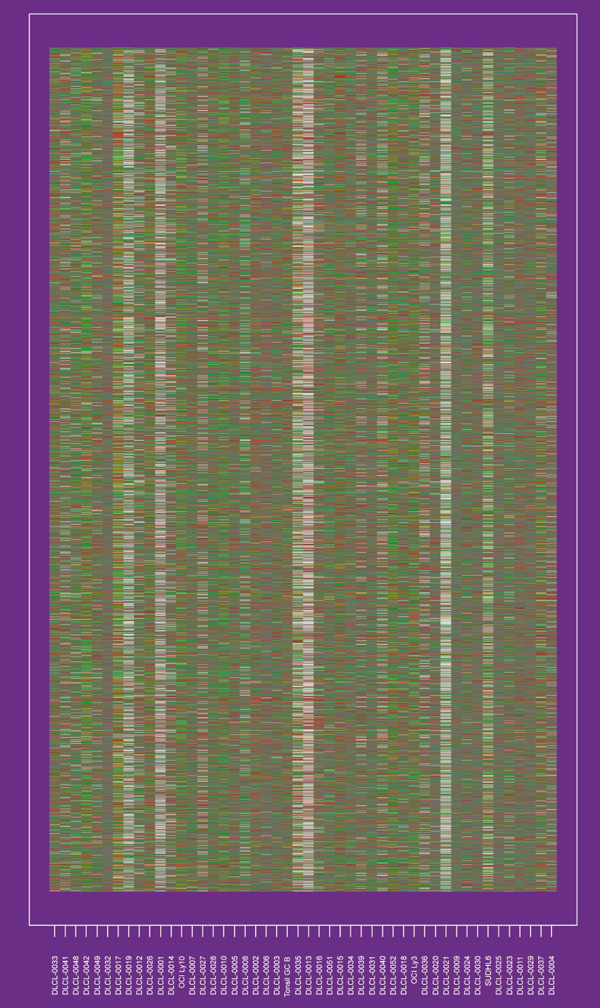
The DLCL expression matrix, in no particular row or column order. The display is a heat map, ranging from bright green (negative, underexpressed) to bright red (positive, overexpressed). The gray cells indicate missing measurements.
Some authors have recently explored the use of clustering methods to arrange the genes in some systematic way, with similar genes placed close together (see [2] for developments and [15] for an overview). In Figure 2, we have applied hierarchical clustering to the genes and samples separately. Each clustering produces a (non-unique) ordering, one that ensures that the branches of the corresponding dendrogram do not cross. Figure 2 displays the original data, with rows and columns ordered accordingly.
Figure 2.
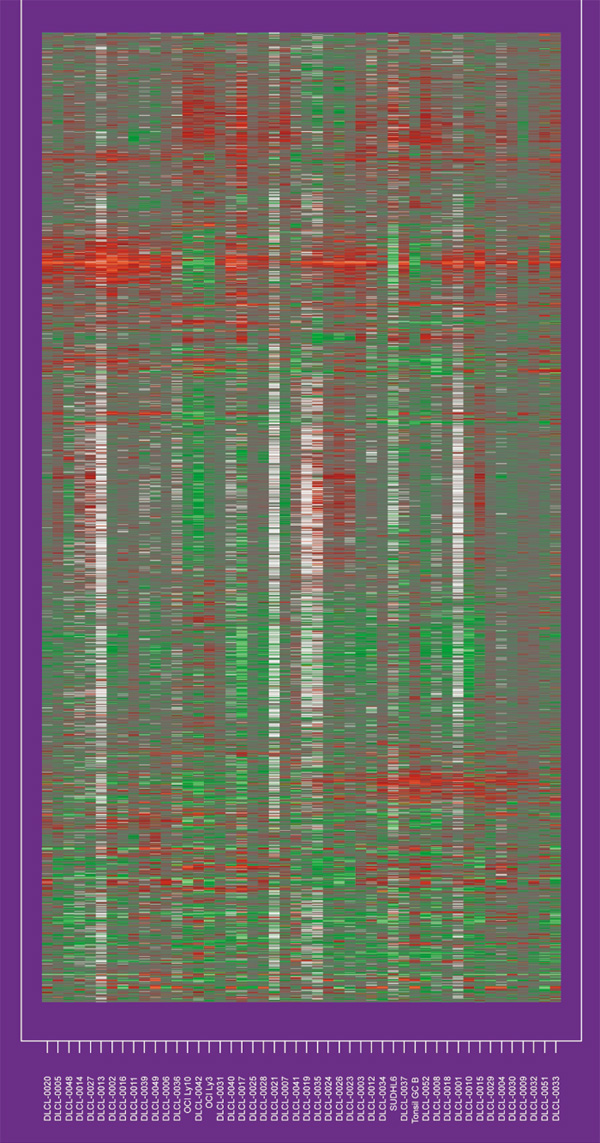
The DLCL expression matrix with rows and columns ordered according to a hierarchical clustering applied separately to the rows and columns.
Some structure is evident in Figure 2, and this method can be used to recognize relationships among the genes and samples.With any method that reduces the dimension of the data, however, finer structure can be lost. For example, suppose the expression of some subset of genes divides the samples in an informative way, correlating with the rate of proliferation of tumor cells, for example, whereas another subset of genes divides the samples a different way, representing the immune response, for example. Then methods such as two-way hierarchical clustering, which seek a single reordering of the samples for all genes, cannot find such structure.
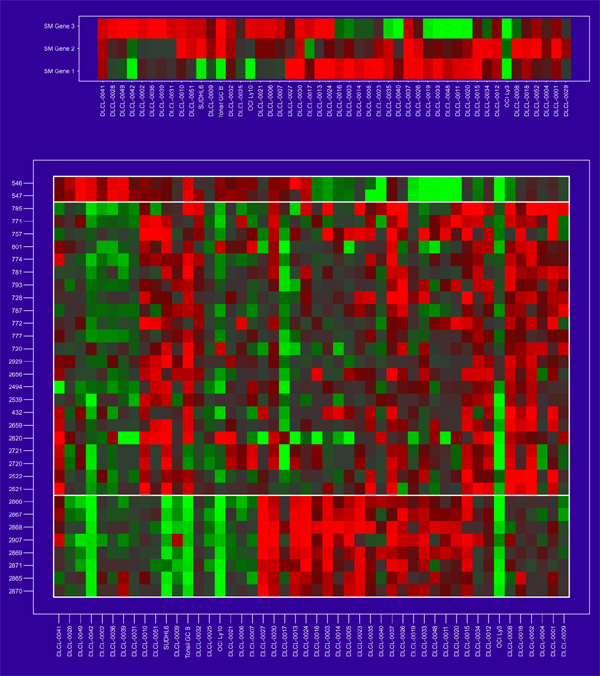
The method of gene shaving we describe here is designed to extract coherent and typically small clusters of genes that vary as much as possible across the samples. Figure 3 shows three gene clusters for the DLCL data, found using shaving. Some of the genes within each cluster lie close to each other in the hierarchical clustering of Figure 2, but others, and the clusters themselves, are quite far apart.
Figure 3.
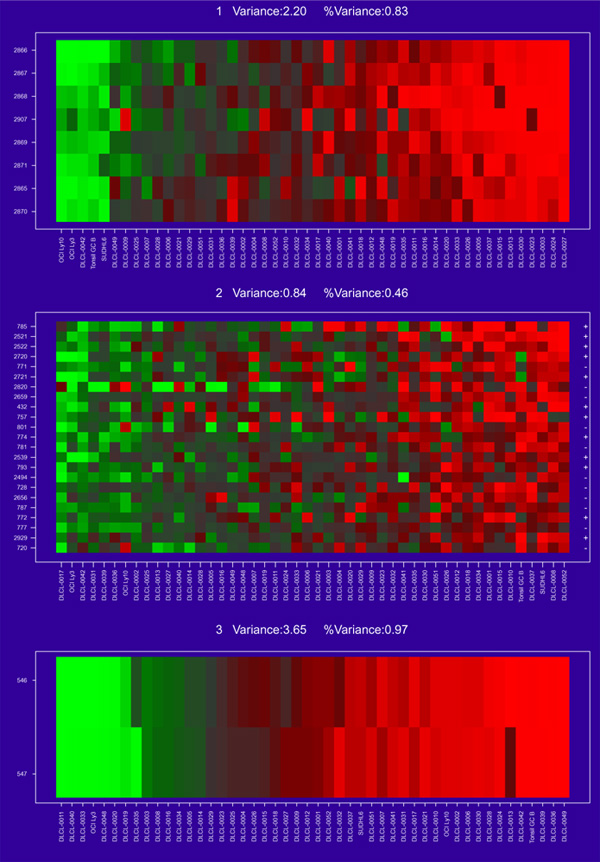
The first three gene clusters found for the DLCL data. Each is a collection of genes showing similar and strong (high variance) expression behavior.
In Figure 3 the samples have been ordered by values of the average gene expression. This average gene is a good representative of the cluster, as all the members are so similar. The variance measures at the top of each cluster are discussed in more detail later. The clusters are all of different sizes. We use an automatic method for determining the size of the clusters, based on a randomization procedure that protects us from looking too hard in the large sea of genes and finding spurious structure. The three cluster-average genes, one from each cluster, are reasonably uncorrelated (see below and Figure 6). This is another aspect of the shaving process - it seeks different clusters, where difference is measured by correlation of the cluster mean. Figure 4 shows the results of a hierarchical clustering applied to the three column-average genes. Whereas hierarchical clustering suggests two main gene groupings, the shaving process may suggest more useful groupings.
Figure 6.
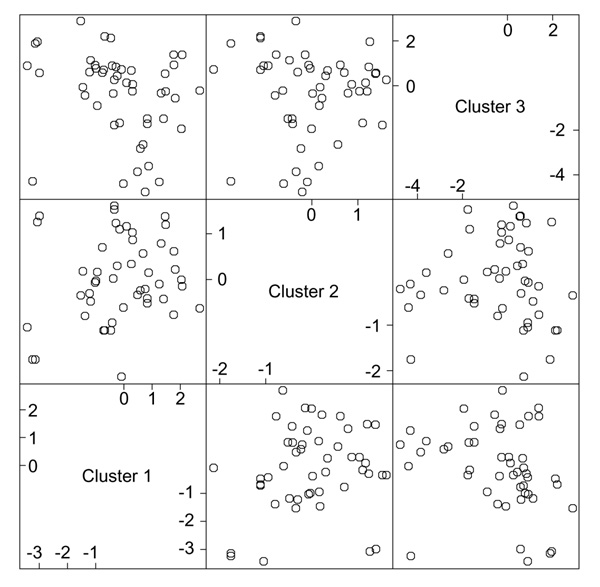
Scatterplot matrix of the three column averages, or `super genes', from each cluster.
Figure 4.
The top panel shows the three signed-mean genes together, and ordered by a hierarchical clustering in this three-dimensional space. The lower panel is similar, except here we show all the genes in each cluster, 33 in all.
This article is organized as follows. In the section 'Gene shaving' we describe the method itself. The section entitled 'The gap estimate of cluster size' outlines the gap test for choosing the cluster size. In the section 'Predicting patient survival' we try to predict patient survival from gene cluster averages. 'Supervised shaving' is discussed in the following section. Finally, in the 'Conclusions' we propose some further generalizations. A more statistical treatment of gene shaving is given in [16].
Results
Gene shaving
In this section we describe in detail our technique for finding clusters like the example in Figure 3. A gene expression matrix is an N × p matrix of real-valued measurements X = xij. The rows are genes, the columns are samples, and xij is the measured (log) expression, relative to a baseline. Typically there are missing entries in X. We use a technique described in [17], an iterative algorithm based on the singular value decomposition, for imputing missing expression values;our analysis here assumes that X has no missing values.
Let Sk be the indices of a cluster of k genes, and
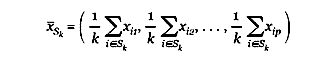
be the collection of p column averages of the expression values for this cluster. Then for each cluster size k, gene shaving seeks a cluster Sk having the highest variance of the column averages:
![]()
The important question of how to choose the cluster size k is addressed in the next section.
Our procedure generates a sequence of nested clusters Sk, in a top-down manner, starting with k = N, the total number of genes, and decreasing down to k = 1 gene. At each stage the largest principal component of the current cluster of genes is computed. This eigen gene is the normalized linear combination of genes with largest variance across the samples. We then compute the inner product (essentially the correlation) of each gene with the eigen gene, and discard ('shave off') a fraction of the genes having lowest (absolute) inner product. The process is repeated on the reduced cluster of genes. The shaving algorithm is depicted in Figure 5 and given in detail in Box 1.
Figure 5.
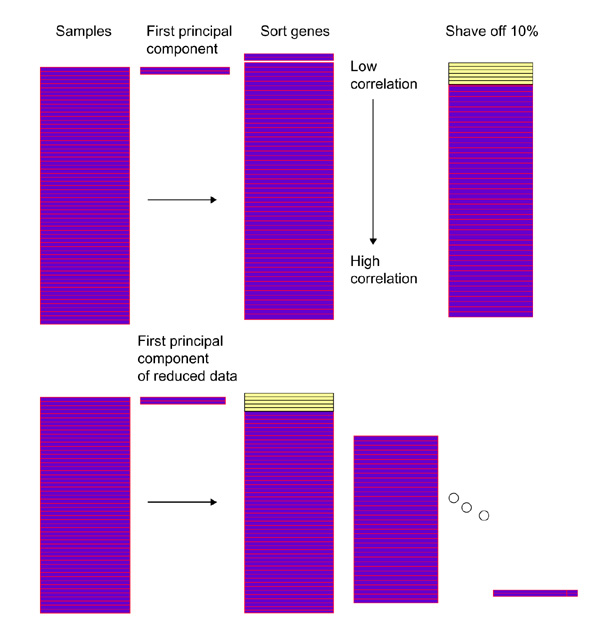
Schematic of the gene shaving process.
Box 1.
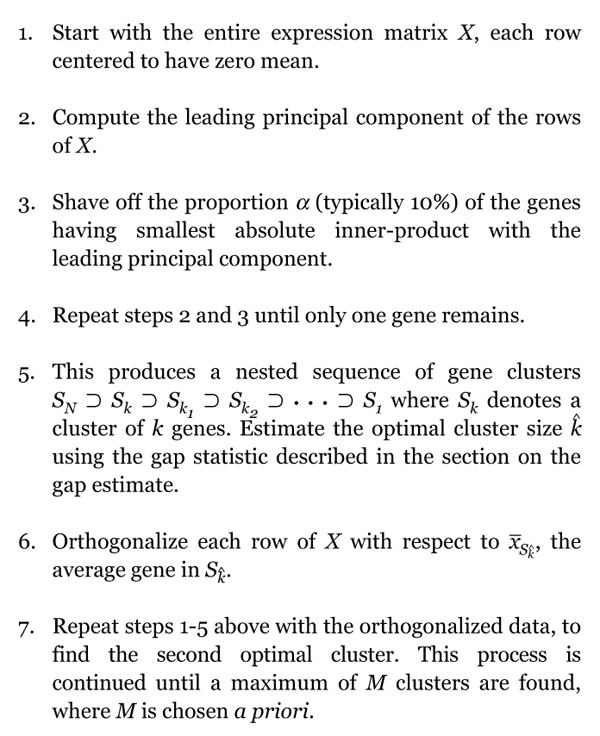
The Shaving algorithm.
There are a number of duplicate genes in the dataset. In some cases the sequence for a given gene appears on the microarray more than once, either by design or by accident. In other cases, more than one different EST (expressed sequence tag) is present for the same gene. Gene shaving can be affected by duplicate genes, since they are highly correlated with each other. We therefore averaged expression profiles for the duplicate genes, leaving 3624 unique gene profiles.
The sequence of operations 1-5 in Box 1 gives the first cluster of rows - the first ribbon in Figure 3. Step 6 orthogonalizes the data to encourage discovery of a different (uncorrelated) second cluster. Note that although we work with the orthogonalized matrix in the shaving process for the second and subsequent clusters, the derived clusters and their averages involve the original genes.
The shaving process requires repeated computation of the largest principal component of a large set of variables. If naively implemented, this requires the construction of a N × N sample covariance matrix ∑ of the genes, and the computation of its largest eigenvector. We can avoid the computational burden by working in the dual space, where the matrices have dimension p × p. Furthermore, as we require only the largest eigenvector, the computations can be reduced even further by using the power method, using the eigenvector of the previous cluster as a starting value.
The three resulting clusters are shown in Figure 3 and again in Figure 4. Figure 6 shows the pairwise scatterplots of each of the three column averages ('super genes') from the clusters. The absolute correlations range from 0.27 to 0.68. The full gene names for the members of the first three clusters are given in Table 1.
Table 1.
The three gene clusters from unsupervised shaving
| Gene number | Clone ID | Description |
| Cluster 1 | ||
| 2866 | "139009" | "Fibronectin I" |
| 2867 | "358168" | "Unknown UG Hs. 106127 ESTs, Highly similar to (defline not available 4689136) [H. sapiens]" |
| 2868 | "323656" | "MMP-2=Matrix metalloproteinase 2=72 kD type IV collagenase precursor=72 kD gelatinase=gelatinase A=TBE-1" |
| 2907 | "897910" | "OSF-2os=osteoblast-specific factor=putative bone adhesion protein with homology with the insect protein fasciclin 1" |
| 2869 | "359412" | "Cyclin D2/KIAK0002=overlaps with middle of KIAK0002 cDNA" |
| 2871 | "754106" | "TIMP-3=Tissue inhibitor of metalloproteinase 3" |
| 2865 | "526335" | "MMP-9=Matrix metalloproteinase 9=92 kD Gelatinase B=92 KD type IV collagenase" |
| 2870 | "487878" | "osteonectin=SPARC=basement membrane protein" |
| Cluster 2 | ||
| 2820 | "753794" | "BLC=BCA-1=B lymphocyte chemoattractant BLC=CXC chemokine" |
| 785 | "1334260" | "Unknown UG Hs. 120716 ESTs" |
| 2521 | "713158" | "Unknown UG Hs.89104 ESTs" |
| 801 | "701361" | "Similar to FXI-TI =FX-induced thymoma transcript" |
| 2720 | "814655" | "Similar to retinol dehydrogenase type 1 (RODH 1)" |
| 2721 | "701122" | "Unknown UG Hs.119410 Homo sapiens cytokine receptor related protein 4 (CYTOR4) mRNA, complete cds" |
| 2522 | "1272196" | "IRF-4=LSIRF=Mum l=homologue of Pip=Lymphoid-specific interferon regulatory factor =Multiple myeloma oncogene 1" |
| 2659 | "685177" | "PTP-1 B=phosphotyrosyl-protein phosphatase" |
| 774 | "701606" | "CD 10=CALLA=Neprilysin=enkepalinase" |
| 771 | "1305913" | "Unknown UG Hs.106771 ESTs" |
| 432 | "417048" | "Similar to human endogenous retrovirus-4" |
| 781 | "1367994" | "myb-related gene A=A-myb" |
| 2539 | "182764" | "EB12=Epstein-Barr virus induced G-protein coupled receptor=Putative chemokine receptor" |
| 757 | "683405" | "SA3=nuclear protein" |
| 793 | "1353041" | "Unknown 166" |
| 2494 | "1357360" | "Cyclin D2/KIAK0002=3\325 end of KIAK0002 cDNA" |
| 2929 | "469297" | "DECI=basic helix-loop-helix protein" |
| 728 | "1338981" | "Unknown UG Hs.l 37038 EST" |
| 2656 | "814768" | "Unknown UG Hs.l 93857 ESTs" |
| 787 | "1338448" | "Unknown UG Hs.224323 ESTs, Moderately similar to alternatively spliced product using exon 13A [H. sapiens]" |
| 720 | "815539" | "JAWI=lymphoid-restricted membrane protein" |
| 772 | "700718" | "Unknown UG Hs.202588 ESTs" |
| 777 | "1352112" | "FMR2=Fragile X mental retardation 2=putative transcription factor=LAF-4 and AF-4 homologue" |
| Cluster 3 | ||
| 546 | "725263" | "immunoglobulin kappa light chain" |
| 547 | "1172268" | "HKG7=cell surface protein in NK and T cells=G-CSF-induced gene" |
The first value given is the gene number in the set of 3624. The second value is the ClonelD. Cross-referencing of this Clone ID with the Accession number is available in the data tables at http://llmpp.nih.gov/lymphoma/data.shtml
It is useful to evaluate how much of the dimensionality of the gene expression variation is captured by the clusters derived from gene shaving. We can approximate the expression profile for each gene in the complete dataset as a linear combination of the three super genes from each cluster (which are simple averages of the genes in each cluster). The percent variance explained by the first j = 1,2, ... 10 super genes is shown in Figure 7.
Figure 7.
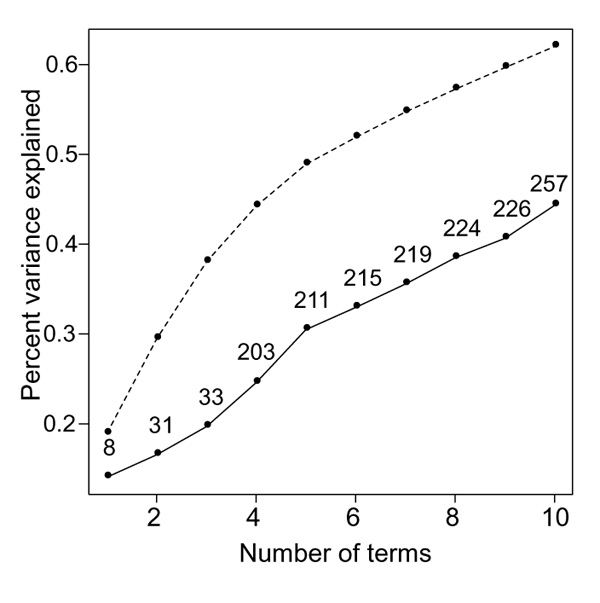
Percent of gene variance explained by first j gene shaving column averages (j = 1,2,... 0) (solid curve), and by first j principal components (broken curve). For the shaving results, the total number of genes in the first j clusters is also indicated.
Thus the three gene clusters, involving a total of 33 genes, explain about 20% of the variation. The percent variance explained by the first j principal components (broken curve) exceeds that from gene shaving. Each principal component gives a non-zero weight to all 3624 genes, however.
The gap estimate of cluster size
Each shaving sequence produces a nested set of gene clusters Sk, starting with the entire expression matrix and then proceeding down to some minimum cluster size (typically 1). If we applied this procedure to null data, in which the rows and columns were independent of each other, we could still find some interesting-looking patterns in the resulting blocks. Hence, we need to calibrate this process so that we can differentiate real patterns from spurious ones. Even in the case of real structure, it is unlikely that a distinct set of genes is correct for a cluster, and the rest not. More likely there is a graduation of membership eligibility, and we have to decide where to draw the line. Here we describe a procedure based on randomization that helps us select a reasonable cluster size.
Our method requires a quality measure for a cluster. We favor both high-variance clusters, and high coherence between members of the cluster. As the generation of the cluster sequence was driven strongly by the former, we focus on the latter in selecting a good cluster. By analogy with the analysis of variance for grouped data, we define the following measures of variance for a cluster Sk:
![]()
![]()
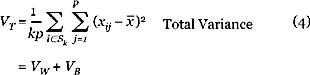
The between variance is the variance of the (signed) mean gene. The within variance measures the variability of each gene about the cluster average, also averaged over samples. As this can be small if the overall variance is small, a more pertinent measure is the between-to-within variance ratio VB/VW, or alternatively, the percent variance explained
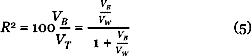
A large value of R2 implies a tight cluster of coherent genes. This is the quality measure we use to select a cluster from the shaving sequence Sk.
Let Sk index the clusters of a given shaving sequence (with k being the number of genes). Let Dk be the R2 measure for the kth member of sequence. We wish to know whether Dk is larger than we would expect by chance, if the rows and columns of the data were independent.
Let X*b be a permuted data matrix, obtained by permuting the elements within each row of X. We form B such matrices, indexed by b = 1,2,... B. Let Dk*b be the R2 measure for cluster Sk*b. Denote by 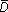 k* the average of Dk*b over b. The Gap function is defined by
k* the average of Dk*b over b. The Gap function is defined by
![]()
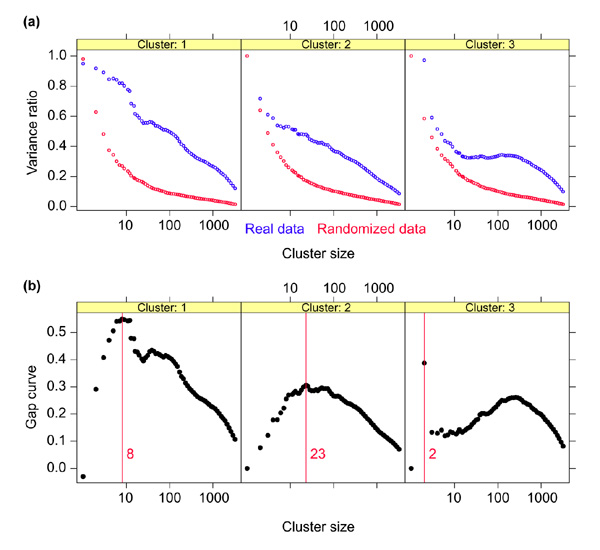
We then select as the optimal number of genes that value of k producing the largest gap:
![]()
The idea is that at the value 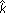 the observed variance is the most ahead of expected. Multiple clusters are produced for the randomized data just like for the original data, and the gap test is used repeatedly to select the cluster size at each stage.
the observed variance is the most ahead of expected. Multiple clusters are produced for the randomized data just like for the original data, and the gap test is used repeatedly to select the cluster size at each stage.
For the DLCL data, the maximum for the first cluster occurs at eight genes. Figure 8 shows the percent-variance curves, Dk, for both the original and randomized tumor data as a function of size, and the gap curves used to select the specific cluster sizes in Figure 3.
Figure 8.
(a) Variance plots for real and randomized data. The percent variance explained by each cluster, both for the original data, and for an average over three randomized versions. (b) Gap estimates of cluster size. The gap curve, which highlights the difference between the pair of curves, is shown.
Predicting patient survival
One important motivation for developing gene shaving was the wish to identify distinct sets of genes whose variation in expression could be related to a biological property of the samples. In the present example, finding genes whose expression correlates with patient survival is an obvious challenge. Group factors g1, g2, g3 were created by splitting each gene cluster in Figure 3 into two groups of 24 patients. We used each of these groupings as a factor in Cox's proportional hazards model for predicting overall survival [18]. Of the group factors only g2 was significant, at the 0.05 level (p = 0.04).
In [14], a cluster of 380 genes was chosen on the basis of their large variation over samples, and their 'germinal center B-like' or 'activated B-like' expression profiles. Using these 380 genes, a hierarchical clustering produced two groups of patients which were (just) statistically different in survival. Close inspection shows that 18 of the 23 genes in the second cluster above also fall into this cluster of 380 genes. Hence, gene shaving can find clinically and biologically relevant subdivisions in gene expression data in an unsupervised fashion.
It may be fortuitous that one of these groupings correlates with survival, as the clusters were not chosen with survival in mind. We next describe a modification of gene shaving that explicitly looks for clusters that are related to patient survival.
Supervised shaving
The methods discussed so far have not used information about the columns to 'supervise' the shaving of rows. Here we generalize gene shaving to incorporate full or partial supervision.
As in Equation (1), we consider a cluster of genes Sk having column average vector  . Let y = (y1, y2, ... yp) be a set of auxiliary measurements available for the samples. For example each yj, might be a survival time for the patient corresponding to sample j or a class label for each sample, such as a diagnosis category. Supervised shaving maximizes a weighted combination of column variance and an information measure J(
. Let y = (y1, y2, ... yp) be a set of auxiliary measurements available for the samples. For example each yj, might be a survival time for the patient corresponding to sample j or a class label for each sample, such as a diagnosis category. Supervised shaving maximizes a weighted combination of column variance and an information measure J( , y):
, y):
![]()
for fixed 0 ≤ α ≤ 1. The value α = 1 gives full supervision; values between o and 1 provide partial supervision.
Choice of the measure J(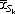 , y) depends on the nature of the auxiliary information y. If the y codes class labels, J(
, y) depends on the nature of the auxiliary information y. If the y codes class labels, J(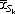 , y) can be taken as the sum of squared differences between the category averages
, y) can be taken as the sum of squared differences between the category averages 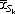 . For censored survival times y, think of
. For censored survival times y, think of 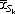 as a covariate in a Cox (proportional hazards) model. If the score vector from this model is g, we set J(
as a covariate in a Cox (proportional hazards) model. If the score vector from this model is g, we set J(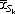 , y) = ggT, a p × p matrix. Computationally we first scale the genes so that the within-risk set variance is 1.
, y) = ggT, a p × p matrix. Computationally we first scale the genes so that the within-risk set variance is 1.
When fully supervised, the shaving procedure reduces to simply ranking the genes from largest to smallest Cox model score test. Thus there is no role for principal components or top-down shaving in this case. However, to encourage coherence within the gene clusters, it can be better to use a partially supervised criterion, which does use principal components and top-down shaving. This is demonstrated in the example below. One can also include other prognostic factors in the model, and direct shaving to find a gene cluster whose column average is a strong predictor in the model. This can be done with other models, for example a linear regression model for a quantitative measurement. Operationally, all of these choices for J are quadratic functions of the column averages 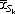 , and gene shaving can be carried out via principal components of a suitably modified variance matrix.
, and gene shaving can be carried out via principal components of a suitably modified variance matrix.
We applied supervised shaving to the set of 3624 genes from the DLCL samples. Figure 9 examines the effect of different values of the supervision weight α, showing the average (absolute) gene correlation and Cox model p value for each choice. From this plot we chose the value α = 0.10, which gives a good mix of high gene correlation and low p value. Partially supervised gene shaving produced a cluster with 234 genes, shown in Figure 10 and in Table 2.
Figure 9.
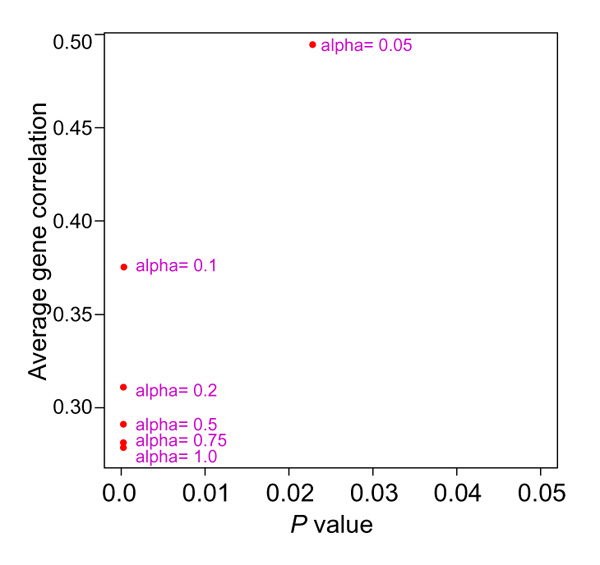
Average (absolute) gene correlation and Cox model p value, for clusters of size 200 from supervised shaving and for different values of α. The value of Qa = 0.1 seems best, and is used in the gene shaving procedure.
Figure 10.
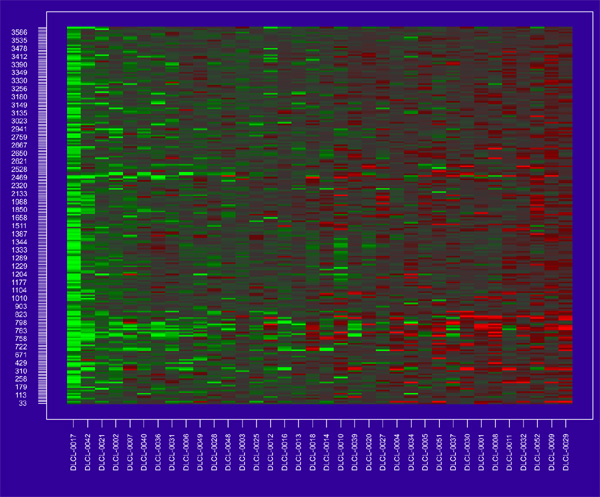
Cluster of 234 genes from supervised shaving.
Table 2.
Cluster from supervised shaving applied to full set of 3624 genes
| Position | ClonelD | Description |
| "-685" | "712937" | "hPMSI=DNA mismatch repair protein=mutL homologue" |
| "-3531" | "1186043" | "Unknown UG Hs.134746 ESTs," |
| "1661" | "352820" | "Unknown UG Hs.231825 ESTs" |
| "-2667" | "1356433" | "Unknown 645" |
| *"798" | "814622" | "Unknown UG Hs.49614 ESTs" |
| "-3545" | "713080" | "CLK-2=cdc2/CDC28-like protein kinase-2" |
| *"-153" | "1339106" | "XE7=B-lymphocyte surface protein" |
| *"824" | "1356501" | "Unknown UG Hs.130721 ESTs" |
| "-3414" | "1319801" | "Similar to non-erythropoietic porphobilinogen deaminase (hydroxymethylbilane synt EC4.3.1.8)" |
| "-1577" | "1353785" | "Unknown UG Hs.119769 ESTs" |
| "-3242" | "376942" | "Ro ribonucleoprotein autoantigen (Ro/SS-A)=autoantigen calreticulin" |
| *"-3535" | "1336373" | "Similar to High mobility group (nonhistone chromosomal) protein isoforms I and Y" |
| "-3412" | "344219" | "5'-terminal region of UMK" |
| "-673" | "279363" | "Adenosine kinase" |
| "920" | "1355987" | "Unknown UG Hs.180836 EST" |
| *"800" | "1358163" | "Phosphatidylinositol 3-kinase p1 10 catalytic, gamma isoform" |
| *"823" | "1319062" | "WIP/HS PRPL-2=WASP interacting protein" |
| *"799" | "1339726" | "Unknown 168" |
| *"788" | "825199" | "Unknown 164" |
| "-3544" | "1285581" | "Similar to myb-related gene A-myb 5'-region" |
| "-68" | "589589" | "homolog of Drosophila splicing regulator suppressor-of-white-apricot" |
| *"759" | "1333557" | "Unknown 161" |
| "339" | "1336946" | "Unknown 80" |
| "-178" | "1354703" | "Unknown UG Hs.150458 ESTs" |
| "-933" | "1184133" | "CASPASE-3=CPP32 isoform alpha=yama=cysteine protease" |
| "-2714" | "149994" | "B12 protein=tumor necrosis factor-alpha-induced endothelial primary response gene |
| "-3364" | "271976" | "ACYI =aminoacylase-I" |
| "-118" | "145409" | "Low-affinity IgG Fc receptor II-B and C isoforms (multiple orthologous genes)" |
| *"-671" | "1317098" | "tyrosine kinase (TnkI)" |
| "-2623" | "324973" | "9G8 splicing factor" |
| *"783" | "814601" | "Unknown UG Hs.161905 EST" |
| "2421" | "1370055" | "Unknown 602" |
| "1855" | "1358160" | "Unknown 428" |
| *"813" | "23173" | "JNK3=Stress-activated protein kinase" |
| "-1412" | "22438" | "RYK receptor-like tyrosine kinase" |
| "1104" | "1336779" | "Unknown 221" |
| "1521" | "1670861" | "Unknown UG Hs.32533 ESTs" |
| "2568" | "1184568" | "Unknown UG Hs.120785 ESTs" |
| "-3161" | "365358" | "pM5 protein=homology to conserved regions of the collagenase gene family" |
| "279" | "1367883" | "KIAA0430" |
| "338" | "1336591" | "Unknown UG Hs.180644 ESTs" |
| *"63" | "746300" | "Unknown UG Hs.136345 ESTs" |
| *"-2661" | "1302032" | "Deoxycytidylate deaminase" |
| *"787" | "1338448" | "Unknown UG Hs.224323 ESTs, Moderately similar to alternatively spliced product exon 13A [H.sapiens]" |
| "2567" | "1354788" | "Unknown 627" |
| *"758" | "1333558" | "Unknown 160" |
| "-3264" | "704732" | "Unknown 699" |
| "-2654" | "724397" | "lymphopain=C 1 peptidase expressed in natural killer and cytotoxic T cells" |
| "1132" | "1354522" | "Unknown UG Hs.125285 ESTs, Highly similar to (defline not available 4200446) [Mlus]" |
| *"-1595" | "1186040" | "Unknown UG Hs.136589 ESTs" |
| "-2320" | "241481" | "CASPASE-10=Mch4=FLICE2" |
| "-3345" | "502761" | "Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase phoribosylaminoimidazole synthetase" |
| "-33" | "268727" | "MYH=DNA mismatch repair protein=mutY homologue" |
| *"774" | "701606" | "CDIO=CALLA=Neprilysin=enkepalinase" |
| "-533" | "276483" | "(2'-5') oligoadenylate synthetase E" |
| "1388" | "1350824" | "Unknown UG Hs.163773 ESTs" |
| "-3244" | "488754" | "DAP-1 =putative mediator of the gamma interferon-induced cell death" |
| "3097" | "686331" | "DCHT=Similar to rat pancreatic serine threonine kinase" |
| "-2641" | "1355868" | "Unknown 643" |
| "-3135" | "199018" | "P120=proliferating-cell nudeolar protein" |
| "-1578" | "713301" | "Unknown UG Hs.32218 ESTs, |
| "-2502" | "153355" | "LD78 beta=almost identical to MIP-1 alpha=chemokine" |
| "2328" | "1341026" | "yotiao=protein of neuronal and neuromuscular synapses that interacts with specific variants of NMDA receptor subunit NR 1" |
| "1863" | "1357676" | "Unknown UG Hs.191211 ESTs" |
| "1399" | "1356420" | "Unknown UG Hs.207995 ESTs" |
| "-3401" | "844479" | "Pig8=p53 inducible gene=etoposide-induced mRNA=Similar to El 24 = p53 responsive (sculus)" |
| "-3040" | "1368740" | "Unknown UG Hs.125307 EST" |
| "-3193" | "152653" | "C-I-Tetrahydrofolate Synthase, cytoplasmic" |
| "-3437" | "814765" | "kinase A anchor protein" |
| "1387" | "1318821" | "Unknown UG Ms. 108614 Homo sapiens mRNAfor KIAA0627 protein, partial cds" |
| "-2527" | "1357085" | "Acidic 82 kDa protein" |
| *"1400" | "682995" | "Unknown 298" |
| *"724" | "1286796" | "Unknown UG Hs.61506 ESTs" |
| "413" | "1334297" | "Unknown 98" |
| *"789" | "825217" | "Unknown UG Hs.169565 ESTs, |
| "-2754" | "1318136" | "5'-AMP-activated protein kinase, gamma-I subunit" |
| "1052" | "1240803" | "Unknown 211" |
| "278" | "815671" | "Unknown UG Hs.101340 ESTs" |
| "-2501" | "346550" | "MIP-1 alpha=LD78 alpha=pAT464=Small inducible cytokine A3=macrophage inflammatory in (GOS 19-1)=chemokine" |
| "1988" | "1320268" | "Unknown 480" |
| "-903" | "704637" | "Unknown UG Hs.5354 ESTs" |
| "-2649" | "181998" | "NFAT3=NFATc4" |
| "-2648" | "171693" | "Lst-1 =IC7=interferon-gamma-inducible gene present in lymphoid tissues, T cells, macrophages, and histiocyte cell lines |
| encoding a transmembrane protein" | ||
| "2373" | "1338072" | "Unknown 592" |
| "223" | "1352327" | "Unknown 52" |
| "1269" | "1339210" | "Unknown 261" |
| "-3004" | "1289545" | "Unknown UG Hs.187869 ESTs" |
| "1177" | "700949" | "Similar to myosin-IXb" |
| *"779" | "703735" | "Unknown UG Hs.28355 ESTs" |
| *"464" | "685761" | "Unknown III" |
| "1229" | "700643" | "Unknown UG Hs.104492 ESTs" |
| "-3482" | "51058" | "E2F-4=pRB-binding transcription factor" |
| "-3584" | "1358191" | "Similar to DNA polymerase beta=DNA alkylation repair protein" |
| *"-429" | "35356" | "Neurotrophic tyrosine kinase, receptor, type 3 (TrkC)" |
| "-3136" | "265590" | "NFI =Neurofibromin" |
| "956" | "1289384" | "Unknown 198" |
| "2491" | "814251" | "SLAM=signaling lymphocytic activation molecule" |
| "2083" | "1353083" | "Unknown UG Hs.136972 EST" |
| "1102" | "1372068" | "KIAA0603=Similar to TBCI" |
| "-1010" | "595474" | "Pak 1 =p21-activated protein kinase" |
| "-3594" | "1269836" | "BCL-7B" |
| "-2270" | "265267" | "HSP70" |
| "-944" | "1337124" | "Unknown UG Hs.81248 CUG triplet repeat, RNA-binding protein 1" |
| "-3330" | "1301224" | "Elongin B=RNA polymerase II transcription factor SIII pi 8 subunit" |
| "1658" | "1241118" | "Unknown 346" |
| "-3140" | "841361" | "GR02=GRO beta=MIP2 alpha=macrophage inflammatory protein-2 alpha=chemokine" |
| "-2651" | "525540" | "BCL-3" |
| "-3350" | "1186114" | "Unknown UG Hs.116447 EST" |
| "-2990" | "1289569" | "Unknown UG Hs.146165 ESTs" |
| *"809" | "1270618" | "Unknown UG Hs.208970 EST, Weakly similar to neuronal thread protein AD7c-NTP [ens]" |
| "-3160" | "703707" | "Protein disulfide isomerase-related protein (PDIR)" |
| "874" | "1320313" | "Unknown UG Hs.132458 ESTs" |
| "-3390" | "1339763" | "Unknown 710" |
| "1343" | "1318717" | "LOK=lymphocyte oriented kinase=STE20-like protein kinase that is expressed predominantly in lymphocytes" |
| "-179" | "301551" | "Integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)" |
| *"723" | "824754" | "Unknown UG Hs.145058 ESTs" |
| "-3406" | "1300230" | "Unknown UG Hs.56421 ESTs, Weakly similar to Similarity to H.influenza ribonucl H [C.elegans]" |
| "-573" | "1341161" | "Similar to rhoGap protein" |
| *"722" | "1341225" | "Unknown UG Hs.186709 ESTs,! [H.sapiens]" |
| "2212" | "1350784" | "Unknown UG Hs.163202 EST" |
| "-3478" | "417897" | "cleavage stimulation factor 77kDa subunit=polyadenylation factor subunit=homolog the Drosophila suppressor of forked protein" |
| "-887" | "756965" | "RGS14=regulator of G protein signaling" |
| "1344" | "825333" | "Unknown UG Hs.193017 ESTs, Highly similar to (defline not available 4220898) [ens]" |
| *"743" | "1358192" | "Unknown UG Hs.228205 EST, |
| "1850" | "1353072" | "Unknown 426" |
| "-3391" | "1340604" | "Unknown UG Hs.127121 ESTs" |
| "-236" | "686771" | "tubulin-gamma" |
| "-3343" | "293934" | "CAS=chromosome segregation gene homolog" |
| "2566" | "1350728" | "Unknown 626" |
| "-2984" | "955354" | "putative cell surface ligand for T1/ST2 receptor (related to IL-1 receptors)" |
| "-3149" | "366713" | "GSK3=glycogen synthase kinase 3" |
| *"720" | "815539" | "JAWI=lymphoid-restricted membrane protein" |
| "-3177" | "378364" | "PRODH=proline dehydrogenase/proline oxidase=p53-induced gene" |
| "1268" | "1339305" | "Unknown 260" |
| "-3616" | "1302092" | "Unknown UG Hs.214428 ESTs" |
| "1210" | "685368" | "Unknown 243" |
| "2330" | "1240688" | "Unknown 577" |
| "259" | "1369262" | "KIAAOO 19=similar to transforming protein tre"-2528" "1184411" "MINOR=mitogen induced nuclear orphan receptor=NOR-l=Nur77 orphan nuclear receptor family member" |
| "-3586" | "1309295" | "Unknown UG Hs.136985 ESTs" |
| "2045" | "1352570" | "Unknown 494" |
| "2067" | "1320316" | "Unknown 508" |
| "-3533" | "298303" | "TECK chemokine" |
| "-3530" | "1355240" | "Unknown UG Hs.130849 ESTs" |
| *"-2469" | "417226" | "c-myc" |
| "1784" | "1355354" | "Unknown 394" |
| "-3023" | "700772" | "Smad2=Madr2=JV 18-1 =Homologue of Mothers Against Decapentaplegic (MAD)=Activated beta" |
| *"793" | "1353041" | "Unknown 166" |
| "-3162" | "1289546" | "Similar to arginine/aspartate-rich 37.3 K protein" |
| *"-2669" | "1186215" | "Unknown UG Hs.190288 EST" |
| "-113" | "1337185" | "KIAA0037" |
| "-3434" | "1338032" | "CPR2=cell cycle progression 2" |
| "-2621" | "1338456" | "c-myc binding protein" |
| "1333" | "824376" | "Similar to (AFO 16450) Similar to acyltransferase" |
| "-3405" | "1334813" | "Unknown UG Hs.17883 protein phosphatase IG (formerly 2C), magnesium-dependent, isoform" |
| "2301" | "300051" | "myosin light chain-2" |
| "1144" | "1372011" | "Unknown UG Hs.209146 ESTs" |
| "-3436" | "485171" | "methionine adenosyltransferase alpha subunit" |
| "1339" | "1355713" | "Unknown 277" |
| "1156" | "1351290" | "Similar to (Z49125) C47G2.4" |
| *"721" | "1353015" | "Unknown 154" |
| "-3125" | "86040" | "Cytochrome P450, subfamily 1, polypeptide 2 (aromatic compound-inducible)" |
| "258" | "1367988" | "Unknown 61" |
| "-3258" | "1304523" | "APRT=adenine phosphoribosyltransferase" |
| "-3548" | "1340120" | "Unknown 733" |
| "151 1" | "1351701" | "Unknown UG Hs.124230 ESTs" |
| "-3280" | "826594" | "replication factor C" |
| "-3363" | "293035" | "APEX=apurinic endonuclease=DNA alkylation repair protein" |
| "1190" | "1371313" | "Similar to G-protein coupled receptor pH218" |
| "1321" | "1309301" | "Unknown UG Hs.136987 EST" |
| "-3180" | "591683" | "GADD45 alpha=growth arrest and DNA-damage-inducible protein alpha" |
| "1748" | "1371159" | "Unknown 377" |
| "-2781" | "1288183" | "BAK=BCL-2 family member" |
| "108" | "1370125" | "Unknown 22" |
| "-2941" | "742132" | "Interferon-induced 17 KD protein" |
| "-2994" | "1271685" | "Unknown UG Hs.176669 ESTs" |
| "1287" | "1353226" | "Unknown UG Hs.30209 Homo sapiens mRNA for KIAA0854 protein, complete cds" |
| "1039" | "1671442" | "Unknown UG Hs.171096 ESTs, Weakly similar to (defline not available 4456988) [ens]" |
| *"83" | "52408" | "ABR=guanine nucleotide regulatory protein" |
| "3624" | "1355859" | "Similar to myosin IE heavy chain" |
| "-2746" | "1350736" | "IRF-3=interferon regulatory factor-3" |
| "1303" | "665682" | "Jnkk2=JNK kinase 2=MAP kinase kinase" |
| "877" | "1367968" | "Unknown UG Hs.105072 ESTs" |
| "-3344" | "1341245" | "CD73=5' nudeotidase" |
| "1191" | "1371317" | "Similar to arylacetyltransferase" |
| *"-310" | "154493" | "HNPP=nuclear phosphoprotein" |
| "1976" | "1334933" | "Unknown UG Hs.144684 ESTs" |
| "-2609" | "1670958" | "SRF=c-fos serum response element-binding transcription factor" |
| "405" | "701689" | "putative tumor suppressor (LUCA15)" |
| "-3319" | "1307997" | "Similar to bromodeoxyuridine-sensitive transcript protein=px 19" |
| "-3255" | "810743" | "MLF2=myelodysplasia/myeloid leukemia factor 2" |
| "2150" | "1353466" | "Unknown UG Hs.124360 EST" |
| "-2650" | "511407" | "69 kDa 2'5' oligoadenylate synthetase (P69 2-5A synthetase)" |
| "252" | "1356345" | "Unknown UG Hs.49500 Homo sapiens mRNA for KIAA0746 protein, partial cds" |
| "1337" | "1367875" | "Unknown UG Hs.128127 ESTs" |
| "1302" | "1351266" | "Unknown UG Hs.134197 ESTs, Moderately similar to FAM [M.musculus]" |
| "1386" | "815165" | "Unknown UG Hs.188732 ESTs" |
| "-3147" | "549277" | "cell cycle protein p38-2G4 homolog (hG4-l)" |
| "-3349" | "1355524" | "Similar to rapamycin-binding protein (FKBP25)" |
| "-173" | "1287032" | "Similar to Drosophila female sterile homeotic (FSH) homologue" |
| *"777" | "1352112" | "FMR2=Fragile X mental retardation 2=putative transcription factor=LAF-4 and AF-4 ogue" |
| "-3334" | "346948" | "nm23-H2=NDP kinase B=Nucleoside dephophate kinase B" |
| "-3256" | "1303575" | "Unknown UG Hs.123304 ESTs" |
| "1289" | "704690" | "Dyrk6=Ser/Thr protein kinase" |
| "1133" | "1351498" | "Unknown UG Hs.189063 ESTs" |
| "2058" | "1339890" | "Unknown 503" |
| "-2927" | "342647" | "MAPKAP kinase (3pK)" |
| "1324" | "687198" | "Unknown UG Hs.125860 ESTs" |
| "-3047" | "203704" | "flavin-containing monooxygenase (FMO 1)" |
| "-2662" | "1288102" | "Similar to nuclear-encoded mitochondrial NADH-ubiquinone reductase 24Kd subunit" |
| "1852" | "1371200" | "Similar to (Z78012) C52E4.6" |
| "1383" | "1319529" | "Unknown 293" |
| "-3360" | "1671396" | "Similar to friend of GATA-1 (FOG)=zinc finger GATA-I coactivator in erythroid and megakaryocyte lineages" |
| "1228" | "1336501" | "Unknown 249" |
| "1353" | "1356762" | "Unknown UG Hs.127480 ESTs" |
| *"-575" | "490387" | "zinc finger protein 42 MZF-1" |
| "1242" | "1031754" | "Protein-tyrosine phosphatase 2C" |
| "1211" | "1372274" | "Unknown UG Hs.208983 ESTs, |
| "-2759" | "489438" | "MyD88=myeloid differentiation primary response protein=death domain-containing p |
| "1227" | "1334962" | "Similar to KIAA0437" |
| "260" | "1341211" | "Unknown UG Hs.191209 ESTs" |
| "-3137" | "1250770" | "Purine nucleoside phophorylase=lnosine phosphorylase=PNP" |
| "1385" | "1371029" | "Unknown 295" |
| "1808" | "1372833" | "Unknown 403" |
| "-2762" | "1184153" | "Unknown UG Hs.230206 EST" |
| "1046" | "1352940" | "Unknown 208" |
| "-2766" | "756452" | "tyk2=non-receptor protein tyrosine kinase" |
| "1204" | "1370570" | "Lamin B receptor (LBR)" |
| "1201" | "1241671" | "Similar to (AE000860) conserved protein [Methanobacterium thermoautotrophicum]" |
| *"735" | "686893" | "Unknown UG Hs.226955 ESTs" |
| "1338" | "1370103" | "Unknown 276" |
| "255" | "1338624" | "Unknown UG Hs.192864 ESTs" |
| "1200" | "1352335" | "Unknown UG Hs.99701 ESTs" |
| "2133" | "1340880" | "Cancer associated surface antigen (RCASI)" |
Genes are ordered from strongest to weakest correlation with survival. The first number is the position in the hierarchical clustering ordering (a minus sign indicates the sign of the gene is to be flipped before averaging); * indicates a gene that also falls in the 380 gene cluster from Alizadeh et al. [14].
Some of the genes are close together in the hierarchical clustering order (indicated by the first number in Table 2), many are not. Some genes have a negative sign, and others have no sign. We will call these 'negative' and 'positive' genes respectively. The negative genes have their expression values flipped before being averaged with other gene expression profiles. Figure 11a shows the gap curve, suggesting a cluster size of 35. However, further analysis below suggests the better cluster size of 234.
Figure 11.
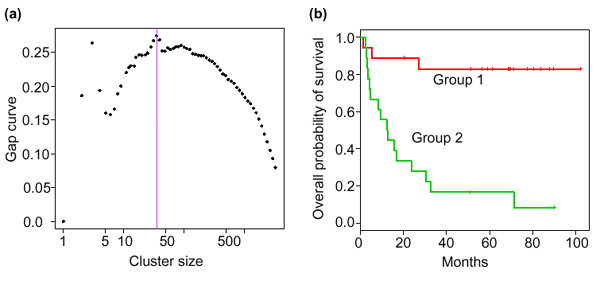
(a) Gap curve for supervised shaving. (b) Survival curves in the two groups defined by the low or high expression of the 234 genes. Group I has high expression of positive genes, and low expression of negative genes; group 2 has low expression of positive genes, and high expression of negative genes. Negative genes are those preceded by a minus sign in Table 2.
The cluster of 234 genes contains many of the strongest individual genes for predicting survival. For example, 130 of the strongest 200 genes are in the cluster. Some weaker genes are, however, also in the cluster, the weakest being the 1332nd strongest gene among the full list of 3624. Figure 11b shows the survival curves stratified by the low and high expression of this gene cluster, using the median of the cutoff point. The two resulting groups are shown in Figure 12.
Figure 12.

The two groups of samples that showed highest and lowest expression of the gene cluster associated with survival.
Using this grouping as a predictor in the Cox model for survival gave a highly significant p value (0.00042). However, this p value must be scrutinized. Figure 13a,b shows the Cox model p value as a function of the cluster size, for both partially and fully supervised shaving. We will call these the 'training set p values'. As the gene shaving process was supervised by the survival times, the training set p values will be biased downward, and it is important to adjust them appropriately. We permuted the survival times, re-ran the shaving process and computed the corresponding p values. This was repeated 100 times, and for each cluster size we computed the proportion of times the permutation p values were less than or equal to the training set p values. These proportions are shown in Figure 13c,d, and are unbiased estimates of the true p values. Fully supervised shaving is too aggressive, and the upward adjustment of the p values is large. As a result the p value is around 0.05 for the full set of genes, but none of the smaller clusters is significant at the 0.05 level. For partially supervised shaving, many of the p values are below 0.05, and from this we chose the cluster size of 234 near the minimum.
Figure 13.
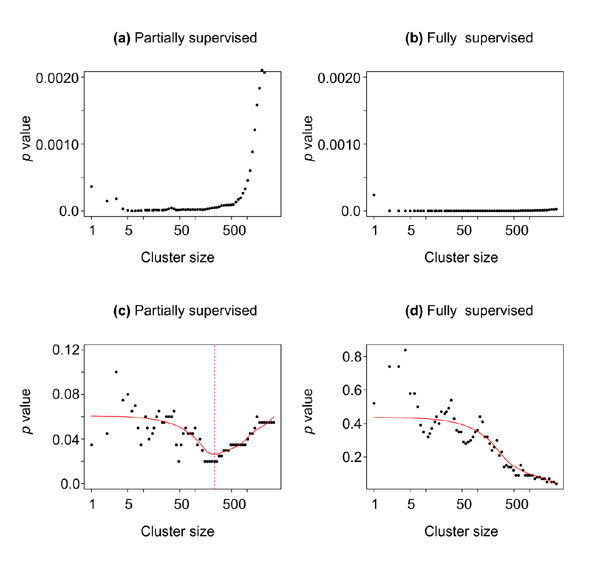
Supervised gene shaving from full gene set. (a,c) Partially supervised with α = 0.10; (b,d) fully supervised (α = 1). (a,b) Training set p values; (c,d) permutation p values for the cluster average as a function of cluster size. The chosen cluster size of 234 is indicated.
Using the full set of genes, flipping each to have positive correlation with survival, averaging their expression values and finally cutting at the median, gave a grouping nearly the same as Groups 1 and 2 in Figure 12. The only change was a swap between DLCL-oo14 and DLCL-oo18, and these two samples are right at the median cutpoint between the two groups in Figure 10.
The international prognostic index (IPI) A score was also available for these patients. Components of the IPI include age, level of the enzyme lactate dehydrogenase (LDH) and the number of extranodal sites. As in [14], we dichotomized IPI scores into low (0, 1 or 2) and high (3, 4 or 5). The resulting grouping seems to be about as predictive as the IPI index, and is quite independent from it, as Table 3 indicates.
Table 3.
Cross-tabulation of gene shaving groups with IPI index
| IPI | ||
| Low | High | |
| Gene shaving groups | ||
| 1 | 7 | 7 |
| 2 | 11 | 7 |
When added to a Cox model containing IPI, this grouping had a training set p value of 0.0006. Figure 14 shows the Kaplan-Meier survival curves for each group, stratified by low and high IPI.
Figure 14.
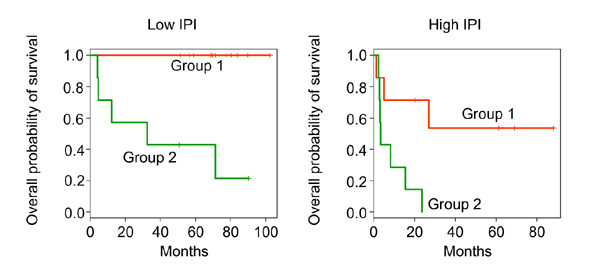
Kaplan-Meier survival curves in the two groups defined by the cluster of 234 genes shown in Figure 10, stratified by IPI. Group I has high expression of positive genes and low expression of negative genes in Figure 9, and vice-versa for Group 2.
In [14], two patient groups were defined from a hierarchical clustering tree grown from a 38o-gene cluster. As a predictor, the grouping was just significant in the low IPI group only, at the 0.05 level. Table 4 gives a cross-tabulation of that grouping with the one used in this paper in Figure 10.
Table 4.
A comparison of the patient groups obtained by gene shaving with those obtained previously [14]
| Patient groups of Alizadeh et al. [14] | ||
| Low | High | |
| Gene shaving groups | ||
| 1 | 13 | 5 |
| 2 | 6 | 12 |
Thus 25/36 = 69% of the patients are classified the same way by both groupings. The patient grouping of Alizadeh et al. [14] was based on a cluster of 380 genes, chosen for their large variation over the samples. Our cluster of 234 genes has 38 genes in common with this cluster of 380, and they are indicated by an asterisk in Table 2. Five of the 234 genes also appear in the unsupervised clusters found earlier, in the second of the three clusters.
Conclusions
We have proposed a set of 'shaving' methods for isolating interesting clusters of genes from a set of DNA microarray experiments. The methods may be unsupervised, or may be supervised - that is, use information available about the samples such as a class label or survival time. The proposed shaving methods search for clusters of genes showing both high variation across the samples, and coherence (correlation) across the genes. Both of these aspects are important and cannot be captured by simple clustering of the genes, or thresholding of individual genes based on the variation over samples.
With our model-based approach for supervised shaving, one can incorporate other prognostic factors in the search for interesting gene clusters. If an outcome such as survival time is available for each sample, the method searches for a gene cluster whose column average gene  has a significant effect, possibly the presence of other prognostic factors, for predicting the outcome.
has a significant effect, possibly the presence of other prognostic factors, for predicting the outcome.
The microarray data xij we have considered are real-valued expression levels. However, other kinds of arrays produce different kinds of data. In particular, some arrays detect the presence or absence of single-nucleotide polymorphisms (SNPs), so that the xij values take on one of k ≥ 2 unordered values. The shaving methods described can be easily modified to handle this kind of data. In detail, we construct k data matrices X1, X2 ... Xk, each of size n × m. The ijth element of Xj is 1 if xij falls in class j, and zero otherwise. Letting ∑jj = 1, 2, ... k be the n × n covariance matrices of the genes in each Xj, we simply apply principal component shaving, using 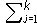 ∑j, as the variance matrix for the penalty. This can be done unsupervised, or a supervision term can also be added.
∑j, as the variance matrix for the penalty. This can be done unsupervised, or a supervision term can also be added.
Acknowledgments
Acknowledgements
We thank Mark Segal for helpful discussions, and Nick Fisher and Jerome Friedman for their work on 'bump-hunting' which inspired some of the ideas in this paper. The first two authors were supported by grants from The National Science Foundation and the National Institutes of Health. W.C.C. was supported by NIH grant UOI-CA 84967. R.L and collaborators were supported by LPPG grant CA 34233.
References
- Weinstein J, Myers T, O'Connor P, Friend S, Fornace A, Jr, Kohn K, Fojo T, Bates S, Rubinstein L, Anderson N, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Eisen M, Spellman P, Brown P, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth FP, Hughes J, Estep P, Church G. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Golub T, Slonim D, Tamayo P, Huard C, Gassenbeek M, Mesirov JP, Coller H, Loh M., Downing J, Caligiuri M, Bloomfield C, Lander E. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–538. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Perou C, Jeffrey S, van de Rijn M, Rees C, Eisen M, Ross D, Perga-menschikov A, Williams C, Zhu S, Lee J, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U, Barkai N, Notterman D, Gish K, Ybarra S, Mack D, Levine A. Broad patterms of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Jonaxsen I, Vilo J, Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998;8:1202–1215. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'haeseleer P, Wen X, Fuhrman S, Somogyi R. Inferring gene relationships from large-scale gene expression data. In Information Processing in Cells and Tissues Edited by Holcombe M, Paton R New York: Plenum; 1998:203–212. [Google Scholar]
- Ewing R, Kahla A, Poirot O, Lopez F, Audic S, Claverie J. Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res. 1999;10:950–959. doi: 10.1101/gr.9.10.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjolsness E, Mann T, Castano R, Wold B. From co-expression to co-regulation: an approach to inferring transcriptional regulation among gene classes from large scale expression data. Technical report JPL-ICTR-99-4 Pasadena: Jet Propulsion Laboratory, Section 365; 1999.
- Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- Walker M, Volkmuth W, Sprinzak E, Hodgson D, Klinger T. Prediction of gene function by genome scale expression analysis: prostate cancer associated genes. Genome Res. 9:1198–1203. doi: 10.1101/gr.9.12.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim T, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E. Interpreting patterns of gene expression with self-organizing maps: Methods and applications to hematopoietic diferentation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh A, Eisen M, Davis RE, Ma C, Losses I, Rosenwal A, Boldrick J, Sabet H, Tran T, Yu X, et al. Identification of molecularly and clinically distinct substypes of diffuse large B-cell lymphoma by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Eisen M, Ross D, Botstein D, Brown P. Clustering methods for the analysis of DNA microarray data. Technical report Stanford: Department of Statistics, Stanford University; 1999.
- Hastie T, Tibshirani R, Eisen M, Brown P, Scherf U, Weinstein J, Alizadeh A, Staudt L, Botstein D. Gene shaving: a new class of clustering methods for expression arrays. Technical report Stanford: Stanford University; 2000.
- Hastie T, Alter O, Sherlock G, Eisen M, Tibshirani R, Botstein D, Brown P. Imputation of missing values in DNA microarrays. Technical report Stanford: Stanford University;
- Cox D. Regression models and lifetables. J Roy Statist Soc B. 1972;74:187–220. [Google Scholar]


