Abstract
In most flowering plants, fertilization is necessary for development of the central cell into endosperm, but in the fie-1 mutant of Arabidopsis, the central cell can proliferate autonomously. However, autonomous fie-1 endosperms do not develop completely: They have fewer nuclei than sexually produced endosperms, cellularization does not take place, and no clear distinction is seen between the different endosperm compartments. Here, we show that autonomous endosperm develop much further in hypomethylated than normally methylated fie-1 mutants, undergoing cellularization and regional specification to resemble endosperm in sexually produced wild-type seeds. Therefore, the combination of maternal hypomethylation and loss of FIE function enables formation of differentiated endosperm without fertilization. A maternal fie-1 mutation is also lethal to sexual seeds, even if the pollen donor is wild type. We report that sexual mutant fie-1 endosperms fail to cellularize and overproliferate, consistent with the hypothesis that embryo abortion may be due, at least in part, to a defect in endosperm development. Finally, we show that pollen from hypomethylated plants rescues fie-1 mutant seeds provided that it also donates a wild-type paternal FIE allele. These results are discussed in light of models for parent-of-origin effects on seed development.
INTRODUCTION
A unique feature of reproduction in flowering plants is that fertilization produces two types of offspring (Friedman, 1995). Each pollen grain transmits two sperm to the embryo sac; one fuses with the haploid egg to give rise to the embryo, and the other fuses with the diploid central cell (like the egg, a derivative of the female meiotic product) to form the triploid endosperm. The role of endosperm during seed development is to support embryogenesis, for example, by acquiring resources from the seed parent or signaling to the embryo (Lopes and Larkins, 1993; Berger, 1999).
In most flowering plants (except for apomictic species [Nogler, 1984]), fertilization is necessary for development of the egg into an embryo and of the central cell into an endosperm. Arabidopsis thaliana is not a natural apomict, but several mutants have been described in which the central cell nucleus can proliferate without fertilization. So far, three loci have been identified in which mutations confer some degree of autonomous endosperm development: MEA (MEDEA)/FIS1 (FERTILIZATION-INDEPENDENT SEED 1)/F644/EMB173; FIS2; and FIE (FERTILIZATION-INDEPENDENT ENDOSPERM)/FIS3 (Castle et al., 1993; Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999). These results indicate that FIE, MEA, and FIS2 function to suppress proliferation of the central cell before fertilization. However, none of these mutants appears to undergo full endosperm development in the absence of fertilization, suggesting that additional genes and processes may be involved in the control of endosperm development.
MEA and FIE encode Polycomb group proteins similar to the Drosophila polypeptides Enhancer of Zeste and extra sex combs, respectively, and both are expressed in flowers before fertilization and in developing siliques afterwards (Grossniklaus et al., 1998; Kiyosue et al., 1999; Ohad et al., 1999). In Drosophila, the best understood function of Polycomb proteins is to maintain transcriptional repression of homeotic genes through many rounds of cell division by forming complexes that modulate chromatin configuration or prevent access of transcription factors (Pirrotta, 1997). The phenotypes of the mea and fie mutants, along with the function of their animal orthologs, suggest models in which the wild-type gene products cooperate to prevent endosperm-specific gene transcription in the central cell until the repression is relieved by fertilization (Kiyosue et al., 1999; Luo et al., 1999; Ohad et al., 1999).
Mutations in the MEA, FIS2, and FIE genes also have parent-of-origin effects on seed viability after pollination. That is, maternal mutations at the MEA, FIS2, and FIE autonomous endosperm loci are lethal to sexually produced seeds. When an embryo sac carrying defective alleles is fertilized, the embryo arrests by the time of the heart or torpedo stage even if the pollen donor is wild type (Ohad et al., 1996, 1999; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999). Several studies suggest that the parent-of-origin effects of mea mutations on endosperm development and seed viability are caused at least in part by imprinting of the MEA gene. Vielle-Calzada et al. (1999) found a MEA probe hybridized to only two chromosomes in the primary endosperm nucleus and detected no paternal MEA transcripts after reverse transcription–polymerase chain reaction (RT-PCR) amplification in whole seedpods containing seeds with globular-stage embryos plus endosperm. They concluded from this evidence that MEA is paternally silenced in the young embryo and endosperm. Working with isolated embryo, endosperm, or endosperm plus seed coat from torpedo stage onwards, Kinoshita et al. (1999) showed that MEA is expressed only from maternal alleles in the endosperm but is biparentally expressed in the embryo, which suggests that the paternal allele is silenced specifically in the endosperm during the mid and late stages of seed development.
A striking aspect of endosperm development is its sensitivity to parental imprinting. In flowering plants, several endosperm-imprinted genes have been identified (Kermicle, 1970; Chaudhuri and Messing, 1994; Lund et al., 1995; Kinoshita et al., 1999; Vielle-Calzada et al., 1999), although the presence of more is inferred from classical genetics, including the analysis of progeny from reciprocal interploidy crosses (Kermicle and Alleman, 1990; Haig and Westoby, 1991). As a result of imprinting, a mother and father contribute different sets of potentially active alleles to the next generation, and therefore disturbance to the normal ratio of parental genomes (two maternal:one paternal in the endosperm) causes abnormalities in developing offspring. For example, interploidy crosses in Arabidopsis show that growth of the seed and, in particular, of endosperm is inhibited by an increased maternal-to-paternal genome ratio but is promoted by a decreased maternal-to-paternal genome ratio (Scott et al., 1998). This is in agreement with a history of strong though circumstantial evidence that imprinting directly affects endosperm development, with mainly indirect consequences for embryo growth (Kermicle, 1970; Lin, 1984; Haig and Westoby, 1991).
DNA methylation, which is usually associated with transcriptional repression (Eden and Cedar, 1994), is known to be an essential component of the imprinting mechanism in mammals (Neumann and Barlow, 1996; Tilghman, 1999), and evidence also suggests that methylation is associated with silencing of imprinted alleles in some late-acting genes in maize endosperm (Lund et al., 1995; Finnegan et al., 1998). The methylation patterns of MEA have not been reported, but in seeds homozygous for a decrease in DNA methylation 1 (ddm1) mutation, which reduces overall cytosine methylation by 70% (Vongs et al., 1993)—most probably indirectly through changes to chromatin configuration (Jeddeloh et al., 1999)—a wild-type paternal MEA allele can rescue seeds that are carrying a normally lethal maternal mea mutation (Vielle-Calzada et al., 1999). This implies that hypomethylation or chromatin remodeling (or both) may have activated the silenced paternal copy. Alternatively, the ddm1 mutation may rescue seeds by activating genes downstream from MEA.
DNA methyltransferase I is thought to be the enzyme primarily responsible for maintaining methylation patterns in the Arabidopsis genome (Genger et al., 1999). In transgenic plants with a METHYLTRANSFERASE I antisense (METI a/s) gene, global cytosine methylation is reduced to 13% of the wild-type amount (Finnegan et al., 1996). Recently we showed that uniparental hypomethylation associated with the METI a/s gene phenocopies interploidy crosses in Arabidopsis, from which we concluded that DNA methylation plays an important role in the parent-of-origin effect and, by inference, in parental imprinting in plants (Adams et al., 2000). For example, crosses between a hypomethylated diploid seed parent and a wild-type diploid pollen parent resembled crosses between a diploid seed parent and a tetraploid pollen parent with regard to increased seed size and weight, greater extent of endosperm proliferation, and delayed onset of endosperm cellularization. This is consistent with a model in which hypomethylation of the seed parent prevented silencing of maternal alleles that promote endosperm and seed growth, which normally (according to theory) are expressed from the paternal genome only. Likewise, the reciprocal cross between a diploid wild-type seed parent and a hypomethylated pollen parent resembled crosses between a tetraploid seed parent and a diploid pollen parent with regard to reduced seed size and endosperm proliferation (Scott et al., 1998). This is consistent with a model in which hypomethylation of the pollen parent prevented silencing of paternal alleles that suppress endosperm and seed growth, which normally are expressed from the maternal genome only.
To understand how genome methylation and Polycomb proteins control seed development, we crossed mutants heterozygous for the fie-1 allele (Ohad et al., 1996, 1999) with plants carrying a METI a/s construct (Finnegan et al., 1996). In pollinated, normally methylated fie-1 mutants, we found that aborted seeds contained overgrown, uncellularized endosperms with phenotypes similar to those produced by interploidy crosses that provided a lethal overdose of paternal genomes (Scott et al., 1998). However, if the pollen donor was hypomethylated by the METI a/s transgene and donated a wild-type paternal FIE allele, fie-1 mutant seeds survived, and their phenotypes were consistent with a nonlethal overdose of paternal genomes in seed size and endosperm morphology.
In addition, consistent with previous reports (Ohad et al., 1996, 1999), we observed that the autonomous endosperm in unfertilized fie-1 mutant seedlike structures did not cellularize; we also saw no morphological signs of regional differentiation of the endosperm. Strikingly, in fie-1 mutants with reduced methylation because of the METI a/s transgene, we found that autonomous endosperm could develop much further than in normally methylated fie-1 plants, undergoing cellularization and regional specification to resemble sexually produced, wild-type endosperm—although the unfertilized seedlike structures contained no embryos. Therefore, the combination of maternal hypomethylation and loss of FIE function enabled formation of differentiated endosperm without fertilization. This phenomenon not only sheds light on the biological problem described by Haig and Westoby (1991) of how autonomous apomicts produce endosperm without a paternal contribution, but it also suggests a method of engineering apomixis into sexual species.
RESULTS
Sexually Produced fie-1 Mutant Seeds Abort with a Paternal Excess Phenotype
In Arabidopsis, as in many other flowering plants, the primary endosperm nucleus undergoes several rounds of mitosis without cytokinesis, producing daughter nuclei that migrate to different regions of the embryo sac before the zygote begins to divide. The endosperm proliferates as a syncytium until the embryo reaches heart stage and then begins to cellularize from the micropylar pole (Mansfield and Briarty, 1990a, 1990b; Scott et al., 1998; Brown et al., 1999). Several days before cellularization, three regions of endosperm are identifiable: central peripheral (composed of regularly spaced nuclei with associated cytoplasm lining the central region of the embryo sac), micropylar peripheral (nuclei embedded in a common cytoplasm surrounding the suspensor), and chalazal (a dense multinucleate tissue at the chalazal pole). After cellularization, the endosperm is gradually consumed by the growing embryo, and only one or two endosperm cell layers persist in the mature seed. A seed with a torpedo stage embryo and cellularizing endosperm is shown in Figure 1.
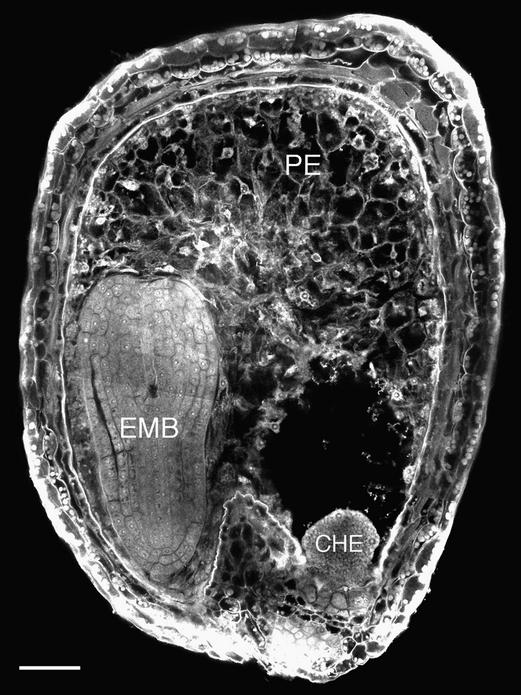
Figure 1.
Confocal Microscopy Image of a Feulgen-Stained Wild-Type Seed at 8 DAP.
The seed contains a torpedo stage embryo (EMB), cellularized peripheral endosperm (PE), and free-nuclear chalazal endosperm (CHE).  .
.
Seeds inheriting a maternal fie-1 mutation do not complete the developmental program described above. Instead, the embryo arrests at heart stage and the seeds collapse (Ohad et al., 1996). Before investigating the effects of hypomethylation on fie-1 mutant seeds, we crossed fie-1/FIE heterozygotes as seed parents with wild-type pollen parents as a control; the results are shown in Table 1 and Figure 2. This cross produced two classes of seed, 99 shriveled and 120 plump (fie-1/FIE and FIE/FIE, respectively) (Ohad et al., 1996), consistent with a 1:1 ratio (χ2 = 2.0, P > 0.1) (Table 1 and Figure 2A). Confocal microscopy of developing seeds showed that differences between the two classes were detectable by 6 to 7 days after pollination (DAP). Figures 2B to 2G show confocal micrographs of the two classes of seeds produced by the [female fie-1/FIE × male FIE/FIE] cross. In plump seeds, embryos reached maturity, and endosperm development appeared identical to that in wild-type seeds (Table 1 and Figures 2B to 2D; compare with Figure 1). In shriveled seeds, the embryo became vacuolate and did not develop past late heart–early torpedo stage, the endosperm failed to cellularize, and chalazal endosperm underwent massive overproliferation (Table 1 and Figures 2E to 2G). Thus, in addition to the previously reported embryo abortion, we observed defects in endosperm development in fie-1 mutant seeds. Moreover, the phenotype of sexually produced fie-1 seeds closely resembled that of seeds from [2x × 6x] crosses (crosses between wild-type diploid seed parents and hexaploid pollen parents), which inherit a lethal overdose of paternal genomes (Scott et al., 1998).
Table 1.
Seed Viability and Endosperm Development after Crosses between fie-1/FIE Heterozygous Seed Parents and Wild-Type Pollen Parents Compared with Crosses between Wild-Type Plants
| Progeny Characteristics | fie-1/FIEa | FIE/FIEa | FIE/FIEb |
|---|---|---|---|
| Expected frequency of embryo genotypes (%) | 50 | 50 | 100 |
| Observed seed phenotypes | 45% shrivelled (n = 219) | 55% plump (n = 219) | 98% plump (n = 126) |
| Seed weight (μg) | No data | 26.0 (n = 120) | 20.2 (n = 60) |
| Germination (%) | 0 (n = 99) | 92 (n = 120) | 100 (n = 20) |
| Maximum no. of PE nuclei (±se) | 408 ± 33 (n = 3) | 447 ± 33 (n = 3) | 429 ± 31 (n = 3) |
| Endosperm cellularization | None at 10 DAP (n = 10) | 5–6 DAP (n = 5) | 5–6 DAP (n = 10) |
| Chalazal endospermc | 10–15X (n = 10) | 1X (n = 5) | 1X |
Cross: fie-1 × FIE/FIE.
Cross: FIE/FIE × FIE/FIE.
Area of maximum cross-section relative to the wild type (1×).
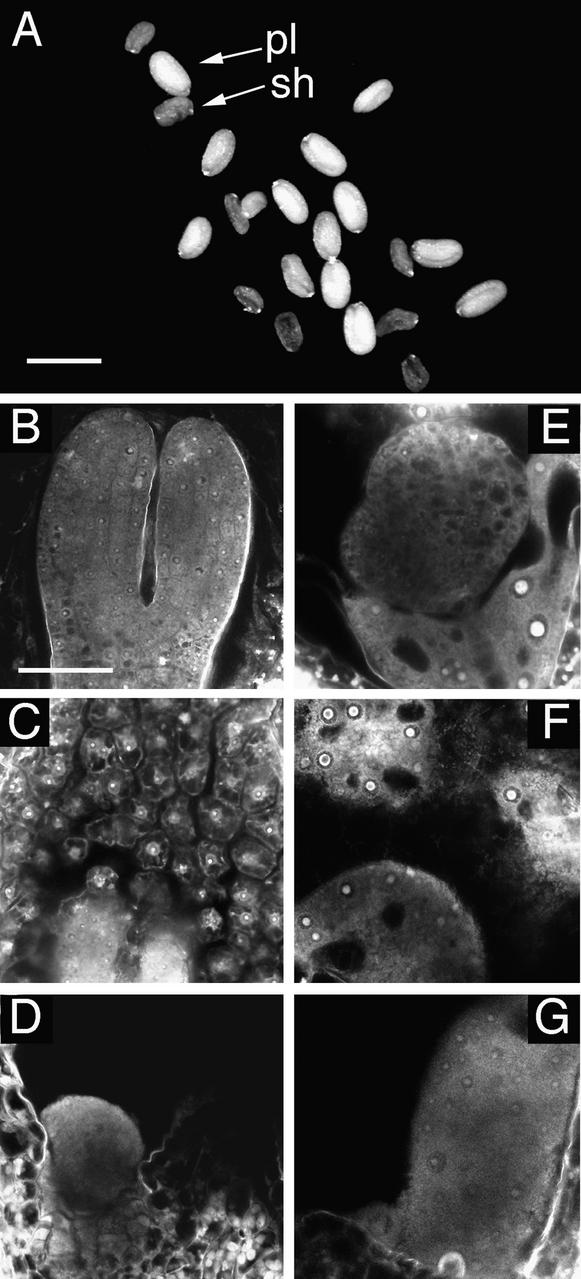
Figure 2.
Seeds from a [fie-1/FIE × FIE/FIE] Cross.
(A) Light microscopy image showing the two classes of seeds, plump (pl) and shriveled (sh).
(B) to (G) Confocal microscopy images of normal (B to D) and aborting (E to G) seeds at 8 DAP, centered on micropylar (B and E), central (C and F), or chalazal (D and G) regions of the embryo sac. The endosperm in (E) to(G) is overgrown and has not cellularized.
 ;
; 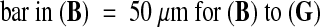 .
.
Hypomethylated Pollen Rescues fie-1 Mutant Seeds
The evidence given above suggests that a maternal fie-1 mutation has a lethal “paternalizing” effect on seed development. Previously, we used uniparental demethylation (Adams et al., 2000) to phenocopy the viable paternal excess phenotype of [2x × 4x] crosses, that is, overgrown endosperms with delayed cellularization and large embryos that germinate (Scott et al., 1998). The phenocopy was achieved by crossing seed parents carrying the METI a/s construct (Finnegan et al., 1996) with wild-type pollen donors (Adams et al., 2000). We interpreted this result as showing that hypomethylation of the seed parent prevented repression of imprinted genes that would normally be silenced in the maternal genome, so that in the endosperm, maternally transmitted genomes expressed paternal- as well as maternal-specific alleles to some extent. We described this as paternalization because, according to our model, the maternal genomes lost their maternal methylation state and expressed maternally silenced genes that are normally expressed only from the paternal genome to promote endosperm development. In contrast, the reciprocal cross between a wild-type seed parent and METI a/s pollen donor resulted in small, viable seeds with small endosperms that cellularized early. These signs of maternal excess (Scott et al., 1998) suggested that hypomethylation of the pollen parent has a “maternalizing” effect (that is, loss of the paternal methylation state) on seed development (Adams et al., 2000). For the present study, we crossed fie-1/FIE heterozygotes as seed parents with METI a/s plants as pollen parents to determine whether the paternalizing effect of a maternal fie-1 mutation and the maternalizing effect of paternal genome hypomethylation could counteract each other.
In the cross [fie-1/FIE × FIE/FIE; METI a/s/METI a/s] all seeds were plump, although half were expected to carry a maternal fie-1 mutation, which is normally lethal. The seeds fell into two classes of approximately equal numbers, small and large (the former were somewhat more variable in size), and most seeds in each class germinated (Table 2 and Figure 3A). Using PCR followed by restriction enzyme digestion to distinguish between fie-1 and FIE alleles, we determined that all plants grown from small seeds had the genotype FIE/FIE, whereas all plants grown from large seeds were fie-1/FIE (Table 2 and Figure 3B). We also confirmed by PCR that all seedlings contained the METI a/s transgene (data not shown). Therefore, hypomethylated pollen rescued the seed lethality of pollinated fie-1 mutants.
Table 2.
Seed Viability and Endosperm Development after Crosses between fie-1/FIE Heterozygous Seed Parents and Pollen Parents Homozygous for the METI a/s Construct
| Progeny Characteristics | fie-1/FIE; METI a/sa | FIE/FIE; METI a/sa |
|---|---|---|
| Expected frequency of embryo genotypes (%) | 50 | 50 |
| Observed seed phenotypes (%) | 51 large (n = 428) | 49 small (n = 428) |
| Observed embryo genotypes (confirmed by PCR) | fie-1/FIE; METI a/s (n = 24) | FIE/FIE; METI a/s (n = 17) |
| Seed weight (μg) | 30.0 (n = 73) | 13.2 (n = 72) |
| Germination (%) | 95 (n = 40) | 85 (n = 40) |
| Maximum no. of PE nuclei (±se) | 673 ± 33 (n = 4) | 192 ± 24 (n = 4) |
| Endosperm cellularization | 7–8 DAP (n = 7) | 3–4 DAP (n = 10) |
| Chalazal endospermb | 3–4X (n = 5) | 0.05–0.1X (n = 5) |
Cross: fie-1/FIE × FIE/FIE; MET1 a/s/MET1 a/s.
Area of maximum cross-section relative to the wild type (1×).
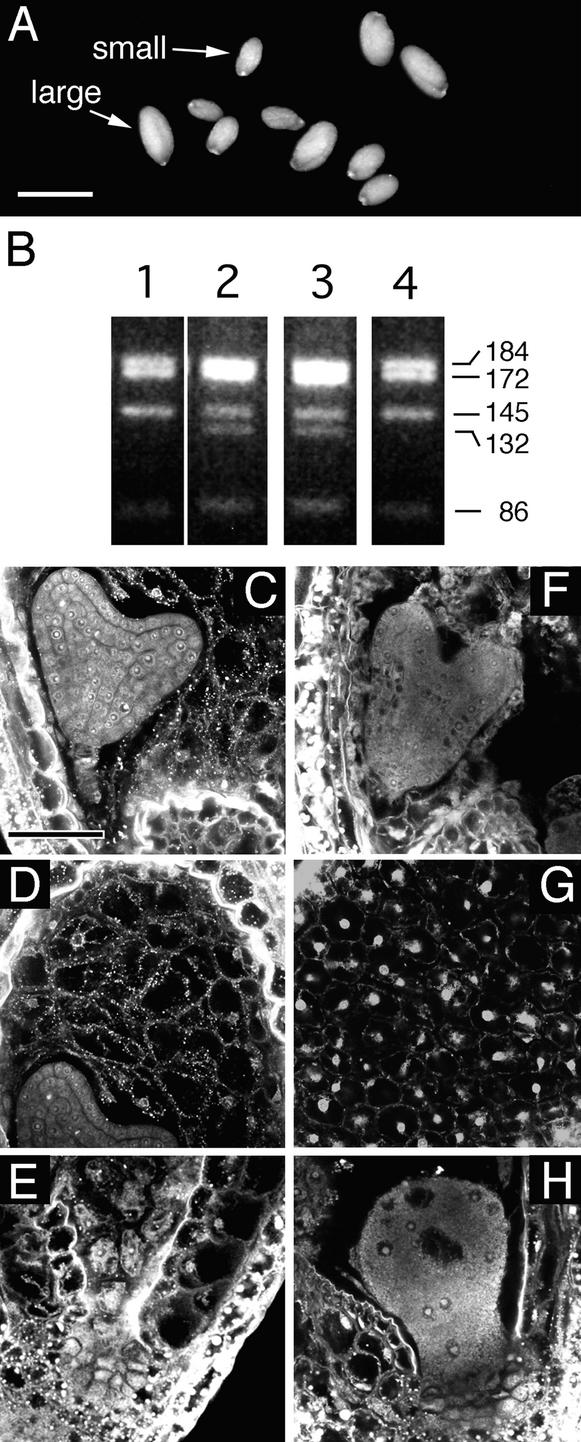
Figure 3.
Seeds from a [fie-1/FIE × FIE/FIE; METI a/s/METI a/s] Cross.
(A) Light microscopy image showing the two classes of seeds. All seeds are plump, indicating that a pollen parent hypomethylated by the METI a/s transgene can rescue fie-1 mutant seeds.
(B) Identification of the fie-1 and FIE alleles by PCR and restriction enzyme analysis (see Methods). The wild-type FIE allele produces four bands (lane 1), whereas fie-1/FIE heterozygotes (lane 2) have an extra band. All large seeds scored had the heterozygous pattern (lane 3), whereas all small seeds were wild type (lane 4).
(C) to (H) Confocal microscopy images of seeds at 8 DAP. The seed in (C) to (E) has a phenotype similar to seeds from interploidy crosses generating maternal genomic excess, whereas the seed in (F) to (H) shows characteristics of paternal excess (see text; Scott et al., 1998).
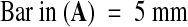 ;
; 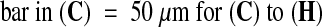 .
.
The small seeds were maternally wild type and paternally hypomethylated, and their phenotypes were similar to those previously reported for [2x × METI a/s] crosses (Adams et al., 2000): small peripheral endosperms that cellularized early and tiny chalazal endosperms (Table 2 and Figures 3C to 3E). In contrast, the large seeds, resulting from fertilization of a fie-1 mutant embryo sac by METI a/s pollen, produced overgrown endosperms. However, unlike the case when fie-1 mutants are fertilized by normally methylated pollen, the endosperm eventually cellularized, and the seeds were mainly viable (Table 2 and Figures 3F to 3H). Both of these results are consistent with a maternalizing effect of hypomethylated pollen (Adams et al., 2000). Wild-type seeds produced by a fie-1/FIE seed parent developed with a maternal excess phenotype (Scott et al., 1998), whereas in pollinated fie-1 mutant seeds, the lethal paternal excess phenotype was reversed to some extent.
Rescue of Maternal fie-1 Mutants by METI a/s–Mediated Hypomethylation Requires a Wild-Type Paternal FIE Allele
One explanation for rescue of fie-1 mutant seeds by pollen from a METI a/s father is that FIE is normally silenced in the paternal genome. Similar to MEA, the silencing could be long-term and extend essentially throughout seed development (Kinoshita et al., 1999). Alternatively, the silencing could be short-term and associated with a transitory delay in activation of the entire paternal genome reported by Vielle-Calzada et al. (2000). In either case, hypomethylation might allow expression of the paternal FIE allele, so that the seed is rescued. To test whether a wild-type FIE allele is necessary for seed rescue, we crossed fie-1/FIE heterozygotes as seed parents with fie-1/FIE pollen parents, which were also hemizygous for the METI a/s transgene (hemiMETI a/s). We reasoned that if seeds inheriting a maternal fie-1 mutation must contain a paternally transmitted wild-type FIE allele to survive, then we would expect two classes of viable embryos, FIE/FIE and fie-1/FIE, and the fie-1/fie-1 seeds (25% of the total) would abort. However, if rescue did not depend on a wild-type paternal FIE allele, then all seeds would complete development, and fie-1/fie-1 homozygous seedlings would be recovered.
To generate hypomethylated fie-1/FIE pollen parents, we germinated large seeds from the [fie-1/FIE × FIE/FIE; METI a/s/METI a/s] cross and confirmed the genotype of the seedlings by PCR followed by restriction enzyme digestion (Table 2). However, because these plants are hemizygous for the antisense construct, we had to establish that pollen from a FIE/FIE; METI a/s plant can still rescue fie-1 mutant seeds, even though half of the pollen does not carry the transgene. HemiMETI a/s plants maintain reduced methylation (Finnegan et al., 1996), and we have found that pollen from hemiMETI a/s plants still has a maternalizing effect on seed development, even when the METI a/s construct does not segregate to the pollen (Adams et al., 2000). To test whether pollen from a hemiMETI a/s plant can rescue fie-1 mutant seeds, we examined progeny of the cross [fie-1/FIE × FIE/FIE; METI a/s]; all of these formed plump seeds, which fell into two size classes, as before (Figure 4A). From this we inferred that all pollen from a hemiMETI a/s plant can compensate for a maternal fie-1 mutation. Next we ruled out the possibility that hypomethylation inhibits paternal transmission of fie-1 mutant alleles by crossing FIE/FIE seed parents with fie-1/FIE; METI a/s pollen parents and scoring the progeny by PCR and enzyme digestion. Of 24 seedlings tested, 13 were FIE/FIE and 11 were fie-1/FIE, consistent with a 1:1 segregation (χ2 = 0.17, P > 0.5).
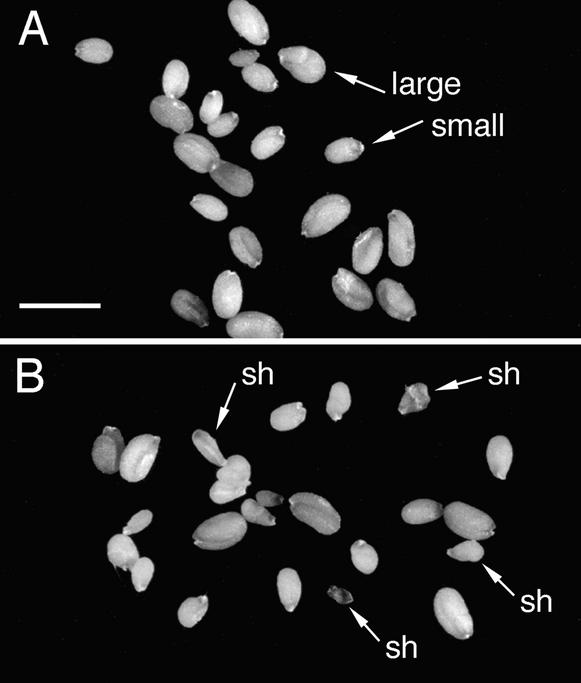
Figure 4.
Light Microscopy Images of Seeds from Crosses with hemiMETI a/s Pollen Parents.
(A) Seeds from a [fie-1/FIE × FIE/FIE; METI a/s] cross. All seeds are plump, indicating that fie-1 mutant seeds are rescued by a pollen parent hemizygous for the METI a/s construct. The two size classes are shown.
(B) Seeds from a [fie-1/FIE × fie-1/FIE; METI a/s] cross. The presence of many shriveled seeds (sh) suggests that some fie-1 mutant seeds have not been rescued.
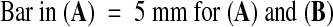 .
.
Once we established that a hemiMETI a/s pollen donor could rescue fie-1 mutant seeds and that mutant fie-1 alleles could be transmitted by hypomethylated pollen at the expected frequency, we crossed fie-1/FIE seed parents with fie-1/FIE; METI a/s pollen parents. This cross produced both plump and shriveled seeds of different sizes (Figure 4B). Plants grown from these seeds were scored for wild-type and mutant alleles. Of 52 seedlings, 11 (all grown from small plump seeds) were FIE/FIE. The remaining 41 seedlings, grown from a mixture of small and large seeds, were fie-1/FIE. No fie-1/fie-1 homozygotes were recovered, suggesting that that class of seeds aborted. Seeds from siliques at 10 DAP were examined either by clearing or by confocal microscopy. Seeds containing maturing and arrested embryos occurred in a 3:1 ratio, as would be expected if those with fie-1/fie-1 embryos aborted (Table 3). Seeds with maturing embryos had either maternal or paternal excess phenotypes (presumably reflecting the presence of a wild-type or mutant maternal copy of FIE, respectively), whereas aborting seeds bore similarities to those from fie-1/FIE × FIE/FIE crosses, containing embryos arrested at heart–torpedo stage and overproliferated chalazal endosperm (Table 3 and Figures 5A to 5E; see also Figures 2E to 2G). We concluded from this evidence that a wild-type paternal FIE allele is needed for METI a/s–mediated rescue.
Table 3.
Seed Viability and Endosperm Development after Crosses between fie-1/FIE Heterozygous Seed Parents and fie-1/FIE Heterozygous Pollen Parents Hemizygous for the METI a/s Construct
| Progeny Characteristics (with respect to FIE) | FIE/FIEa FIE/fie-1a | fie-1/FIEa | fie-1/fie-1a |
|---|---|---|---|
| Expected frequency of embryo genotypes (%) | 25 25 | 25 | 25 |
| Observed seed phenotypes at 10 DAP (%)b | 49 Maternal excess (n = 123) | 25 Paternal excess (n = 123) | 26 Lethal paternal excess (n = 123) |
| Seed weight (μg) | Small seeds 17.9 (n = 96) | Large seeds 32.9 (n = 125) | Shrivelled seeds 10.6 (n = 59) |
| Latest embryo stage observed | Mature | Mature | Heart–torpedo |
| Chalazal endosperm at 12 DAP | None visible | None visible | Persistent and filling seed |
Cross: fie-1/FIE × fie-1/FIE; METI a/s.
Segregation of maturing and aborted seeds consistent with a 3:1 ratio, χ2 = 0.068, P > 0.1.
Figure 5.
Seeds from a [fie-1/FIE × fie-1/FIE;METI a/s] Cross.
(A) to (C) Light microscopy images of embryos isolated from seeds 12 DAP representing the three phenotypic classes (see Table 3). (A) Plump, maternal excess (predicted genotypes FIE/FIE and maternal FIE/paternal fie-1); (B) plump, paternal excess (predicted genotype fie-1/FIE); (C) embryo from shriveled seed, with lethal paternal excess (predicted genotype fie-1/fie-1). Seeds expected to constitute the fie-1/fie-1 fraction contain arrested heart–torpedo stage embryos, which suggests that FIE is not required for embryogenesis.
(D) and (E) Confocal microscopy images of endosperm in (D) a plump seed with a paternal excess phenotype (predicted genotype fie-1/FIE) and embryo from (E) a collapsed seed with a lethal paternal excess phenotype (predicted genotype fie-1/fie-1). Both are shown 12 DAP. The seed shown in (D) contains abundant cellularized peripheral endosperm; in the seed shown in (E), this is replaced by persistent chalazal endosperm.
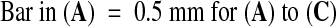 ;
; 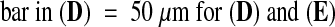 .
.
Relationship of Methylation and Autonomous Endosperm in fie-1 Mutants
Normally the central cell in Arabidopsis will not proliferate without fertilization, but in fie-1/FIE heterozygotes, central cells in as many as half of the ovules, presumably those with a fie-1 embryo sac, undergo division and sometimes generate large free-nuclear structures (Ohad et al., 1996, 1999). To further investigate the unpollinated fie-1 mutant phenotype, we emasculated floral buds from fie-1/FIE heterozygotes and used confocal microscopy to image Feulgen-stained siliques. Results and representative images are shown in Table 4 and Figures 6A to 6C. We found that in nearly half of the ovules, the central cell proliferated without cytokinesis, producing ∼200 nuclei (Table 4). There was no development of a morphologically distinct chalazal endosperm, although occasionally clusters of nuclei were found in a common cytoplasm at the embryo sac periphery (Figure 6B). We saw no autonomous embryo development, consistent with previous reports of the unpollinated fie-1 mutant phenotype (Ohad et al., 1996).
Table 4.
Autonomous Endosperm Development in Unpollinated fie-1/FIE Heterozygotes with Normal and Reduced DNA Methylation
| Embryo Sac Genotypes (with Respect to FIE) |
fie-1a | FIEa | fie-1b | FIEb | |
|---|---|---|---|---|---|
| Expected frequency of embryo sac genotypes |
50% | 50% | 50% | 50% | |
| Observed phenotypes | 47% Autonomous endosperm (n = 186) |
53% No development (n = 186) |
51% Autonomous endosperm (n = 233) 60% Type 1c 40% Type 2d (n = 30) (n = 30) |
49% No development (n = 233) |
|
| Maximum no. of PE nuclei (±se) |
199 ± 44 (n = 4) |
—e | 532 ± 117 (n = 3) |
697 ± 42 (n = 3) |
— |
| Endosperm cellularization | None at 10 DAE f (n = 20) | — | 5–6 DAE (n = 25) |
7–8 DAE (n = 20) |
— |
| Chalazal endospermg | 0 (n = 20) | — | 0.01–0.05X (n = 5) |
5–10X (n = 5) |
— |
Parent (emasculated): fie-1/FIE.
Parent (emasculated): fie-1/FIE; MET1 a/s.
Cellularized peripheral endosperm; chalazal endosperm absent (see text for details).
Cellularized peripheral endosperm; large chalazal endosperm present (see text for details).
Dash indicates no development.
DAE, days after emasculation.
Area of maximum cross-section relative to the wild type (1×).
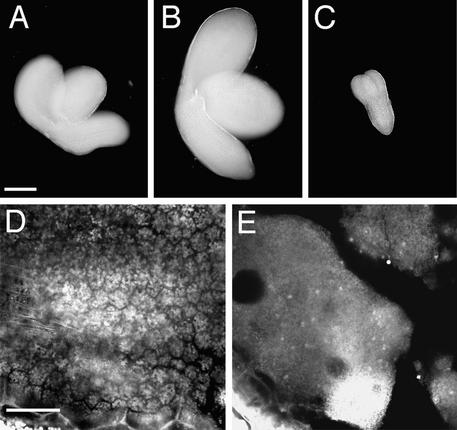
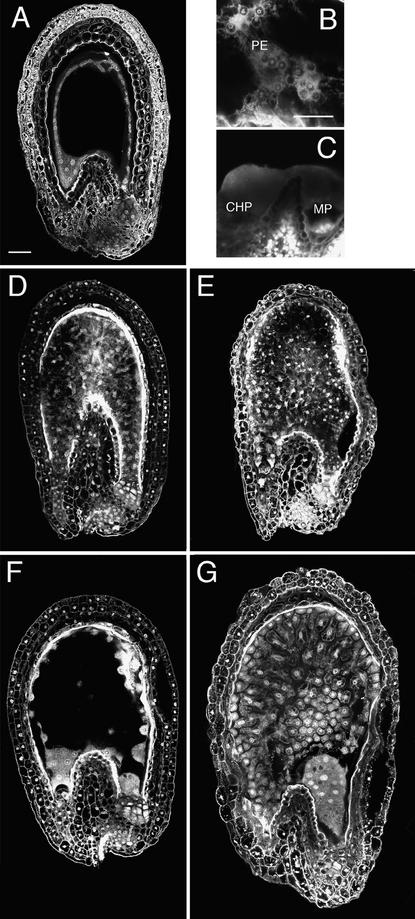
Figure 6.
Confocal Microscopy Images of Fertilization-Independent Seedlike Structures Produced by Emasculated fie-1/FIE Heterozygotes with Normal and Reduced Methylation.
(A) to (C) Seedlike structure from a plant with normal methylation. (A) An optical section showing peripheral endosperm but no well-differentiated chalazal endosperm. (B) Clustered endosperm nuclei at periphery (PE, peripheral endosperm). (C) Endosperm at micropylar (MP) and chalazal (CHP) poles.
(D) to (G) Seedlike structures from fie-1/FIE; METI a/s plants. (D) and (E) Type 1 seedlike structures at 7 (D) and 10 (E) days after emasculation (DAE); in these, the endosperm cellularizes and fills the interior of the embryo sac. (F) and (G) Type 2 seedlike structures at 7 (F) and 10 (G) DAE; these produce micropylar and chalazal in addition to peripheral endosperm.
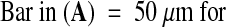 (A)
(A)
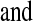 (D)
(D)
 (G)
(G) (B) =
(B) =  μ
μ (B)
(B)
 (C).
(C).
Although fie-1 mutant central cells undergo some degree of autonomous development, they do not produce differentiated, cellularized endosperms without fertilization. Therefore, some block to full endosperm development must be present in fie-1 mutants. To investigate the role of methylation in this block, we emasculated fie-1/FIE; METI a/s plants (see above) and looked for autonomous endosperm development. As shown in Table 4 and Figures 6D to 6G, half of the ovules underwent autonomous endosperm development as before but progressed much further than in fie-1 mutants with normal methylation, producing more nuclei and exhibiting more features of normal endosperm development. We found two phenotypic classes of autonomous endosperms in emasculated fie-1/FIE; METI a/s plants. In one class, which we designated type 1, the free-nuclear endosperm cellularized, but there was still little regional differentiation (Table 4 and Figures 6D and 6E). Type 2 seedlike structures resembled sexually produced seeds (except for absence of an embryo), containing more peripheral endosperm nuclei (which cellularized later than did type 1) and large chalazal and micropylar endosperms (Table 4 and Figures 6F and 6G). The chalazal endosperms in this class were also larger than in sexual seeds from wild-type [2x × 2x] crosses. No embryos were found in either type, indicating that no accidental fertilization occurred and that hypomethylation promotes fertilization-independent development of fie-1 mutant endosperms but not of embryos.
DISCUSSION
FIE as a Regulator of Endosperm Proliferation
Ohad et al. (1996)(1999) previously reported that when fie-1/FIE heterozygotes are pollinated, half the seeds arrest with the embryo at heart stage. We have shown here that in such seeds the endosperm proliferates without cellularizing, and the chalazal endosperm is massively overgrown (Table 1 and Figure 2), a phenotype similar to that seen in seeds inheriting a loss-of-function mea-3 allele from the seed parent (Kiyosue et al., 1999) and in seeds from [2x × 6x] crosses in wild-type Arabidopsis (Scott et al., 1998). One interpretation is that important functions of FIE and MEA are to directly or indirectly inhibit proliferation of the central cell and endosperm, explaining why these undergo extra divisions in both unpollinated and pollinated fie-1 and mea-3 mutants (Figures 2 and 6) (Kiyosue et al., 1999). According to this model, the seed phenotypes of pollinated fie-1 and mea-3 mutants on the one hand and wild-type [2x × 6x] crosses on the other all result from failure to inhibit endosperm proliferation. In the first case, according to the model, the loss of wild-type FIE or MEA product causes derepression of gene activity that promotes endosperm growth. In the second case, the overdose of paternal genomes contributes to the seed extra active alleles of genes that promote endosperm growth.
Our results lead to the hypothesis that FIE and MEA normally participate in repression of genes (including, perhaps, promoters of endosperm growth) in the maternal genome. In Drosophila and mammals, FIE orthologs are thought to recruit other Polycomb proteins—including MEA orthologs—to repressive complexes (Simon, 1995; Pirrotta, 1997; Jones et al., 1998). Perhaps FIE also serves such a recruitment role in Arabidopsis. If so, the maternal fie-1 mutation could allow expression of alleles that would normally be silenced in the maternal genome.
METI a/s Suppresses Parent-of-Origin Effects of the fie-1 Mutation on Seed Viability
The fie-1 mutation causes parent-of-origin effects on seed viability. When a maternal mutant allele is inherited, the seed aborts even when the pollen donor contributes a wild-type paternal FIE allele. Two models have been proposed to explain this phenomenon. The first model requires very early expression of the maternal wild-type FIE allele. This in turn could reflect a requirement for expression either before fertilization in the female gametophyte, as Springer et al. (2000) described for the PROLIFERA locus, or very soon after fertilization, before the general activation of the paternal genome reported by Vielle-Calzada et al. (2000). The second model includes a specific, long-term silencing of the paternal wild-type FIE allele. For example, the paternal FIE allele, like the paternal MEA allele, may be silenced specifically in the endosperm during mid and late stages of seed development (Kinoshita et al., 1999). Recent analysis of maternal and paternal FIE gene expression in dissected seeds at the mid to late stages of development (torpedo stage and later) has revealed biallelic FIE gene expression in both endosperm and embryo (T. Kinoshita and R.L. Fischer, unpublished results). Thus, we favor the first model, which emphasizes an early essential role of maternal FIE gene expression for seed viability.
The seed-lethal phenotype of maternal fie-1 mutants can be rescued if the pollen donor is hypomethylated by the METI a/s construct (Table 2) and if the pollen transmits a wild-type FIE allele. There are several possible explanations for this result. First, METI a/s may affect the global activation of the paternal genome, as reported by Vielle-Calzada et al. (2000). Reduction in a general transitory delay in paternal genome expression could allow for earlier expression of a wild-type paternal FIE allele and subsequent rescue of the seed. Alternatively, it is formally possible that METI a/s rescues the seed by altering expression of genes downstream of FIE. Although a wild-type paternal FIE allele is required for rescue, we cannot rule out the possibility that FIE may have other essential functions during seed development (i.e., in addition to controlling endosperm proliferation). Additional analysis of FIE gene function is required to resolve these points.
We found that rescued fie-1 mutant seeds showed many features of paternal genomic excess, including large size and weight and overproliferation and delayed cellularization of endosperm (Table 2). Vielle-Calzada et al. (1999) also reported that rescued mea-1/MEA seeds were enlarged, containing large embryos and sometimes excess persistent and partially cellularized endosperm. We interpret these phenotypes as indicating that reactivation of paternally silenced alleles—which in effect maternalizes the sperm genome—can compensate partly but not fully for the paternalizing effect of maternal fie-1 or mea-3 mutations (Table 1 and Figures 2 and 3) (Kiyosue et al., 1999).
One reason why wild-type FIE and MEA (Vielle-Calzada et al., 1999) could be essential to seed rescue by METI a/s and ddm1, respectively, is that without them the genes promoting seed growth are overexpressed. Rescued seeds might retain an attenuated paternal excess phenotype of overgrown but viable endosperms because postfertilization rescue cannot fully compensate for loss of gene function in the female gametophyte. An alternative explanation is dosage sensitivity of Polycomb proteins (e.g., Campbell et al., 1995): The single copy of each activated FIE or MEA gene from the haploid sperm might not be as effective as the two copies normally transmitted by the diploid central cell. However, the requirement for FIE and MEA wild-type alleles for rescue might also reflect distinct biological processes. The fact that homozygous mutant mea seedlings can be generated (Chaudhury et al., 1997; Kiyosue et al., 1999; Vielle-Calzada et al., 1999), but not fie homozygous mutant seedlings (Ohad et al., 1996; Chaudhury et al., 1997), suggests that, compared with MEA, FIE may have additional essential functions during seed development.
The failure to obtain fie-1 homozygotes indicates an essential role for FIE during seed development, in the endosperm, the embryo, or both. Putative fie-1/fie-1 seeds (the abortive fraction from a [fie-1/FIE × fie-1/FIE; METI a/s] cross) contained large amounts of endosperm with a chalazal morphology at 12 DAP (like seeds from a [2x × 6x] cross) (Scott et al., 1998), whereas putative fie-1/FIE seeds at that same time did not. This suggests that embryo abortion of fie-1/fie-1 seeds may result, at least in part, from paternalization of the endosperm.
Global DNA Hypomethylation Promotes Autonomous Endosperm Development in fie-1 Mutants
In plants heterozygous for the loss-of-function fie-1 allele, proliferation of the central cell without pollination is seen in half of the ovules (Table 4 and Figure 6) (Ohad et al., 1996, 1999). This phenotype has been interpreted as showing that wild-type FIE represses endosperm development before fertilization. However, much of the endosperm developmental program in fie-1 mutants does not take place—for example, there is no regional specification of micropylar and chalazal endosperm, and no cellularization (Table 4 and Figures 6A and 6C). Therefore, unpollinated fie-1 mutants are blocked from complete endosperm development. Aside from a wild-type FIE allele, several components of normal seed development are missing from unpollinated fie-1 ovules: not only pollination and fertilization themselves, and therefore any gene expression they might trigger, but also a paternally transmitted genome, which is not equivalent to maternal genomes because of parental imprinting (Haig and Westoby, 1991).
Demethylating fie-1/FIE heterozygotes by using the METI a/s construct allowed autonomous endosperm to develop much further than previously reported (Table 4 and Figure 6); therefore, demethylating the maternal genome appears to relieve the block on endosperm development in fie-1 mutants. In half of the ovules—presumably those carrying wild-type FIE alleles—central cells did not proliferate without pollination, so hypomethylation on its own did not promote fertilization-independent seed development. The half that did develop fell into two classes. In type 1, the central cell usually underwent more rounds of mitosis than is seen in normally methylated fie-1 mutants; the endosperm did cellularize but still had little or no regional differentiation (Table 4 and Figures 6D and 6E). In contrast, type 2 ovules had full endosperm development, at least judging by morphology (Table 4 and Figures 6F and 6G). However, no embryos were found in either type. The presence of two types of autonomous endosperm in proportions consistent with a 1:1 ratio ( , P > 0.1) could reflect segregation of the METI a/s transgene. In contrast, we found previously that seed phenotypes subsequent to reciprocal crosses between hemiMETI a/s and wild-type plants were apparently determined by the methylation status of the parents and not by the presence of the transgene in the seed (Adams et al., 2000). Here, an effect of the transgene might be uncovered in fie-1 mutants, or some other effect could be operating, independently of the presence of the transgene: For example, hypomethylation might affect different DNA sequences in different embryo sacs.
, P > 0.1) could reflect segregation of the METI a/s transgene. In contrast, we found previously that seed phenotypes subsequent to reciprocal crosses between hemiMETI a/s and wild-type plants were apparently determined by the methylation status of the parents and not by the presence of the transgene in the seed (Adams et al., 2000). Here, an effect of the transgene might be uncovered in fie-1 mutants, or some other effect could be operating, independently of the presence of the transgene: For example, hypomethylation might affect different DNA sequences in different embryo sacs.
Hypomethylation of the maternal genome could relieve the endosperm block in fie-1 mutants in several ways. For example, it might allow expression of genes that would normally be transcribed only at pollination or fertilization (e.g., Dong et al., 1998). Or perhaps hypomethylation might prevent repression of imprinted genes that are normally silenced in the maternal genome, thus in effect supplying the missing paternal genome in the absence of fertilization. However, evidence presented above suggests that a maternal fie-1 mutation could itself derepress maternally silenced genes. The additive effects of the fie-1 mutation and maternal hypomethylation on autonomous endosperm development could indicate that FIE does not regulate all maternal-specific genes required for endosperm development. In this scenario, Polycomb proteins and DNA methylation would mediate repression of overlapping (or different) sets of targets. An alternative explanation is that FIE participates in repression of all maternal-specific loci required for endosperm development, but the fie-1 mutation alone does not completely release their expression, implying that Polycomb proteins and DNA methylation participate in repressive complexes at the same loci.
In type 2 seedlike structures, the endosperm shows several features characteristic of paternal genomic excess (Table 4 and Figures 6F and 6G): a high maximum number of peripheral endosperm nuclei (697 ± 42) (mean ±se; 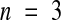 ) compared with 429 ± 31 (
) compared with 429 ± 31 (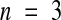 ) for [2x × 2x] seeds and in particular a large chalazal endosperm. The cellularized peripheral endosperm in type 2 seedlike structures fills the space that would otherwise be occupied by an embryo (cf. Figures 6G and 1), so the absence of an embryo could explain the increased number of peripheral endosperm nuclei. However, we interpret the large chalazal endosperm as probably reflecting an overdose of paternal-specific genes, because this region of the endosperm seems to be particularly sensitive to changes in parental genome balance. A paternal excess phenotype for type 2 seedlike structures is consistent with a model in which the combination of the fie-1 mutation and hypomethylation activates alleles in the central cell genome that would normally be expressed only from a paternally derived genome. This would give the central cell genome a rough equivalent of 2m:2p genomes, the same ratio as seen in [2x × 4x] crosses.
) for [2x × 2x] seeds and in particular a large chalazal endosperm. The cellularized peripheral endosperm in type 2 seedlike structures fills the space that would otherwise be occupied by an embryo (cf. Figures 6G and 1), so the absence of an embryo could explain the increased number of peripheral endosperm nuclei. However, we interpret the large chalazal endosperm as probably reflecting an overdose of paternal-specific genes, because this region of the endosperm seems to be particularly sensitive to changes in parental genome balance. A paternal excess phenotype for type 2 seedlike structures is consistent with a model in which the combination of the fie-1 mutation and hypomethylation activates alleles in the central cell genome that would normally be expressed only from a paternally derived genome. This would give the central cell genome a rough equivalent of 2m:2p genomes, the same ratio as seen in [2x × 4x] crosses.
Conclusion
Our results show that FIE and DNA methylation are likely to participate in regulation of endosperm proliferation both before and after fertilization. Maternal fie-1 mutant seeds are rescued if the pollen parent is hypomethylated and if it contributes a wild-type FIE allele. The effect of hypomethylation on unpollinated fie-1 ovules is particularly dramatic: Autonomous endosperm develops further than in fie, fis2, or mea mutants with normal methylation, in many cases resembling endosperm found in sexually produced wild-type seeds. Our results show that neither fertilization nor embryogenesis is necessary to make endosperm, which should be of great importance in developing apomictic crop plants such as cereals, in which the endosperm is the major product. Haig and Westoby (1991) described the following problem in understanding the evolution of autonomous apomicts: How is the requirement for a paternal genome in the endosperm circumvented? Our observation that endosperm can develop without any paternal contribution in a normally sexual species suggests a solution: Autonomous apomicts could overcome the need for a paternal genome through the loss of FIE expression and a decrease in DNA methylation.
METHODS
Plant Material
Arabidopsis thaliana plants were grown for 3 to 4 weeks at 22°C with 16 hr of light in a Fisons growth cabinet and then transferred to a green house and grown at 24 ± 2°C. fie mutant plants were heterozygous for the fie-1 allele (Ohad et al., 1996, 1999); these were originally in the Landsberg erecta background but had been introgressed into Columbia for two generations. Hypomethylated plants were from the T3 generation of family 10.5, homozygous for the Arabidopsis METI a/s construct under control of the cauliflower mosaic virus 35S promoter, in the C24 background (Finnegan et al., 1996). Wild-type plants were in the C24 background. For cross-pollinations, flower buds were emasculated 1 day before anthesis and pollinated 2 days later. Developing siliques were collected after pollination and processed for confocal microscopy as described below. Mature seeds were collected when pods were desiccated. Seeds were weighed with a Mettler UMT 2 microbalance (Mettler-Toledo, Leicester, UK). For investigation of unpollinated fie-1 ovules, anthers were dissected from flower buds of fie-1/FIE heterozygotes; the carpels were collected 0 to 15 days after emasculation and processed for confocal microscopy.
Seed Clearing
To clear seeds for light microscopy, seeds were dissected from siliques in a drop of 1 M KOH on a microscope slide under a dissecting microscope. The pod walls were removed and a cover slip applied. Embryos were released from the seed coat by applying gentle pressure to the cover slip. Preparations were observed by using an Olympus BH-2 microscope (Olympus, Southall, UK) with dark-field illumination and photographed with a Nikon CoolPix 990 digital camera (Nikon Ltd., UK).
Confocal Laser Scanning Microscopy
Samples were prepared as described by Braselton et al. (1996) and imaged at the University of Bath with an Axiovert 100M Zeiss LSM510 laser scanning microscope (Carl Zeiss Ltd., Wellwyn Garden City, UK). Feulgen-stained samples were excited with an argon ion laser at 458 or 488 nm, and emissions were detected at ⩾515 nm. Images measuring 1024 × 1024 pixels were collected with a C-Apochromat 63×/1.2 water lens (Zeiss), saved in PSD format, and processed by using Adobe Photoshop 4.0.1.
Detection of the fie-1 and FIE Alleles by Polymerase Chain Reaction
Genomic DNA was extracted from 0.1 g of leaf tissue according to the small-scale method described by Edwards et al. (1991). DNA template (10 ng) was added to a 20-μL reaction mix containing 1.8 μL of 11× buffer (500 mM Tris-HCl, pH 8.8, 120 mM NH4SO4, 50 mM MgCl2, 75 mM β-mercaptoethanol, 0.05 mM EDTA, 11 mM dNTPs, 1.25 mg/mL DNase-free BSA), 10 pmol of each primer (see below), and 1 U of Taq polymerase (Advanced Biotechnologies, Surrey, UK). The primers used were 5′-GTCCAATCGGCAATGAGTAGAG-3′ and 5′-CACTGTTGACGTCAATGACTCGG-3′, which amplify a 587-bp genomic fragment spanning the intron1–exon2 boundary of the FIE gene. Cycling conditions were 95°C for 1 min followed by 30 cycles of 94°C for 30 sec, 62°C for 1 min, 72°C for 1 min, and a final extension step of 72°C for 4 min, performed in an MJ Research PTC-200 Peltier Thermal Cycler (GRI Ltd., Braintree, UK). Gel-purified polymerase chain reaction fragments (Qiaex II purification kit; Qiagen, Crawley, UK) were digested with the restriction enzyme Tsp509I (New England Biolabs, Hitchin, UK) at 65°C for 30 min and separated on a 4% agarose gel in TAE buffer (Tris-acetate EDTA; 40 mM Tris-acetate, 1 mM EDTA, pH 8.0). The wild-type FIE allele gave a pattern of four visible bands at 86, 145, 172, and 184 bp; the fie-1 allele had an extra band at 132 bp because of a G-to-A transition at the intron border (Ohad et al., 1999).
Acknowledgments
We gratefully acknowledge Jean Finnegan for METI a/s seed and Linda Margossian for primer sequences. R.V. was supported by Biotechnology and Biological Sciences Research Council (BBSRC) Grant No. P08575; S.A. was supported by a BBSRC Case studentship and Biogemma UK Ltd.; and M.S. was supported by BBSRC Grant No. P12018. The confocal microscope was purchased by the Wellcome Trust (Grant No. 049452).
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127 2493–2502. [DOI] [PubMed] [Google Scholar]
- Berger, F. (1999). Endosperm development. Curr. Opin. Plant Biol. 2 28–32. [DOI] [PubMed] [Google Scholar]
- Braselton, J.P., Wilkinson, M.J., and Clulow, S.A. (1996). Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem. 71 84–87. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H., and Olsen, O.-A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12 32–42. [Google Scholar]
- Campbell, R.B., Sinclair, D.A.R., Couling, M., and Brock, H.W. (1995). Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 245 291–300. [DOI] [PubMed] [Google Scholar]
- Castle, L.A., Errampalli, D., Atherton, T.L., Franzmann, L.H., Yoon, E.S., and Meinke, D.W. (1993). Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 241 504–514. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, S., and Messing, J. (1994). Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc. Natl. Acad. Sci. USA 91 4867–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y.-H., Kvarnheden, A., Yao, J.-L., Sutherland, P.W., Atkinson, R.G., Morris, B.A., and Gardner, R.C. (1998). Identification of pollination-induced genes from the ovary of apple (Malus domestica). Sex. Plant Reprod. 11 277–283. [Google Scholar]
- Eden, S., and Cedar, H. (1994). Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 4 255–259. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Genger, R.K., Peacock, W.J., and Dennis, E.S. (1998). DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 223–247. [DOI] [PubMed] [Google Scholar]
- Friedman, W.E. (1995). Organismal duplication, inclusive fitness theory, and altruism: Understanding the evolution of endosperm and the angiosperm reproductive syndrome. Proc. Natl. Acad. Sci. USA 92 3913–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genger, R.K., Kovac, K.A., Dennis, E.S., Peacock, W.J., and Finnegan, E.J. (1999). Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol. Biol. 41 269–278. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb-group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Haig, D., and Westoby, M. (1991). Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. London Ser. B 333 1–14. [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22 94–97. [DOI] [PubMed] [Google Scholar]
- Jones, C.A., Ng, J., Peterson, A.J., Morgan, K., Simon, J., and Jones, R.S. (1998). The Drosophila Esc and E(z) proteins are direct partners in polycomb group–mediated repression. Mol. Cell. Biol. 18 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L. (1970). Dependence of the R-mottled phenotype in maize on mode of sexual transmission. Genetics 66 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L., and Alleman, M. (1990). Gametic imprinting in maize in relation to the angiosperm life cycle. Development (suppl.), 9–14. [PubMed]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J., Goldberg, R.B., and Fischer, R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, G., Ciceri, P., and Viotti, A. (1995). Maternal-specific hypomethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 8 571–581. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990. a). Development of the free-nuclear endosperm in Arabidopsis thaliana. Arab. Inf. Serv. 27 53–64. [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990. b). Endosperm cellularization in Arabidopsis thaliana L. Arab Inf. Serv. 27 65–72. [Google Scholar]
- Neumann, B., and Barlow, D.P. (1996). Multiple roles for DNA methylation in gametic imprinting. Curr. Opin. Genet. Dev. 6 159–163. [DOI] [PubMed] [Google Scholar]
- Nogler, G.A. (1984). Gametophytic apomixis. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 475–518.
- Ohad, N., Margossian, L., Hsu, Y.-C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta, V. (1997). PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 7 249–258. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Simon, J. (1995). Locking in stable states of gene expression: Transcriptional control during Drosophila development. Curr. Opin. Cell Biol. 7 376–385. [DOI] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during G1 phase and is required maternally for early Arabidopsis development. Development 127 1815–1822. [DOI] [PubMed] [Google Scholar]
- Tilghman, S.M. (1999). The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell 96 185–193. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.-P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.-P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404 91–94. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260 1926–1928. [DOI] [PubMed] [Google Scholar]