Abstract
We examined the promoter hypermethylation of tumor-suppressor genes RASSF1A and TSLC1, quantitated EBV DNA load in nasopharyngeal carcinoma (NPC) tissues (T tissues), and matched tumor-adjacent tissues outside 0.5 cm (P tissues) and outside 1.0 cm (Z tissues) to evaluate the role of promoter hypermethylation of RASSF1A and TSLC1 as well as viral load in the pathogenesis of NPC. Methylation-specific polymerase chain reaction (PCR) for RASSF1A and TSLC1 and quantitative real-time PCR analysis of EBV DNA were performed on matched T, P, and Z tissues (n = 28) as well as chronic nasopharyngitis tissues (n = 8). Hypermethylated RASSF1A was frequently detected in the T (82%) and P tissues (75%), but less frequently in Z tissues (46%). The average quantities of EBV DNA (copies/µg DNA) in matched T, P, and Z tissues were 673,000, 90,000, and 7000. The differences of promoter hypermethylation of RASSF1A and EBV viral load among T, P, and Z tissues were statistically significant, with more frequent methylation and higher viral load detected when tissues examined were nearer to the NPC tissues. Our results suggest that aberrant hypermethylation of RASSF1A and high EBV load might be important events in NPC pathogenesis, and they may be useful molecular diagnostic markers for this cancer.
Keywords: NPC, RASSF1A, TSLC1, EBV, hypermethylation
Introduction
Nasopharyngeal carcinoma (NPC) is a malignancy particularly prevalent in Southern China and Southeast Asia. Various studies indicate that the development of NPC is a multistage process involving multiple factors. Genetic predisposition and epigenetic alterations, Epstein-Barr virus (EBV), and environmental factors including dietary habits have been suggested to play roles in the development of NPC [1,2].
3p21.3 is a chromosome region of particular significance to NPC [3,4]. Loss of heterozygosity (LOH) and comparative genomic hybridization (CGH) analyses indicate a high incidence of allelic loss of 3p in NPC. Microsatellite marker deletion and familial NPC pedigree analyses suggest that the NPC-related locus is 3p21.3 [4–7]. Many tumor-suppressor genes locate on the 3p21.3 region, including the NPC-related tumor-suppressor gene RASSF1A [8,9]. Lost or altered expression of this gene has been associated with the pathogenesis of a variety of cancers. Aberrant promoter hypermethylation of RASSF1A occurs frequently in NPC [10–13]. Inactivation of RASSF1A was found to be related with the hypermethylation of CpG island in its promoter region. 11q22–23 is another important tumor-suppressive region in NPC, involving candidate tumor-suppressor gene TSLC1. Expression of TSLC1 was downregulated by promoter hypermethylation in NPC cell lines [14].
EBV, also referred to as the human herpesvirus-4 (HHV-4), is a double-strand DNA gamma herpesvirus with a genome of 172 kb [15]. EBV is directly associated with human malignancies. EBV infection occurs worldwide and most people become infected during their lifetime. Infection with EBV usually occurs at a very early age, particularly in developing countries, and is closely associated with NPC and Burkitt's lymphoma. In contrast, in developed countries, most people are infected during adolescence or young adulthood, associated with infectious mononucleosis (IM). Basically, there exist two cell types susceptible to EBV infection: epithelial cells located in human oropharynx/nasopharynx and B-lymphocytes. EBV is strongly associated with NPC, an epithelial-derived cancer [15]. Many evidences indicate that EBV may be one of the most important factors involved in the tumorigenesis of NPC, but the exact role of EBV in NPC pathogenesis is not clear yet.
In this study, we detected the promoter hypermethylation of RASSF1A and TSLC1 genes, quantitatively analyzed the EBV DNA load in NPC tissues, and matched tumor-adjacent tissues to evaluate the role of promoter hypermethylation of RASSF1A and TSLC1 as well as EBV load in the development of NPC.
Materials and Methods
Specimens
Twenty-eight matched tumor and tumor-adjacent tissues as well as eight chronic nasopharyngitis tissues were obtained from the Pathology Department of Xiangya Hospital (Hunan, China), with the consent of patients according to the regulation of university policy. Detailed information of these patients was summarized in Tables 1 and 2. Specimens were snap-frozen in liquid nitrogen and subsequently stored at -80°C. Hematoxylin-eosin (HE)-stained sections were examined for the presence or absence of tumor cells, as well as tumor cell density. Only samples containing more than 70% of tumor cells were selected for T tissues. P and Z tissues were defined as tissues located 0.5 and 1.0 cm outside of visible NPC lesions, respectively. Histopathologic examination indicated that P tissues had mild, moderate, to severe presence of atypical hyperplastic cells, and were infiltrated by a number of lymphocytes. The Z tissues had a mild presence of atypical hyperplastic cells and were also infiltrated by inflammatory cells [16].
Table 1.
Clinical Information of Patients with Chronic Nasopharyngitis; Summary of MSP Results of RASSF1A and TSLC1, and EBV DNA Quantities in Chronic Nasopharyngitis Tissues.
| Patient Number | Sex | Age (years) | Methylation Status at RASSF1A | Methylation Status at TSLC1 | EBV (copies/µg DNA) |
| CN1 | F | 62 | - | - | 5.040 x 102 |
| CN2 | M | 38 | - | - | 6.493 x 102 |
| CN5 | M | 19 | - | - | 10.95 x 102 |
| CN10 | F | 13 | - | - | 0 |
| CN20 | F | 52 | - | - | 20.86 x 102 |
| CN2a | F | 51 | - | - | 0.089 x 102 |
| CN21 | F | 60 | - | - | 0 |
| CN50 | M | 50 | - | - | 2.564 x 102 |
CN: patients with chronic nasopharyngitis; (-) no hypermethylation detected.
Table 2.
Clinical Information of Patients with NPC; Summary of MSP Results of RASSF1A and TSLC1, and EBV DNA Quantities in Matched T, P, and Z Tissues.
| Patient Number | Sex | Age (years) | TNM | Methylation Status at RASSF1A | Methylation Status at TSLC1 | EBV (copies/µg DNA) | ||||||
| T | P | Z | T | P | Z | T | P | Z | ||||
| 29 | M | 64 | IVa | - | - | - | + | - | - | 3.110 x 102 | 4.211 x 102 | 19.63 x 102 |
| 30 | M | 42 | IIb | + | + | + | + | + | + | 395.2 x 102 | 93.80 x 102 | 0 |
| 31 | F | 31 | IIb | + | + | + | - | - | - | 9307 x 102 | 4634 x 102 | 210.8 x 102 |
| 32 | M | 45 | IIb | + | + | + | + | + | - | 868.0 x 102 | 917.0 x 102 | 353.8 x 102 |
| 36 | M | 68 | IVa | + | + | + | + | + | - | 665.0 x 102 | 483.8 x 102 | 30.78 x 102 |
| 40 | M | 40 | IIb | + | + | - | + | + | - | 8480 x 102 | 2188 x 102 | 19.27 x 102 |
| 44 | F | 50 | IVb | + | - | - | + | + | + | 2149 x 102 | 4.350 x 102 | 0 |
| 46 | M | 30 | I | + | + | + | + | + | + | 29190 x 102 | 3.247 x 102 | 0.816 x 102 |
| 48 | M | 37 | III | + | + | - | - | - | - | 37580 x 102 | 331.4 x 102 | 0 |
| 49 | M | 28 | III | + | + | - | - | - | - | 4650 x 102 | 105.4 x 102 | 17.22 x 102 |
| 50 | M | 26 | IIb | - | - | - | - | - | - | 9580 x 102 | 22.23 x 102 | 3.723 x 102 |
| 51 | M | 32 | IIb | + | - | - | + | + | + | 6880 x 102 | 10.62 x 102 | 8.970 x 102 |
| 54 | M | 54 | IIa | - | - | - | + | + | + | 17.92 x 102 | 2.615 x 102 | 0 |
| 56 | M | 37 | IIb | + | + | + | + | + | + | 21720 x 102 | 889.0 x 102 | 192.8 x 102 |
| 58 | F | 42 | IIb | - | - | - | + | - | - | 430.5 x 102 | 37.59 x 102 | 3.738 x 102 |
| 59 | F | 35 | IIb | - | - | - | + | + | + | 434.0 x 102 | 10.38 x 102 | 3.620 x 102 |
| 60 | M | 41 | IIb | + | + | - | + | + | + | 153.3 x 102 | 29.31 x 102 | 7.110 x 102 |
| 62 | M | 52 | IIa | + | + | + | - | - | - | 5620 x 102 | 373.0 x 102 | 2.201 x 102 |
| 64 | M | 55 | IIa | + | + | - | + | + | + | 1142 x 102 | 878.0 x 102 | 0.256 x 102 |
| 61 | F | 62 | IVb | + | + | + | - | - | - | 6.960 x 102 | 3.982 x 102 | 0 |
| 66 | M | 60 | IIa | + | + | - | + | + | + | 10190 x 102 | 0 | 0 |
| 70 | M | 55 | III | + | + | + | + | + | + | 98.00 x 102 | 29.41 x 102 | 0.107 x 102 |
| 71 | F | 45 | III | + | + | - | + | + | + | 2851 x 102 | 2283 x 102 | 23.97 x 102 |
| 72 | F | 38 | IIb | + | + | + | - | - | - | 10510 x 102 | 712.0 x 102 | 3.460 x 102 |
| 73 | F | 48 | I | + | + | + | + | + | + | 12330 x 102 | 1310 x 102 | 920.0 x 102 |
| 76 | M | 45 | III | + | + | + | - | - | - | 421.0 x 102 | 0.268 x 102 | 0 |
| 77 | M | 55 | IVb | + | + | + | + | + | + | 1807 x 102 | 1031 x 102 | 210.8 x 102 |
| 79 | F | 55 | IVa | + | + | - | - | - | - | 11040 x 102 | 760.0 x 102 | 40.30 x 102 |
TMN: tumor-node-metastasis stage; (+) hypermethylation; (-) no hypermethylation detected.
EBER-ISH (EBV-Encoded Small RNA-In Situ Hybridization)
Five cases of matched T, P, and Z tissues were randomly selected. ISH was performed using the EBER-ISH kit (PanPath Co., Amsterdam, The Netherlands) according to the manufacturer's instruction. Briefly, after the sections were dewaxed with dimethyl benzene, dehydrated with alcohol, and predigested with pepsin, hybridization solution containing the fluorescein-conjugated EBV nucleic acid probe was applied. After application of alkaline phosphatase-conjugated antibody against fluorescein isothiocyanate, nitroblue tetrazolium chloride/bromochloroindolylphosphate (NBT/BCIP) was applied as the chromogen. Dark brown nuclear staining was interpreted as a positive hybridization signal.
Cell Lines and Culture Conditions
The NPC cell lines were cultured in RPMI 1640 supplemented with 10% FCS (Invitrogen, Carlsbad, CA).
DNA Preparation
Genomic DNA from matched T, P, and Z tissues as well as chronic nasopharyngitis tissues were prepared using an improved method of extracting high-molecular-weight DNA with phenol/chloroform as described in Molecular Cloning [17]. Briefly, tissues were ground in liquid nitrogen, dissolved in 500 µl of Tris/NaCl/EDTA/SDS with proteinase K (100 µg/ml), incubated at 55°C for 2 hours, heated at 65°C for 15 minutes to inactivate the enzyme, and then extracted by phenol/chloroform followed by precipitation with 70% of ethanol. Finally, pellets of DNA were dissolved in DNase-free water and the concentrations were determined before being stored at -80°C.
Bisulfite Treatment
DNA was subjected to bisulfite treatment [18] according to the protocol of Tao et al. [19], with a little modification. Briefly, 2 µg of genomic DNA was denatured in 0.3 M NaOH for 10 minutes at 37°C. The denatured DNA were diluted in 300 ml of freshly prepared solution of 10 mM hydroquinone and 4.8 M sodium bisulfite, and incubated for 4 hours at 55°C in darkness. After the incubation, the DNA samples were desalted through a column provided by genome DNA clean-up system (TaKaRa Bio Inc., Shiga, Japan). The eluted DNA was treated with 0.3 M NaOH for 8 minutes at room temperature, and precipitated with acetate ammonia and ethanol. The bisulfite-modified genomic DNA was resuspended in 20 µl of TE buffer and stored at -20°C.
Methylation Analysis
Methylation-specific polymerase chain reaction (MSP) was performed according to the TaKaRa HS Taq kit protocol. Primers for MSP were described by Qiu et al. [11] for RASSF1A, and redesigned for TSLC1 according to the reports by Kuramochi et al. [20], Fukuhara et al. [21], and Lung et al. [14]. For each reaction, 1 µl of sodium bisulfite-treated DNA was added to 24 µl of reaction buffer (1.25 mM dNTPs, 16.6 mM (NH4)2SO4, 6.7 mM MgCl2, and 1.5 U of HS Taq polymerase; TaKaRa Bio Inc.) containing 100 ng of forward and reverse primers specific to the methylated or unmethylated DNA sequences. Conditions for amplification of DNA were 94°C for 4 minutes, followed by 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds (53°C for TSLC1), and 72°C for 30 seconds, with a final extension of 72°C for 5 minutes.
Sequencing of MSP Products of RASSF1A and TSLC1
Both U (reactions using primers specific for unmethylated CpG sites) and M (reactions using primers specific for methylated CpG sites) products of the RASSF1A and TSLC1 MSP from one T tissue were excised from agarose gels and purified by the PCR product purification system (TaKaRa). Subsequently, sequencing reactions were performed separately on the purified U and M products by 377 ABI PRISM DNA sequencer (Shanghai BoYa Company, Shanghai, China).
RT-PCR
The expression of RASSF1A and TSLC1 in NPC tissues and NPC cell lines as well as chronic nasopharyngitis tissues was examined by RT-PCR analysis. Total RNA was isolated by TRIZOL reagent. Two micrograms of total RNA was subjected to cDNA synthesis using a Reverse Transcription System according to the manufacturer's protocol (Promega, Madison, WI). PCR analysis of RASSF1A expression was carried out with primers according to Qiu at al. [11], and primer sequences of TSLC1 are 5′-ATGAAATGCCTCAACACGCCGTAC-3′ and 5′-TCTGCTGCGTCATCGGCTC CT-3′.
Real-Time PCR
EBV DNA copies in matched T, P, and Z tissues as well as chronic nasopharyngitis tissues were measured by realtime quantitative PCR analysis. The PCR primers and DNA probe with fluorescent dye (FAM) and TAMRA were designed according to Lo et al. [22] and synthesized by Dalian Baosheng Life Technologies, Inc. (Dalian, China), and used according to the manufacturer's protocol. In brief, PCR was set up in a total volume of 25 µl, including 2.5 µl of 10x buffer, 300 nM of each of the primers, 25 nM of the fluorescent probe, 200 nM dNTPs, 3 mM MgCl2, 2.5 µl of DNA template, and 1 U of Taq DNA polymerase (Promega). The reactions were performed and analyzed using the Double Fluorescent Quantitative PCR System (Light Cycler; Roche, Morristown, NJ). Parameters for PCR were as follows: 95°C for 5 minutes, followed by 40 cycles of 94°C for 15 seconds and 56°C for 1 minute.
Statistical Analysis
For comparison of hypermethylation level and EBV quantities in matched samples of T, P, and Z tissues, odds ratios were calculated and evaluated using chi-square analysis for hypermethylation data and analysis of variance (ANOVA) for EBV data. We used the logarithm of EBV data for statistical analysis in this study. P < .05 was considered as statistically significant. The statistical analyses were performed using the Excel software (Microsoft Corp., Redmond, WA) and statistical software SPSS Version 11.0 (SPSS Inc., Chicago, IL).
Results
Detection of EBV Infection in NPC and Matched Adjacent Tissues
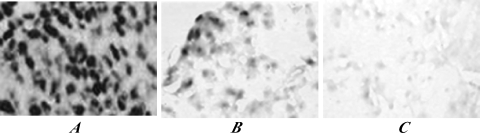
EBER-ISH detected many EBV+ tumor cells in all five cases of T tissues with strong positive signals, but only scattered EBV+ cells in all five cases of P tissues. However, all five cases of Z tissues were EBER- (Figure 1).
Figure 1.
Nuclear staining for EBER by ISH in NPC and matched tumor-adjacent tissues (original magnification, x400). (A) Strong positive signals were detected in many carcinoma nuclei of T tissues. (B) Weak to strong positive signals were detected in the nuclei of some atypical hyperplasic cells of P tissues. (C) No positive signal was detected in any cell of Z tissues.
Methylation Status and Expression of RASSF1A and TSLC1 in NPC Cell Lines
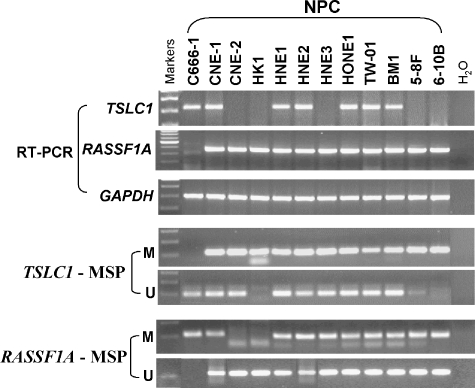
The methylation status of RASSF1A and TSLC1 in all currently available NPC cell lines (n = 11) was examined by MSP. Promoter hypermethylation of RASSF1A and TSLC1 was detected in the majority of cell lines. RT-PCR results showed that the expression levels of both genes were inversely correlated with their promoter methylation status (Figure 2).
Figure 2.
Methylation status and expression of RASSF1A and TSLC1 in NPC cell lines. Promoter hypermethylation of RASSF1A and TSLC1 was detected in the majority of cell lines with expression downregulated or silenced.
Hypermethylation of RASSF1A in Matched T, P, and Z Tissues
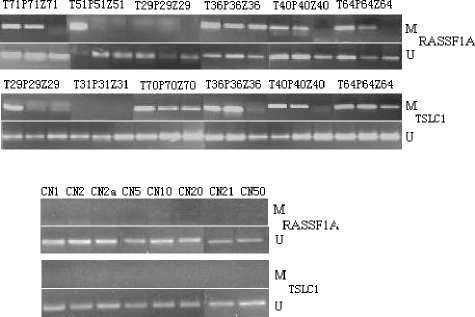
Because the aberrant methylation of RASSF1A and TSLC1 was frequently detected in NPC cell lines, we further investigated the methylation of these two tumor-suppressor genes in NPC tumors and the tumor-adjacent tissues. By MSP, the frequencies of promoter hypermethylation of RA SSF1A in matched T, P, and Z tissues were 82% (23 of 28), 75% (21 of 28), and 46% (13 of 28), respectively. No promoter hypermethylation of RASSF1A was detected in chronic nasopharyngitis tissues (zero of eight). All P tissues with hypermethylation of RASSF1A also had methylation in their matched T tissues. All Z tissues with hypermethylated RASSF1A also had methylation in matched T and P tissues. The results are summarized in Tables 1 and 2, and the representative MSP results are shown in Figure 3.
Figure 3.
Detection of hypermethylation of RASSF1A and TSLC1 in matched T, P, and Z tissues as well as chronic nasopharyngitis tissues. MSP analysis was performed using primers specific for both promoters. PCR products were visualized after electrophoresis on a 2% agarose gel (T: NPC tissues; P: tumor-adjacent tissues outside 0.5 cm; Z: tumor-adjacent tissues outside 1.0 cm; CN: chronic nasopharyngitis tissues).
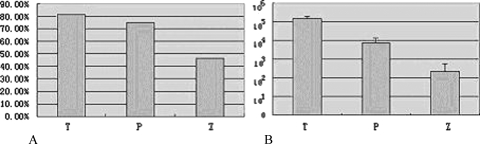
A statistically significant difference in hypermethylation levels among matched T, P, and Z tissues was found, and there were a significant difference between P and Z tissues, but no significant difference between T and P tissues (Figure 4A). Hypermethylation levels in T and P tissues were significantly higher than that in matched Z tissues (P = .002 and .029, respectively), and the hypermethylation level in Z tissues was significantly higher than that in chronic nasopharyngitis tissues (P = .016).
Figure 4.
Histogram of frequencies of aberrant promoter hypermethylation of RASSF1A (A) and geometric mean of EBV quantities (B) in T, P, and Z tissues of NPC patients. They followed a similar trend from high to low in these samples.
Hypermethylation of TSLC1 in Matched T, P, and Z Tissues
The frequencies of TSLC1 promoter hypermethylation in matched T, P, and Z tissues were 68% (19 of 28), 61% (17 of 28), and 50% (14 of 28), respectively. No promoter hypermethylation of TSLC1 was detected in chronic nasopharyngitis tissues (zero of eight). All P tissues with hypermethylation of TSLC1 also had methylation in their matched T tissues. All Z tissues with hypermethylation TSLC1 also had methylation in the matched Tand P tissues. Three cases exhibited aberrant promoter hypermethylation in T tissues and matched P tissues, but not in Z tissues. Two cases exhibited aberrant promoter hypermethylation in T tissues, but not in matched P and Z tissues. Fourteen cases exhibited aberrant promoter hypermethylation in T, P, and Z tissues. The results were listed in Tables 1 and 2, and representative MSP results are shown in Figure 3.
No statistically significant difference in TSLC1 hypermethylation among matched T, P, and Z tissues was found.
Sequencing Confirmation of Methylated and Unmethylated PCR Products
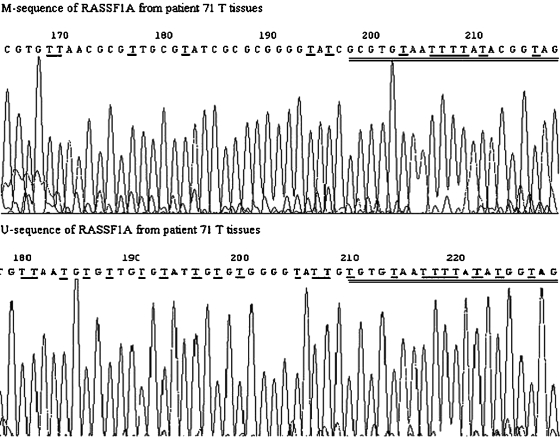
The M-MSP and U-MSP products amplified from one T tissue (patient 71) for either M or U primers of RASSF1A and TSLC1 were sequenced. The sequencing result showed that all cytosine residues in the sequence were converted to thymines, except for cytosine residues in CpG dinucleotides, indicating the presence of methylated cytosines in these CpG dinucleotides; the sequencing result of U-PCR product showed that all cytosine nucleotides, including those within CpG dinucleotides, were converted to thymines, indicating complete conversion of all cytosines to uracils by sodium bisulfite and the presence of unmethylated CpG sites. The representative sequencing results of M-and U-sequences of RASSF1A were shown in Figure 5.
Figure 5.
Sequencing analysis of M-MSP and U-MSP products for RASSF1A from NPC tissues (patient 71). For M-products, all cytosines were changed to thymines, except cytosines in CpG dinucleotide. The changes are underlined and M-and U-primers of MSP are double-underlined.
EBV DNA Quantities in Matched T, P, and Z Tissues as Well as Chronic Nasopharyngitis Tissues
Quantitative analysis of EBV DNA was performed on all matched T, P, and Z tissues. EBV DNA was detectable in 28 (100%) T tissues, and the average EBV DNA quantity was 6730 x 102 copies/µg DNA. EBV DNA was detectable in 27 (96%) matched P tissue samples of NPC patients, and the average EBV DNA quantity was 900 x 102 copies/µg DNA. EBV DNA was detectable in 21 (75%) matched Z tissues, and the average EBV DNA quantity was 70 x 102 copies/µg DNA. EBV DNA was also detectable in six (75%) chronic nasopharyngitis tissues, but the average EBV DNA quantity was only 6 x 102 copies/µg DNA (Tables 1 and 2).
A statistically significant difference of EBV DNA quantities among T, P, and Z tissues and chronic nasopharyngitis tissues was discovered (Figure 4B). The EBV DNA quantities in P tissues were significantly lower than those in matched T tissues (P = .027), whereas the EBV DNA quantities in Z tissues were significantly lower than those in matched P tissues (P < .01). The EBV DNA quantities in chronic nasopharyngitis tissues were lower than those matched tissues in NPC patients (P < .01).
Expression of RASSF1A and TSLC1 in T Tissues and Chronic Nasopharyngitis Tissues
Expression of RASSF1A and TSLC1 in T tissues and chronic nasopharyngitis tissues was examined by RT-PCR. Figure 6 shows that chronic nasopharyngitis tissues expressed a relatively higher level of both RASSF1A and TSLC1 mRNA when compared with T tissues.
Discussion
NPC is an endemic cancer with high incidence in Southeast Asia and Southern China. Both physical mapping and functional studies have assigned NPC-related tumor-suppressor gene(s) to 3p21.3. RASSF1A is a critical tumor-suppressor gene located at this locus, which has been shown to be frequently inactivated by promoter hypermethylation in Hong Kong and Singaporean NPC, both in NPC cell lines and primary tumors [9–11]. Although these studies demonstrated that promoter hypermethylation inactivated RASSF1A expression in NPC tissues and cell lines, they could not elucidate whether promoter hypermethylation is indeed involved in NPC tumorigenesis, or just a passenger phenomenon after the development of NPC.
Although our locality (Hunan Province, China) is not located in the highly prevalent area of NPC like Hong Kong, we still detected aberrant promoter methylation of RASSF1A in 82% of NPC tissues, similar to the previous reports of NPC tumors from Hong Kong and Singapore. We also detected aberrant methylation in 75% of P tissues and 46% of Z tissues of NPC patients. These results demonstrated the lowest level of RASSF1A methylation in Z tissues, a moderate level of methylation in P tissues, the highest methylation level in T tissues, and no aberrant methylation in chronic nasopharyngitis tissues. There are statistically significant differences in hypermethylation levels among T, P, and Z tissues. Our results suggest that: 1) aberrant promoter methylation of RASSF1A is a critical event in the development of NPC because no aberrant methylation was detected in chronic nasopharyngitis tissues; and 2) aberrant promoter methylation of RASSF1A is an early event during the pathogenesis of NPC because aberrant methylation of RASSF1A exists in both Z and P tissues.
In our studies, RASSF1A and TSLC1 genes were both frequently inactivated by promoter hypermethylation in NPC tissues and NPC cell lines. The results confirmed that promoter methylation plays a direct role in silencing both genes in NPC and contributes to NPC pathogenesis.
EBV is a member of the herpesvirus family, widely associated with human tumors. Although the association of EBV with the development of NPC is well-established, the precise role of EBV in the carcinogenic process remains unknown. Humans appear to be the only reservoir for EBV. Infection with the virus can lead to three different outcomes: 1) replication of the virus in epithelial cells permissive for EBV proliferation; 2) latent infection of B-lymphocytes in the presence of competent T-lymphocytes; and 3) stimulation and immortalization of B-cells [15]. Previous studies have demonstrated that EBV appears in many healthy subjects as well as patients with other cancers [23–25]. In our studies, the difference of EBV DNA levels among T, P, and Z tissues and chronic nasopharyngitis tissues was statistically significant. Further analyses showed that there were significant differences in EBV levels between either of the two abovementioned tissues. The EBER-ISH results also showed the different numbers of latent EBV-infected cell among T, P, and Z tissues. The results indicated that EBV copies were more abundant in regions nearer to the NPC tissues, suggesting either an infiltration of EBV tumor cells to adjacent area or a continuous malignant transformation of nearby epithelial cells. Our study also demonstrated that EBV is present in both NPC and healthy nasopharyngeal tissues, but the EBV load in healthy subjects was far less than that in NPC patients.
Aberrant promoter hypermethylation of RASSF1A and EBV quantities in T, P, and Z tissues followed a similar trend from high to low, suggesting a possible correlation between them. The promoter hypermethylation levels of TSLC1 in matched T, P, and Z tissues are not statistically significant, although a larger-scale study is needed to confirm this observation. There was no evidence for possible correlation between RASSF1A and TSLC1 methylation in this study.
In summary, our results indicate that promoter hypermethylation of both RASSF1A and TSLC1, and high EBV DNA load were commonly found in NPC tissues. The differences of aberrant promoter hypermethylation of RASSF1A and EBV levels between matched T, P, and Z tissues of NPC patients were statistically significant, with both levels being higher in tissues nearer to the NPC tissues. The results suggest that both promoter hypermethylation of RASSF1A and EBV infection might have occurred in the early phase of the development of NPC, and there may be a direct correlation between these two events.
Figure 6.
Expression of RASSF1A and TSLC1 in chronic nasopharyngitis tissues and T tissues. Chronic nasopharyngitis tissues expressed a relatively higher level of both genes compared with T tissues (CN: chronic nasopharyngitis tissues; T: NPC tissues).
Acknowledgements
We are grateful to the molecular pathology laboratory of the Cancer Research Institute in Central South University for technical assistance and encouragement.
Abbreviations
- CN tissues
chronic nasopharyngitis tissues
- EBER
EBV-encoded small RNA
- ISH
in situ hybridization
- HE
hematoxylin-eosin
- M
PCR reaction using primers specific for methylated CpG sites
- U
PCR reaction using primers specific for unmethylated CpG sites
- NPC
nasopharyngeal carcinoma
- P tissues
matched tumor-adjacent tissues outside 0.5 cm
- T tissues
nasopharyngeal carcinoma tissues
- Z tissues
matched tumor-adjacent tissues outside 1.0 cm
References
- 1.Choi PH, Suen MW, Huang DP, Lo KW, Lee JC. Nasopharyngeal carcinoma: genetic changes, Epstein-Barr virus infection, or both clinical and molecular study of 36 patients. Cancer. 1997;72:2873–2878. doi: 10.1002/1097-0142(19931115)72:10<2873::aid-cncr2820721003>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Yao KT. Epidemiological characteristics and presumed carcino-genesis of nasopharyngeal carcinoma in Hunan province—based on 1973–1975 cancer mortality survey. Bull Hunan Med Coll. 1982;7:10–17. [Google Scholar]
- 3.Cheng Y, Poulos NE, Lung ML, Hampton G, Ou B, Lerman MI, Stanbridge EJ. Functional evidence for a nasopharyngeal carcinoma tumor suppressor gene that maps at chromosome 3p21. Proc Natl Acad Sci USA. 1998;95:3042–3047. doi: 10.1073/pnas.95.6.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, Hu DX, Tan C, Xiang JJ, Zhou J, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972–1974. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- 5.Huang DP, Lo KW, Choi PH, Ng AY, Tsao SY, Yiu GK, Lee JC. Loss of heterozygosity on the short arm of chromosome 3 in nasopharyngeal carcinoma. Cancer Genet Cytogenet. 1991;54:91–99. doi: 10.1016/0165-4608(91)90035-s. [DOI] [PubMed] [Google Scholar]
- 6.Huang DP, Lo KW, van Hasselt CA, Woo JK, Choi PH, Leung SF, Cheung ST, Cairns P, Sidransky D, Lee JC. A region of homozygous deletion on chromosome 9p21–22 in primary nasopharyngeal carcinoma. Cancer Res. 1994;54:4003–4006. [PubMed] [Google Scholar]
- 7.Lo K, Tsao SW, Leung SF, Choi PHK, Lee JCK, Huang DP. Detailed deletion mapping on the short arm of chromosome 3 in nasopharyngeal carcinomas. Int J Oncol. 1994;4:1359–1364. doi: 10.3892/ijo.4.6.1359. [DOI] [PubMed] [Google Scholar]
- 8.Peng H, Zhao T, Yao KT. Expression of RASSF1A gene in nasopharyngeal carcinoma. Di Yi Jun Yi Da Xue Xue Bao. 2003;23:376–673. [PubMed] [Google Scholar]
- 9.Chow LS, Lo KW, Kwong J, To KF, Tsang KS, Lam CW, Dammann R, Huang DP. RASSF1A is a target tumor suppressor from 3p21. in nasopharyngeal carcinoma. Int J Cancer. 2004;109:839–847. doi: 10.1002/ijc.20079. [DOI] [PubMed] [Google Scholar]
- 10.Lo KW, Kwong J, Hui AB, Chan SY, To KF, Chan AS, Chow LS, Teo PM, Johnson PJ, Huang DP. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61:3877–3881. [PubMed] [Google Scholar]
- 11.Qiu GH, Tan LK, Loh KS, Lim CY, Srivastava G, Tsai ST, Tsao SW, Tao Q. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21., is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene. 2004;23:4793–4806. doi: 10.1038/sj.onc.1207632. [DOI] [PubMed] [Google Scholar]
- 12.Kwong J, Lo KW, To KF, Teo PM, Johnson PJ, Huang DP. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res. 2002;8:131–137. [PubMed] [Google Scholar]
- 13.Hesson L, Bieche I, Krex D, Criniere E, Hoang-Xuan K, Maher ER, Latif F. Frequent epigenetic inactivation of RASSF1A and BLU genes located within the critical 3p21. region in gliomas. Oncogene. 2004;23:2408–2419. doi: 10.1038/sj.onc.1207407. [DOI] [PubMed] [Google Scholar]
- 14.Lung HL, Cheng Y, Kumaran MK, Liu ET, Murakami Y, Chan CY, Yau WL, Ko JM, Stanbridge EJ, Lung ML. Fine mapping of the 11Q22-23 tumor suppressive region and involvement of TSLC1 in nasopharyngeal carcinoma. Int J Cancer. 2004;112:628–635. doi: 10.1002/ijc.20454. [DOI] [PubMed] [Google Scholar]
- 15.Bisson Theresa M. The Epstein-Barr virus. Infect Dis Update. 2004;11:34–39. [Google Scholar]
- 16.Jiang WH, Xiao XZ. Study on infectious status and molecular mechanism of Epstein-Barr virus in nasopharyngeal poorly differentiated squamous cell carcinoma. China: PhD. Thesis Changsha, Hunan; 2003. [Google Scholar]
- 17.Sambrool J, Fritsch EF, Maniatis T. Molecular Cloning. New York, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;11:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao Q, Swinnen LJ, Yang J, Srivastava G, Robertson KD, Ambinder RF. Methylation status of the Epstein-Barr virus major latent promoter C in iatrogenic B cell lymphoproliferative disease. Application of PCR-based analysis. Am J Pathol. 1999;155:619–625. doi: 10.1016/S0002-9440(10)65157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, et al. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 21.Fukuhara H, Kuramochi M, Fukami T, Kasahara K, Furuhata M, Nobukuni T, Maruyama T, Isogai K, Sekiya T, Shuin T, et al. Promoter methylation of TSLC1 and tumor suppression by its gene product in human prostate cancer. Jpn J Cancer Res. 2002;93:605–609. doi: 10.1111/j.1349-7006.2002.tb01297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo YM, Chan LY, Chan AT, Leung SF, Lo KW, Zhang J, Lee JC, Hjelm NM, Johnson PJ, Huang DP. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 23.You SL, Yao KT. Latency of Epstein-Barr virus and its relationship to nasopharyngeal carcinomas. Zhong Huazhong Liu Za Zhi. 1996;8:23–26. [PubMed] [Google Scholar]
- 24.Hausen zur H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. Epstein-Barr virus DNA in biopsies of Burkitt-tumor and anaplastic carcinoma of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- 25.Anderson-Anvert M, Fosby N, Klein G, Henle W. Relationship between the Epstein-Barr virus and undifferentiated nasopharyngeal carcinoma: correlated nuclear acid hybridization and histopathological examination. Int J Cancer. 1977;20:486–494. doi: 10.1002/ijc.2910200403. [DOI] [PubMed] [Google Scholar]