Abstract
ClpXP is an ATP-dependent protease that denatures native proteins and translocates the denatured polypeptide into an interior peptidase chamber for degradation. To address the mechanism of these processes, Arc repressor variants with dramatically different stabilities and unfolding half-lives varying from months to seconds were targeted to ClpXP by addition of the ssrA degradation tag. Remarkably, ClpXP degraded each variant at a very similar rate and hydrolyzed ∼150 molecules of ATP for each molecule of substrate degraded. The hyperstable substrates did, however, slow the ClpXP ATPase cycle. These results confirm that ClpXP uses an active mechanism to denature its substrates, probably one that applies mechanical force to the native structure. Furthermore, the data suggest that denaturation is inherently inefficient or that significant levels of ATP hydrolysis are required for other reaction steps. ClpXP degraded disulfide-cross-linked dimers efficiently, even when just one subunit contained an ssrA tag. This result indicates that the pore through which denatured proteins enter the proteolytic chamber must be large enough to accommodate simultaneous passage of two or three polypeptide chains.
Keywords: Arc repressor/chaperone/Clp/Hsp100/energy-dependent proteolysis/ssrA
Introduction
ClpXP is a bipartite protease responsible for degrading certain key regulatory proteins and aberrant translation products bearing the ssrA degradation tag (Levchenko et al., 1995; Kruklitis et al., 1996; Laachouch et al., 1996; Gottesman et al., 1998; Jenal and Fuchs, 1998; Karzai et al., 2000; Liu and Zuber, 2000). The ClpX component of ClpXP is a hexameric ring ATPase, belonging to the Clp/Hsp100 subfamily of the AAA+ ATPases (Schirmer et al., 1996). ClpX, by itself, has the capacity to recognize specific substrates and to denature and/or remodel the tertiary structures of these proteins in an ATP-dependent reaction (Levchenko et al., 1995; Wawrzynow et al., 1995; Kruklitis et al., 1996; Konieczny and Helinski, 1997; Jones et al., 1998). The ClpP component of ClpXP is a serine peptidase with broad sequence specificity. ClpP consists of two, stacked heptameric rings, which enclose a central chamber containing the 14 active sites of the enzyme (Wang et al., 1997). Because access to this proteolytic chamber requires passage through small axial pores, ClpP alone is able to degrade small peptides but not larger peptides or native proteins (Thompson and Maurizi, 1994). This mechanism serves to protect cellular proteins from cleavage unless they are recognized, denatured and translocated into ClpP by an associated ATPase, such as ClpX. In Escherichia coli, ClpP also associates with ClpA, an ATPase related to ClpX, to form the ClpAP protease.
In bacteria, incomplete polypeptides associated with stalled ribosomes are marked for degradation by co-translational addition of the ssrA peptide (AANDEN YALAA) to their C-terminal ends (Karzai et al., 2000). The ssrA tag is sufficient to target proteins for degradation by ClpXP (Gottesman et al., 1998; Kim et al., 2000). ClpAP also recognizes and degrades ssrA-tagged proteins but, in addition, interacts with a set of target substrates distinct from those recognized by ClpXP. Recent studies have shown that ClpA and ClpX catalyze denaturation of ssrA-tagged green fluorescent protein (GFP) and transfer of the resulting unfolded protein into the ClpP chamber (Weber-Ban et al., 1999; Hoskins et al., 2000; Kim et al., 2000; Singh et al., 2000). How ClpXP and ClpAP denature and translocate substrates is not understood. Similarly, the effect of substrate stability on these protein processing steps is not known, although denaturation appears to be the rate-limiting step in the overall degradation cycle for GFP–ssrA, a very stable protein (Kim et al., 2000).
Here, we use variants of a model substrate, P22 Arc repressor bearing a C-terminal ssrA tag, to probe the influence of protein stability on degradation and ATP utilization by the ClpXP machine. Native Arc is a homodimer consisting of two intertwined but unlinked subunits, each composed of a β-strand and two α-helices (Breg et al., 1990); denatured Arc is monomeric (Bowie and Sauer, 1989). Arc variants with a wide range of thermodynamic stabilities have been characterized in previous studies, including molecules in which the dimer subunits are covalently connected by a disulfide bond across the β-sheet (Sauer et al., 1996; Robinson and Sauer, 2000). We show that changing Arc-ssrA stability over a range that spans 15 kcal/mol changes the rate of ClpXP degradation by less than a factor of two. Thus, enzyme-catalyzed denaturation is largely insensitive to the intrinsic stability of the substrate. During a degradation cycle, ClpXP hydrolyzed significantly more ATP than would be needed for efficient denaturation of even the most stable substrate tested, but the hydrolysis rate was slower for hyperstable Arc-ssrA mutants than for unstable variants. Finally, we find that the ClpXP machine degraded both subunits of a disulfide-linked dimer without reducing the covalent linkage, even when just one subunit contained an ssrA tag. These results have important implications for the mechanism of denaturation and for the minimum size of the pore through which denatured polypeptides enter ClpP.
Results
Protein substrates
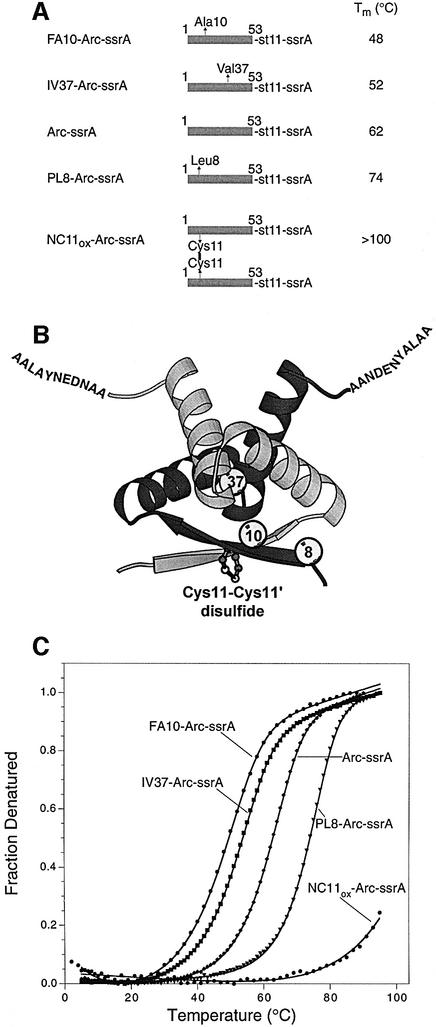
Figure 1 shows a ribbon model of the Arc repressor dimer, schematic representations of the Arc-ssrA variants used in the studies reported here and melting curves for each variant. Arc-ssrA contains residues 1–53 of wild-type Arc, followed by an 11 residue sequence containing a His6 affinity tag, and then the ssrA tag sequence AADENY ALAA (Levchenko et al., 1997). The four variants are identical to Arc-ssrA except for the introduction of destabilizing substitution mutations (FA10 or IV37) or stabilizing mutations (PL8 or NC11). The FA10 and IV37 substitutions decrease Arc stability by removing portions of the hydrophobic core (Milla et al., 1994). The PL8 mutation increases stability by introducing an extra hydrogen bond at either end of the wild-type β-sheet (Schildbach et al., 1995). The NC11 substitution allows formation of a native disulfide bond between Cys11 and Cys11′ in the two subunits of the Arc dimer (Robinson and Sauer, 2000). Disulfide-bonded NC11-Arc (NC11ox-Arc) is extremely stable, with a Tm in excess of 100°C (Figure 1).
Fig. 1. (A) Arc-ssrA variants used for degradation studies. Mutations in residues 1–53 of Arc are indicated by arrows. The st11 and ssrA sequences are H6KNQHE and AANDENYALAA, respectively. The reported Tm values are taken from the fits in (C). (B) Ribbon model of the Arc dimer. Spheres mark the sites of individual mutations in one subunit. A model of the disulfide bond in the NC11ox-Arc-ssrA dimer is shown as a ball-and-stick representation. (C) Thermal denaturation of 10 µM Arc-ssrA or variants (monomer equivalents) in 0.1 M NaH2PO4 pH 6.8 followed by changes in circular dichroism at 222 nm. The data were fit assuming an equilibrium between unfolded monomers and folded dimers (Bowie and Sauer, 1989).
Degradation by ClpXP
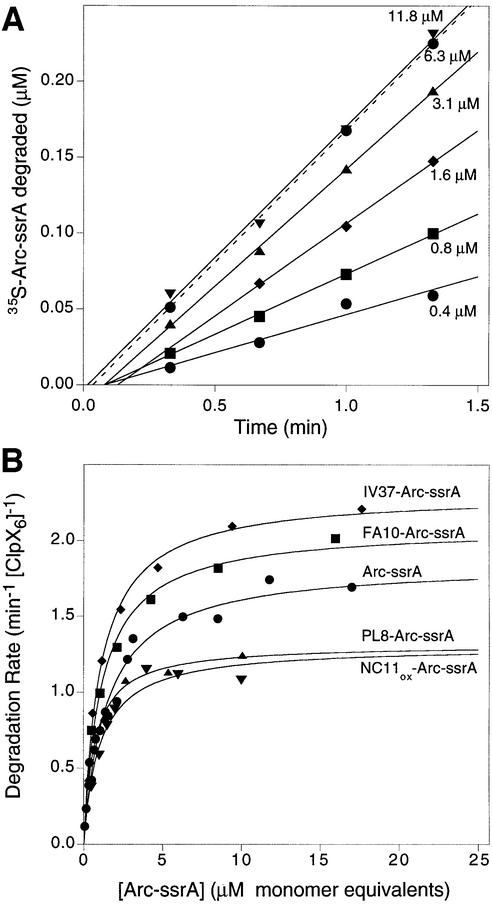
The Arc-ssrA variants with differing intrinsic stabilities allowed us to ask how protein stability affects the rate of degradation by ClpXP. Using 35S-labeled proteins, rates of ClpXP degradation at different concentrations of each Arc-ssrA variant were determined from the linear portions of kinetic experiments like those shown in Figure 2A. Control experiments confirmed that degradation required ClpX, ClpP and ATP (data not shown). For each variant tested, the dependence of the degradation rates on substrate concentration fit well (R >0.95) to a simple Michaelis–Menten model (Figure 2B). The kcat values (units per min per ClpX6 hexamer), obtained from non-linear least squares fitting, ranged from 1.3/min for the hyperstable PL8 and disulfide-bonded NC11 variants, to 2.3/min for the destabilized IV37 variant (Table I). As expected from previous experiments comparing degradation of different ssrA-tagged proteins (Kim et al., 2000), the KM values for the Arc-ssrA variants were also very similar (1–1.5 µM; Table I).
Fig. 2. (A) ClpXP degrades different concentrations of [35S]Arc-ssrA with linear kinetics as assayed by release of TCA-soluble counts. (B) Variation of ClpXP degradation rates with substrate concentration for Arc-ssrA and four mutant variants. The lines are non-linear least-squares fits to the Michaelis–Menten equation. The fitted values for KM and kcat are listed in Table I. All Arc concentrations are calculated in terms of subunit equivalents. The concentrations of ClpX6 and ClpP14 were 0.1 and 0.5 µM, respectively, except in the NC11ox-Arc-ssrA degradations, which were performed with 0.05 µM ClpX6.
Table I. Stability parameters for Arc-ssrA variants and steady-state kinetic parameters for ClpXP degradation.
| Variant | ΔGD at 1 µM (kcal/mol)a | Unfolding rate constant (/min) | ClpXP, KM (µM) | ClpXP, kcat (/min/[ClpX6]) |
|---|---|---|---|---|
| Arc-ssrA | 1.3 | 8.4 | 1.5 ± 0.1 | 1.8 ± 0.1 |
| PL8-Arc-ssrA | 2.2 | 0.12 | 1.0 ± 0.2 | 1.3 ± 0.1 |
| FA10-Arc-ssrA | –0.4 | 184 | 1.2 ± 0.1 | 2.1 ± 0.1 |
| IV37-Arc-ssrA | 0.2 | 44 | 1.1 ± 0.1 | 2.3 ± 0.1 |
| NC11ox-Arc-ssrA | 14.6 | 4.8 × 10–6 | 1.0 ± 0.2 | 1.3 ± 0.1 |
aFree energy changes of denaturation (ΔGD) at 25°C and a standard-state concentration of 1 µM were calculated from Ku values reported in Milla et al. (1994), Milla and Sauer (1995), Schildbach et al. (1995) and Robinson and Sauer (2000). Values for the unfolding rate constants were taken from the same references and from Milla et al. (1995). The KM and kcat values for ClpXP degradation are from non-linear least squares fits of the data shown in Figure 2B.
The similar KM values observed for Arc-ssrA variants having vastly different stabilities indicate that ClpXP must be able to bind to the ssrA tag sequence in native substrates. This result was expected, because the ssrA tag peptide is not part of the native Arc structure, and thus should be equally accessible to ClpXP in both the native and denatured substrates. Although some correlation between substrate stability and kcat was evident, the striking result of these experiments was that substrates that differed dramatically in thermodynamic stability were degraded at remarkably similar rates. For example, at 30°C and a concentration of 1 µM, roughly half of the FA10-Arc-ssrA molecules are denatured in solution, whereas for disulfide-bonded NC11-Arc-ssrA, only 1 in 1010 molecules is denatured (Milla et al., 1994; Robinson and Sauer, 2000). Nevertheless, ClpXP degraded both proteins at rates that differed by less than a factor of two. Assuming that denaturation is rate limiting, this result indicates that ClpXP does not depend on spontaneous denaturation of native Arc-ssrA substrates but rather actively unfolds these molecules at rates that do not depend strongly on intrinsic substrate stability. These data also show that ClpXP efficiently degrades disulfide-bonded proteins, a feature of the reaction that is explored further below.
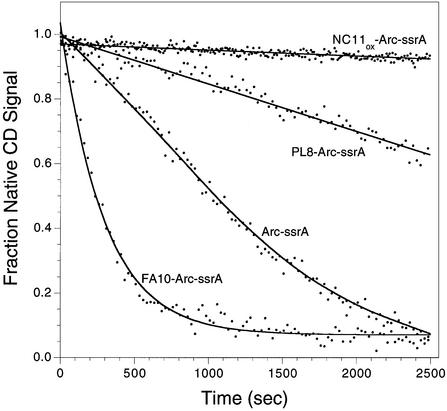
To compare the influence of substrate stability on the activity of an energy-independent protease, we examined degradation of four Arc-ssrA variants by the Arg-C endoproteinase. As assayed by loss of circular dichroism signal (Figure 3), the initial rates of Arg-C proteolysis were highly dependent on the stabilities of the Arc-ssrA substrates. For example, Arg-C degraded the unstable FA10-Arc-ssrA variant at a rate at least 1000-fold faster than the hyperstable, disulfide-bonded NC11-Arc-ssrA variant. These results eliminate the possibility that the native and denatured states of these proteins are equally resistant to degradation by any protease. It appears extremely likely, therefore, that the ability of ClpXP actively to unfold substrates in an ATP-dependent reaction allows it to degrade substrates with dramatically different native stabilities at similar rates.
Fig. 3. Endoproteinase Arg-C degradation of Arc-ssrA variants assayed by circular dichroism. Reactions were carried out as described in Materials and methods.
ATP consumption during substrate degradation
How much ATP hydrolysis is associated with ClpXP degradation of protein substrates with different stabilities? Using a coupled spectrophotometric assay, we determined ATP hydrolysis rates during degradation of the Arc-ssrA variants, using concentrations of ATP (2.5 mM; KM ∼0.3 mM) and protein substrate (20 µM; KM ∼1 µM) that ensured ∼90% saturation of ClpXP. The results listed in Table II show ATP hydrolysis rates ranging from 170 to 380/min per ClpX6P14 complex depending on the presence and identity of different Arc substrates. Several controls and observations indicate that these ATPase activities derive from ClpX and not from contaminating enzymes. First, a single peak of ATPase activity co-eluted with ClpX protein (Figure 4A) during ion-exchange chromatography on a column (MonoQ) that had not been used to purify ClpX. Secondly, no ATP hydrolysis was observed in the presence of ClpP alone or Arc variants alone (data not shown). Thirdly, the basal rate of ATPase hydrolysis by ClpX alone (260/min) was reduced in the presence of ClpP (170/min; Table II). Repression of ATPase activity by ClpP is a known property of ClpX (Kim et al., 2001). Finally, the presence of specific protein substrates stimulated ATPase hydrolysis significantly (Table II), consistent with previous results using ClpX purified by a different procedure (Wawrzynow et al., 1995).
Table II. Rates of ClpXP-catalyzed ATP hydrolysis stimulated by Arc-ssrA variants.
| Protein substrate | ATP turnover (/min) |
|---|---|
| None | 170 ± 10 |
| Arc-ssrA | 340 ± 20 |
| FA10-Arc-ssrA | 380 ± 30 |
| IV37-Arc-ssrA | 370 ± 30 |
| PL8-Arc-ssrA | 240 ± 10 |
| NC11ox-Arc-ssrA | 240 ± 10 |
Fig. 4. (A) Co-elution of ClpX protein and ATPase activity during cation-exchange chromatography. ClpX (90 µg), purified as described in Materials and methods, was applied to a MonoQ column and eluted with a linear gradient from 150 to 500 mM KCl in buffer pH 7.6 containing 50 mM HEPES-KOH, 15% glycerol, 3 mM DTT, 2 mM MgCl2, 0.1 mM ATP, 0.1 mM ZnCl2 and 0.01% Triton. Fractions of 200 µl were collected and assayed for ATPase activity and total protein by the Bradford assay. SDS–PAGE analysis of column fractions is shown in the inset. (B) Correlation between rates of ClpXP-mediated Arc-ssrA degradation and ATP hydrolysis. Data are from Tables I and II, and the error bars reflect the standard deviations of at least three independent measurements. The solid line is a linear fit to the data (slope = 150 ± 20, intercept = 45 ± 39, r2 = 0.94).
For the set of Arc variants, the rates of ATP hydrolysis were highly correlated with the maximal rates of ClpXP-mediated degradation (Figure 4B), with hydrolysis rates being slower for the most stable variants and vice versa. The slope of the linear fit to the Figure 4B data indicated that 150 ± 20 molecules of ATP were hydrolyzed during the time taken to degrade a single molecule of Arc-ssrA, regardless of its intrinsic stability. Two conclusions emerge. First, a large amount of ATP is consumed during a single degradation cycle. Secondly, when the ClpXP machine engages hyperstable substrate proteins, the rate of ATP hydrolysis slows, a result consistent with strong coupling between the chemical and mechanical reaction steps.
Proteolytic fate of untagged subunits in Arc heterodimers
The ssrA peptide tag is both necessary and sufficient to target Arc for degradation by ClpXP (Levchenko et al., 1997). Because Arc is a dimeric protein, however, the proteolytic fate of a heterodimer containing one tagged and one untagged subunit is uncertain. Are both subunits degraded, is just the tagged subunit degraded or are two tags required for degradation? To address these questions, we examined the degradation of both covalent and non-covalent Arc dimers.
The experiments discussed above showed that ClpXP can degrade Arc dimers that are covalently linked by an intersubunit disulfide bond. To determine whether both subunits needed to carry the ssrA tag for this degradation to occur, we mixed the reduced forms of tagged NC11- Arc-ssrA and excess untagged 35S-labeled NC11-Arc and allowed disulfide bond formation to occur. This resulted in a mixture of three disulfide-bonded molecules: a tagged homodimer, a singly tagged heterodimer and an untagged homodimer in a ratio of 1:6:11 as judged by densitometry of stained SDS–gels (Figure 5A, lane 4). Because only the untagged subunits were 35S-labeled in this experiment, the tagged homodimer was not detected by autoradiography (Figure 5A, lanes 1–3).
Fig. 5. (A) ClpXP degrades a disulfide-bonded heterodimer in which just one subunit contains a ssrA tag. Three disulfide-bonded species were present in the experiment, i.e. untagged homodimers, singly tagged heterodimers and tagged homodimers, but only the untagged subunits were 35S-labeled and thus the tagged homodimer was not detected following SDS–PAGE and autoradiography. Lane 4 shows SDS–PAGE of the protein mixture used in the degradation experiment, stained for total protein with SYPRO Orange. (B) Quantitation of the autoradiogram in (A). (C) Specific trapping of the singly tagged heterodimer in ClpPDFP as measured by gel filtration chromatography The top panel shows the absorbance profile at 214 nm of samples eluting from the column. The elution positions of molecular weight standards are shown above the chromatogram. Fractions 10–35 were analyzed by SDS–PAGE and quantified by PhosphorImager analysis. The ClpP14 and ClpX peaks were identified by staining for total protein with SYPRO Orange. The intensities of the heterodimer band (middle panel) and untagged homodimer band (bottom panel) are shown.
ClpXP was added to this mixture of disulfide-linked dimers and degradation was assayed by SDS–PAGE followed by autoradiography. This experiment showed no degradation of the untagged homodimer, as expected, but revealed that the heterodimer was degraded efficiently (Figure 5A and B). No accumulation of degradation intermediates was observed. If the disulfide-linked portion of the polypeptide could not enter the proteolytic chamber and be degraded, then an intermediate roughly the size of an Arc monomer should have been observed. We conclude that a single ssrA tag is sufficient to target both subunits of a disulfide-cross-linked dimer for degradation.
Further evidence that ClpXP can process both subunits of the covalent heterodimer effectively was obtained in experiments using ClpP that had been inactivated by chemical modification with diisopropylfluorophosphate (ClpPDFP). This version of ClpP efficiently accepts substrate proteins from ClpX, where they become ‘trapped’ rather than degraded (Kim et al., 2000). When the same mixture of disulfide-linked dimers was incubated with ClpXPDFP and analyzed by gel filtration chromatography under conditions that separated ClpX and ClpP, the labeled heterodimer was found to be quantitatively associated with the inactivated ClpPDFP. In contrast, none of the untagged dimers co-eluted with the ClpP oligomer (Figure 5C). The heterodimer associated with ClpP remained covalently linked, suggesting that the ClpXP machine translocated the disulfide-linked heterodimer directly into the proteolytic chamber of ClpP without reduction. Finally, in experiments designed to detect ClpXP-catalyzed exchange of subunits between disulfide-bonded Arc dimers, no exchange was observed, indicating that disulfide-bonded substrates are not reduced and then reoxidized during the denaturation and translocation reactions (data not shown).
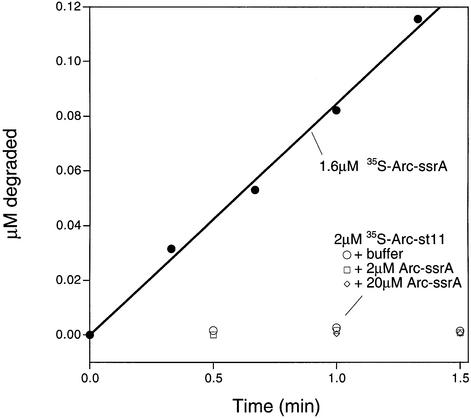
To address whether ClpXP degrades untagged subunits in non-covalent Arc heterodimers, we mixed 35S-labeled Arc lacking the ssrA tag (Arc-st11) with unlabeled Arc-ssrA. The formation of heterodimers between these species was confirmed by SDS–PAGE after cross-linking with bis(sulfosuccinimidyl) suberate (data not shown). In a control reaction, efficient ClpXP degradation of 35S-labeled Arc-ssrA was observed (Figure 6). In contrast, no ClpXP degradation of the 35S-labeled, untagged Arc subunit was detected by itself, in the presence of equimolar unlabeled Arc-ssrA or in the presence of a 10-fold excess of unlabeled Arc-ssrA (Figure 6). Under the last set of conditions, >90% of the 35S-labeled, untagged subunits should be present in heterodimers with tagged subunits. The experiment presented above using disulfide-linked dimers rules out the possibility that Arc dimers require two ssrA tags to be bound or degraded by ClpXP. We conclude, therefore, that ClpXP does not engage the untagged subunit in the non-covalent heterodimer, even though it binds and degrades the ssrA-tagged subunit. Taken together, these data suggest that the mechanism of protein processing by ClpXP involves some degree of tracking along a covalently linked protein chain.
Fig. 6. ClpXP does not degrade untagged 35S-labeled Arc monomers in non-covalent heterodimers with ssrA-tagged monomers. Degradation mixtures were prepared with 2 µM 35S-labeled Arc-st11 plus buffer, 2 µM cold Arc-ssrA or 20 µM unlabeled Arc-ssrA, and proteolysis was followed by the appearance of TCA-soluble counts as described in Materials and methods. The degradation of a sample of 35S-labeled Arc-ssrA is shown for comparison.
Discussion
ClpX-mediated degradation is nearly independent of substrate stability
The results presented here show that the rate of ClpXP-mediated degradation of a set of Arc-ssrA variants with radically different thermodynamic stabilities is scarcely affected by substrate stability. The ability of ClpXP to degrade proteins efficiently regardless of their inherent thermodynamic stability makes biological sense in terms of its role in the SsrA protein quality control system. SsrA-tagging occurs when ribosomes stall during translation, and tagged substrates would thus have a wide range of stabilities depending on both the protein and the position of tagging (Keiler et al., 1996; Roche and Sauer, 1999; Abo et al., 2000). At one extreme, ssrA tagging early in a protein sequence would generate a denatured fragment. At the other extreme, the tagging of some endogenous proteins occurs at positions corresponding to their stop codons, resulting in full-length native proteins fused to the ssrA peptide (E.Roche and R.Sauer, unpublished). ClpXP also degrades many native protein substrates that are not ssrA tagged, and ClpX, by itself, catalyzes energy-dependent disassembly of extraordinarily stable macromolecular complexes (Levchenko et al., 1995; Kruklitis et al., 1996; Jones et al., 1998). The capacity of ClpX or ClpXP to denature very stable proteins is likely, therefore, to be important in both its protein degradation and disassembly activities.
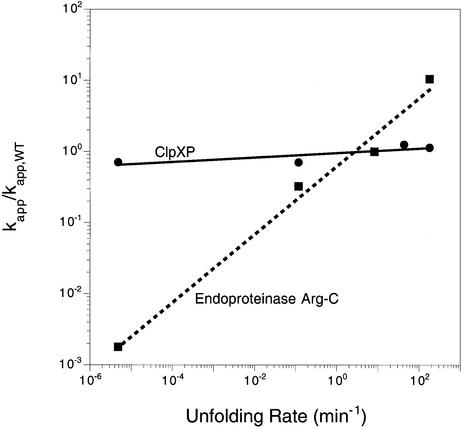
Although ClpX can trap globally denatured ssrA-tagged proteins (Singh et al., 2000), this does not appear to be the normal mechanism of denaturation. Native GFP–ssrA, for example, is denatured by ClpXP 107-fold faster than the rate at which spontaneous global unfolding and trapping would lead to unfolding (Kim et al., 2000). The results presented here reinforce the idea that ClpX does not function by trapping denatured substrates, a model that predicts that the rate of substrate degradation by ClpXP should depend on the equilibrium concentration of the denatured species. In contrast, we found that ClpXP degrades Arc-ssrA variants for which spontaneously denatured protein concentrations vary over 10 orders of magnitude at rates that differed by less than a factor of two. The trapping model also requires that the rate of uncatalyzed denaturation be as fast or faster than the degradation rate. Although this condition is satisfied for wild-type Arc-ssrA and the unstable variants, it is clearly inadequate for the hyperstable mutants (Table I). For example, kcat for ClpXP degradation of disulfide-bonded NC11-Arc-ssrA is 1.3/min, but this molecule unfolds spontaneously at a rate almost 300 000-fold slower (4.8 × 10–6/min; Robinson and Sauer, 2000). Whatever the mechanism, catalysis of active denaturation of Arc-ssrA variants by ClpXP is not significantly correlated with the spontaneous unfolding dynamics of these substrates (Figure 7).
Fig. 7. Variation of normalized degradation rates with the rate constants for spontaneous denaturation of Arc-ssrA and variants. ClpXP degradation (squares); Arg-C degradation (circles).
Denaturation of GFP–ssrA is the rate-determining step in ClpXP-mediated degradation (Kim et al., 2000). Moreover, acid-denatured GFP–ssrA is bound more tightly by ClpX than native GFP–ssrA and is degraded more rapidly when ClpP is added (Singh et al., 2000). Why then does the stability of Arc-ssrA variants, even those in which a substantial fraction of the molecules are already denatured, have such a small effect on their degradation rates by ClpXP? The simplest possibility is that the reaction step that leads to denaturation is both the slowest in the overall reaction and is also required for subsequent steps whether or not the substrate is native. For example, the conformational change that drives denaturation might also be needed to initiate translocation. In this case, what accounts for the observation that ClpXP degrades more stable Arc-ssrA variants slightly more slowly than less stable proteins (Figure 7)? An attractive possibility is that hyperstable proteins provide increased resistance to denaturation, which in turn slows the ATPase cycle of ClpXP and the overall rate of degradation. This model provides an explanation for the correlation between the observed rates of ClpXP-mediated ATP hydrolysis and substrate degradation (Figure 4).
Models for the mechanism of ClpX-mediated denaturation
The emerging view is that ClpX probably denatures proteins by mechanical destabilization of the native structure. This process could occur, for example, by binding the ssrA tail and pulling or translocating it through a cleft, channel or pore smaller than the native protein in a reaction coupled to ATP binding or hydrolysis. Continued pulling, once the native protein was lodged in the channel opening, could then provide the destabilizing force and ultimately result in unfolding. This model is attractive because it requires only a single site of initial contact with substrates, and results presented here show that Arc-ssrA dimers need just one ssrA tag for degradation. Alternatively, ClpX might catalyze denaturation by wrenching, squeezing or deforming the native proteins. These models, however, require ClpX binding to two or more distinct regions of the substrate, followed by a conformational change in the enzyme that leads to the application of force. The additional binding site(s) in this model would have to be sufficiently common to occur in most, if not all, native proteins.
Models that do not utilize mechanical force for ClpX-dependent denaturation are difficult to rationalize with the current data. Traditional transition state stabilization seems improbable because different proteins would have structurally distinct transition states for denaturation. Could ClpX tether substrates via the ssrA tag and then bind and trap a region of polypeptide that becomes locally denatured because of transient breathing motions, leading to increased breathing, more trapping, etc.? Arc, for example, is known to undergo a prominent breathing motion in which the two strands of the antiparallel β-sheet separate and open transiently (Burgering et al., 1995). One problem with this mechanism, however, is that substitutions in this region (e.g. FA10, PL8 and NC11) should dramatically affect the rate of this opening transition, but ClpXP degrades the ssrA-tagged forms of each of these proteins at similar rates, arguing against a role for this particular protein breathing motion in ClpXP-mediated denaturation. In addition, ClpXP degrades GFP–ssrA at a rate similar to the Arc-ssrA proteins and yet these substrates undoubtedly undergo highly distinct dynamic motions that occur at different rates.
In solution, the kinetic barrier for unfolding of disulfide-bonded NC11-Arc-ssrA must be >14 kcal/mol, the free energy change upon denaturation, and is probably closer to 20 kcal/mol. If a similar energetic barrier occurs for the ClpXP-bound substrate, then hydrolysis of a minimum of 2–3 molecules of ATP (ΔG° ∼ –7.3 kcal/mol ATP) would appear to be needed to drive the denaturation reaction. It also seems likely that this energy is expended in a single power stroke, because the spontaneous unfolding of Arc, like the unfolding of many small single-domain proteins, is highly cooperative (Bowie and Sauer, 1989; Milla and Sauer, 1994). Unless the mechanism of ClpX-mediated unfolding is significantly different from that observed in solution, there would be no way to unfold just a small part of the native structure when one ATP molecule was hydrolyzed, unfold a bit more when the next ATP was hydrolyzed, and so on. Although the cooperativity of ATP hydrolysis has not been investigated, as many as six molecules of ATP could, in principle, be hydrolyzed at once by the ClpX hexamer.
Why are 150 molecules of ATP hydrolyzed during ClpXP-mediated denaturation, translocation and degradation of a single molecule of Arc-ssrA or its variants? One explanation for this seemingly wasteful use of ATP is that the chance of denaturing Arc-ssrA during any given ATPase cycle is relatively low. In fact, because ClpXP does not bind native ssrA-tagged proteins tightly (Singh et al., 2000), it might be difficult for it to apply enough force to overcome a large denaturation barrier without slippage or substrate dissociation being the normal outcome. It is also possible that some of the excess ATP hydrolysis is required for other steps in the reaction, including translocation or substrate release.
Some of the substrate transactions of ClpXP are similar to those mediated by the mitochondrial import machinery. In both cases, native proteins are denatured in an ATP-dependent reaction and translocated through a small pore into another compartment. The effects of substrate stability on mitochondrial import have been studied using barnase with an attached signal sequence for import (Huang et al., 1999). When the unstructured linker between barnase and the signal peptide was sufficiently long to reach through the pore and engage the ATPase on the inner mitochondrial membrane, the observed import rates were independent of the thermodynamic and kinetic stability of the barnase variants. Interestingly, proteins containing bovine pancreatic trypsin inhibitor fusions to the signal sequence could not be imported into mitochondria unless the disulfide bonds in the native protein were reduced (Jascur et al., 1992).
The entrance pore into ClpP must be larger than 10 Å
Our finding that ClpXP efficiently degrades disulfide-bonded NC11-Arc-ssrA has important implications for the size of the pore required to allow polypeptide passage into the proteolytic chamber of ClpP. Figure 8 shows that import of a disulfide-cross-linked protein would, at some point, require two or three denatured chains to pass simultaneously through the pore. If the disulfide bond portion of the [35S]NC11-Arc-st11–NC11-Arc-ssrA heterodimer could not enter ClpP, then translocation would stall at a position similar to those depicted in Figure 8B or C. Moreover, because the active sites of ClpP are some distance from the entrance portal, only some of the polypeptide chain within the peptidase chamber would then be accessible for degradation. As a result, one of the degradation products would have a molecular weight slightly larger than an Arc monomer, and this product would retain most or all of the radioactivity present in the initial heterodimer. Figure 5A shows no evidence for such a degradation product, suggesting that the entire heterodimer is transferred into ClpP and degraded. In the ClpP crystal structure, the axial portals are ∼10 Å in diameter, dimensions that would barely allow passage of a single polypeptide chain. Hence, a dramatic opening of this pore would be required to accommodate entrance of disulfide-bonded NC11-Arc-ssrA into the proteolytic chamber of ClpP. If protein denaturation by ClpX also involves pulling the polypeptide chain through a cleft or pore, then the dimensions of these structures would also have to be sufficient to permit concurrent passage of two or three polypeptide chains. Examination of the Arc structure suggests that the pore would have to be ∼20–25 Å in diameter to accommodate three polypeptide chains at the point of the disulfide cross-link (Figure 8). Thus, we suggest that in the context of the functional ClpXP machine, the entry pore into ClpP must expand substantially to allow ClpX and ClpP to collaborate efficiently in promoting protein degradation.
Fig. 8. Models for import into ClpP of denatured proteins either as a single polypeptide chain (A) or as a disulfide-bonded pair of polypeptides (B and C). The presence of the disulfide cross-link requires that two or three polypeptide chains must pass simultaneously through the axial pore of the ClpP tetradecamer.
Materials and methods
Buffers
PD buffer pH 7.6 contained 25 mM HEPES-KOH, 5 mM MgCl2, 200 mM KCl, 15 mM NaCl, 0.032% (v/v) NP-40, 10% (v/v) glycerol, 5 mM ATP, 2.5 mM creatine phosphate and 0.05 mg/ml creatine kinase. Ni2+-NTA buffer A pH 8.0 was composed of 10 mM Tris–HCl, 100 mM NaH2PO4, and 6 M GdnHCl. Ni2+-NTA wash buffer pH 8.0 included 50 mM NaH2PO4, 300 mM NaCl and 20 mM imidazole. Ni2+-NTA elution buffer pH 8.0 contained 50 mM NaH2PO4, 300 mM NaCl and 250 mM imidazole. SP buffer pH 7.5 contained 10 mM Tris–HCl, 100 mM NaCl and 0.1 mM EDTA. Arc storage buffer pH 7.5 consisted of 10 mM Tris–HCl and 50 mM NaCl.
Plasmids
An arc-st11 gene with a C-terminal ssrA tag (Arc-ssrA) gene was amplified by PCR from pET3a-Arc-ssrA (Levchenko et al., 1997) and ligated into the NcoI–XhoI backbone fragment of pET28b (Novagen) to construct pET-28b-Arc-ssrA. Mutations encoding the PL8, FA10 and IV37 amino acid substitutions were introduced into the latter vector by cross-over PCR. The NC11-Arc gene was amplified by PCR from pLA11B-NC11 (Robinson and Sauer, 2000) and ligated into NcoI–XhoI-digested pET28b (Novagen). A gene encoding NC11-Arc-ssrA was constructed by introducing the NC11 mutation into pET-28b-Arc-ssrA using the Quik-Change™ Site-Directed Mutagenesis Kit protocol (Stratagene). The DNA sequences of the mutant genes were determined to confirm the presence of the appropriate mutation and the absence of any other changes in the amino acid sequence.
Protein purification and assays
Arc-ssrA and variants were expressed in E.coli strain SG1146a (clpP::cat, Δlon, λDE3), which was a gift from S.Gottesman (NIH). NC11-Arc-st11 was expressed in BL21(DE3) cells (Novagen). Cells were grown at 30°C to an OD600 of 1.0, and expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. After 90 min, cells were harvested by centrifugation, resuspended in buffer A and frozen at –80°C. The frozen suspension was thawed, centrifuged at 15 000 r.p.m. in a Sorvall SA600 rotor for 30 min, and the supernatant was decanted and mixed with Ni2+-NTA resin (Qiagen; 2 ml of resin per liter of cell culture) for 30 min. The slurry was poured into a column and washed with buffer A plus 10 mM imidazole (15 ml/l of culture). The column was washed with 15 ml of wash buffer, and bound protein was eluted with elution buffer until there was no detectable absorbance at 280 nm in the eluant. Fractions containing protein were pooled, dialyzed against three changes of SP buffer, centrifuged for 30 min at 10 000 r.p.m. in an SA600 rotor and applied to a 5/5 MonoS FPLC column (Pharmacia) equilibrated in SP buffer. A linear gradient from 100 mM to 2 M NaCl in SP buffer was applied, and fractions containing Arc-ssrA or variants were pooled and dialyzed against three changes of Arc storage buffer. NC11-Arc-ssrA and NC11-Arc-st11 were purified as described previously (Robinson and Sauer, 2000). Purified proteins were judged to be >95% pure by SDS–PAGE, and the molecular weight of each variant was confirmed by MALDI-TOF mass spectroscopy using a Perspective Biosystems Voyager instrument.
Arc-ssrA and variants were radioactively labeled by growth of cells in media containing [35S]methionine as described previously (Gottesman et al., 1998), incorporating the radiolabel at positions 1, 4, 7 and 42. ClpXP-mediated degradation of 35S-labeled Arc protein substrates was performed in PD buffer at 30°C and was assayed by the release of trichloroacetic acid (TCA)-soluble peptides (Kim et al., 2000). ClpX and ClpP were purified as described (Kim et al., 2000).
Oxidation to form disulfide-bonded NC11-Arc-ssrA was performed as described (Robinson and Sauer, 2000). An excess of [35S]NC11-Arc was mixed with NC11-Arc-ssrA in 4.5 M GuHCl, 10 mM dithiothreitol (DTT). This mixture was dialyzed against buffer containing 50 mM Tris–HCl pH 7.5, 250 mM KCl, 0.2 mM EDTA, resulting in oxidation and three disulfide-bonded dimers: NC11-Arc-ssrA–NC11-Arc-ssrA, [35S]NC11-Arc–NC11-Arc-ssrA and [35S]NC11-Arc–[35S]NC11-Arc. Degradation of this mixture of covalent dimers was performed in a buffer identical to PD buffer except with 5 mM KCl at 30°C by addition of 0.2 µM ClpX6 and 0.5 µM ClpP14. The initial substrate concentrations in these reactions were 0.1 µM untagged homodimer, 0.04 µM singly tagged heterodimer and 0.006 µM tagged homodimer.
The ClpPDFP-trapping experiments were performed by mixing 0.19 µM untagged dimer, 0.07 µM singly tagged heterodimer, 0.3 µM ClpX and 0.8 µM ClpPDFP in buffer identical to PD buffer except with 5 mM KCl, and incubating for 30 min at 30°C. The reaction mixture was then subjected to gel filtration chromatography on a Superdex 200 PC 3.2/30 column in Clp buffer. Fractions were analyzed by SDS–PAGE followed by autoradiography and quantitation using ImageQuant.
Degradation at 30°C of Arc-ssrA and variants by the endoproteinase Arg-C was assayed by changes in circular dichroism ellipticity at 222 nm in a buffer containing 90 mM Tris–HCl pH 7.6, 8.5 mM CaCl2, 5 mM DTT and 0.5 mM EDTA. For proteolysis of disulfide-bonded NC11-Arc-ssrA, the DTT was replaced with 1 mM β-mercaptoethanol to prevent reduction of the disulfide bond. SDS–PAGE analysis confirmed that the disulfide-bonded NC11-Arc-ssrA remained dimeric throughout the reaction, and the rate of Arg-C degradation of wild-type Arc-ssrA was found to be identical in 5 mM DTT and 1 mM β- mercaptoethanol.
ATP hydrolysis by ClpXP complexes was measured using a coupled assay (Nørby, 1988). A 40 ml mixture of 0.1 mM ClpX6, 0.5 mM ClpP14, 2.5 mM ATP, 1.0 mM NADH, 2.0 mM phosphoenolpyruvate, 3.0 U/ml lactate dehydrogenase and 3.0 U/ml pyruvate kinase in PD buffer was prepared and incubated at 30°C for 2 min. Arc-ssrA protein stock (10 ml) plus buffer that had been pre-warmed to 30°C was added to bring to the total Arc-ssrA concentration to 20 mM in monomer equivalents. This mixture was incubated for 2 min at 30°C, and the absorbance at 340 nm was recorded for the next 5 min. The rate of ADP formation was calculated assuming a 1:1 correspondence between ATP regeneration and NADH oxidation and a Δε340 nm of 6.23/mM/cm (Nørby, 1988).
Acknowledgments
Acknowledgements
We thank members of the Sauer and Baker labs for advice and help. Supported by NIH grant AI-15706 and the Howard Hughes Medical Institute. R.E.B. was supported by an NIH postdoctoral fellowship. T.A.B. is an employee of the Howard Hughes Medical Institute.
References
- Abo T., Inada,T., Ogawa,K. and Aiba,H. (2000) SsrA-mediated tagging and proteolysis of lacI and its role in the regulation of the lac operon. EMBO J., 19, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J.U. and Sauer,R.T. (1989) Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry, 28, 7139–7143. [DOI] [PubMed] [Google Scholar]
- Breg J.N., van Opheusden,J.H., Burgering,M.J., Boelens,R. and Kaptein,R. (1990) Structure of Arc repressor in solution: evidence for a family of β-sheet DNA-binding proteins. Nature, 346, 586–589. [DOI] [PubMed] [Google Scholar]
- Burgering M.J.M., Hald,M., Boelens,R., Breg,J.N. and Kaptein,R. (1995) Hydrogen exchange studies of the Arc repressor: evidence for a monomeric folding intermediate. Biopolymers, 35, 217–226. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Roche,E., Zhou,Y. and Sauer,R.T. (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the ssrA-tagging system. Genes Dev., 12, 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J.R., Singh,S.K., Maurizi,M.R. and Wickner,S. (2000) Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl Acad. Sci. USA, 97, 8892–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Ratliff,K.S., Schwartz,M.P., Spenner,J.M. and Matouschek,A. (1999) Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nature Struct. Biol., 6, 1132–1138. [DOI] [PubMed] [Google Scholar]
- Jascur T., Goldenberg,D.P., Vestweber,D. and Schatz,G. (1992) Sequential translocation of an artificial precursor protein across the two mitochondrial membranes. J. Biol. Chem., 267, 13636–13641. [PubMed] [Google Scholar]
- Jenal U. and Fuchs,T. (1998) An essential protease involved in bacterial cell-cycle control. EMBO J., 17, 5658–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.M., Welty,D.J. and Nakai,H. (1998) Versatile action of Escherichia coli ClpXP as protease or molecular chaperone for bacteriophage Mu transposition. J. Biol. Chem., 273, 459–465. [DOI] [PubMed] [Google Scholar]
- Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The ssrA–smpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol., 7, 449–455. [DOI] [PubMed] [Google Scholar]
- Keiler K.C., Waller,P.R.H. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of protein synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- Kim Y.-I., Burton,R.E., Burton,B.M., Sauer,R.T. and Baker,T.A. (2000) Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell, 5, 639–648. [DOI] [PubMed] [Google Scholar]
- Kim Y.-I., Levchenko,I., Fraczkowska,K., Woodruff,R.V., Sauer,R.T. and Baker,T.A. (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nature Struct. Biol., 8, 230–233. [DOI] [PubMed] [Google Scholar]
- Konieczny I. and Helinski,D.R. (1997) The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc. Natl Acad. Sci. USA, 94, 14378–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruklitis R., Welty,D.J. and Nakai,H. (1996) ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. EMBO J., 15, 935–944. [PMC free article] [PubMed] [Google Scholar]
- Laachouch J.E., Desmet,L., Geuskens,V., Grimaud,R. and Toussaint,A. (1996) Bacteriophage Mu repressor as a target for the Escherichia coli ATP-dependent Clp protease. EMBO J., 15, 437–444. [PMC free article] [PubMed] [Google Scholar]
- Levchenko I., Luo,L. and Baker,T. (1995) Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev., 9, 2399–2408. [DOI] [PubMed] [Google Scholar]
- Levchenko I., Smith,C.K., Walsh,N.P., Sauer,R.T. and Baker,T.A. (1997) PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell, 91, 939–947. [DOI] [PubMed] [Google Scholar]
- Liu J. and Zuber,P. (2000) The ClpX protein of Bacillus subtilis indirectly influences RNA polymerase holoenzyme composition and directly stimulates σH-dependent transcription. Mol. Microbiol., 37, 885–897. [DOI] [PubMed] [Google Scholar]
- Milla M.E. and Sauer,R.T. (1994) P22 Arc repressor: folding kinetics of a single domain dimeric protein. Biochemistry, 33, 1125–1133. [DOI] [PubMed] [Google Scholar]
- Milla M.E. and Sauer,R.T. (1995) Critical side-chain interactions at a subunit interface in the Arc repressor dimer. Biochemistry, 34, 3344–3351. [DOI] [PubMed] [Google Scholar]
- Milla M.E., Brown,B.M. and Sauer,R.T. (1994) Protein stability effects of a complete set of alanine substitutions in Arc repressor. Nature Struct. Biol., 1, 518–523. [DOI] [PubMed] [Google Scholar]
- Milla M.E., Brown,B.M., Waldburger,C.D. and Sauer,R.T. (1995) P22 Arc repressor: transition state properties inferred from mutational effects on the rates of protein unfolding and refolding. Biochemistry, 34, 13914–13919. [DOI] [PubMed] [Google Scholar]
- Nørby J.G. (1988) Coupled assay of Na+,K+-ATPase activity. Methods Enzymol., 156, 116–119. [DOI] [PubMed] [Google Scholar]
- Robinson C.R. and Sauer,R.T. (2000) Striking stabilization of Arc repressor by an engineered disulfide bond. Biochemistry, 39, 12494–12502. [DOI] [PubMed] [Google Scholar]
- Roche E.D. and Sauer,R.T. (1999) SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J., 18, 4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R.T., Milla,M.E., Waldburger,C.D., Brown,B.M. and Schildbach,J.F. (1996) Sequence determinants of folding and stability for the P22 Arc repressor dimer. FASEB J., 10, 42–48. [DOI] [PubMed] [Google Scholar]
- Schildbach J.F., Milla,M.E., Jeffrey,P.D., Raumann,B.E. and Sauer,R.T. (1995) Crystal structure, folding and operator binding of the hyperstable Arc repressor mutant PL8. Biochemistry, 34, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Glover,J.R., Singer,M.A. and Lindquist,S. (1996) HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci., 21, 289–296. [PubMed] [Google Scholar]
- Singh S.K., Grimaud,R., Hoskins,J.R., Wickner,S. and Maurizi,M.R. (2000) Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl Acad. Sci. USA, 97, 8898–8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.W. and Maurizi,M.R. (1994) Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J. Biol. Chem., 269, 18201–18208. [PubMed] [Google Scholar]
- Wang J., Hartling,J.A. and Flanagan,J.M. (1997) The structure of ClpP at 2.3Å resolution suggests a model for ATP-dependent proteolysis. Cell, 91, 447–456. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A., Wojtkowiak,D., Marszalek J., Banecki,B., Jonsen,M., Graves,B., Georgopoulos,C. and Zylicz,M. (1995) The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP–ClpX protease, is a novel molecular chaperone. EMBO J., 14, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban E.U., Reid,B.G., Miranker,A.D. and Horwich,A.L. (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature, 401, 90–93. [DOI] [PubMed] [Google Scholar]