Abstract
The general transcription factor TFIID, composed of the TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs), is important for both basal and regulated transcription by RNA polymerase II. Although it is well known that the TAF N-terminal domain (TAND) at the amino-terminus of the TAF1 protein binds to TBP and thereby inhibits TBP function in vitro, the physiological role of this domain remains obscure. In our previous study, we screened for mutations that cause lethality when co-expressed with the TAF1 gene lacking TAND (taf1-ΔTAND) and identified two ΔTAND synthetic lethal (nsl) mutations as those in the SPT15 gene encoding TBP. In this study we isolated another nsl mutation in the same screen and identified it to be a mutation in the histone fold domain (HFD) of the TAF12 gene. Several other HFD mutations of this gene also exhibit nsl phenotypes, and all of them are more or less impaired in transcriptional activation in vivo. Interestingly, a set of genes affected in the taf1-ΔTAND mutant is similarly affected in the taf12 HFD mutants but not in the nsl mutants of TBP. Therefore, we discovered that the nsl mutations of these two genes cause lethality in the taf1-ΔTAND mutant by different mechanisms.
INTRODUCTION
In eukaryotes, transcriptional initiation and activation of protein-coding genes involve a large number of proteins, such as general transcription factors (GTFs; TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH), upstream factor stimulatory activity (USA)-derived negative and positive cofactors, mediators and other types of coactivators and corepressors, including histone acetyltransferase (HAT) complexes, ATP-dependent nucleosome remodelers, RNA polymerase II (PolII) and gene-specific activators (reviewed in 1–5). TFIID, composed of the evolutionarily conserved TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs) (6), is required for specific recognition of core promoter elements such as the TATA box, the initiator element and the downstream promoter element (DPE) (reviewed in 7,8). TFIID nucleates the assembly of the preinitiation complex (PIC) around the transcription initiation site by recruiting other GTFs and RNA PolII, either sequentially or as part of a pre-assembled holoenzyme complex (reviewed in 9,10). The efficiency of PIC assembly is regulated by gene-specific activators that are usually bound to upstream activating sequences (UAS) (9,11). One of the most crucial molecular roles of activators is postulated to be the binding of TFIID to the core promoter (12–17). A number of biochemical studies using in vitro transcription reconstitution systems demonstrated that suboptimal core promoter binding by TFIID might provide an energetic blockade for initiating transcription, and activators can overcome this rate-limiting step by inducing conformational changes in TFIID (13,18,19). In fact, basal transcriptional activity mediated by TFIID was significantly lower than that mediated by TBP, for example, on the adenovirus E1b and HIV core promoters, even when both factors added to the reaction were adjusted to include the same amount of TBP (20). Importantly, gene-specific activators such as NF-κB can stimulate TFIID-mediated transcriptional activity to a much greater extent than TBP-mediated activity (20). Therefore, TAFs appear to have a dual function; that is, they inhibit TBP-mediated basal transcription in the absence of activators and stimulate transcription in response to the presence of activators (20–22). At least a part of the former function can be attributed to TAF1 since a binary complex comprised of human TAF1 (hsTAF1) and TBP showed lower basal activity than that of TBP alone, whereas transcription was restored to the level of TBP-mediated basal activity when activators were present (20,22) [PolII TAFs are named according to the new nomenclature (6)].
Consistent with the observation described above, the amino-termini of hsTAF1 and its orthologues carry an inhibitory activity for TBP-TATA interactions which can be suppressed by the coordinated action of TFIIA and certain types of activators (e.g., c-Jun, Zta and VP16) (23–30). The structural organization of this inhibitory domain, which was designated as TAF N-terminal domain (TAND), has been extensively characterized, especially for the yeast TAF1 (scTAF1) and Drosophila TAF1 (dmTAF1) (23,26,31). In both species, TAND is composed of two functionally distinct subdomains, TAND1 and TAND2, which bind to the concave and convex surface of TBP, respectively (31). Interestingly, acidic activation domains (ADs) of some activators derived from yeast cells and mammalian viruses were demonstrated to be functionally exchangeable with scTAND1 in vitro as well as in vivo, indicating that these two distinct classes of molecules might be similar in structure at least when bound to a common target, i.e., the concave surface of TBP (29). These observations encouraged us to build a two-step hand-off model depicting an early stage of the activation process in which TAND1, bound to the concave surface of TBP, could first be displaced by AD, and AD could be successively displaced by the TATA element (29). However, the precise molecular events involving TFIID during activation remain in dispute. For instance, another mechanism has been proposed for the action of AD in dissociating TAND-TBP interactions; c-Jun binds to hsTAND directly so as to cancel the TAND inhibition (30). This direct binding model sharply contrasts with our two-step hand-off model (indirect competitive model) in which AD and TAND1 bind to the overlapping surface of TBP, thereby allowing AD to compete out the inhibitory effect of TAND1 (29). More recent studies have demonstrated that the interaction between the convex surface of TBP and the high-mobility group (HMG) box-like region of hsTAF1 is also crucial for activation by several activators, including GAL4-E1A and GAL4-VP16 in mammalian cells (32). Although one molecule of TBP per TFIID complex is present (33), multiple potential binding sites for TBP were identified on the scTAF1 protein (four molecules of TBP could be bound to one molecule of scTAF1) (34). Interestingly, at both binding sites (TAND of scTAF1 and the HMG box-like region of hsTAF1) TFIIA appears to play a key role in dissociating TAF1-TBP interactions (22,26,27,31,32). Considering that this same action of TFIIA was not shown to be involved in c-Jun-mediated hsTAND-TBP dissociation (30), the molecular mechanisms triggering the initial step of TFIID activation may differ depending on the sort of activator and/or promoter structure of target genes.
Although accumulating evidence suggests that TAND is involved in transcriptional activation, it is hard to obtain direct proof of such involvement in vivo, probably because there are multiple and redundant pathways to support transcriptional activation (35). Indeed, in vitro transcription systems reconstituted with factors derived from either yeast or mammalian cells revealed that TAFs are dispensable for transcriptional activation, for example, when mediator components are added to the reaction (33,36–39). Genetic studies consistently show that at least a subset of TAFs can be deleted without affecting general activator function in yeast (reviewed in 40). Furthermore, mutations of TAFs included in both TFIID and Spt-Ada-Gcm5-acetyltransferase (SAGA) tend to affect transcription more broadly than those of TAFs specifically included in TFIID (reviewed in 35). These observations suggest that mediators and SAGA might be functionally redundant with TFIID in regulated transcription.
To obtain further evidence indicating that TAND is indeed involved in regulated transcription in vivo, we screened ΔTAND synthetic lethal (nsl) mutations that cause lethality in combination with a taf1 gene lacking TAND (taf1-ΔTAND) in Saccharomyces cerevisiae (41). As reported previously, the NSL1 gene we isolated in this screen was found to be allelic with the SPT15 gene encoding TBP (41). Intriguingly, our studies demonstrated that nsl1/spt15 alleles encoding TBP mutants, which are activation-deficient and especially defective at the post-TBP recruitment step, showed stronger nsl phenotypes (41). In the present study, we used the same screen to isolate a temperature-sensitive (TS) mutation of another gene designated as NSL2. Further analyses reveal that NSL2 is allelic with the TAF12 gene that encodes a common subunit of TFIID and SAGA (42). The nsl2/taf12 allele we isolated has a missense mutation (L420S) in the histone fold domain (HFD). Several other previously characterized nsl2/taf12 alleles harboring HFD mutations (W486stop, L446A, L464A) (43–45) also show nsl phenotypes. All of these nsl2/taf12 mutants are more or less deficient in transcriptional activation. Interestingly, further analyses suggest that nsl1/spt15 and nsl2/taf12 alleles cause lethality in the taf1-ΔTAND mutant, apparently by different molecular mechanisms. Based on these observations, we discuss how TAND is involved in regulating transcription in vivo.
MATERIALS AND METHODS
Yeast strains, media and genetic analyses
Standard techniques were used for yeast growth, transformation and tetrad dissection (46–48). Yeast strains used in this study are listed in Table 1.
Table 1. S.cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| CH1305 | MATa ade2 ade3 leu2 ura3 lys2 can1 | Kranz and Holm (49) |
| TMY4-2 | MATa ade2 ade3 leu2 ura3 lys2 can1 taf1-ΔTAND | Kobayashi et al. (41) |
| A22 | MATa ade2 ade3 leu2 ura3 lys2 can1 taf1-ΔTAND taf12-L420S pTM17/TAF1-ADE3-URA3 | This study |
| YAK278 | MATα ade2 ade3 leu2 ura3 lys2 can1 taf12-L420S | This study |
| YAK983 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pTM138/TAF12 | This study |
| YAK985 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pM1240/TAF12 | This study |
| YAK986 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pM1241/TAF12(L420S) | This study |
| YAK987 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pM1523/TAF12(W486stop) | This study |
| YAK1019 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pM3350/TAF12(391-539) | This study |
| YAK1156 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pM3852/TAF12(L446A) | This study |
| YAK1157 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf12::His3MX6 pYN1/TAF1 pM3853/TAF12(L464A) | This study |
| YAK1008 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pTM138/TAF12 | This study |
| YAK1010 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pM1240/TAF12 | This study |
| YAK1011 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pM1241/TAF12(L420S) | This study |
| YAK1012 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pM1523/TAF12(W486stop) | This study |
| YAK1021 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pM3350/TAF12(391-539) | This study |
| YAK1154 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pM3852/TAF12(L446A) | This study |
| YAK1155 | MATa ura3-52 trp1-63 leu2,3-112 ade2 Δtaf12::His3MX6 pM3853/TAF12(L464A) | This study |
| H2450 | MATa ura3-52 trp1-63 leu2,3-112 ade2 | Kokubo et al. (26) |
| Y13.2 | MATa ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 pYN1/TAF1 | Kokubo et al. (26) |
The host strain, TMY4-2, used for the synthetic lethal screen was constructed from CH1305 (49), as described previously (41). The YAK983 strain was generated from Y13.2 by targeted disruption of the TAF12 gene using a PCR-based gene deletion method (50). A His3MX6 marker module derived from Saccharomyces kluveri was amplified by PCR from the pFA6a-His3MX6 plasmid (kindly provided by Dr Mark Longtine) using the primer pair of TK1337 and TK1338 (oligonucleotides used in this study are listed in Supplementary Material Table S1). The resulting 1.5 kb PCR fragment was used to transform Y13.2 that had been transformed with the LEU2 marked plasmid carrying the TAF12 gene. The structure of the disrupted gene was confirmed by Southern blotting and genomic PCR using the TK1499 and TK1915 primer pair. The resulting Leu+ Trp– His+ strain in which both TAF1 and TAF12 genes were deleted was designated as YAK983. The YAK985, 986, 987, 1019, 1156, 1157 strains (Leu– Trp+ His+) were then generated from YAK983 by plasmid segregation.
YAK983 (Δtaf1 Δtaf12 strain) was crossed with H2450 (wild-type strain) and then dissected to obtain the haploid strain, YAK1008, carrying a single deletion of the TAF12 gene. The YAK1008 strain was grown on 5-fluoroorotic acid (5FOA)-containing plates to be completely deprived of the pYN1/TAF1 (URA3 marker) plasmid (26). The YAK1010, 1011, 1012, 1021, 1154, 1155 strains (Trp+ Leu–) were then generated from YAK1008 (Trp– Leu+) by plasmid segregation.
Cloning of a gene that complements the A22 nsl mutant
The screen for synthetic lethality with the taf1-ΔTAND gene was performed as described previously (41). The A22 mutant isolated in this screen was found to carry a single recessive mutation responsible for the synthetic lethal phenotype. The A22 strain showed the TS phenotype. Since multiple backcrosses revealed that the TS phenotype was linked to synthetic lethality, mutant segregants were transformed with a low-copy-number plasmid library (ATCC77162) yielding ∼180 000 transformants on SD-Leu plates when grown at 25°C for 12–18 h and then shifted to 36°C and incubated for 7 days. Plasmids containing complementing genomic DNA fragments were recovered from the positive colonies and amplified in Escherichia coli DH5α. These plasmids were retransformed into the A22 strain to confirm the complementation of the red/white sectoring, 5FOA lethality and the TS phenotype. Insert DNA boundaries were sequenced and compared to the yeast genome database. Overlapping regions from chromosome IV were obtained in all cases, and subcloning indicated that the presence of the TAF12 open reading frame (ORF) was sufficient to complement all mutant phenotypes shown by A22.
Identification of amino acid substitutions in the TAF12 gene of A22
The mutation in the TAF12 gene of the A22 strain responsible for synthetic lethality was identified by sequencing. The 2.6 kb DNA fragment, including the entire TAF12 gene, was amplified by PCR using the primer pairs TK678 and TK679 from the isolated genomic DNA of the A22 mutant. Direct sequencing of amplified DNA fragments using TK680, TK681, TK682, TK683, TK684, TK685, TK687 and TK741 as sequence primers revealed a single T→C point mutation at 1259 bp, which results in the amino acid substitution L420S. Proof that this mutation conferred synthetic lethality was obtained by testing the nsl phenotype of the taf12 allele bearing this mutation produced by site-specific mutagenesis (51), as described below.
Construction of plasmids encoding taf12 mutants
pTM138 was constructed by ligating the 3.8 kb PstI–PstI fragment including the entire TAF12 gene from the genomic insert (obtained as described above) into the PstI site of pRS315 (52). pM1240 was subsequently constructed by ligating the 3.0 kb PstI–BglII fragment into the PstI/BamHI sites of pRS314 (52). pM1240 was subjected to site-specific mutagenesis to create various taf12 alleles. Oligonucleotides TK765, TK1096, TK2658 and TK2659 were used to generate the plasmids pM1241 (L420S), pM1523 (W486stop), pM3852 (L446A) and pM3853 (L464A), respectively. To express truncated TAF12 protein (amino acids 391–539) in yeast cells, pM3350 was constructed by ligating two PCR fragments, namely, the 0.7 kb PstI–BamHI fragment including the promoter region of the TAF12 gene amplified by the primer pair of TK1978 and 1979, and the 1.2 kb BamHI–NotI fragment including the coding region of amino acids 391–539 amplified by the primer pair of TK1980 and 1981 into the PstI/BamHI and BamHI/NotI sites of pRS314, respectively.
To prepare GST-fused TAF12 derivatives expressing only the 391–539 amino acid C-terminal region, pM3405 (wild type), pM3924 (L420S), pM3926 (L446A) and pM3927 (L464A) were constructed by ligating the 0.4 kb BamHI–EcoRI fragments amplified from pM1240, pM1241, pM3852 and pM3853, respectively, with the same set of PCR primers (TK2063 and TK2064) into the bacterial expression vector, pGEX2T (Amersham Biosciences). To prepare shorter forms (amino acids 414–490) of these mutant proteins, pM3854 (wild type), pM3855 (L420S), pM3857 (L446A) and pM3858 (L464A) were similarly constructed by ligating the 0.2 kb BamHI–EcoRI fragments amplified with the primer set of TK2900 and TK2901 into pGEX2T. In addition, pM3925 and pM3856 were constructed by ligating the 0.3 and 0.2 kb BamHI–EcoRI fragments amplified from pM1240 with the primer sets of TK2064/2065 and TK2900/2065 into pGEX2T to express the 391–485 and 414–485 amino acid C-terminal regions, respectively, both of which correspond to the taf12-W486stop mutant.
Construction of the plasmid encoding TAF4
For in vitro binding studies, pM3869 was constructed by ligating the 1.2 kb NdeI–BamHI fragment corresponding to the TAF4 ORF (amplified with the primer pair of TK2708 and TK2709) into the NdeI/BamHI sites of the pACYC184 (New England Biolabs) based bacterial expression vector, which was originally designed to express scADA1 (amino acids 259–359) (kindly provided by Dr Irwin Davidson) (53).
Phenotypic analyses
To confirm the presence of the synthetic lethal phenotype in different general genetic backgrounds, YAK985, YAK986, YAK987, YAK1019, YAK1156 and YAK1157 were transformed with pRS315-based plasmids encoding TAF1 (pTM26) (41) and taf1-ΔTAND (pM3217) (41) and then incubated on 5FOA plates at 30°C for 5 days. To test the complementing activities of various plasmids using a red/white sectoring assay, colonies transformed with these plasmids were streaked onto YPD plates and then incubated at 25°C for 8–10 days. Recovery of the TS phenotype was assayed by comparing the growth rates at 25 and 35°C of yeast transformants incubated on YPD plates for 3–4 days.
Plasmids encoding activation domains or TAF12 derivatives fused with the GAL4 DNA binding domain
Expression plasmids encoding GAL4 (amino acids 842–874), GCN4 (amino acids 107–144), ADR1 TADIV (amino acids 642–704), EBNA2 (amino acids 426–462), VP16 (amino acids 457–490) and scTAND1 (amino acids 10–42) ADs fused with the GAL4 DNA binding domain were constructed as described previously (41,54). To express GAL4-TAF12 derivatives in yeast cells, pM385 (wild type), pM1247 (L420S), pM3320 (W486stop), pM3321 (amino acids 391–539), pM3860 (L446A) and pM3861 (L464A) were constructed similarly by ligating EcoRI–BamHI, PCR-amplified fragments into pM471 (41). The primer pairs used were TK1088/TK772 (wild type, L420S, L446A and L464A), TK1088/TK1955 (W486stop) and TK1976/TK772 (amino acids 391–539).
In vivo activation as measured by β-galactosidase activity
For the artificial recruitment experiments, plasmids encoding GAL4-TAF12 derivatives were introduced into the CH1305 strain containing pB20, a multicopy URA3 plasmid with the GAL1 promoter upstream of the LacZ structural gene (kindly provided by Dr A. G. Hinnebusch). The resulting strains were grown to an OD600 of 0.7 in YPD medium and then treated with repeated freeze/thaw cycles. The β-galactosidase activity was measured as described previously (29). To measure activation by classical ADs, yeast strains bearing TAF12 derivatives (YAK1010, YAK1011, YAK1012, YAK1021, YAK1154 and YAK1155) were transformed with pB20 and plasmids expressing various activators.
Co-expression of HFD proteins in E.coli and GST pulldown assays
Plasmid pairs encoding scADA1 (amino acids 259–359) (kindly provided by Dr Irwin Davidson) (53) or the entire region of TAF4 and GST-fused TAF12 derivatives (the longer form of amino acids 391–539 or the shorter form of amino acids 414–490) were introduced into E.coli BL21(DE3) (Novagen), and double transformants were selected on plates containing ampicillin (200 mg/l) and chloramphenicol (35 mg/l). Bacteria were grown to an OD600 of 0.45 and induced for 4 h at 25°C with 1 mM IPTG in 10 ml of LB medium. Cells were harvested and washed once with lysis-wash buffer (25 mM Tris–HCl, pH 6.0/0.4 M NaCl) and then resuspended in 250 µl of lysis-wash buffer (53). After repeated cycles of sonication, cell debris was removed by centrifugation (15 000 r.p.m. for 10 min; Hitachi himac CF15R centrifuge with T15AP21 rotor), and the supernatant was stored at –30°C until use.
To study interactions of GST-TAF12, supernatants (200 µl) obtained from cells co-expressing ADA1 (amino acids 259–359) or TAF4 were prepared as described above, incubated with 50 µl of glutathione–Sepharose 4B (Amersham Biosciences) at 4°C for 30 min, then washed four times with 500 µl of the lysis-wash buffer. The complexes on the beads were eluted by boiling in 100 µl of SDS sample buffer, and one-fifth of the eluted fraction was analyzed by SDS–PAGE and Coomassie brilliant blue (CBB) staining.
Northern blot analyses
Northern blot analyses were performed as described previously (54). To prepare the probes, DNA fragments surrounding the initiating methionine were amplified by PCR from yeast genomic DNA, purified and 32P-labeled using a random priming method. The PCR primer pairs used were as follows: TK245/TK246 for CTT1, TK1043/TK1044 for PHO84, TK1879/TK1880 for YJR078W, TK1881/TK1882 for HIS4, TK1889/TK1890 for YJR079W, TK1895/TK1896 for YDR539W, TK2111/TK2112 for GNA1, TK2113/TK2114 for HTA3, TK2115/TK2116 for CAM1, TK2117/TK2118 for PMT4, TK2119/TK2431 for YPL019C, TK2121/TK2432 for PHO5, TK2125/TK2126 for YDL124W, TK2294/TK2295 for TRX1, TK2296/TK2297 for WTM2 and TK2298/TK2299 for YHB1.
RESULTS
Isolation and identification of the nsl2/taf12 mutant, which is synthetically lethal to taf1-ΔTAND
In our previous study (41), we attempted to identify genes involved in parallel pathway(s) of transcriptional activation to the one in which TAND function might be essential. We screened nsl genes displaying synthetic lethal interactions with the taf1 gene that lacks TAND (taf1-ΔTAND), and 14 mutants were isolated as candidates carrying nsl mutations (41). Two of them, C40 and D7, referred to as nsl1-1 and nsl1-2, respectively, were previously characterized and found to carry different amino acid substitutions (S118L and P65S, respectively) in the SPT15 gene encoding TBP (41). In this study, another nsl mutant, A22, was characterized as described below.
Genetic crosses and tetrad dissection showed that the A22 mutant carries a recessive mutation belonging to a different complementation group from nsl1/spt15 mutants (data not shown). The gene responsible for the A22 nsl phenotype was designated as NSL2. We also refer to a mutant allele of A22 as nsl2-1. Since linkage analysis indicated that the nsl2-1 allele exhibits a TS growth phenotype, we tried to isolate the wild-type NSL2 gene by complementing the TS phenotype of the nsl2-1 mutant. The nsl2-1 mutant was transformed with a partial Sau3A yeast genomic library, and several complementing colonies were isolated. Retransformation and sequencing analyses revealed that genomic inserts in the plasmids all encoded entire regions of the SAN1, MKC7 and TAF12 genes. We subcloned each gene into a centromeric vector and tested it in the complementation assay. Only the plasmid carrying the TAF12 gene rescued the TS growth defects of the nsl2-1 mutant, suggesting that NSL2 might be allelic to TAF12.
To identify possible mutations in the TAF12 gene of the nsl2-1 mutant, we sequenced PCR-amplified genomic fragments encompassing the entire ORF plus 5′- and 3′-adjacent DNA regions (∼500 bp each) of the TAF12 gene. We found a single amino acid substitution, L420S, in the coding region of the TAF12 gene of the nsl2-1 mutant (Fig. 1A). We next asked whether this mutation was sufficient to reproduce the nsl phenotype and whether such a phenotype depended on a particular genetic background. To address these questions, we reconstructed the taf12-L420S allele on a centromeric TRP1 plasmid by site-directed mutagenesis to exclude any other possible mutations. Then we constructed yeast strains containing either the wild-type TAF12 gene (YAK985) or the taf12-L420S mutant allele (YAK986) on a centromeric TRP1 plasmid as well as the wild-type TAF1 gene on a centromeric URA3 plasmid in combination with double deletions of chromosomal TAF12 and TAF1 genes. These strains have different general genetic backgrounds from the one used in the original genetic screen for the nsl mutants. The centromeric LEU2 plasmid harboring either the wild-type TAF1 gene or taf1-ΔTAND mutant allele was transformed into the YAK985 and YAK986 strains described above, and strains were tested for their growth on 5FOA plates. We reasoned that if taf12-L420S is responsible for the nsl phenotype, strains derived from YAK986 (i.e., those carrying the taf12-L420S gene on a TRP1 plasmid) would be viable on 5FOA plates, which select for cells that have lost the URA3- and TAF1-containing plasmid, only when the LEU2- and TAF1-containing plasmid has been introduced into cells as a substitute. Consistent with this expectation, yeast strains carrying the taf12-L420S gene grew well on 5FOA plates only when they expressed the wild-type TAF1 gene but not when they expressed the taf1-ΔTAND gene (Fig. 1B). Also as expected, yeast strains carrying the wild-type TAF12 gene grew well on 5FOA plates whether they expressed the TAF1 gene or the taf1-ΔTAND gene (Fig. 1B). We also confirmed that the taf12-L420S allele was recessive and showed a TS phenotype in the genetic background of the YAK986 strain just as in the original mutant strain A22 (data not shown). These observations support the notion that the taf12-L420S gene is synthetically lethal with the taf1-ΔTAND gene even when expressed in a different general genetic background.
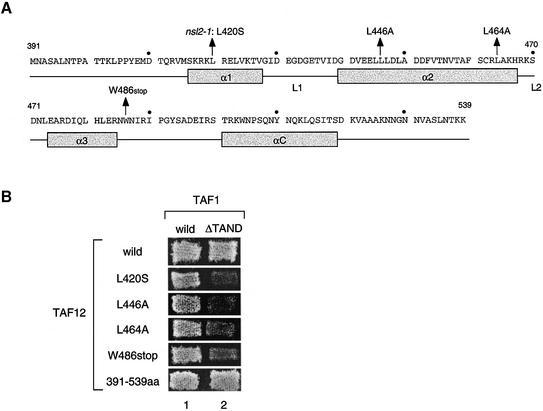
Figure 1.
Positions of nsl2/taf12 mutations and their nsl phenotypes. (A) Schematic representation of primary sequence and proposed secondary structures of the HFD of the TAF12 protein (53). Four α-helices are depicted as grey boxes and two linker regions between α-helices 1 and 2 or 2 and 3 are represented as L1 and L2 (53). The positions of the nsl2/taf12 mutations tested in this study are indicated with arrows above the primary sequence. The 410, 430, 450, 470, 490, 510 and 530 amino acids are marked with a dot. (B) The nsl phenotypes are shown by several nsl2/taf12 mutants. The LEU2-marked plasmid encoding either the wild-type TAF1 gene or the taf1-ΔTAND gene, as indicated at the top, was individually introduced into the strains with double deletions of TAF1 and TAF12 genes containing the TRP1-marked plasmid encoding each TAF12 derivative, as indicated on the left, in addition to the URA3 marked plasmid encoding wild-type TAF1. The resulting transformants were grown on 5FOA plates at 30°C for 5 days.
The integrity of the HFD of the TAF12 protein is crucial for cell growth of the taf1-ΔTAND mutant
Recent studies demonstrated that TFIID contains at least nine TAF proteins containing a HFD (reviewed in 55,56). Sequence comparison and biochemical analyses suggest that TAF9, TAF6, TAF4 and TAF12 are homologous in their HFD to the H3, H4, H2A and H2B components of the histone octamer, respectively (44,55,57,58). Leucine at the 420 amino acid position of scTAF12, which was substituted with serine in the nsl2-1 mutant, corresponds to isoleucine at the 36 amino acid position of Xenopus laevis H2B located on the α1 helix, which directly contacts a deoxyribose in a nucleosome (Fig. 1A) (59).
To determine whether the integrity of the HFD of the TAF12 protein is important for growth of the taf1-ΔTAND mutant, we decided to test several other HFD mutations that were previously characterized (43–45). Leucines at positions 446 and 464 on the α2 helix of TAF12 (Fig. 1A) (equivalent to methionine at position 59 and leucine at position 77 of X.laevis H2B) are assumed to be located on the interaction surface with TAF4 and TAF6, respectively (45). On the other hand, a truncated mutation, W486stop, which lacks the αC helix, was isolated as a taf12 TS mutant by two groups independently (43,44). We examined nsl phenotypes of these three taf12 alleles (L446A, L464A and W486stop) in the same assay as conducted for taf12-L420S, and all of them appeared to be synthetically lethal to the taf1-ΔTAND gene (Fig. 1B). However, synthetic effects were found to be milder in the L446A and L464A mutants than those in the other two mutants after longer incubation (data not shown). Importantly, the taf12 mutant expressing only HFD (amino acids 391–539) did not display any nsl phenotype and showed normal growth (Fig. 1B), as reported previously (60). These observations indicate that the integrity of the TAF12 HFD is necessary and sufficient for growth of the taf1-ΔTAND mutant, although the severity of the nsl phenotype represented by each allele depends on the position and/or the type of amino acid substituted.
Transcriptional activation is impaired in taf12/nsl2 mutants
In previous studies, the taf12-W486stop mutation was shown to affect transcription of most, if not all, promoters of PolII-transcribed genes (43,44). After incubation at 37°C for 1 h, induction of the RNR2 gene by DNA damage and derepression of the SUC2 gene by transfer to the low dextrose medium were prevented in the taf12-W486stop mutant (44). In addition, the reduced expression of the TAF12 C-terminal segment (amino acids 391–539) impaired both GCN4-dependent and -independent transcription of the HIS3 gene (60). These observations indicate that the HFD function of the TAF12 protein is required for both basal and activated transcription of at least a subset of genes. Considering that two activation-defective TBP mutants, P65S and S118L, were isolated in the same screen (41), we reasoned that the taf12-L420S mutant might also be deficient in its response to some activators. Thus, we examined activation efficiencies in the taf12-L420S mutant (YAK278) which had been backcrossed to an isogenic wild-type strain more than three times so as to avoid the effect of other unrelated mutations. The β-galactosidase activity from the Gal4 UAS-dependent reporter plasmid was measured when ADs of VP16 and TAND1 fused to the Gal4 DNA binding domain were expressed in the cell (Fig. 2A). As expected, activation efficiencies of both activator proteins were substantially lower in the taf12-L420S mutant than in the wild type (Fig. 2A).
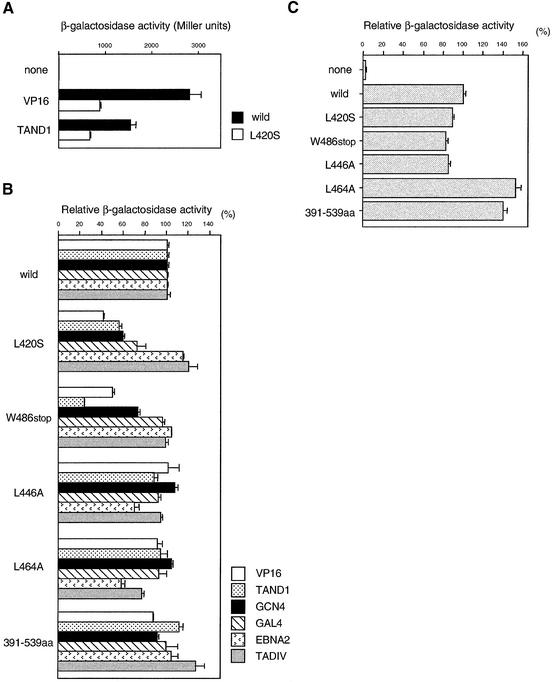
Figure 2.
Reporter analyses measuring transcriptional activities of the nsl2/taf12 mutants. (A) GAL4-dependent transcriptional activation in the wild-type (solid bars) and nsl2-1 (taf12-L420S) mutant strains (open bars). Expression vectors encoding VP16 and the TAND1 ADs fused to the GAL4 DNA binding domain were transformed into yeast, and transcription activity was determined by measuring the lacZ reporter activity expressed from a reporter plasmid (29,41). (B) Activation by various activators in the nsl2/taf12 mutants. β-galactosidase activities of a GAL4-dependent reporter system were measured in strains containing the indicated TAF12 derivatives and one of six activators: GAL4DBD-VP16AD, GAL4DBD-TAND1AD, GAL4DBD-GCN4AD, GAL4DBD-GAL4AD, GAL4DBD-EBNA2AD or GAL4DBD-TADIVAD (41,54). The values are represented as a percentage of the value obtained in the wild-type strain containing the corresponding activators. Note that the host strain used here is different from that in (A). (C) Artificial recruitment assay of GAL4DBD-TAF12 derivatives. The relative β-galactosidase activities of a GAL4-dependent reporter plasmid were measured in the wild-type strain expressing the indicated GAL4DBD-TAF12 derivatives. Each value represents the average of three different isolates of each strain (A, B and C).
To see the correlation between the degree of the nsl phenotype and that of activation defects, we compared activation efficiencies of several taf12 HFD mutants described above, under the same conditions where VP16, TAND1, GCN4, GAL4, EBNA2 and TADIV (ADR1) ADs were used as activators (Fig. 2B). Consistent with the results in Figure 2A, activation efficiencies of VP16 and TAND1 were significantly decreased in the L420S mutant. Additionally, activation efficiencies of GCN4 and GAL4 also decreased in this mutant, whereas those of EBNA2 and TADIV did not (Fig. 2B). A similar differential response to activators was observed for the W486stop mutant, where the activation efficiencies of VP16, TAND1 and GCN4 were specifically reduced (Fig. 2B). In contrast, in the weaker nsl mutants like L446A and L464A, activation efficiencies of EBNA2 and/or TADIV were somewhat lower, whereas those of VP16, TAND1, GCN4 and GAL4 were almost normal (Fig. 2B). Importantly, no similar activation defect was observed for the 391–539 amino acid mutant, which did not exhibit any nsl phenotypes (Figs 1B and 2B). This is consistent with previous experiments showing that transcription was affected only when the 391–539 amino acid mutant was expressed at low levels (60). Collectively, the degree of the nsl phenotype is apparently correlated with that of the activation defect and/or the type of ADs in the various taf12 mutants we tested.
The post-recruitment step appears to be normal in nsl2/taf12 mutants
Previous studies demonstrated that transcription could be activated in the absence of activators by artificial recruitment of TBP and/or TAFs physically connected to a heterologous DNA binding domain (61–64). This simple in vivo recruitment assay could roughly predict which step(s) is impaired in each activation-defective TBP/TAF mutant (65). Namely, if an activation-defective TBP/TAF can activate transcription when it is recruited to the template DNA, its defect is probably involved in the step(s) before its recruitment to the core promoter. Conversely, if the TBP/TAF mutant fails to activate transcription under the same conditions, it probably lacks the ability to function at a post-recruitment step. We previously found that a number of nsl mutants of TBP (e.g., P65S, S118L, K138T/Y139A, N159D, N159L, V161A, E236P and F237D) showed lower activities in this assay (<40%), suggesting that the defects in the post-TBP (apparently post-TFIID) recruitment step might cause a stronger nsl phenotype (41).
To test whether nsl2/taf12 mutants are also defective at a post-recruitment step, we transformed wild-type strains with the reporter plasmid harboring the GAL1 promoter-driven LacZ gene as well as the effector plasmid expressing each taf12 mutant fused to the GAL4 DNA binding domain (Fig. 2C). As already shown for other scTAFs (e.g., TAF5, TAF6, TAF9 and TAF10) (61–63,66), artificial recruitment of scTAF12 to the promoter activates transcription (Fig. 2C). This parallels the effect of LexA-hsTAF12 in mammalian cells, which activates transcription from chromosomally integrated promoters (64). Unexpectedly, however, all taf12 mutants we tested activated transcription at the same level or more efficiently than the wild type when recruited to the promoter (Fig. 2C), indicating that they were not impaired in a post-recruitment step. Therefore, nsl mutants of TBP and TAF12 probably cause lethality in the taf1-ΔTAND mutant by different mechanisms, despite the fact that both are affected in activated transcription.
The expression of TFIID- and SAGA-dependent genes are affected in the nsl2/taf12 mutants
TFIID and SAGA have five subunits in common: TAF5, TAF6, TAF9, TAF10 and TAF12 (42). Genome-wide expression analysis using high-density oligonucleotide arrays demonstrated that the sum of the effects seen with mutations in each complex-specific subunit was substantially less than the sum of effects of the shared TAFs, suggesting redundant roles for TFIID and SAGA complexes in global transcription (35).
Since TAF12 was one of the shared TAFs, we decided to examine whether nsl2/taf12 mutations affect the function of either TFIID or SAGA or both. The expression of a subset of genes depends on either TFIID or SAGA function (35). We examined transcriptional defects of those genes that require complex-specific function to assess which complex, TFIID or SAGA, is more severely damaged in the nsl2/taf12 mutants. We used northern blotting to examine the expression levels of TFIID-dependent genes listed by Lee et al. (35): GNA1, CAM1, PMT4, HTA3, TRX1 and WTM2 genes in the taf12-L420S and -W486stop TS mutants as well as in the 391–539 amino acid mutant (Fig. 3A, left panel). Consistent with the difference of restrictive temperatures of the W486stop and L420S mutants (36 and 37°C, respectively), the expression of these genes was most severely damaged in the W486stop mutant when cells were incubated at 37°C for 2 h (Fig. 3A). A significant reduction was observed for only the TRX1 gene in the taf12-L420S mutant (Fig. 3A). Unexpectedly, the expression of TRX1 was slightly reduced even in the 391–539 amino acid mutant. These observations suggest that TFIID function was more or less impaired in these three taf12 mutants and that the extent of transcriptional defects appeared to correlate with that of the nsl phenotypes (Figs 1 and 3). Next, we examined the expression levels of SAGA-dependent genes, that is, PHO84, YPL019C, PHO5, YHB1 and YDL124W (35), in these three taf12 mutants (Fig. 3A, right panel). The expression of PHO84 and YPL019C was significantly reduced at 37°C in the taf12-W486stop mutant, whereas other genes were expressed at near normal levels. In contrast, the expression of PHO5 and YHB1 increased in the taf12-L420S and 391–539 amino acid mutants, while the expression of other genes was only slightly affected. These results suggest that SAGA function is also affected in a mutation-specific manner in these three taf12 mutants. The transcriptional defects of the W486stop mutant were rather different from those of the other two mutants, implying that the impaired function of the SAGA complex might be less closely related to the nsl phenotype than that of TFIID.
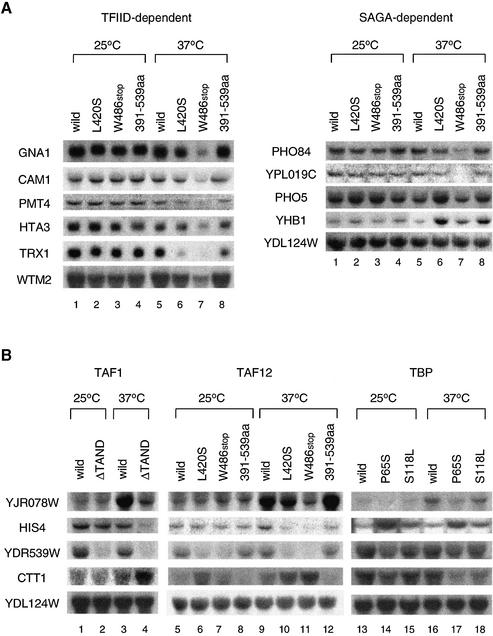
Figure 3.
Transcription analyses of the nsl2/taf12 mutants. (A) The expression of TFIID- and SAGA-dependent genes (35) was compared in three taf12 mutants. Cultures were grown in YPD media to log phase at 25°C; a portion of each culture was shifted to 37°C and incubation continued for 2 h. Total RNA was isolated from wild-type or mutant strains 2 h after a temperature shift to 37°C (lanes 5–8) or continuous incubation at 25°C over the same time period (lanes 1–4). The same amounts of total RNA were blotted onto a nylon membrane and hybridized with the probes indicated. (B) The expression of TAND-dependent genes was compared in the taf1-ΔTAND, nsl2/taf12 and nsl1/spt15 (TBP) mutants. Northern blot analysis was conducted as described in (A) for wild-type or various mutant strains as shown above the lanes using probes of TAND-dependent genes (YJR078W, HIS4, YDR539W and CTT1) and a TAND-independent gene (YDL124W). Although YDL124W was listed as a SAGA-dependent gene by Lee et al. (35), its expression was not changed in the nsl2/taf12 mutants (right panel in A). Thus it serves as an RNA loading control.
A comparison of expression of TAND-dependent genes in the nsl2/taf12 and nsl1/spt15 mutants
Expression analyses of TFIID- and SAGA-dependent genes described above imply that TFIID, which appears to be more substantially damaged than SAGA in the activation-defective nsl2/taf12 mutants, might be predominantly responsible for the nsl phenotypes. This supports our previous results demonstrating that activation-defective nsl1/spt15 (i.e., TBP) mutants also display strong nsl phenotypes (41). However, an artificial recruitment assay implies that molecular mechanisms yielding nsl phenotypes are different in these two mutants (Fig. 2C). To further pursue this point, we decided to examine the expression level of TAND-dependent genes (YJR078W, HIS4, YDR539W and CTT1), which were selected by our DNA chip experiments (K.Ohtsuki, Y.Tsukihashi, K.Shirahige and T.Kokubo, unpublished observations). Each of these four genes was expressed differently in the taf1-ΔTAND mutant (Fig. 3B). For instance, expression of the YJR078W gene was induced at 37°C in the wild-type strain but not in the taf1-ΔTAND mutant (Fig. 3B, left panel). In contrast, the expression of the CTT1 gene was induced at 37°C specifically in the taf1-ΔTAND mutant (Fig. 3B, left panel). The expression of HIS4 and YDR539W genes was reduced at 37°C specifically in the taf1-ΔTAND mutant; however, expression of the latter gene (YDR539W) was reduced even at 25°C (Fig. 3B, left panel). Intriguingly, careful inspection of other data obtained for the taf12 and spt15 (TBP) mutants revealed that only the taf12-L420S and -W486stop mutants recapitulated transcriptional defects of the taf1-ΔTAND mutant. Since the expression of the SAGA-dependent YDL124W gene remained unchanged in all lanes (Fig. 3B, the lowest column), we believe that the changes in other genes are not due to differences in the amount of RNA loading. In the spt15-P65S and -S118L mutants, distinct expression profiles were observed (Fig. 3B, right panel). For instance, the expression of the CTT1 gene was reduced in both spt15 mutants, whereas it was induced in the taf1-ΔTAND and taf12 mutants. It is notable that the enhanced expression of the HIS4 gene as well as the slightly reduced expression of the YJR078W and YDR539W genes was specific to the P65S mutant but not the S118L mutant. Only the latter finding is also observed in the taf1-ΔTAND and taf12 mutants. Taken together, transcription of TAND-dependent genes appears to be similarly impaired in the taf1-ΔTAND and the taf12 mutants but not in the spt15 mutants. Moreover, the extent of transcriptional defects of these genes was closely related to that of nsl phenotypes in the taf12 mutants but not in the spt15 mutants. These observations further support the idea that nsl phenotypes of taf12 and spt15 mutants are generated by different molecular mechanisms. We speculate that impairment of overlapping and distinct functions of TFIID may cause synthetic lethality in the taf1-ΔTAND/taf12 and taf1-ΔTAND/spt15 mutants, respectively.
Physical interaction of TAF12 HFD mutants with ADA1 and TAF4
We showed that the integrity of the HFD of TAF12 is important for cell growth of the taf1-ΔTAND mutant (Fig. 1). Previous studies demonstrated that direct interacting partners of this domain are different in SAGA and TFIID com plexes, i.e., ADA1 and TAF4, respectively (44,45,53). Gene expression analyses suggest that the damage to TFIID is more closely related to the nsl phenotypes of the taf12 mutants than that of SAGA (Fig. 3). Hence, the interaction between TAF12 and TAF4 might be more substantially weakened than that between TAF12 and ADA1 in these taf12 mutants.
To test for this possibility, we used bacterial cells to co-express HFD portions in the following combinations: TAF12 (amino acids 391–539) and TAF4 (amino acids 1–388) or TAF12 (amino acids 414–490) and ADA1 (amino acids 259–359). As previously described, unless HFD of TAF4 or ADA1 was co-expressed together with the HFD of TAF12, complexes were not efficiently recovered in the soluble fraction (53). Binary complexes of TAF12 (amino acids 391–539)-TAF4 (amino acids 1–388) and TAF12 (amino acids 414–490)-ADA1 (amino acids 259–359) could be recovered by glutathione Sepharose resin via a GST molecule fused to TAF12 derivatives. Trapped complexes were eluted and visualized with CBB staining after fractionation by SDS–PAGE (Fig. 4). Note that GST-TAF12 (amino acids 414–490) could form a stable complex with ADA1 (amino acids 259–359) (Fig. 4B) (53) but not with TAF4 (amino acids 1–388) (data not shown), suggesting that the αC helix of TAF12 may be crucial only for forming a complex with TAF4.
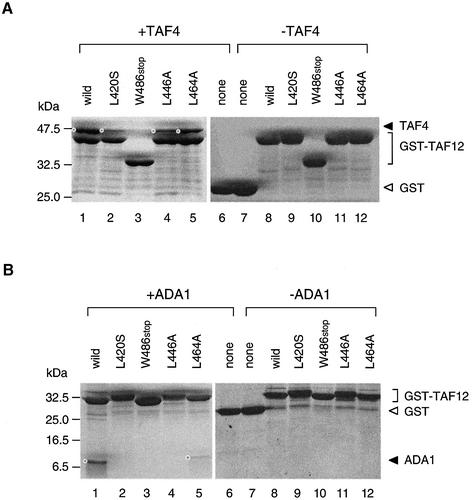
Figure 4.
GST-pulldown assay to test for protein–protein interactions between TAF12 derivatives and TAF4 or ADA1. (A) Interaction of TAF4 (full-length; amino acids 1–388) with GST-TAF12 derivatives (amino acids 391–539). Bacteria were transformed concurrently with two plasmids expressing TAF4 and GST-TAF12 derivatives, respectively. After selection with ampicillin and chloramphenicol, bacteria were grown to an OD600 of 0.45 and induced for 4 h at 25°C with 1 mM IPTG in LB medium. Cell lysates containing TAF4-TAF12 complexes were chromatographed on glutathione Sepharose resin. Complexes bound to the resin were washed extensively with the lysis-wash buffer and then analyzed by SDS–PAGE and CBB staining (lanes 1–6). The bands corresponding to TAF4 are marked with a dot on the left (lanes 1, 2, 4 and 5). The identity of the bands was judged by the molecular size as well as the results obtained without a plasmid encoding TAF4 (lanes 7–12). (B) Interaction of ADA1 (amino acids 259–359) with GST-TAF12 derivatives (amino acids 414–490) (53). Co-expression and GST-pulldown assays were conducted as described in (A) except that shorter forms (amino acids 414–490) of TAF12 derivatives were used according to the protocol of Gangloff et al. (53). ADA1 bound to the beads is also marked with a dot on the left (lanes 1 and 5). The identity of these bands was judged as described in (A).
As shown in Figure 4A, TAF12 (amino acids 391–539) could form a stable complex with TAF4 (amino acids 1–388) (lane 1). The band apparently corresponding to TAF4 (amino acids 1–388) was not visible when it was not added to the reaction (lane 8) or when the GST molecule alone was used for the pulldown assay (lane 6). The two TAF12 mutant proteins, L420S (lane 2) and L446A (lane 4), were significantly impaired in their ability to form a complex with TAF4 (amino acids 1–388). Additionally, the W486stop mutant could not form any detectable complex with TAF4 (amino acids 1–388) under the same conditions (lane 3). This is not consistent with a previous study demonstrating that in vitro translated TAF4 (amino acids 1–388) could interact with both GST-TAF12 (amino acids 330–539) and GST-TAF12 (amino acids 330–485) (44). Although the reason for this discrepancy is uncertain, differences in experimental procedures for protein preparation (e.g., an in vitro translation system versus a bacterial co-expression system) might be responsible. In any case, the inability of the W486stop mutant to form any detectable complex with TAF4 in our assays again underscores the importance of the αC helix of the TAF12 protein in the formation of a binary complex with TAF4. In contrast to these three TAF12 mutants, the L464A mutant appeared to be normal in this assay (lane 5). These results are consistent with previous work demonstrating that the L446A and L464A mutants disrupt the TAF12-TAF4 and TAF12-TAF6 interfaces of the TAF octamer, respectively (45).
We next examined the interaction between TAF12 (amino acids 414–490) and ADA1 (amino acids 259–359) (Fig. 4B). Somewhat unexpectedly, all of the taf12 mutants were strongly impaired in their ability to form a complex with ADA1 (Fig. 4B). A weakly stained band apparently corresponding to ADA1 (amino acids 259–359) was visible for only the L464A mutant (lane 5) but not for the others (lanes 2–4). Thus, the integrity of the HFD of TAF12, except for the αC helix, seems to be more critical for the association with ADA1 than TAF4. Furthermore, these observations imply that the HFDs of TAF4 and ADA1 interact with the HFD of TAF12 in different ways. Nevertheless, it remains unclear whether nsl phenotypes of the taf12 mutants could be ascribed to these weakened interactions between TAF12 and ADA1 and/or TAF12 and TAF4.
DISCUSSION
In this study, we screened nsl genes that have a genetic interaction with TAND of TAF1. The NSL2 gene isolated in our screen was found to be allelic to the TAF12 gene encoding one of the five shared subunits between TFIID and SAGA (42). In contrast to the NSL1/SPT15 gene encoding TBP that was previously isolated in the same screen (41), only a very limited number of taf12 mutants have been characterized in vivo (43,44,60). Conditional (TS) taf12 alleles characterized by two groups contain nonsense or frameshift mutations around W486, which generate C-terminal truncated proteins of ∼50 residues (43,44). A non-conditional taf12 allele characterized by another group carries a transposon insertion between amino acids 278 and 279, which generates an amino-terminal truncated 17 kDa polypeptide apparently corresponding to the 391–539 amino acid fragment initiated at the first in frame ATG codon downstream of the inserted transposon (60). Phenotypes of the latter allele [e.g., slow growth and 3-aminotriazole (3-AT)-sensitivity on synthetic media] result from reduced expression of the C-terminal region of amino acids 391–539 (60). This region encoding HFD, comprises four α-helices (α1, α2, α3 and αC) and is essential and sufficient for wild-type yeast cell growth (Fig. 1) (60,67). The previously characterized conditional alleles, such as taf12-W486stop, encode proteins truncated for the αC helix, thereby affecting the stability of TFIID and transcription of a set of genes at restrictive temperatures (43,44). The nsl2-1/taf12-L420S allele we isolated here also carries a mutation in HFD, indicating that the integrity of HFD is crucial for growth when TAND is removed from TFIID. Consistent with this idea, taf12-W486stop, -L446A and -L464A, all of which harbor mutations within HFD, exhibit nsl phenotypes in our assay; however, the phenotypes of the L446A and L464A mutants were weaker than those of W486stop and L420S mutants (Fig. 1; data not shown). HFD of TAF12 was shown to be important in forming a nucleosome-like structure with TAF4, TAF6 and TAF9 in TFIID, and it can form a similar structure with ADA1, TAF6 and TAF9 in SAGA (45). Indeed, mutant proteins encoded by severe nsl alleles such as taf12-W486stop and -L420S exhibit reduced binding to both of their direct partners, i.e., TAF4 and ADA1 (Fig. 4). However, the integrity of TFIID and SAGA complexes containing these mutant proteins appears to be unaffected, at least at lower temperatures (25°C), even when TAND is absent (A.Kobayashi and T.Kokubo, unpublished observations). This excludes the possibility that a weakened nucleosome-like structure in TFIID and SAGA could be stabilized by TAND.
It is important to understand whether mutations in shared TAFs result in transcriptional defects in the TFIID complex, the SAGA complex, or both. The taf12-W486stop allele was shown to yield a partially disrupted SAGA complex that lacks TAF12 and contains lower amounts of TAF5 and SPT3 when the mutant was cultured at 37°C for a few hours (42). In the in vitro experiments, this defective SAGA complex could not acetylate nucleosome histones nor could it stimulate transcription from chromatin templates in an acetyl-CoA-dependent manner (42). On the other hand, immunoblot analysis of lysates prepared from this mutant at different time periods after the temperature shift to 37°C showed that unique TAFs are more quickly degraded than TAFs shared between TFIID and SAGA (44). This suggests that the taf12-W486stop mutation probably destabilizes TFIID more substantially than SAGA at higher temperatures. Four sets of 10 genes were recently listed as representative of TFIID-dependent, SAGA-dependent, TFIID- and SAGA-dependent, and either TFIID- or SAGA-dependent genes, respectively (35). The expression of these genes was examined to assess TFIID and SAGA specificities for various taf10 mutants (68,69). We employed the same approach to determine which function of TFIID and/or SAGA was impaired in taf12 mutants (Fig. 3). The results showed that the W486stop mutation affects the function of these two complexes, whereas it appears to impair TFIID more extensively than SAGA. This confirms previous results of immunoblot analyses as described above (42,44). On the other hand, the effect of the L420S mutation on transcription of these representative genes was not evident (Fig. 3). Intriguingly, the effects of the W486stop and L420S mutations on SAGA function appeared to be different since only the former decreased the expression of PHO84 and YPL019C genes and only the latter increased the expression of PHO5 and YHB1 genes (Fig. 3). Although the reason for this differential effect on SAGA function remains unknown, the interaction between TAF12 and TBP might be related to this effect since only the W486stop mutant protein was severely defective in TBP binding (44) (A.Kobayashi and T.Kokubo, unpublished observations).
Previously, we conducted an artificial recruitment assay to determine which step, i.e., pre- or post-TBP recruitment step, is more severely damaged by nsl1 mutations (41). The results showed that nsl phenotypes were closely correlated to the defects in a post-TBP recruitment step. This is consistent with our two-step hand-off model in which TAND may be involved in the initial step of activation, i.e., stable binding of TFIID to the promoter (pre-TBP recruitment step) (29,41). The combined defects of the pre- and post-TBP recruitment steps can be expected to yield the lowest activation and thereby inhibit cell growth (41). In contrast, a similar approach, in which TAF12 instead of TBP was recruited to the promoter by a GAL4 DNA binding domain, revealed that the post-TAF12 recruitment step was not impaired for any of the nsl2 mutations we tested (Fig. 2). Thus, we speculate that the pre-TAF12 recruitment step must be impaired in the nsl2 mutants. It is known that TFIID as well as SAGA can be recruited to the promoter by activators (70–72). In fact, GCN5, a HAT component of SAGA, can activate transcription when it is artificially recruited to the promoter (73). At present, it is not yet clear whether both the pre-TFIID and SAGA recruitment steps are affected by nsl2 mutations. Nevertheless, the fact that a post-TFIID (and SAGA) recruitment step was intact in the nsl2 mutants is in stark contrast to the results obtained for the nsl1 mutants. Consistently, the expression of TAND-dependent genes was changed similarly in the nsl2 mutants and in the taf1-ΔTAND mutant but not in the nsl1 mutants (Fig. 3). Taken together, these observations indicate that nsl1 and nsl2 mutations affect distinct steps of activation, and only the step(s) targeted by the latter should overlap with those that require TAND function. We conclude that TAND may be involved in, but is not essential for, the pre-TFIID recruitment step; however, the combined defects of ΔTAND and nsl2 mutations debilitate transcriptional activation, thereby preventing yeast cell growth.
Recently, non-classical activators connecting components of the mediator complex with the Zif DNA binding domain were demonstrated to work much better on CYC1 and GAL1 promoters in the taf1-ΔTAND1 mutant (74). At present, this effect is hard to explain since both promoters are believed to be TAF-independent (54,75). In fact, chromatin immunoprecipitation analyses showed that TBP and several mediator components, but not TAF1, are recruited to the GAL1 promoter during activation by GAL4 (75,76). However, to achieve any stimulatory effect on transcription, TAND1-deleted TFIID must be recruited on the promoter together with non-classical activators. Thus, one possibility is that non-classical activators (i.e., artificially recruited mediator complexes) could stimulate TFIID binding to the promoter, especially when TAND1 is removed. Consistently, such cooperative recruitment between TFIID and the mediator was observed for human factors (77). If this is the case, TAND1 might negatively regulate such a cooperative interaction, thereby minimizing the function of non-classical activators in normal cells. Alternatively, TAND1-deleted TFIID might increase the amount of free TBP and thereby help non-classical activators to overcome a critical rate-limiting step in transcription, e.g., TBP binding to the TATA element. The latter possibility is supported by recent observations demonstrating that TBP quite dynamically associates with the 14-subunit holo-TAF complex (33). The TAND1 deletion may move this equilibrium dramatically to favor the TBP-free state. In either case, it is of significance that the presence or absence of TAND1 in TFIID could effectively change the function of non-classical activators. Isolation of other NSL genes and their characterization may provide additional insight with regards to the function of TAND during transcriptional activation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank K. Kasahara for constructing yeast deletion strains and for his helpful input. We thank K. Kasahara, Y. Tsukihashi, Y. Ohyama, H. Ohta and S. Takahata for sharing antibodies and Y. Ohyama for his assistance in preparing yeast strains for TS assays. We also thank A. G. Hinnebusch, Y. Nakatani, C. Holm, I. Davidson and M. Longtine for yeast strains and plasmids and Y. Ohya and K. Matsumoto for yeast genomic libraries. This study was supported by grants from the Ministry of Education, Science, and Culture of Japan; the Mitsubishi Foundation; the Asahi Glass Foundation; the NAITO Foundation; the Sumitomo Foundation and the NOVARTIS Foundation (JAPAN) for the Promotion of Science.
REFERENCES
- 1.Roeder R.G. (1998) Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb. Symp. Quant. Biol., 63, 201–218. [DOI] [PubMed] [Google Scholar]
- 2.Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naar A.M., Lemon,B.D. and Tjian,R. (2001) Transcriptional coactivator complexes. Annu. Rev. Biochem., 70, 475–501. [DOI] [PubMed] [Google Scholar]
- 4.Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- 5.Woychik N.A. and Hampsey,M. (2002) The RNA polymerase II machinery: structure illuminates function. Cell, 108, 453–463. [DOI] [PubMed] [Google Scholar]
- 6.Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., 16, 673–675. [DOI] [PubMed] [Google Scholar]
- 7.Kutach A.K. and Kadonaga,J.T. (2000) The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol., 20, 4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smale S.T. (2001) Core promoters: active contributors to combinatorial gene regulation. Genes Dev., 15, 2503–2508. [DOI] [PubMed] [Google Scholar]
- 9.Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- 10.Lee T.I. and Young,R.A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet., 34, 77–137. [DOI] [PubMed] [Google Scholar]
- 11.Koleske A.J. and Young,R.A. (1995) The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem. Sci., 20, 113–116. [DOI] [PubMed] [Google Scholar]
- 12.Klein C. and Struhl,K. (1994) Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science, 266, 280–282. [DOI] [PubMed] [Google Scholar]
- 13.Chi T. and Carey,M. (1996) Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev., 10, 2540–2550. [DOI] [PubMed] [Google Scholar]
- 14.Lieberman P.M., Ozer,J. and Gursel,D.B. (1997) Requirement for transcription factor IIA (TFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol. Cell. Biol., 17, 6624–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- 16.Li X.Y., Virbasius,A., Zhu,X. and Green,M.R. (1999) Enhancement of TBP binding by activators and general transcription factors. Nature, 399, 605–609. [DOI] [PubMed] [Google Scholar]
- 17.Wu S.Y. and Chiang,C.M. (2001) TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem., 276, 34235–34243. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman P.M. and Berk,A.J. (1994) A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev., 8, 995–1006. [DOI] [PubMed] [Google Scholar]
- 19.Shykind B.M., Kim,J. and Sharp,P.A. (1995) Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev., 9, 1354–1365. [DOI] [PubMed] [Google Scholar]
- 20.Guermah M., Malik,S. and Roeder,R.G. (1998) Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell. Biol., 18, 3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burley S.K. and Roeder,R.G. (1998) TATA box mimicry by TFIID: autoinhibition of pol II transcription. Cell, 94, 551–553. [DOI] [PubMed] [Google Scholar]
- 22.Guermah M., Tao,Y. and Roeder,R.G. (2001) Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription. Mol. Cell. Biol., 21, 6882–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokubo K., Yamashita,S., Horikoshi,M., Roeder,R.G. and Nakatani,Y. (1994) Interaction between the N-terminal domain of the 230 kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA box-binding. Proc. Natl Acad. Sci. USA, 91, 3520–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa J., Kokubo,T., Horikoshi,M., Roeder,R.G. and Nakatani,Y. (1997) Drosophila TAF(II)230 and the transcriptional activator VP16 bind competitively to the TATA box-binding domain of the TATA box-binding protein. Proc. Natl Acad. Sci. USA, 94, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Y., Perez,G.M., Beechem,J.M. and Weil,P.A. (1997) Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAF(II)130. Mol. Cell. Biol., 17, 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kokubo T., Swanson,M.J., Nishikawa,J.I., Hinnebusch,A.G. and Nakatani,Y. (1998) The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol., 18, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozer J., Mitsouras,K., Zerby,D., Carey,M. and Lieberman,P.M. (1998) Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J. Biol. Chem., 273, 14293–14300. [DOI] [PubMed] [Google Scholar]
- 28.Liu D., Ishima,R., Tong,K.I., Bagby,S., Kokubo,T., Muhandiram,D.R., Kay,L.E., Nakatani,Y. and Ikura,M. (1998) Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell, 94, 573–583. [DOI] [PubMed] [Google Scholar]
- 29.Kotani T., Banno,K., Ikura,M., Hinnebusch,A.G., Nakatani,Y., Kawaichi,M. and Kokubo,T. (2000) A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc. Natl Acad. Sci. USA, 97, 7178–7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lively T.N., Ferguson,H.A., Galasinski,S.K., Seto,A.G. and Goodrich,J.A. (2001) c-Jun binds the N terminus of human TAF(II)250 to derepress RNA polymerase II transcription in vitro. J. Biol. Chem., 276, 25582–25588. [DOI] [PubMed] [Google Scholar]
- 31.Kotani T., Miyake,T., Tsukihashi,Y., Hinnebusch,A.G., Nakatani,Y., Kawaichi,M. and Kokubo,T. (1998) Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem., 273, 32254–32264. [DOI] [PubMed] [Google Scholar]
- 32.Martel L.S., Brown,H.J. and Berk,A.J. (2002) Evidence that TAF-TATA box-binding protein interactions are required for activated transcription in mammalian cells. Mol. Cell. Biol., 22, 2788–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders S.L., Garbett,K.A. and Weil,P.A. (2002) Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol., 22, 6000–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banik U., Beechem,J.M., Klebanow,E., Schroeder,S. and Weil,P.A. (2001) Fluorescence-based analyses of the effects of full-length recombinant TAF130p on the interaction of TATA box-binding protein with TATA box DNA. J. Biol. Chem., 276, 49100–49109. [DOI] [PubMed] [Google Scholar]
- 35.Lee T.I., Causton,H.C., Holstege,F.C., Shen,W.C., Hannett,N., Jennings,E.G., Winston,F., Green,M.R. and Young,R.A. (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature, 405, 701–704. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y.-J., Bjorklund,S., Li,Y., Sayre,M.H. and Kornberg,R.D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell, 77, 599–608. [DOI] [PubMed] [Google Scholar]
- 37.Oelgeschlager T., Tao,Y., Kang,Y.K. and Roeder,R.G. (1998) Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell, 1, 925–931. [DOI] [PubMed] [Google Scholar]
- 38.Wu S.Y., Kershnar,E. and Chiang,C.M. (1998) TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J., 17, 4478–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fondell J.D., Guermah,M., Malik,S. and Roeder,R.G. (1999) Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl Acad. Sci. USA, 96, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi A., Miyake,T., Ohyama,Y., Kawaichi,M. and Kokubo,T. (2001) Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae. J. Biol. Chem., 276, 395–405. [DOI] [PubMed] [Google Scholar]
- 42.Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates,J.R. and Workman,J.L. (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- 43.Michel B., Komarnitsky,P. and Buratowski,S. (1998) Histone-like TAFs are essential for transcription in vivo. Mol. Cell, 2, 663–673. [DOI] [PubMed] [Google Scholar]
- 44.Reese J.C., Zhang,Z. and Kurpad,H. (2000) Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem., 275, 17391–17398. [DOI] [PubMed] [Google Scholar]
- 45.Selleck W., Howley,R., Fang,Q., Podolny,V., Fried,M.G., Buratowski,S. and Tan,S. (2001) A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol., 8, 695–700. [DOI] [PubMed] [Google Scholar]
- 46.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sherman F. and Hicks,J. (1991) Micromanipulation and dissection of asci. Methods Enzymol., 94, 21–37. [DOI] [PubMed] [Google Scholar]
- 48.Lundblack V. (1998) Saccharomyces cerevisiae. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols In Molecular Biology. John Wiley and Sons, New York, NY, Vol. 2, pp. 13.10.11.–13.13.19.
- 49.Kranz J.E. and Holm,C. (1990) Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl Acad. Sci. USA, 87, 6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 51.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 52.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gangloff Y.G., Werten,S., Romier,C., Carre,L., Poch,O., Moras,D. and Davidson,I. (2000) The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol., 20, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukihashi Y., Miyake,T., Kawaichi,M. and Kokubo,T. (2000) Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol., 20, 2385–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gangloff Y.G., Romier,C., Thuault,S., Werten,S. and Davidson,I. (2001) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci., 26, 250–257. [DOI] [PubMed] [Google Scholar]
- 56.Leurent C., Sanders,S., Ruhlmann,C., Mallouh,V., Weil,P.A., Kirschner,D.B., Tora,L. and Schultz,P. (2002) Mapping histone fold TAFs within yeast TFIID. EMBO J., 21, 3424–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanders S.L. and Weil,P.A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem., 275, 13895–13900. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan S., Sink,D.W., Trout,K.L., Makalowska,I., Taylor,P.M., Baxevanis,A.D. and Landsman,D. (2002) The Histone Database. Nucleic Acids Res., 30, 341–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 60.Natarajan K., Jackson,B.M., Rhee,E. and Hinnebusch,A.G. (1998) yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell, 2, 683–692. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Couto E., Klages,N. and Strubin,M. (1997) Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc. Natl Acad. Sci. USA, 94, 8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keaveney M. and Struhl,K. (1998) Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell, 1, 917–924. [DOI] [PubMed] [Google Scholar]
- 63.Gaudreau L., Keaveney,M., Nevado,J., Zaman,Z., Bryant,G.O., Struhl,K. and Ptashne,M. (1999) Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc. Natl Acad. Sci. USA, 96, 2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorris D.R. and Struhl,K. (2000) Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol., 20, 4350–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stargell L.A. and Struhl,K. (1996) A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol. Cell. Biol., 16, 4456–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Apone L.M., Virbasius,C.M., Reese,J.C. and Green,M.R. (1996) Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev., 10, 2368–2380. [DOI] [PubMed] [Google Scholar]
- 67.Moqtaderi Z., Yale,J.D., Struhl,K. and Buratowski,S. (1996) Yeast homologues of higher eukaryotic TFIID subunits. Proc. Natl Acad. Sci. USA, 93, 14654–14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirchner J., Sanders,S.L., Klebanow,E. and Weil,P.A. (2001) Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol., 21, 6668–6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirschner D.B., vom Baur,E., Thibault,C., Sanders,S.L., Gangloff,Y.G., Davidson,I., Weil,P.A. and Tora,L. (2002) Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol., 22, 3178–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Utley R.T., Ikeda,K., Grant,P.A., Cote,J., Steger,D.J., Eberharter,A., John,S. and Workman,J.L. (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature, 394, 498–502. [DOI] [PubMed] [Google Scholar]
- 71.Larschan E. and Winston,F. (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev., 15, 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhaumik S.R. and Green,M.R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev., 15, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Candau R., Zhou,J.X., Allis,C.D. and Berger,S.L. (1997) Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J., 16, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng J.X., Nevado,J., Lu,Z. and Ptashne,M. (2002) The TBP-inhibitory domain of TAF145 limits the effects of nonclassical transcriptional activators. Curr. Biol., 12, 934–937. [DOI] [PubMed] [Google Scholar]
- 75.Li X.Y., Bhaumik,S.R. and Green,M.R. (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science, 288, 1242–1244. [DOI] [PubMed] [Google Scholar]
- 76.Pokholok D.K., Hannett,N.M. and Young,R.A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell, 9, 799–809. [DOI] [PubMed] [Google Scholar]
- 77.Johnson K.M., Wang,J., Smallwood,A., Arayata,C. and Carey,M. (2002) TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev., 16, 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.