Abstract
Using the extensive segmental duplications of the Arabidopsis thaliana genome, a comparative study of homoeologous segments occurring in chromosomes 1, 2, 4 and 5 was performed. The gene-by-gene BLASTP approach was applied to identify duplicated genes in homoeologues. The levels of synonymous substitutions between duplicated coding sequences suggest that these regions were formed by at least two rounds of duplications. Moreover, remnants of even more ancient duplication events were recognised by a whole-genome study. We describe a subchromosomal organisation of genes, including the tandemly repeated genes, and the distribution of transposable elements (TEs). In certain cases, evidence of the possible mechanisms of structural rearrangements within the segments could be found. We provide a probable scenario of the rearrangements that took place during the evolution of the homoeologous regions. Furthermore, on the basis of the comparative analysis of the chromosomal segments in the Columbia and Landsberg erecta accessions, an additional structural variation in the A.thaliana genome is described. Analysis of the segments, spanning 7 Mb or 5.6% of the genome, permitted us to propose a model of evolution at the subchromosomal level.
INTRODUCTION
The Arabidopsis genome has been explored by an extensive sequencing effort and several in silico studies (1–3) have shown that its five chromosomes (n = 5) are built up prevalently from several dozens of duplicated segments ranging in size from several kilobases to 4.6 Mb. According to these reports, the duplications most probably resulted from a single tetraploidisation event, since >60% of the chromosomal sequences are duplicated and there are no overlapping segments. It was estimated that this tetraploidisation event occurred some 65 million years ago (4). At present, it is not clear whether the tetraploidisation was a result of the merging of genomes from two different species (allotetraploidisation), or by the doubling of a genome within the species (autotetraploidisation). However, it has been accepted that the unusual segmental duplication was due to breakage and reshuffling of chromosomal segments that followed the whole-genome duplication event. This process is also known as diploidisation and prompts a primary tetraploid genome to evolve towards the differentiation of chromosomes until an independent diploid inheritance.
The duplicated nature of Arabidopsis thaliana chromosomal segments does not imply that they are identical. Actually, only approximately a quarter of the genes are still preserved in the pairs of duplicated segments (1–3). This suggests that the duplicated segments could be considered as homoeologous (also used as homeologous), as was proposed by Grant et al. (5) for partially homologous chromosomal regions. The loss of redundant gene copies may be due to the divergent evolution of each copy of a chromosome segment, in which some genes may change their structure and function while others may be inactivated and deleted. Moreover, homoeologous segments evolve by many other mechanisms such as tandemly repeated gene (i.e. tandem arrays) expansion, accumulation of transposable elements (TEs) or small-scale rearrangements. The mechanisms of these processes are known; however, their occurrence and contribution to genome evolution is still unclear (6).
Another unexpected insight concerning the present Arabidopsis genome structure comes from the analyses of Ku et al. (7) and Vision et al.(3). They suggested that there were several rounds of duplication (i.e. several duplication events, each followed by the diploidisation process) in the history of the Arabidopsis species. Although the number of those rounds and their dating may be controversial (6), it has become clear that at least some of them must have taken place.
Identification of the chromosomal segments that originated from different large-scale duplication events, such as duplication of the whole or of a substantial part of the genome, could be limited by many factors (reviewed in 6). The initial structure of the duplicated regions in an ancestor degenerated because of divergence of a sequence and gene loss, obscuring the relationships between older homoeologous regions. The lengths of collinear fragments, i.e. those with conserved gene content and order, could also decrease, as ancestral segments became progressively fragmented and were distributed throughout the genome by chromosome translocations. Hence, some older rounds of large-scale duplications may be undetectable in the contemporary A.thaliana genome. Indeed, the number of ancient duplications, their scale and date remain unknown.
In this context, the identification of gross ancient events such as the duplication rounds and the subsequent extensive process of diploidisation can be crucial in the understanding of the present-day Arabidopsis genome. To date, the genome-wide approaches (1–3) are insufficiently precise, because they are unable to offer insights into the numerous structural events that have taken place in each chromosome arm. Even detailed comparative studies between Arabidopsis and other dicots based on a few hundred kilobase pair-long fragments may miss the well conserved regions and the nearest breakpoints of segmental rearrangements. While such studies can provide important observations on gene organisation at the nucleotide level (for example see 8–10), they are limited for a wider understanding of the mechanisms involved in genome evolution. Therefore, there is a need for high resolution bioinformatic analyses of large segments that will describe the structure and allow insight to various aspects of evolution of the plant genome, particularly those connected to ancient polyploids.
In this study, we attempt to shed light on the level of genome evolution that we refer to as subsegmental. We report on the analysis of four segments in chromosomes 1, 2, 4 and 5 that have been identified to be of common origin (3) and we have found even more ancient duplicated regions using a histogram-based approach. Extrapolation of our findings to the architecture of the whole genome suggests that Arabidopsis was involved in at least three rounds of duplication events. We refer to the most recent one as the α duplication event, the middle one as the β duplication event, and the earliest one as the γ duplication event. We describe the subchromosomal organisation of genes, including the tandemly repeated genes (tandem arrays), the distribution of TEs, and we try to describe the phenomenon of gene loss. On the basis of the structure and organisation of the homologous genes detected we were able to establish a probable scenario of chromosomal rearrangements that took place within the segments under study over the dozens of million years of their co-evolution. The analysis of the rearrangement breakpoints allowed us to recognise the probable mechanisms involved. Furthermore, by comparing the DNA sequences available for two accessions of Arabidopsis, we have investigated the present-day variation of insertions and deletions within A.thaliana. A general model of molecular evolution at the subchromosomal level is proposed.
MATERIALS AND METHODS
Characterisation of duplicated segments and identification of successive duplication events
Identification of homoeologous regions within segments 1A, 2A, 4B and 5B derived from the β and α duplications. This identification was based on the analysis of homology of all genes among the segments in a pair-wise manner. To establish the homology of genes among the segments, formed as a result of the β and α duplication events, the BLASTP (http://www.arabidopsis.org/Blast) (11) analysis was performed using P-values of 10–10 and 10–30, respectively, chosen after preliminary testing. Sequences of proteins coded by the genes from the chromosomal segments 2A and 4B were used as queries for this study.
Identification of homoeologous regions derived from the γ duplication: a histogram-based approach. Since the classical BLAST search for ancient events may give unsatisfactory results, we developed an approach based on a histogram analysis of BLASTP data (see Fig. 3). It is based on the following assumption: taking into account a minor role of gene transposition during evolution, all genes present in the four homoeologous segments (1A, 2A, 4B and 5B) can be considered as derivatives from the same ancestral region. Therefore, the search for possible older duplication events may be improved by integrating BLAST data obtained from them.
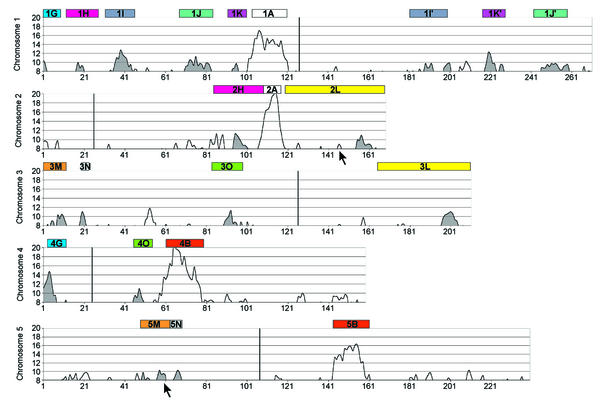
Figure 3.
The histogram-based approach describing the distribution of subsegments that originated from the γ duplication event within the Arabidopsis chromosomes. The x-axis represents individual chromosomes divided into 25-gene-long sections. The y-axis represents numbers of BLASTP hits for genes from segments 1A, 2A, 4B and 5B. The centromere positions are shown as vertical bars within the charts. Colour boxes above the charts depict physical positions of the segments, which arose by the α duplication event [according to maps in Arabidopsis Genome Initiative (1) and Blanc et al. (2)]; corresponding segments are marked by the same colour. Regions defined as significant, and thus being descendants from the γ duplication event, are shaded. Two γ subsegments illustrated in Figure 2 (2L and 5M) are marked by arrows.
For each segment (1A, 2A, 4B and 5B) a BLASTP analysis was performed separately using the P-value of 10–5 as a cutoff, BLOSUM45 as a matrix and the filter option switched off. Reanalysis of the data using the 10–10 cutoff did not bring a significant improvement. Genes that produced more than 50 significant hits (a hit is defined as a gene similar to that of BLASTP query according to specific criteria) were excluded from the analysis as they produced high background. Some genes that could have originated from tandem array expansion or ancient tandem duplications would affect data interpretation. To avoid these discrepancies for individual genes, hits separated by less than 30 genes on a chromosome were considered as a single hit. Similarly, if the same hits were produced for several different genes in a segment of question, they were also considered as single hits. All repeated element-related genes were excluded, which reduced the background level. The chromosomes were divided arbitrarily into 25-gene-long sections producing an approximate resolution of ∼120 kb/section (average gene density of 4.6 kb/gene). This procedure produced 271, 169, 211, 159 and 240 sections for chromosomes 1, 2, 3, 4 and 5, respectively. The hits for segments 1A, 2A, 4B and 5B were added and assigned to individual sections creating the distribution chart. The Moving Average option with a period of 4 (which corresponds to 100 genes) was used to recalculate data points; this period was chosen after a preliminary testing. All regions that generated picks >8 hits for both homoeologous segments from the α duplication [as defined on the map in Arapdopsis Genome Initiative (1) and Blanc et al. (2)], and >10 hits for at least one of them, were considered as significant and originating from the γ duplication event. The lengths of the selected regions were not considered as crucial since even for the α duplication event they could be very short (2).
For a more accurate comparative analysis of the four duplicated segments, a distribution of sequence elements such as tandem arrays and repeated elements was determined (see below).
Analysis of tandemly repeated genes. The presence and distribution of tandem arrays was determined using the BLASTP program with the P-value of <10–20; up to 10 unrelated genes between members (i.e. genes) of an array were tolerated.
Identification of repeated elements. Transposon and satellite-based repeats were identified by the RepeatMasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; A.F.A. Smit and P. Green, unpublished data) against the Repbase, version 03/31/00 (12,13) using the default setting. For a study of the distribution of repeat units along the segments, they were divided arbitrarily into 50-gene-long sections. For each section, a separate RepeatMasker analysis was performed.
Estimation of the level of synonymous substitutions (Ks)
An accurate homology relationship between genes present in tandem arrays was established by the use of a phylogenetic tree-based analysis (Clustal W, version 1.7) (14) of the complete family and defined as proteins showing the shortest phylogenetic distances. Protein sequences corresponding to the duplicated genes were aligned using the Clustal W program (14) and the resulting alignment used as a guide to align the corresponding nucleotide sequences. After removing the gaps, the level of synonymous substitutions between the nucleotide sequences was estimated by the use of the maximum likelihood method implemented in the codeml program (http://abacus.gene.ucl.ac.uk/software/paml.html) (15) under the F3 × 4 model of codon substitution (16). We kept all Ks values ≤ 10, which allows reliable phylogenetic analyses (17,18).
Analysis of structural rearrangements within the segments
Detected rearrangements were classified according to the following definitions. (i) Inversions were defined as regions of reoriented gene pairs within reversed blocks. (ii) Duplications were defined as doubled blocks of genes. (iii) Deletions were defined as missing gene blocks within a given segment disclosed by a comparison with its homoeologous segment. For the identification of deletions the minimal size of missed blocks was defined arbitrarily at 11 genes for α and at 31 genes for β duplication events. These values seemed to be sensitive with minimal possibility to mistake small deletions for random gene loss. (iv) Translocations were defined as changes of a subsegment position in the homoeologous segment. All structural changes, except for deletions, were considered as authentic when they were represented by at least two pairs of genes preserved in two segments, and separated in each segment by up to 10 unrelated genes. Note, the sizes of structural changes are only estimates, especially for the ancient events. Some deletions could be confused with small translocations.
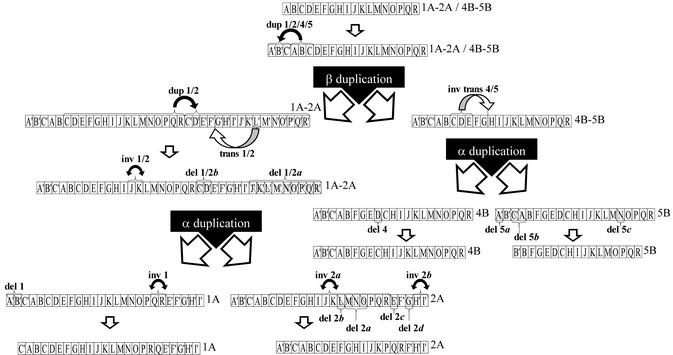
The sequence of rearrangements within the analysed segments 1A, 2A, 4B and 5B was established in the following consecutive steps. (i) The more recent rearrangements that took place after the α duplication event (independent analyses of segment pairs 1A–2A and 4B–5B) were identified from Figure 2. To establish in which segment a structural change occurred, the gene order and orientation were compared with segments from the β and γ duplication events, if applicable. (ii) A comparison of segment 1A with segments 4B–5B allowed the identification of rearrangements that occurred before the α duplication. Because of its dramatically rearranged structure, segment 2A was used only to verify and to correct the results. (iii) For the intrachromosomal duplication (dup 1/2 in Fig. 5), a tandem origin and consecutive partial deletion were concluded, as the probability of these changes appears the highest (19). (iv) All identified rearrangements were incorporated into Figure 5, and illustrated as boxes within a segment. For each section, a label was used (A–R). A few small sections spanning less than 10 genes and not involved directly in the segment rearrangements, were excluded from the analysis to reduce the number of sections. (v) Finally, a figure representing the sequence of chromosome rearrangements was built, in which individual rearrangements are depicted as letter-labelled boxes (Fig. 5). The duplicated gene blocks were indicated as boxes labelled additionally with prime (′). The estimate of the extents of more ancient events may be imprecise.
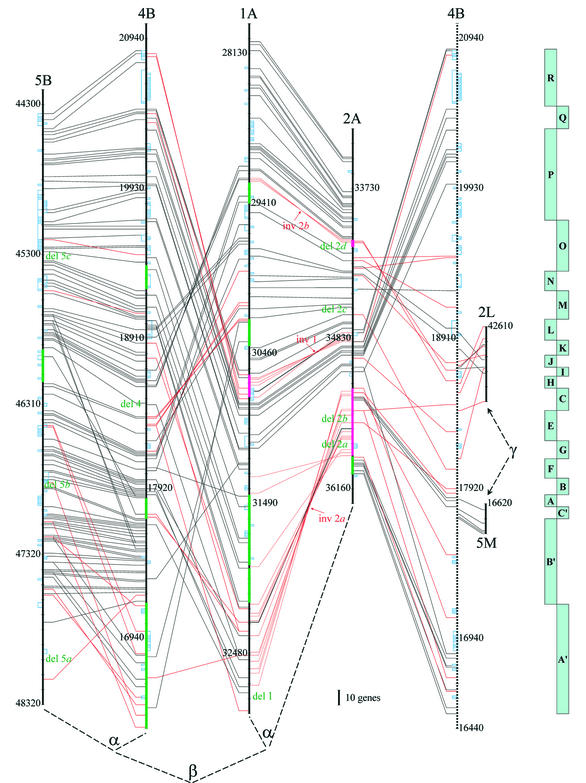
Figure 2.
Schematic representation of four segments of the Arabidopsis genome and their interrelation. Individual segments are depicted as vertical black bars. Segment 4B is reoriented relative to the other segments. Every 100th gene on the map of segments is numbered according to the TAIR nomenclature. Each link represents a conserved pair of genes between two corresponding segments, whereas links for genes with reversed orientation are shown in red. Tandem arrays are depicted as blue boxes on the segment bars. The structural rearrangements are identified on the basis of the presence and order of conserved genes among segments. The rearrangements that occurred after the α duplication are marked: deletions in green and inversions in pink. To illustrate the homology relationship between segments 2A and 4B, the latter one is depicted twice with the second copy as a dashed line. The phylogenetic relations of segments in the light of successive duplications β and α are also indicated. Two short fragments of γ duplication events are shown on the right. Turquoise broken bar on the right depicts sections for 4B described in Figure 5.
Figure 5.
The model of chromosomal structural changes that occurred during the evolution of the segments. Each section of the segments shown in Figure 2 (turquoise broken bar) is represented by a box with a letter. The segments are numbered on the right (also the original segments that occurred before duplication events). The large β and α duplication events are marked by large blank doubled arrows. Sections that occurred after the tandem duplications are marked by prime (′). All identified rearrangements are depicted by brackets and appropriate description (del, dup, inv and trans for deletion, duplication, inversions and translocation, respectively). For simplicity, there is no size relationship to the segments depicted in Figure 2.
Identification of the rearrangement breakpoints was conducted in two steps. First, an approximate localisation was established as shown in Figures 2 and 5. Then, the precise sites were investigated by a combination of methods available in the BLAST_2 Sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) (20) and PipMaker (http://bio.cse.psu.edu) (21). Verification of the identified sequences was carried out by RepeatMasker and BLASTN.
Estimation of gene loss
The hypothesis of gene loss caused by the accumulation of point mutations was tested by the PipMaker analysis (21). This study allows a comparison of large genomic sequences and produces high-resolution displays of the resulting alignments. It was performed for segment 2A against segment 1A, with default settings. Gene positioning for 2A was retrieved from TIGR (ftp://ftp.tigr.org/pub/data/a_thaliana; version date 03/17/00) and corrected with TAIR (http://www.arabidopsis.org) and MATDB (MIPS; http://mips.gsf.de/proj/thal) databases (versions of 04/11/02). Pips (homologous fragments between two analysed sequences detected and plotted by PipMaker) that did not overlap with annotated genes on segment 2A were extracted from the sequence and aligned against the Arabidopsis proteome using BLASTX. They were classified as inactivated genes if they showed significant homology (P = 10–5) to any Arabidopsis protein. Matches to genes in the corresponding region of the segment 1A were classified as significant even with a higher P-value (P = 10–2), since the probability of their occurrence by chance was very low.
Comparison of sequence variation between two Arabidopsis accessions: large indels analysis
The sequence variation within the species is mainly due to single-nucleotide polymorphisms and insertion/deletion events (indels) between compared accessions. Large (>100 bp) indels were analysed by a comparison of the Arabidopsis Columbia (Col-0) and Landsberg erecta (Ler). The Ler sequence data and the positions of the polymorphic sites for segments 1A, 2A, 4B and 5B were obtained from the Cereon Arabidopsis Landsberg Sequence Database (22). On average, the Ler DNA sequences consisted of 1.5 kb plasmid contigs. For each indel, the corresponding sites were identified in a segment by BLAST_2 Sequences analysis using the oligonucleotide set provided by Cereon Genomics. For all indels, the corresponding sequence from Ler was searched with the use of BLASTN. The sequences from the two accessions were also compared using BLAST_2 Sequences to confirm the variant sites. The positions were verified to determine whether they bordered on repeated elements. If so, the sequence was extracted from the repeats and tested by BLASTX and GENSCAN (http://genes.mit.edu/GENSCAN.html) (23) if it enclosed some genes. Otherwise, if a polymorphic fragment was duplicated in tandem with an identity of >95%, it was considered to be a result of unequal homologous recombination.
RESULTS
Successive duplication events in the Arabidopsis genome
One of the basic problems in the genome analysis of ancient polyploids is to distinguish chromosome segments deriving from different large-scale duplication events. By using gene-by-gene BLASTP search and developing a histogram-based approach we have identified successive genome duplications. Through the study of homoeologous segments, we described three successive duplications that took place during the history of the Arabidopsis genome. The relative chronology of these events was studied by estimating the levels of Ks between preserved genes in homoeologous regions.
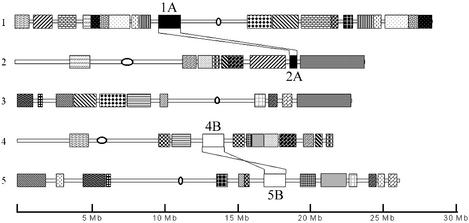
Structure of homoeologous segments: identification of the α and β duplications. We identified duplicated regions in the Arabidopsis genome with a homoeologous relationship to fragment 2A (Fig. 1) using the BLASTP gene-by-gene approach. This fragment was reported to be homoeologous to the 1A segment (1–3) of chromosome 1; our study has confirmed this observation. Further BLAST analysis conducted for both segments 1A and 2A with less stringent threshold parameters identified additional two homoeologous segments, on chromosomes 4 and 5 (4B and 5B) (1,2), which showed lower gene collinearity to 1A and 2A (Fig. 2) (see below for a structural comparison). Moreover, we have found two short internal fragments within the homoeologous segments 1A, 2A, 4B and 5B that have additional copies in the genome (Fig. 2, regions 2L and 5B). They are represented by regions containing sets of genes with higher sequence divergence and more complex rearrangements (see Fig. 2 for details). To differentiate clearly among the sequential duplication events, we referred to the most recent one as the α duplication event [1A–2A split and 4B–5B split; described earlier in Arapdopsis Genome Initiative (1) and Blanc et al. (2)], the middle one as the β duplication event (A–B split), and the earliest one as the γ duplication event. The γ duplication event may comprise more than a single event.
Figure 1.
Localisation of duplicated segments in the Arabidopsis genome. Individual chromosomes are represented as horizontal bars. Centromere positions are marked by black open circles. Blocks within chromosomes (patterned) represent segmental duplications [according to maps in Arapdopsis Genome Initiative (1) and Blanc et al. (2), modified]. Each segment that we refer to in the study is identified by the chromosome number and by the letter describing its relationship to another segment. The duplications between 1A–2A and 4B–5B are shown by bands connecting the segments. Segments 4B–5B are in reverse orientations.
Older events of duplication: identification of the γ duplication in a histogram-based approach. A BLASTP search was performed for all genes from segments 1A, 2A, 4B and 5B, followed by a background elimination step. Each Arabidopsis chromosome was divided into 25-gene-long sections and the numbers of BLAST hits for successive sections were plotted and reanalysed on a histogram. The peaks obtained were treated as significant if they were higher than a threshold of 10 hits. Altogether, we detected 17 such regions (Fig. 3). The authenticity and accuracy of the detection of these regions was confirmed by finding peaks (with a threshold >8 hits) in the corresponding homoeologous segments, which originated from the α duplication event [according to the physical maps in Arapdopsis Genome Initiative (1) and Blanc et al. (2)]. Only in two regions out of the 17 we did fail to find peaks within homoeologous segments; they were excluded from further study (Fig. 3). Taken together, this approach allowed us to identify 18 regions (shaded in Fig. 3) which lie within nine pairs of segments from the α duplication. We believe therefore that they originate from the γ duplication event.
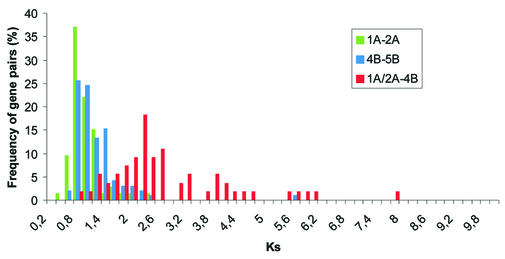
Relative chronology of the duplication events. In order to assess the relative chronology of the three duplication events, we estimated the level of Ks between duplicated genes for each pair of homoeologous regions (Fig. 4). The median Ks values for the pairs 1A–2A and 4B–5B were very similar (0.793 and 0.879, respectively), suggesting that both duplications occurred at contemporaneous dates. Nevertheless, the Mann–Whitney U-test indicates that the small difference observed between these two median values is significant (P < 0.001). The distribution obtained from the pairs of genes duplicated between segments 1A or 2A and 4B shows greater Ks values (median = 2.278), indicating that the β duplication is an older event. The genes duplicated between other subsegments detected by the gene-by-gene approach (2L and 5M in Fig. 2) gave very high Ks values (data not shown). Since these sequences are highly saturated at synonymous sites, they do not allow reliable phylogenetic inferences. However, from their length and the number of rearrangements we can presume they originated from very ancient events.
Figure 4.
Frequency distributions of homologous gene pairs as a function of the estimated level of synonymous substitution per site (Ks). The green and blue distributions correspond to duplicated genes between block pairs 1A–2A and 4B–5B (α duplication event), whereas the red distribution corresponds to 1A or 2A and 4B (β duplication event), respectively.
A structural characterisation of duplicated chromosomal segments
On the basis of the available whole-genome sequence of A.thaliana it was possible to make detailed structural characterisation of identified homoeologues. In this section we describe four homoeologous segments and their comparison at several different levels. We analyse their size, the number of genes within individual segments, the number of conserved genes between segments, gene reorientation events, the frequencies of tandem array, and the distribution of repeated elements. The differences detected allowed the identification of various evolutionary pathways of these segments presumed to be of common origin.
For analyses at the gene and sequence level, we selected four segments: 1A, 2A, 4B and 5B, localised on chromosomes 1, 2, 4 and 5, respectively (Fig. 1). Although these four segments are of common origin, they differ markedly in several respects. The most apparent from the BLASTP analysis was a dissimilarity in size: 1A, 2A, 4B and 5B cover 1.996, 0.980, 1.927 and 1.739 Mb, and comprise 430, 223, 445 and 380 genes, respectively. With similar gene densities within the segments (Table 1), size differences must have resulted from the deletion/insertion events. Since it is known that the pairs of segments 1A–2A and 4B–5B are homoeologous (1,2), the number of preserved genes could be established as 69 for the first pair and 108 for the second pair. A similar comparison between 1A and 4B revealed a conservation of 58 genes, and between 2A and 4B a conservation of 39 genes.
Table 1. The summary of a structural comparison between duplicated chromosomal segments.
| Feature\segment | 1A | 2A | 4B | 5B |
|---|---|---|---|---|
| Size (Mb) | 1.996 | 0.980 | 1.927 | 1.739 |
| Size (gene number) | 430 | 223 | 445 | 380 |
| Gene densities (kb/gene) | 4.64 | 4.39 | 4.33 | 4.58 |
| Tandem arrays | 28 | 9 | 27 | 35 |
| Genes within tandem arrays | 95 (22%) | 21 (9.4%) | 105 (23.6%) | 106 (27.9%) |
| TEs | 6.60% | 1.57% | 4.85% | 5.64% |
| Class I | 2.13% | 0.14% | 2.16% | 1.53% |
| Class II | 4.46% | 1.42% | 2.68% | 4.06% |
Within the segment pairs 1A–2A and 4B–5B, with five exceptions, all homologous genes were in the same orientation (Fig. 2). Of the five exceptions, three reoriented genes were in tandem arrays (At2g34810, At4g19470, At4g19080). Although At4g17230 was present in a single copy, tandem array of this gene is still present as a relic. An attempt to determine the cause of reorientation of the fifth gene pair At1g32270–At2g35460 failed. We detected several examples of reoriented genes for the distal region of the segment pair 4B–5B (Fig. 2, green fragment at the very end of 4B), however, this may be due to an ancient tandem duplication event (see below). The gene orientation was also analysed within the pairs 1A/4B and 2A/4B, revealing a much higher number of repositioned sequences (Fig. 2).
An analysis of tandem arrays showed more evident differences between the segments analysed. We detected tandem arrays enclosing 22, 9.4, 23.6 and 27.9% of genes for segments 1A, 2A, 4B and 5B, respectively (for details see Table 1). Remarkably, the frequency of tandemly repeated genes in segment 2A was distinctly lower (9.4%).
The interspersed repeated element frequency and content is not uniformly distributed among the segments. Segment 1A consists of 6.60% of transposons; 2A, 1.57%; 4B, 4.85%; and 5B, 5.64% (for details see Table 1). The distribution of repeat units along the segments is not uniform in segments 1A and 2A (data not shown); it varies from 2.76 to 7.55% for 1A and from 0 to 3.54% for 2A (counted as 50-gene-long sections). Among all segments analysed, 2A apparently has a lower level of the interspersed repeated elements.
Structural rearrangements within homoeologues segments
One of the main results of the previous whole-genome analyses in Arabidopsis was the evidence of a massive gene loss in the duplicated segments. To give a more accurate evaluation of this phenomenon, a PipMaker-based comparison between two homoeologous segments was performed. This analysis revealed potential remnants of the gene loss process. Moreover, the types of actual rearrangements and their relative sequences could be identified for four homoeologous segments analysed in this study (1A, 2A, 4B and 5B). It was done by comparing gene order and orientation in one fragment relative to the remaining three. As a consequence, we were able to establish precisely the breakpoints of the most recent rearrangements which allowed us to deduce probable mechanisms responsible for these events.
A search for evidence of gene loss in the corresponding segments was conducted. For this purpose, we compared the nucleotide sequence of segment 2A (807 746 bp; 223 genes) to that of 1A using PipMaker (see Supplementary Material). Those pips that were not related to any gene present in segment 2A were analysed further by BLASTX against the Arabidopsis genome. In total, we identified 35 sequence elements; at least 19 of these seemed to have originated from inactivated genes. While unequal recombination was deduced for 11 cases out of 19, the remaining eight may correspond to genes inactivated by the accumulation of point mutations (see Supplementary Material).
The phenomenon of ectopic homologous recombination is the most probable explanation of the two inversions within the segments. One of the recombination events occurred in segment 1A (inv 1 in Fig. 2). It spans 76.8 kb and contains 15 genes (At1g30620 to At1g30780). Most interestingly, remnants of the ATREP2 transposon element are present on each flank of this rearrangement, in the opposite orientation, but highly similar to each other (∼90%, 537 bp). A similar event might have produced inversion 2a (38.7 kb fragment containing nine genes, from At2g34150 to At2g34230) except that it is flanked by an AT-rich sequence (identity ∼75%, 590 bp, A/T content = 71%) and is also in the opposite orientation. This sequence appears low-copy, as it has only a few significant matches in the genome. All tests for a more accurate identification of this sequence by gene-finding programs did not produce any significant hits. For the third inversion that took place on chromosome 2 after the α duplication event (inv 2b), no breakpoints could be identified, mainly because of a large deletion (del 1 in Figs 2 and 5) in the corresponding region on chromosome 1. We failed to find any remnants of the elements responsible for the deletions or intrachromosomal duplications.
Having compared four homoeologous segments and analysed gene copy orientation (Fig. 2), we constructed a probable sequence of each of the 19 rearrangements detected (Fig. 5). In total, two tandem duplications (>150 and >250 genes), two translocations (one of them inversional) (=10 and ∼30 genes), four inversions (9, 15, ∼20, ∼150 genes), and 11 deletions (up to 200 genes) were present. Further analysis revealed that large chromosomal rearrangements were followed by a wave of smaller changes, especially deletions that accumulated at the rearranged site. For example, there are at least three relatively large deletions within inversion 2b in both segments.
Genome divergence resulting from large indels in Arabidopsis accessions
In this part of our work we focused on the mechanisms responsible for the sequence divergence observed in different Arabidopsis lines. For this purpose we compared the sequence data from four segments (1A, 2A, 4B and 5B) between two different Arabidopsis accessions.
To evaluate sequence variation over a relatively short evolutionary distance, we conducted an analysis of large indel (>100 bp) polymorphism between Col-0 and Ler accessions, identified and provided by the Cereon Genomics, for the segments 1A, 2A, 4B and 5B. Altogether, 40 such large indels were localised by Cereon within those segments; however, we excluded six of them as we were unable to re-identify them in the Col-0 segments and/or in the Ler contigs (Table 2). The variations caused by homology-dependent mechanisms such as unequal recombination or replication slippage were recognisable in 15 of these indels. Among these 15, 12 indels involved gene sequences. Moreover, in three out of these 12, the deletion of genes fragments most likely occurred. They resulted from gene truncation in the Ler accession, in comparison with their full-length copies in Col-0 (At1g28700, At1g28760, At4g19930). In 13 other indels, the variation was generated by insertions or excisions of TEs, with the same frequencies of class I (7) and class II (7) elements involved. For the remaining six indels, the mechanisms remain unknown, but three of them probably resulted from deletions in the Ler accession, since they produced truncated genes (At1g30600, At1g31355, At2g34840). It is noteworthy that in five out of the six such cases we detected short direct repeats of 4–8 bp in length at the breakpoints, indicating a recombination-based mechanism.
Table 2. The summary of a comparison between Col-0 and Ler accessions.
| Feature\segment | 1A | 2A | 4B | 5B | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of polymorphic sitesa | 9 | 4 | 16 | 5 | ||||||||
| Indel size (range; mean) (bp) | 172–9364; 2621 | 177–2077; 856 | 117–5171; 1683 | 385–12277; 5547 | ||||||||
| Probable mechanism | c/ob | TEc | und | c/ob | TEc | und | c/ob | TEc | und | c/ob | TEc | und |
| 6 | 1 | 2 | 2 | 0 | 2 | 6 | 8 | 2 | 1 | 4 | 0 | |
| Polymorphic sites within genes | 5 | 0 | 2 | 1 | – | 1 | 6 | 1 | 0 | 0 | 0 | – |
| Indel size (mean) (bp) | 3130 | 2660 | 1470 | 210 | – | 1500 | 690 | 2660 | 370 | 390 | 6840 | – |
aThe values represent only the sites that were detected by Cereon Genomics, Rel. 1–3. All sites analysed are insertions in Col-0 or deletions in Ler, due to the Ler contig size.
bUnequal recombination or replication slippage.
cTE insertion/excision.
dUnknown.
DISCUSSION
Successive duplication events
Identification of homoeologous chromosomal segments derived from separate rounds of duplication is severely limited. Assuming the contribution of several large-scale duplications in the Arabidopsis evolution, one can expect that the older the duplication, the less evidence will remain at the sequence and the gene organisation level. Therefore, a search for some older events may be relatively difficult in the present-day genome of this species. In this study, we uncovered evidence of chromosome segments and subsegments that derived from at least three successive duplication events in the evolutionary past of the Arabidopsis genome. Being most recent and thus the least masked by structural modifications, the α duplication is the best known (1–3). Most probably it was a result of a whole genome duplication (tetraploidisation), because >60% of chromosomal segments are present in the duplicated form (2). Even so, on average only 25% of the genes are still preserved in the homoeologous segments. This implies an extremely high rate of minor rearrangements and gene loss. The β duplication event was discovered and analysed on the basis of a comparison of segments 1A–2A with 4B–5B. This relationship was partially shown by Vision et al. (3). The homoeologous segment pair 1A/4B represents 58 preserved genes corresponding to >15% of genes within these segments. However, only some of them should be considered, since segment 1A was involved in a major tandem duplication (Fig. 5, dup 1/2).
The γ duplication event, the remnants of which were detected using a histogram-based approach, was discovered on the basis of the analysis of 18 regions (shaded peaks in Fig. 3) of the Arabidopsis chromosomes. This event was followed by a flurry of subchromosomal rearrangements during the genome evolution, resulting in many fragmentary gene blocks representing a mosaic of segments of ancestral chromosomes. Thus, the regions described herewith represent short remnants of duplicated blocks rather than entire segments. Some of these regions still exist in a conserved form within segments of the α duplication event (for example regions within 2L–3L in Fig. 3). Two regions depicted in Figure 2, within regions 2L and 5M, are also illustrated in Figure 3. Applying the criteria assumed in this study the number of hits for the segment 2L was insignificant. These observations confirmed the usefulness and the proper stringency of the histogram-based approach.
Synonymous codon positions in a nucleotide sequence are largely free from selection pressure and accumulate changes at a clock-like rate. Thus, we could determine the chronology of the α, β and γ duplication events by estimating the Ks values for homoeologous segments. It is noteworthy that the median values for the α duplication are consistent with the conspicuous secondary peak centred on Ks = 0.8 in the distribution obtained by Lynch and Conery (4). They concluded that this peak reflected a genome duplication event. Interestingly, the median values for 1A–2A and 4B–5B differ slightly and this can imply that either there is a regional variation in the silent substitution rate, similarly to that described in mammals (24,25), or that the α duplication occurred by segmental allopolyploidy, as was found in maize (26). Segmental allopolyploidy arises from hybridisation of species with partially differentiated chromosomes sets. The duplicated regions in the differentiated chromosomes have divergence times corresponding to the separation between the two parental species. In contrast, the segments residing on chromosomes not differentiated enough at the time of hybridisation might have formed quadrivalents at meiosis in the newly formed polyploid and, therefore, diverged only more recently, after the switch from the tetrasomic to disomic inheritance. Further comparative analysis of the duplicated segments should clarify whether segmental allopolyploidy could proceed also in Arabidopsis.
The origin of structural rearrangements: compensation effect
Our work concentrated primarily on a comparison of four segments of the A.thaliana genome and its purpose was to identify different types of local chromosomal rearrangements (Figs 2 and 5). Interestingly, each one of those rearrangements occurred at a short physical distance from the original location on a chromosome. We conclude that most probably all of them were produced by ectopic and/or unequal recombination events. These events could have taken place because of the presence of repeated elements (Figs 2 and 5, inv 1) or duplicated unique sequences (Figs 2 and 5, inv 2a). These mechanisms seem to be similar to those described for Drosophila (27) and yeast (28).
A comparison of four homoeologous segments in terms of gene orientation has prompted us to deduce a probable sequence of their structural modifications (Fig. 5). Their frequency and distribution appear to show a trend towards accumulation of nested rearrangements (del 1, del 2a, del 2b within inv 2b, Fig. 5). On the basis of this finding we propose a hypothesis of compensation of chromosomal rearrangements, the major assumptions of which are as follows: a rearrangement event, a non-deleterious one which can potentially be fixed in a population, may perturb bivalent formation of the wild- and the new-type homologues in heterozygotes. On a large evolutionary scale, natural selection works towards compensation of this effect by the elimination of the non-pairing fragments (both the original and the rearranged ones). This is achieved by small-scale deletions and gene loss within the non-pairing regions that proceed at a higher rate than in intact regions. This effect could work only in a limited time scale after a polyploidisation event, when the majority of genes are duplicated, accelerating gene loss. Moreover, as reduced pairing inhibits homologous recombination in the region, new smaller structural mutations can accumulate. The compensation process works until the elimination of the region (both its rearranged and the wild form) from the chromosomes. However, in the case of an extensive chromosomal rearrangement, the meiotic imbalance could favour the elimination of the modified chromosome from the population, thus preserving its wild form.
While we were able to identify the breakpoints of the segments, we failed to characterise the mechanism by which the segmental reshuffling occurred. We speculate that it was accomplished by a process similar to that detected in the F2 of Brassica napus, newly synthesised from B.oleracea and B.rapa (29). Following hybridisation, major chromosome mutations took place rapidly. A similar mechanism of genome rebuilding was proposed by Wolfe and Shields (30) for yeast, where the original chromosome-sized duplication could break up into smaller blocks by reciprocal translocations.
Dynamics of evolution at the subsegmental level
Despite being of common origin, segments 1A, 2A, 4B and 5B differ markedly. The most evident is the difference in size. The shortest segment, 2A, is half the size of segment 1A, and comprises 1.9 times fewer genes. Although segments 1A, 4B and 5B are similar in size, their internal organisation is quite different, mainly because of a large tandem duplication (Fig. 5, dup 1/2) that took place in 1A preceding the α duplication event (Fig. 2). The gene density of all segments is similar, but major differences are evident in other features, such as the number of tandem gene arrays and repeated elements. Since the distribution of repeated elements is random along the segments analysed in this study, their frequency appears to be segment-specific. In this context, the most conspicuous is segment 2A, because of its relatively low level of interspersed repeats. In this segment, a low level of repeats is associated with a high level of rearrangements, especially deletions (Fig. 5) and a low number of tandem arrays. This implies no evident correlation between the number of repeat units and the number of rearrangements in a fragment. Moreover, as 2A is the penultimate segment on the long arm of chromosome 2 (Fig. 1), we suspect that in the very recent history of the species this segment constituted the end of the chromosome, until the chromosome was elongated by a newly attached fragment. This may imply that the terminal parts of a chromosome tend to evolve towards length contraction, by deletions and gene loss. A similar situation may occur at the end of the short arm of chromosome 1, where three homoeologous regions located more proximally in the chromosomes are much larger. This phenomenon could be related to the telomere position effect responsible for an increased rate of gene silencing towards the ends of the chromosomes (reviewed in 31).
Because of the predominance of deletion events, we speculate that following the α duplication event, the rearrangements in the Arabidopsis genome proceeded towards the reduction of genome size. There were at least seven deletions (more than 10 genes in size, the largest covered ∼70 genes) in the segments analysed, and no local duplications. For the more ancient β and γ duplications, there is also evidence of the prevalence of deletions. We detected only two intrachromosomal duplications that occurred in tandem (Fig. 5). A similar situation was deduced for grasses, in which a slow but steady process of genome size reduction counteracted the amplification of TEs (32,33). It has to be emphasised that this study was based on a relatively small portion, 7 Mb, or <5.6% of the genome.
Gene evolution and sequence divergence
The above analysis revealed only a few examples of gene reorientation in the homoeologous segment pairs derived from the α duplication. Thus, the underlying mechanism was inconsequential for the segment divergence, as shown by Seoighe et al. (34) in yeast. In fact, we detected several examples of reoriented genes for the distal region of the segment pair 4B–5B (Fig. 2, green fragment at the extremity of 4B), but we believe this region originated from a very ancient tandem duplication event and was subsequently deleted from segment 5B (del 5b in Fig. 2). For all identified cases of gene reorientation the most probable mechanism is an expansion of a tandem array assisted by inversion events.
In order to identify the sequence divergence that might have occurred within the A.thaliana, we studied in detail the polymorphism of large indels between the Col-0 and Ler accessions. These two accessions derive from the same wild population and so they must be very closely related. The Ler accession was irradiated after being selected from the population, but the irradiation seems unlikely to influence significantly the outcome of a large indels analysis. The results confirmed unequal recombination (15 cases out of 34 indels) as the mechanism responsible for the loss, duplication and divergence of genes, as these modifications were identified mainly within the coding sequences. On the other hand, the activity of TEs was recognised to be of significance for sequence divergence (12 cases out of 34), but did not play such an important role in the evolution of the gene content. We did not discover any examples of the TEs-mediated gene transposition or inactivation, as was described for large plant genomes (reviewed in 35). Six other indels identified were caused by an unknown process. Interestingly, their breakpoints contained short direct repeats of 4–8 bp in length. Similar observations were reported for deletion events within TE sequences in Drosophila (36) and in Arabidopsis (37). Devos et al. (37) suggested illegitimate recombination as the driving force for these events and found them important for elimination of TEs. In this context, our data indicate that this mechanism can proceed also in the coding regions, and work together with unequal recombination for the evolution of gene content. Identification of other mechanisms of indels generation, such as those acting via double-strand breaks and subsequent repair, would probably need a comparison of more than two accessions, as was shown for RPS5 by Henk et al. (38). Because of their nature, they could be confused with the recombination-based mechanisms. However, there are some premises suggesting a relatively minor role of the mechanisms in the creation of indels (19). Because of the incomplete sequence data available for the Ler accession we have refrained from interpretation of the differences in the number of indels and mechanisms responsible for indels occurrence in segments 1A, 2A, 4B and 5B (Table 2). It should be noted that the Ler plasmid clone contigs were relatively short (average size 1.5 kb) (22) which might have affected the identification of some indels in this accession.
Gene loss initiated through the duplication events was probably executed by several different mechanisms. The best known would be point mutations causing gene knockout, as revealed by our PipMaker analysis (see Supplementary Material). With the active copy of a gene present in the duplicated region and the selective pressure absent, rapid alteration of the inactivated copy is possible (39). As a consequence, the rate of gene diversification after the primary gene damage is the same as for the non-coding sequences. This phenomenon resembles the process identified in yeast (40), though in Arabidopsis it seems to be of minor importance. On the other hand, most recently Bancroft (41) and Schmidt (42) suggested that the duplicated or triplicated genes in Arabidopsis and Brassica lineages could be deleted by unequal recombination. We tested this hypothesis by comparative data analysis of two Arabidopsis accessions (Col-0 and Ler) and detected three instances (deletion of gene copy from a tandem array) that could have resulted from unequal homologous recombination. Moreover, this study revealed three cases of gene loss most probably caused by illegitimate recombination, as proposed by Devos et al. (37). If this was the case, the third mechanism of gene loss would be illegitimate recombination. We postulate that gene loss in Arabidopsis could be mainly due to the loss of gene function mediated by point mutations and deletions via both illegitimate and unequal homologous recombination.
Different levels of Arabidopsis genome evolution
Our study suggests four different levels of the Arabidopsis genome evolution: nucleotide, subsegmental, subchromosomal and genomic. The nucleotide evolution is based on changes in the DNA sequence caused by point mutations, a gene-specific process. Some genes may evolve more rapidly, such as the genes encoding resistance proteins, while others may show very low levels of acceptable change, and the rate of change may not depend on the chromosomal location. The subsegmental evolution involves many different mechanisms, mainly small-scale rearrangements caused by ectopic recombination, accumulation of repeated elements, DNA/gene loss caused by unequal homologous or illegitimate recombination, and tandem array expansion. The rate of evolution at this level depends strongly on the chromosomal location. Segments localised at the ends of a chromosome and far from the centromeres could evolve towards the reduction in size, while the pericentromeric regions could evolve towards the enlargement, mainly by the accumulation of repeat units and their inefficient removal (37). At the subchromosomal level, the evolution is based on large-scale genome reassembling whose mechanism is still unclear. This phenomenon can imply the presence of a selective pressure towards maximum divergence of the initially modified homoeologous chromosomes. Numerous rearrangements would preserve chromosome specificity during cell division. The evolution at the genomic level proceeds by rapid and rare events that can duplicate or eliminate single chromosome, or form auto- or allopolyploids (43).
NOTE ADDED IN PROOF
Since the submission of this paper, a manuscript regarding large-scale duplication events in Arabidopsis by using whole-genome analysis has been published (44). Interestingly, the latter paper reported three successive rounds of duplications that complement our findings.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Steve Rounsley, Michel Delseny and Richard Cooke for providing their Arabidopsis genome databases and stimulating discussions. Special thanks are due to Adam Lukaszewski for critical reading of the manuscript. The financial support within the collaboration program between the Polish Academy of Sciences and CNRS France covering the cost of a 1 month visit of P.A.Z. to Laboratoire Genome et Développement des Plantes, Perpignan, France is gratefully acknowledged. The work was supported by a grant, no. PBZ/KBN/029/PO6/2000, from the State Committee for Scientific Research, Poland to J.S.
REFERENCES
- 1.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Blanc G., Barakat,A., Guyot,R., Cooke,R. and Delseny,M. (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell, 12, 10931–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vision T.J., Brown,D.G. and Tanksley,S.D. (2000) The origins of genomic duplications in Arabidopsis. Science, 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M. and Conery,J.S. (2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- 5.Grant D., Cregan,P. and Shoemaker,R.C. (2000) Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl Acad. Sci. USA, 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe K.H. (2001) Yesterday’s polyploids and the mystery of diploidization. Nature Rev. Genet., 2, 333–341. [DOI] [PubMed] [Google Scholar]
- 7.Ku H.M., Vision,T., Liu,J. and Tanksley,S.D. (2000) Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl Acad. Sci. USA, 97, 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossberg M., Theres,K., Acarkan,A., Herrero,R., Schmitt,T., Schumacher,K., Schmitz,G. and Schmidt,R. (2001) Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis and Capsella genomes. Plant Cell, 13, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quiros C.F., Grellet,F., Sadowski,J., Suzuki,T., Li,G. and Wroblewski,T. (2001) Arabidopsis and Brassica comparative genomics: sequence, structure and gene content in the ABI-Rps2-Ck1 chromosomal segment and related regions. Genetics, 157, 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer K., Murphy,G., Tarchini,R., Wambutt,R., Volckaert,G., Pohl,T., Dusterhoft,A., Stiekema,W., Entian,K.D., Terryn,N. et al. (2001) Conservation of microstructure between a sequenced region of the genome of rice and multiple segments of the genome of Arabidopsis thaliana. Genome Res., 11, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 12.Jurka J. (1998) Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol., 8, 333–337. [DOI] [PubMed] [Google Scholar]
- 13.Jurka J. (2000) Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet., 9, 418–420. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS, 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 16.Goldman N. and Yang,Z. (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol., 11, 725–736. [DOI] [PubMed] [Google Scholar]
- 17.Anisimova M., Bielawski,J.P. and Yang,Z. (2001) Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol., 18, 1585–1592. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z. (1998) On the best evolutionary rate for phylogenetic analysis. Syst. Biol., 47, 125–133. [DOI] [PubMed] [Google Scholar]
- 19.Achaz G., Netter,P. and Coissac,E. (2001) Study of intrachromosomal duplications among the eukaryote genomes. Mol. Biol. Evol., 18, 2280–2288. [DOI] [PubMed] [Google Scholar]
- 20.Tatusova T.A. and Madden,T.L. (1999) Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett., 174, 247–250. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jander G., Norris,S.R., Rounsley,S.D., Bush,D.F., Levin,I.M. and Last,R.L. (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol., 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 24.Smith N.G. and Lercher,M.J. (2002) Regional similarities in polymorphism in the human genome extend over many megabases. Trends Genet. 18, 281–283. [DOI] [PubMed] [Google Scholar]
- 25.Williams E.J. and Hurst,L.D. (2000) The proteins of linked genes evolve at similar rates. Nature, 407, 900–903. [DOI] [PubMed] [Google Scholar]
- 26.Gaut B.S. and Doebley,J.F. (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl Acad. Sci. USA, 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cáceres M., Ranz,J.M., Barbadilla,A., Long,M. and Ruiz,A. (1999) Generation of a widespread Drosophila inversion by a transposable element. Science, 285, 415–418. [DOI] [PubMed] [Google Scholar]
- 28.Ryu S.L., Murooka,Y. and Kaneko,Y. (1998) Reciprocal translocation at duplicated RPL2 loci might cause speciation of Saccharomyces bayanus and Saccharomyces cerevisiae. Curr. Genet., 33, 345–351. [DOI] [PubMed] [Google Scholar]
- 29.Song K., Lu,P., Tang,K. and Osborn,T.C. (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl Acad. Sci. USA, 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfe K.H. and Shields,D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 31.Tham W.H. and Zakian,V.A. (2002) Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene, 21, 512–521. [DOI] [PubMed] [Google Scholar]
- 32.Bennetzen J.L. and Kellogg,E.A. (1997) Do plants have a one-way ticket to genomic obesity? Plant Cell, 9, 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SanMiguel P., Gaut,B.S., Tikhonov,A., Nakajima,Y. and Bennetzen,J.L. (1998) The paleontology of intergene retrotransposons of maize. Nature Genet., 20, 43–45. [DOI] [PubMed] [Google Scholar]
- 34.Seoighe C., Federspiel,N., Jones,T., Hansen,N., Bivolarovic,V., Surzycki,R., Tamse,R., Komp,C., Huizar,L., Davis,R.W. et al. (2000) Prevalence of small inversions in yeast gene order evolution. Proc. Natl Acad. Sci. USA, 97, 14433–14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennetzen J.L. (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol., 42, 251–269. [PubMed] [Google Scholar]
- 36.Petrov D.A., Lozovskaya,E.R. and Hartl,D.L. (1996) High intrinsic rate of DNA loss in Drosophila. Nature, 384, 346–349. [DOI] [PubMed] [Google Scholar]
- 37.Devos K.M., Brown,J.K. and Bennetzen,J.L. (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res., 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henk A.D., Warren,R.F. and Innes,R.W. (1999) A new Ac-like transposon of Arabidopsis is associated with a deletion of the RPS5 disease resistance gene. Genetics, 151, 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno S. (1970) Evolution of Gene Duplication. Springer-Verlag, New York.
- 40.Fischer G., Neuveglise,C., Durrens,P., Gaillardin,C. and Dujon,B. (2001) Evolution of gene order in the genomes of two related yeast species. Genome Res., 11, 2009–2019. [DOI] [PubMed] [Google Scholar]
- 41.Bancroft I. (2001) Duplicate and diverge: the evolution of plant genome microstructure. Trends Genet., 17, 89–93. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt R. (2002) Plant genome evolution: lessons from comparative genomics at the DNA level. Plant Mol. Biol., 48, 21–37. [PubMed] [Google Scholar]
- 43.Soltis P.S. and Soltis,D.E. (2000) The role of genetic and genomic attributes in the success of polyploids. Proc. Natl Acad. Sci. USA, 97, 7051–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simillion C., Vandepoele,K., Van Montagu,M.C., Zabeau,M. and Van de Peer,Y. (2002) The hidden duplication past of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 99, 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.