Abstract
The human β-globin gene is abundantly expressed specifically in adult erythroid cells. Stage-specific transcription is regulated principally by promoter proximal cis-regulatory elements. The basal promoter contains a non-canonical TATA-like motif as well as an initiator element. These two elements have been shown to interact with the TFII-D complex. Here we show that in addition to the TATA and initiator elements, conserved E-box motifs are located in the β-globin downstream promoter. One of the E-box motifs overlaps the initiator and this composite element interacts with USF1 and TFII-I in vitro. Another E-box, located 60 bp 3′ to the transcription initiation site, interacts with USF1 and USF2. Mutations of either the initiator or the downstream E-box impair transcription of the β-globin gene in vitro. Mutations of a putative NF-E2-binding site in the downstream promoter region do not affect transcription in vitro. USF1, USF2, TFII-I and p45 can be crosslinked to a β-globin promoter fragment in MEL cells in vivo, whereas only TFII-I and USF2 crosslink to the β-globin gene in K562 cells. The summary data demonstrate that in addition to the well-characterized interactions of the TFII-D complex with the basal promoter, E-box motifs contribute to the efficient formation of transcription complexes on the adult β-globin gene.
INTRODUCTION
The five genes of the human β-globin locus are organized in sequential order on chromsosome 11, with the embryonic ε-globin gene at the 5′ end and the adult β-globin gene at the 3′ end of the locus (1). Expression of the human β-globin genes is restricted to erythroid cells. A variety of cis-regulatory DNA elements participate in the developmental and tissue-specific control of all of the β-like globin genes. A critical element for high level β-globin gene expression is the locus control region (LCR), located far upstream of the genes and composed of several subregions that have extremely high sensitivity to nucleases in erythroid cells (hypersensitive sites) (2–4). Deletion of the LCR dramatically impairs globin gene expression in both the murine and human globin loci (5,6).
At least two modes of regulation control the stage-specific expression of the globin genes. The first parameter regulating stage-specific expression is the relative location of the genes with respect to the LCR. When the order of the genes relative to the LCR is reversed, the β-globin gene is inappropriately expressed at the embryonic stage (7). The second parameter is the binding of stage-specific trans-acting factors to the individual globin promoter regions leading to activation of transcription at the appropriate times during development. A well-characterized example for this mode of regulation is represented by the transcription factor EKLF, which specifically activates expression of the adult β-globin gene during definitive erythropoiesis (8–11). EKLF exerts part of its activating function by recruiting chromatin-remodeling complexes to the β-globin gene promoter, which leads in turn to a local change in nucleosome organization (12).
The basal promoter of the human adult β-globin gene is composed of a TATA-like element and an initiator sequence, both of which were shown to interact with the TFII-D protein complex (13,14). In addition to components of the TFII-D complex (specifically, TAF250 and TAF150), a diverse group of other proteins can also interact with initiator sequences (15,16). Notable among these are the helix–loop–helix proteins USF and TFII-I, because both proteins have been implicated in the recruitment of transcription complexes to TATA-less promoters and in the stabilization of transcription complexes in TATA-box-containing promoters (17–19).
Regulatory DNA elements located downstream of transcription initiation sites (downstream promoter elements, DPE) that contribute significantly to the formation of stable transcription complexes have been well characterized in a variety of Drosophila genes (20). In some cases these downstream promoter elements function in the absence of TATA elements and cooperate with initiator sequences to recruit transcription complexes. The transcription cofactor NC2 (negative cofactor 2), originally identified as a repressor of RNA Pol II transcription, activates transcription of DPE-containing promoters and inhibits transcription of TATA-box-containing promoters in vitro (21).
The human β-globin gene contains two conserved E-box motifs located 3′ to the initiator sequence. The first E-box overlaps the initiator while the second E-box is located 60 bp 3′ to the transcription initiation site. We analyzed the function of these E-boxes and characterized proteins that interact with these sequences. In these studies we used two erythroid cell lines, MEL, a murine erythroleukemia cell line expressing the adult β-globin gene, and K562, a human erythroleukemia cell line expressing the embryonic β-type globin genes. Our results demonstrate that the human β-globin promoter contains a TATA-like motif, an initiator and a downstream E-box element that are all required for high level transcription in vitro. We further show that USF1, USF2, TFII-I and p45 interact with the β-globin downstream promoter in vitro and in vivo. The interactions of USF1 and p45 are cell type specific. The results indicate that helix–loop–helix proteins contribute to the formation of transcription complexes on the adult human β-globin gene, and suggest that the differential association of HLH proteins with the basal promoter contribute to the stage-specific expression of the gene.
MATERIALS AND METHODS
Cell culture and protein extracts
MEL and K562 cells were grown in RPMI and processed as described previously (22). Protein extracts used in EMSA or in vitro transcription experiments were prepared from MEL cells as described previously (19,22). NF-E2, p45 tethered to mafg, has been previously described (23). The cDNA was subcloned into a pET expression vector and expressed in bacteria. The final concentration of NF-E2 was 0.3 µg/µl.
Plasmid constructs and oligonucleotides
The templates for in vitro transcription were all derived from pRSβ, which contains the human β-globin gene as an ApaI–XbaI fragment cloned into the ApaI and XbaI sites of pRS306 (24) (kindly provided by Doug Engel, Northwestern University). pRSβTATA, pRSβINImut and pRSβCACCmut were generated by replacing the TATA-box, the initiator or the 5′-CACCC site in the promoter with NotI sites by site-directed PCR mutagenesis. For the mutagenesis, primers were designed that amplify a region of the β-globin gene from the SphI site at position 61519 (25) to the individual mutated regions (TATA, INI and 5′-CACC) and from these regions to the EcoRI site at position 63530 (5′SphI: 5′-TTAGCATGCATGAGCAAATTAAG-3′; 3′TATA: 5′-GACGCGGCCGCCCAGCCCTGGCTCCT; 3′INI: 5′-AGCGCGGCCGCAA GCAATAGATGGCT-3′; 5′CACC: 5′-CACGCGGCCGCCTCCACAGGGTGAGG-3′; 5′TATA: 5′-TGCGCGGCCGCGTCAGGGCAGAGCCA-3′; 5′INI: 5′-GCTGCGGCCGCT TCTGACACAACTGT-3′; 3′EcoRI: 5′-CCGGAATTCTTTGCCAAAGTG-3′). The PCR products were digested with SphI and NotI (for the upstream fragments) or NotI and EcoRI (for the downstream fragments) and ligated into the SphI and EcoRI site of pRSβ, replacing the wild-type SphI–EcoRI fragment. The mutant globin templates pRSβ5′INImut, pRSβ3′INImut, pRSβ+20E-boxmut, pRSβ+50mut, pRSβ+60E-boxamut, pRSβ+60E-boxbmut pRSβMAREa and pRSβMAREb were generated by PCR-directed mutagenesis using the Quik-Change mutagenesis kit (Stratagene). The following primers were used for the mutagenesis: +20E-box: 5′-GCTTACATTTGCTTCTGACAGCGCTCTGTTCACTAGCAACCTCAAACAGACACCATGG-3′; +60E-boxa: 5′-CCTC AACAGACACCATGGTGAGCGCTACTCCTGAGGAGAAGTCTGCCGTTACTGCC-3′; +60E-boxb: 5′-CCTCAAA CAGACACCATGGTGAAGCTTACTCCTGAGGAGAAGTCTGCCGTTACTGCC-3′; 5′INI: 5′-GAGCCATCTATTGCGGACATTTGCTTCTGACACAACTGTGTTCACTAGCAACC-3′; 3′INI: 5′-GAGCCATCTATTGCTTACATTGT CTTCTGACACAACTGTGTTCACTAGCAACC-3′; +40NS: 5′-CTAGCAACCTCAAACAAGCGCTATGGTGCACCTGCTCCTGAGGAG-3′; MAREa: 5′-CATTTGCTTCTGACACAACTATGTTCACTAGCAACCTCAAACAGACACC-3′; MAREb: 5′-CATTTGCTTCTGACACAACTGTATTCGACAGCAACCTCAAACAGACACC-3′.
All plasmids were sequenced to verify that the desired mutations were introduced without changing other sequences.
EMSA
For EMSA, ∼100 fmol of an end-labeled 40 bp oligonucleotide corresponding to the wild-type (or mutant) β-globin initiator, or downstream region, was incubated with ∼240 µg/ml of MEL extract in a 1× binding buffer containing 18 mM HEPES (pH 7.9), 80 mM KCl, 20 mM MgCl2 and 10% glycerol, in a final reaction volume of 30 µl. Poly(dI-dC) was included at a concentration of 50 ng/ml to reduce non-specific interactions. Reactions were incubated without the DNA for 20 min at 30°C at which time the DNA was added and the binding reactions allowed to continue for an additional 45 min. Where indicated, reactions were pre- or post-incubated for 20 min at 30°C with 3 µl of antibody against TFII-I, USF1 or USF2. Immediately following incubation, 25 µl of each reaction was loaded onto a native 4% polyacrylamide gel consisting of 0.8% glycerol and the 1× running buffer contained 6.7 mM Tris (pH 7.9), 3.3 mM sodium acetate and 1 mM EDTA. Electrophoresis was allowed to continue for 3–4 h at 4°C, after which time the gel was dried. Complex formation was then visualized by autoradiography. The following double-stranded oligonucleotides were used in the experiments (only the non-coding strand is shown): WT INI: 5′-TCTATTGCTTACATTTGCTTCTGACACAACTGTGTTCACT-3′; INImut: 5′-TCTATTGCGATGCGGCCCTTCTGACACAACTGTGTTCACT-3′; +60 E-box: 5′-GACACC ATGGTGCACCTGACTCCTGAGGAGAAGTCTGCCG-3′; β-MARE: 5′-CTCAAAACTGTGTTCACTAGCAACCTCAAAC-3′; HS2-MARE: 5′-CTCAAAGCAATGCTGAGTCATGATGAGTCAT-3′.
Western blot analysis
Approximately 60 µg of nuclear protein extract was electrophoresed on 10% Ready Gels (Bio-Rad) under denaturing conditions. Electrotransfer was performed with the Mini Protean apparatus from Bio-Rad according to the manufacturer’s instructions. Incubation with antibodies and detection of signals with horseradish peroxidase-conjugated secondary antibody were done with reagents from Amersham according to the manufacturer’s instructions.
Primer extension in vitro transcription
In vitro transcription experiments were performed as previously described using a mixture of HeLa and MEL cell extracts (22). Using MEL cell extract alone led to extremely weak transcription signals, especially for the internal control, suggesting that the MEL extract has limiting amounts of one or more of the basal transcription factors. We therefore decided to supplement the MEL extract with HeLa extract. As templates in these experiments we used either the wild type or mutants of the β-globin gene in combination with the human growth hormone gene under control of the thymidine kinase promoter (26), or a CMV construct (Promega), which served as an internal control. The following primers were used: β-glob90: 5′-CTTGCCCCACAGGGCAGTAACGGCAG-3′; β-glob80: 5′-CGGCAGACTTCTCCTCAGGAGTCAGGTG-3′; CMV: 5′-ACCAAGCCTATGCCTACAGCATCC-3′; Gh: 5′-GTGAGCTGTCCACAGGACCCTGAG-3′.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed as described by Forsberg et al. (27) with minor modifications. MEL and K562 cells (108 cells) were grown in RPMI medium. Crosslinking of proteins and DNA was induced by incubating the cells in 1% formaldehyde with rocking at room temperature for 10 min. The crosslinking was quenched with 0.125 M glycine for 5 min. Cells were washed twice with 1× PBS (including protease inhibitors), resuspended in swelling buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, 11 mg/ml sodium butyrate, protease inhibitors) and incubated on ice for 10 min. Nuclei were lysed by incubation in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1, 11 mg/ml sodium butyrate, protease inhibitors) on ice for 10 min. DNA was sonicated on ice to yield an average size of <500 bp and cleared by centrifugation. The supernatant was diluted in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM Tris–HCl pH 8.1, 167 mM NaCl, 11 mg/ml sodium butyrate, protease inhibitors). Aliquots of 500 µl Protein A–Sepharose beads were added to the diluted lysate and incubation was done for 2 h with rotation at 4°C. The beads were pelleted and 5 µl of appropriate antibody was added to the aliquoted supernatant and incubated overnight with rotation at 4°C. The solutions were rotated for 2 h at 4°C with 60 µl Protein A–Sepharose beads previously blocked and resuspended in blocking buffer (3% BSA, 0.05% Na azide and protease inhibitor in 10 mM Tris–HCl pH 8.1, 1 mM EDTA). The beads were pelleted and supernatants removed. The supernatant of the ‘no antibody’ sample was kept and labeled ‘input’. The pelleted beads were washed with low salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 200 mM Tris–HCl pH 8.1, 150 mM NaCl), high salt wash (0.1% SDS 1% Triton X-100, 2 mM EDTA, 200 mM Tris–HCl pH 8.1, 500 mM NaCl), LiCl wash (0.25 M LiCl, 1% NP-40, 1% Na deoxycholate, 1 mM EDTA, 100 mM Tris–HCl pH 8.1) and two washes with TE (pH 8.0). DNA was eluted with 500 µl 1% SDS and 0.1 M NaHCO3 at 65°C for 30 min. NaCl was added to the eluates to a final concentration of 200 mM and crosslinking was reversed by incubation at 65°C for 5 h. RNA was digested by incubation with 40 µg/ml RNase A for 30 min at 37°C. Proteins were then digested with 40 µg/ml proteinase K in 10 mM EDTA, 40 mM Tris–HCl pH 6.5 at 37°C for 1 h. DNA was purified using a purification kit (Qiagen).
A PCR contained 10% input DNA or 10% immunoprecipitated DNA, 50 pM primers and 10 µl Platinum PCR Supermix (Life Technologies) in a total volume of 25 µl. The following primers were used: mouse HS2 5′flank, US: 5′-CTATTTGCTAACAGTCTGACAATAGAGTAG-3′, DS: 5′-GTTACATATGCAGCTAAAGCCACAAATC-3′; human HS2 5′flank, US: 5′-AGTAGATATTCCTGCCCTGTG GCC-3′, DS: 5′-GAAACTTTGTTGTCAGACCCGGC-3′; mouse β-globin, US: 5′-AAGCCTGATTCCGTAGAGCCACAC-3′, DS; 5′-CCCACAGGCAAGAGACAGCAGC-3′; human β-globin, US: 5′-TATCTTAGAGGGAGGGCTGAGGGTTTG-3′, DS: 5′-CCAACTTCATCCACGTTCACCTTGC-3′. Titration of cycle numbers was performed with the different primer pairs to determine the range of linear amplification. After 30 cycles, DNA was run in a 5% TBE polyacrylamide gel. The gels were stained with SyBr-green and analyzed by fluorescence scanning.
In vivo footprinting
DMS treatments of cells (MEL and 3T3L1) and naked genomic DNA were performed essentially as described by Hornstra and Yang (28). The sequencing ladder was generated as described by Maxam and Gilbert (29).
Ligation-mediated PCR (LMPCR) was essentially performed as described by Hornstra and Yang (30) except that Taq polymerase (Life Technologies) was used in the PCR. PCR included a denaturing step at 95°C for 5 min followed by 20 cycles of PCR under the following conditions: 95°C for 20 s, 65°C for 1 min, 72°C for 1 min with an increase of 5 s/cycle and an additional five cycles of 95°C for 20 s, 65°C for 1 min, 72°C for 2 min 30 s, followed by a final extension at 72°C for 15 min.
Radiolabeled probes were synthesized using the Prime-a-Probe kit (Ambion) from a gel purified PCR product (PCR primers β-maj FWD and MPB 1) containing the region of interest. The labeling reaction was quenched on ice and stopped by adding 35 µl formamide loading dye. The probe was denatured at 95°C for 10 min and purified on a 5% poylacrylamide gel. After exposing the gel to film (Type 57; Polaroid) the probe was excised from the gel, crushed and soaked in 4 ml hybridization buffer (250 mM Na2PO4 pH 7.2, 7% SDS, 1% BSA). The probe was hybridized to the blots at 65°C, overnight. The blots were washed three times at 65°C in washing solution (20 mM Na2PO4 pH 7.2, 1% SDS) and visualized by autoradiography. The following primers were used for LMPCR: MPA 3: 5′-TGAAGGGCCAATCTGCTCACACAGG-3′; MPA 1: 5′-ATGTCCAGGGAGAAATATCG-3′; MPB 1: 5′-TCTGTCTCCAAGCACC CAA-3′; MPB 2: 5′-GGCCTCACCACCAACTTCATCGGA-3′; β-maj FWR: 5′-GACAAACATTATTCAGAGGGAGTACCC-3′.
RESULTS
The human β-globin downstream promoter contains two conserved E-box motifs and a partial NF-E2-binding site
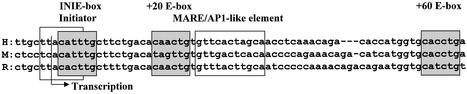
The downstream promoter region of the human β-globin gene contains three E-box motifs (consensus sequence CANNTG). Two of these are well conserved in both position and primary sequence among different species (Fig. 1). The proximal E-box overlaps the initiator sequence whereas the distal E-box is located 60 bp downstream of the transcription initiation site.
Figure 1.
Sequence alignment of the human β-globin downstream promoter region. Shown are three sequences of the adult β-globin downstream promoter region from human (H), mouse (M) and rabbit (R) (26). Shaded boxes highlight the position of E-box motifs (CANNTG). Two of these E-boxes, the one overlapping the initiator and the distal E-box, are conserved in all three species, whereas the E-box located at +20 is only present in the human and rabbit genes. The open box delineates the position of the MARE-like sequence.
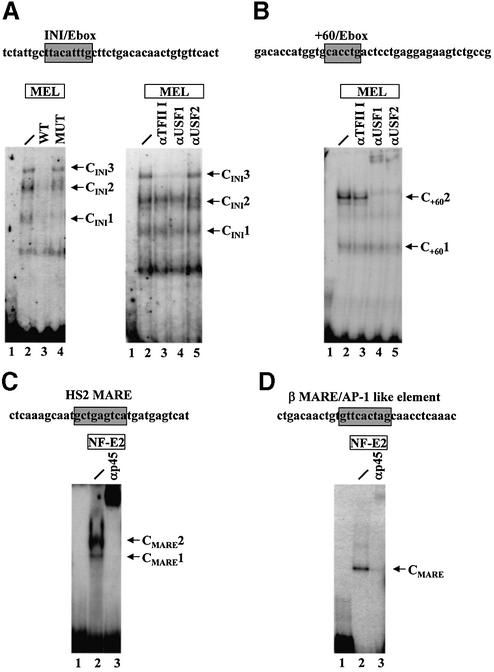
We used EMSA to characterize proteins interacting with the β-globin downstream promoter (Fig. 2). We began these studies by utilizing a DNA fragment that encompasses the initiator/E-box and the E-box at +20. After incubating this fragment with protein extracts from MEL cells, three specific protein–DNA complexes are detectable (Fig. 2A, lane 2, labeled complexes CINI1–CINI3). Pre-incubation with unlabeled wild-type competitor DNA reduced the formation of complexes CINI1, CINI2 and CINI3 (Fig. 2A, lane 3). The band below complex CINI1 is not affected by competition with the wild-type oligonucleotide and is thus non-specific. A mutated oligonucleotide in which the initiator/E-box motif was altered (INImut) did not efficiently compete for formation of complexes CINI2 and CINI3 (Fig. 2A, lane 4), indicating that the initiator/E-box constitutes or is part of the protein-binding site in this fragment. Formation of CINI1 was reduced by competition with the initiator/E-box mutant indicating that this complex represents the binding of proteins outside the initiator.
Figure 2.
Interaction of HLH proteins and NF-E2 with human β-globin downstream promoter sequences in vitro. (A) EMSA was carried out as described in Materials and Methods using a fragment encompassing the initiator/E-box as well as the E-box at +20 (WT). Radiolabeled DNA fragments were incubated with 8 µg MEL protein extracts for 45 min at 30°C (lanes 2–4 and 6–9; lanes 1 and 5, no protein). MEL protein extracts were pre-incubated either without (lane 2) or with a 50-fold excess (compared to the radiolabeled fragment) of unlabeled competitor DNA, either using the wild-type initiator (lane 3) or a mutant fragment in which the initiator/ E-box was mutated (INImut, lane 4). Antibody supershift reactions were post-incubated either without (lane 6) or with specific antibodies raised against TFII-I (αTFII-I, lane 7), USF1 (αUSF1, lane 8) or USF2 (αUSF2, lane 9) for 20 min at 30°C. (B) Characterization of proteins interacting with an E-box motif located 60 bp downstream of the β-globin transcription initiation site. EMSA was carried out with a fragment encompassing the E-box element located 60 bp downstream of the start site of transcription. Radiolabeled fragment was incubated without (lane 1) or with 8 µg MEL protein extract (lanes 2–5). Proteins were post-incubated with specific antibodies raised against TFII-I (αTFII-I, lane 3), USF1 (αUSF1, lane 4) or USF2 (αUSF2, lane 5) for 20 min at 30°C. (C) Interaction of NF-E2 with a consensus MARE sequence from LCR element HS2. A radiolabeled oligonucleotide bearing the NF-E2-binding site from HS2 was incubated with 0.06 µg of a protein fraction enriched for a tethered p45/mafg protein (lane 2; lane 1 depicts the free probe). The binding reaction was pre-incubated with antibodies against p45 (lane 3) for 20 min at 30°C before addition of the radiolabeled fragment. (D) Interaction of NF-E2 with a MARE-sequence from the human β-globin downstream promoter region. A radiolabeled fragment bearing the β-globin downstream MARE like sequence was incubated with 0.2 µg of a protein fraction enriched for the tethered p45/mafg fusion protein (lane 2; lane 1 shows the free probe). Post-incubation with p45 antibodies (lane 3) was done as described in (B).
To characterize the proteins interacting with the β-globin initiator fragment, we employed antibodies specific for USF1, USF2 or TFII-I in further EMSA studies. Formation of complex CINI3 is disrupted by post-incubation with either antibodies specific for USF1 or antibodies recognizing TFII-I (Fig. 2A, lanes 7 and 8, respectively). This result suggests that complex CINI3 is generated by the binding of USF1 and TFII-I to the initiator/E-box binding site. Longer gel runs resolved complex CINI2 into two bands (Fig. 2A, right panel), the lower of which also disappeared after post-incubation with TFII-I and USF1 antibodies (Fig. 2A, lanes 7 and 8), indicating that TFII-I and USF1 are part of this protein–DNA complex as well. Antibodies against USF2 had no effect on complex formation on the initiator/E-box fragment in these experiments (Fig. 2A, lane 9).
We also analyzed the interaction of proteins with the E-box located at +60 bp. Figure 2B shows that two major protein–DNA complexes are formed with a DNA fragment encompassing the +60 E-box when incubated with MEL extract (C+601 and C+602; Fig. 2B, lane 2). Further experiments showed that the mobility of the upper complex (C+602) is affected by incubating the proteins and DNA with antibodies directed against USF1 or USF2 (Fig. 3B, lanes 4 and 5) but not by incubating with anti-TFII-I antibodies (Fig. 2B, lane 3). Mutations in the +60 E-box motif also affected the formation of the upper complex (C+602) in a manner similar to pre-incubation with anti-USF antibodies (data not shown). The combined data show that TFII-I and USF1 interact with the initiator and proximal E-box motifs, whereas USF1 and USF2 interact with the distal E-box.
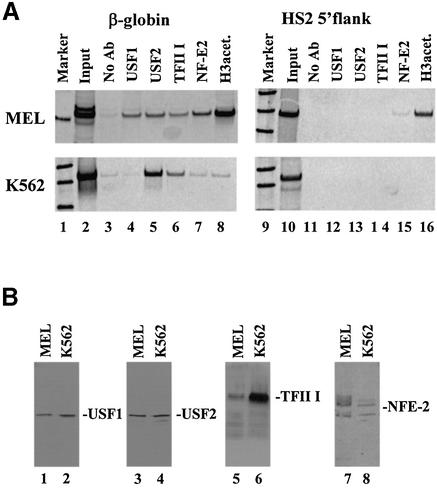
Figure 3.
Characterization of protein–DNA interactions in the human β-globin downstream promoter region in vivo. (A) Chromatin immunoprecipitation (ChIP) experiment analyzing the interaction of proteins with the murine/human β-globin gene or the HS2 5′flanking region in MEL and K562 cells in vivo. Cells were incubated in 1% formaldehyde to crosslink protein–DNA and protein–protein interactions. After sonication, the cells were lysed and the chromatin was precipitated with either no antibody (lanes 3 and 11) or with antibodies specific for USF1 (lanes 4 and 12), USF2 (lanes 5 and 13), TFII-I (lanes 6 and 14), NF-E2 (p45, lanes 7 and 15) or acetylated histone H3 (lanes 8 and 16). DNA was purified from the precipitate and analyzed by PCR for the presence of the murine or human β-globin gene (210 and 321 bp, respectively, lanes 2–8) or the murine or human HS2 5′flank (336 and 565 bp, respectively, lanes 10–16). As positive controls, the input DNA was also analyzed by PCR (lanes 2 and 10). Lanes 1 and 9 show radiolabeled 100 bp markers. (B) Western blot analysis of protein extracts from MEL and K562 cells. Nuclear extracts were prepared from MEL or K562 cells and electrophoresed on denaturing polyacrylamide gels. Proteins were electroblotted to a nitrocellulose membrane, which was then hybridized to antibodies specific for USF1, USF2, TFII-I or NF-E2 (p45) as indicated. Bands were visualized by incubation with horseradish peroxidase-conjugated secondary antibody and autoradiography.
Previous studies have shown that NF-E2 can be crosslinked to the murine β-globin gene in erythroid cells (31). By using a combination of ChIP and DNA footprinting we have demonstrated that p45-selected chromatin fragments reveal a footprint over a partial NF-E2-binding site downstream of the murine βmaj-globin gene (32). The corresponding sequence of the human β-globin gene reveals a poor homology to the consensus NF-E2-binding site. We used EMSA to analyze whether NF-E2 binds to this sequence in vitro. Recombinant NF-E2 (p45 tethered to mafg; kindly provided by G. Blobel) (24) was incubated either with a consensus NF-E2-binding site from LCR element HS2 (Fig. 2C) or with the NF-E2-like binding site downstream of the human β-globin promoter (Fig. 2D). In each case NF-E2 binds specifically to the fragment (Fig. 2C and D, lanes 2) and the protein–DNA complexes are supershifted with antibodies specific for the p45 subunit (Fig. 2C and D, lanes 3). The binding is specific because the recombinant protein did not bind to an unrelated fragment isolated from the erythropoietin promoter region (C. Dame, unpublished results).
USF1 and USF2 interact with the β-globin gene in erythroid cells in vivo
We next examined whether USF1, USF2 or TFII-I interact with the β-globin gene in vivo by performing ChIP experiments in MEL and in K562 cells. The results of these experiments are shown in Figure 3A and demonstrate that both USF1 and USF2 could be crosslinked to the β-globin gene promoter in MEL cells in vivo (Fig. 3A, lanes 4 and 5). TFII-I could also be crosslinked to the globin promoter (Fig. 3A, lane 6), however, the signal for TFII-I was consistently weaker in MEL versus K562 cells. Antibodies recognizing the p45 subunit of NF-E2 and acetylated histone H3 also precipitated the β-globin gene in MEL cells (Fig. 3A, lanes 7 and 8, respectively). The precipitation with acetylated H3 antibodies was expected from previous studies (27). To analyze whether the interaction of USF proteins with the β-globin promoter could be correlated with the transcriptional activity of the gene we carried out ChIP analyses in K562 cells, in which the β-globin gene is not expressed. In contrast to our observations in MEL cells, USF1 does not interact with the β-globin gene in K562 cells, whereas TFII-I and USF2 do interact (Fig. 3A, lanes 5 and 6). The interactions of p45 (NF-E2) and acetylated H3 with the β-globin gene were also less efficient in K562 cells when compared to MEL cells (compare Fig. 3A, lanes 7 and 8, MEL versus K562). In control experiments we amplified a region within the LCR located upstream of HS2 (HS2 5′flank; Fig. 3A, lanes 10–16). The results demonstrate that the observed interactions of USF1, USF2, TFII-I and p45 in erythroid cells are specific for the β-globin gene.
To analyze the possibility that the differential binding of USF1, NF-E2 and, to a lesser extent, TFII-I is due to differential expression of these proteins in the two cell lines, we carried out western blotting experiments using antibodies specific for USF1, USF2, TFII-I and NF-E2 (Fig. 3B). The results show that USF1 and USF2 are expressed at similar levels in K562 and MEL cells (Fig. 3B, lanes 1–4). However, TFII-I is expressed at higher levels in K562 cells (Fig. 3B, lanes 5 and 6) and p45 (NF-E2) is expressed at higher levels in MEL cells (Fig. 3B, lanes 7 and 8). The lower signal for TFII-I in protein extracts from MEL cells is not due to species specificity of the antibody, which was raised against a conserved region of the protein (33).
We next performed DMS in vivo footprinting to determine whether the conserved E-box motifs in the β-globin promoter region are protected in MEL cells. To analyze whether footprinted regions in the β-globin gene are specific for erythroid cells, we also analyzed the DMS cleavage pattern in 3T3L1 cells (a mouse adipocyte cell line). Figure 4 shows that a guanine residue in the E-box located at +60 bp is protected from cleavage by DMS/piperidine in MEL cells (Fig. 4, lane 3). The guanine residue that is protected from modification by DMS is the most 5′ nucleotide of the consensus E-box (CANNTG, opposite strand). The footprint is specific for erythroid cells and not detectable in 3T3L1 cells (Fig. 4, lane 2). Because guanine residues 5′ to this E-box are not protected, the result suggests that the footprint over the +60 E-box is not due to the binding of the transcription complex but reflects the binding of other proteins to this region, most likely USF. We also detected footprints in the initiator region (data not shown). However, because this region is likely bound by a variety of proteins, including the TFII-D complex, we cannot conclude that the protection reflects the consequence of the interaction with E-box-binding proteins. Taken together, the in vivo experiments suggest that helix–loop–helix proteins interact with downstream promoter elements of the adult β-globin gene in vivo.
Figure 4.
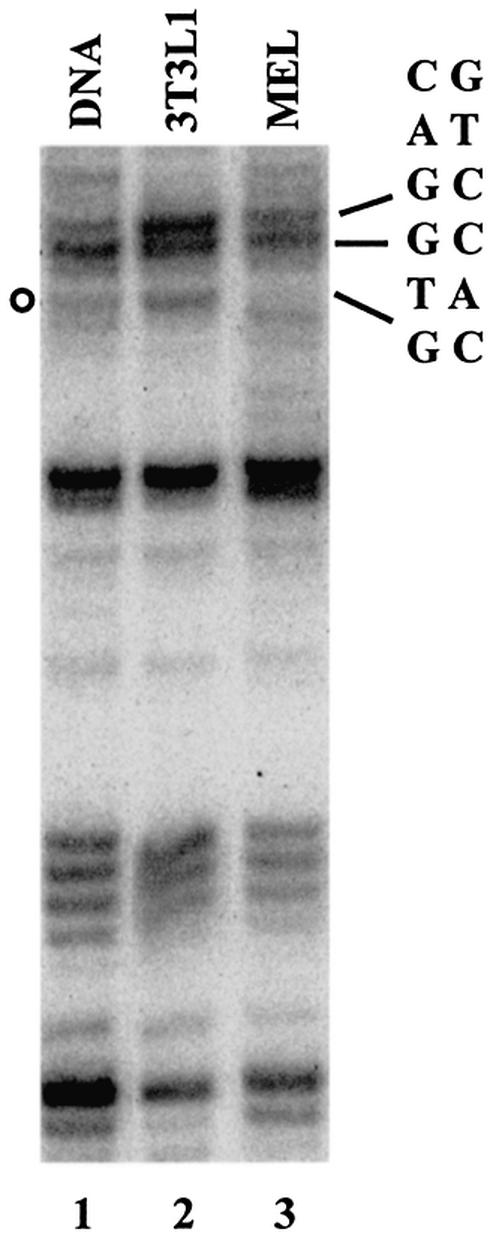
In vivo footprint analysis of a conserved E-box located in the murine adult β-globin downstream promoter region. NIH3T3L1 (lane 2) or MEL (lane 3) cells were treated with DMS. The DNA was purified from these cells and analyzed by linker ligation-mediated PCR as described in Materials and Methods. Lane 1 depicts the G sequencing ladder of the region. The position of the E-box at +60 is indicated on the right. The open circle on the left highlights a G residue that is consistently protected from cleavage by DMS in MEL cells.
The β-globin downstream promoter E-box elements contribute to efficient transcription of the human β-globin gene in vitro
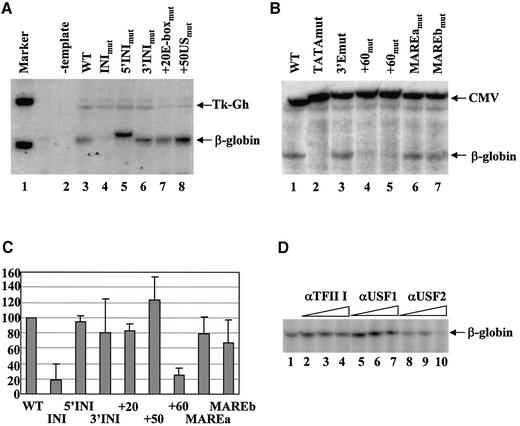
Because previous studies have shown that E-box-binding proteins contribute to the efficient formation of RNA Pol II transcription complexes in the presence or absence of a TATA-box (17–19), we were interested in addressing their possible function in β-globin gene transcription. Therefore, we mutated these elements and analyzed the consequences of these mutations on β-globin gene transcription by primer extension assays in vitro (Fig. 5). As internal controls in these experiments we used the human growth hormone gene under control of the thymidine kinase promoter (Tk-Gh, Fig. 5A) or a CMV construct (CMV, Fig. 5B). Figure 5A shows that using mixed nuclear extracts from HeLa and erythroid cells (22), the human β-globin gene is efficiently transcribed in vitro, whereas transcription of the human growth hormone gene is significantly less efficient (Fig. 5A, lane 3). We first analyzed the consequence of mutating the β-globin initiator on in vitro transcription. Mutation of the initiator (and its overlapping E-box) significantly reduced transcription (Fig. 5A, lane 4). To determine whether the INI mutation affected transcription by interfering with initiator complex or HLH complex formation, we independently mutated both cis motifs. Neither mutation alone significantly reduced transcription efficiency of the β-globin gene (Fig. 5A, lanes 5 and 6, and C). However, mutations in the 5′ sequences of the initiator altered the transcription initiation site (Fig. 5A, lane 5). The mutation changes two nucleotides immediately upstream of the start site; our results show that these nucleotides are involved in start site selection. Mutation of sequences between the initiator and the E-box at +60 had no effect (Figs. 5A, lane 8, and C) nor did a mutation introduced into the species non-conserved E-box at +20 (Fig. 5A, lane 7, and C). We next analyzed the role of the +60 E-box and of the MARE-like element on in vitro transcription of the β-globin gene (Fig. 5B). Because the primer used for the experiment in Figure 5A interfered with the mutated E-box at +60 we had to change the primer sequences for both the β-globin gene and for the internal control (CMV instead of TK-Gh). We generated two different mutations of the +60 E-box (a and b, see Materials and Methods). Both mutants revealed a significant reduction in β-globin gene transcription in vitro (25–30% of WT; Fig. 5B, lanes 4 and 5, and C and data not shown). Mutations within the β-globin TATA-box completely abolished transcription (Fig. 5B, lane 2) suggesting that in vitro the initiator and +60 E-box do not function in the absence of the TATA-box. Mutations within the partial NF-E2-binding site did not significantly alter transcription efficiency of the β-globin gene in vitro (Fig. 5B, lanes 6 and 7, and C) nor did the deletion of the β-globin 3′ enhancer. Taken together, the data suggest that the biologically active basal promoter of the β-globin gene consists of a TATA-box, an initiator and a downstream promoter element that is composed of an E-box motif.
Figure 5.
Mutations of the initiator and E-boxes in the downstream promoter region impair transcription of the β-globin gene in vitro. In vitro transcription was monitored by primer extension using wild-type and mutant constructs of the β-globin gene and, as an internal control, the human growth hormone gene under control of the thymidine kinase promoter (Tk-Gh) (A) or a CMV construct (CMV) (B) was used. (A) Transcription analysis of mutations in the initiator (INImut), the 5′ region of the initiator (5′INImut), the 3′ region of the initiator (3′INImut), the E-box located 20 bp downstream of the initiation site (+20E-boxmut) and a region 50 bp downstream of the initiation site (+50USmut). Transcription was compared to that of a negative control lacking a DNA template (–template) and to that of the wild-type β-globin gene (WT). The first lane shows a radiolabeled marker (φX174/HinfI, denatured 100 bp marker). β-globin transcription was monitored using the β-glob80 primer. (B) Transcription of the wild-type (WT) and various β-globin gene templates carrying the following mutations: 5′TATAmut, mutation of the β-globin TATA-box; 3′Emut, deletion of the β-globin 3′ enhancer; +60amut, mutation of the E-box at +60; MAREamut and MAREbmut, mutations in the MARE-like element. Transcription of the β-globin gene was monitored using the β-glob90 primer and compared to transcription of the CMV internal control. (C) Quantitative analysis of in vitro transcription of the various promoter mutants shown in (A) and (B). Radiolabeled samples were electrophoresed on sequencing gels and the gels subjected to phosphorimaging. Graphs represent the relative expression levels of each β-globin mutant. RNA levels were normalized using the internal control and WT expression was set as 100%. Results shown are the average over two or three separate experiments. Error bars represent +1 SD from the mean. (D) Analysis of the effect of antibodies against HLH proteins on transcription of the β-globin gene in vitro. In vitro transcription of the wild-type β-globin gene was carried out as described in Figure 2. Protein extracts were pre-incubated with increasing concentrations of antibodies (2, 4 and 8 µl) against TFII-I (lanes 2–4), USF1 (lanes 5–7) and USF2 (lanes 8–10).
The transcription studies have shown that both the initiator/E-box as well as the E-box located at +60 are required for efficient transcription of the β-globin gene in vitro. The EMSA analysis revealed that these sequences interact in vitro with USF1 and TFII-I (initiator/E-box) or USF1 and USF2 (+60 E-box). To establish a link between these observations we performed in vitro transcription experiments with protein extracts that had been pre-incubated with antibodies directed against TFII-I, USF1 and USF2 (Fig. 5D). The results of multiple experiments show that antibodies against USF2 strongly reduced transcription efficiency of the β-globin gene in vitro (Fig. 5D, lanes 8–10), whereas antibodies against USF1 inhibited transcription only weakly (Fig. 5D, lanes 5–7). Antibodies against TFII-I had no effect on transcription efficiency of the β-globin gene in vitro (Fig. 5D, lanes 2–4). These data indicate that the interaction of USF with the E-box element at +60 is required for high level transcription of the β-globin gene in vitro.
DISCUSSION
The promoter of the human β-globin gene is characterized by the presence of a non-canonical TATA-box (CATAAA) located 25–30 bp upstream of the transcription start site (34). Deviation from consensus TATA sequences often weakens promoters and leads to the requirement of additional elements for the efficient recruitment or stabilization of transcription complexes. For example, a recent report demonstrated that the TBP/TFII-A pre-initiation complex interacts only weakly with the CATAA motif (35). An additional sequence that contributes to high level β-globin gene transcription is an initiator element located at the transcription start site. Lewis et al. (13,14) have shown that this element interacts with the TFII-D complex in vitro and that mutations in this sequence reduce transcription efficiency.
In addition to the initiator element, the β-globin gene contains several E-box motifs downstream of the transcription start site (25) (Fig. 1). Two of these elements are conserved across species. Because previous studies have shown that E-box-binding proteins can participate in the formation of transcription complexes on both TATA-containing and TATA-less genes (17–19), we were interested in examining the functional role of these sequences in β-globin gene transcription and in determining the proteins that interact with these elements. Mutations of the E-box at +60 reduced transcription to a level comparable to that observed after mutation of the initiator sequence, indicating that the E-box motif and the initiator significantly contribute to the formation of transcription complexes on the β-globin gene in vitro (Fig. 4). Mutation of the β-globin gene TATA-box completely abolished transcription indicating that the initiator and downstream E-boxes are not functioning independently from the TATA-box. On the other hand, the TATA-box does not appear to function efficiently in the absence of either the initiator or the distal E-box suggesting that all three elements act together to establish functional transcription complexes on the β-globin gene. Our results are consistent with a recent study showing that mutations of either a non-canonical TATA-box or a promoter-proximal E-box motif abolished transcription of the vasopressin promoter (36). Together these data suggest that specific promoters use non-canonical TATA-boxes in combination with initiator and/or E-box motifs for the efficient recruitment of transcription complexes.
The β-globin initiator/E-box interacts with TFII-I and USF1, whereas the distal E-box at +60 interacts with USF1 and USF2 (Fig. 2). There are a number of reports in the literature documenting the interaction of USF and TFII-I with sequence elements in the vicinity, mostly downstream, of transcription initiation sites (37–40). Hsieh et al. (41) recently identified an E-box motif located downstream of the APEG-1 gene from +39 to +44. This element interacts with USF proteins and mutation of this E-box dramatically reduces transcription.
TFII-I interacts with the β-globin initiator in vitro and in vivo and may play a role in activating transcription of the β-globin gene in MEL cells. However, several observations suggest that TFII-I is not required for β-globin gene transcription. First, antibodies against TFII-I were not able to inhibit transcription of the β-globin gene in vitro (Fig. 5D). Secondly, crosslinking of TFII-I to the β-globin gene in adult erythroid cells is inefficient compared to USF1 and USF2 (Fig. 3A and unpublished data). The ChIP assay does not provide information as to what fraction of cells have a specific protein bound at a specific site. It is thus possible that the weak PCR signal from TFII-I-selected MEL chromatin fragments indicates that only a small fraction of cells have TFII-I bound at the initiator. This fraction of cells could represent non-expressing cells. This is consistent with our observation that TFII-I crosslinks more efficiently to the β-globin gene in K562 cells (Fig. 3A), suggesting that TFII-I may only be able to interact with the β-globin gene in the absence of transcription complex formation. Whether the binding of TFII-I to the β-globin promoter in erythroid cells with embryonic phenotype contributes to repression of the β-globin gene or simply reflects the absence of TFII-D binding remains to be determined. However, a recent report shows that TFII-I interacts with histone deacetylase 3 (42). It is thus possible that TFII-I interacts with the β-globin gene in embryonic erythroid cells, recruits histone deacetylases and contributes to the repression of β-globin gene expression by rendering the chromatin structure inaccessible. Our ChIP data clearly show a strong reduction of acetylated H3 binding to the β-globin promoter in K562 cells compared to MEL cells (Fig. 3A, lane 8).
We have performed RT–PCR (not shown) and western blotting experiments to characterize the expression pattern of TFII-I in K562 versus MEL cells. The RT–PCR analysis revealed that TFII-I is transcribed with equal efficiency in both cell types. However, western blots of nuclear extracts consistently showed that full-length TFII-I protein is present at higher levels in K562 than it is in MEL nuclear extracts. The reason for the lower amount of TFII-I in MEL nuclear extracts may be due to post-transcriptional mechanisms.
Our data led us to formulate a hypothesis on the function of proteins interacting with the β-globin downstream promoter during development (Fig. 6). We suggest that TFII-D, NF-E2, USF1 and USF2 interact with the globin promoter in adult erythroid cells and provide a platform for the efficient recruitment of transcription complexes (13,14,32,43; this study). The functional role of NF-E2 in this process remains to be established. Although our data show that mutation of the putative NF-E2-binding site in the downstream promoter has no effect on in vitro transcription of the β-globin gene, NF-E2 could still play a role in the context of the whole locus in vivo. We recently demonstrated that NF-E2-selected crosslinked chromatin from MEL cells revealed a footprint over a partial MARE-binding site in the mouse β-globin downstream promoter region (32). In addition, Johnson et al. (43) demonstrated that MEL cells lacking NF-E2 show a reduction in Pol II recruitment to the murine β-globin gene in vivo. The mechanisms of transcription complex recruitment are likely to be similar between the mouse and human locus, so it is possible that NF-E2 also plays a role in recruiting Pol II to the human β-globin gene. Consistent with this hypothesis is our observation that NF-E2 is capable of interacting with the MARE-like sequence of the human β-downstream promoter in vitro (Fig. 2D).
Figure 6.
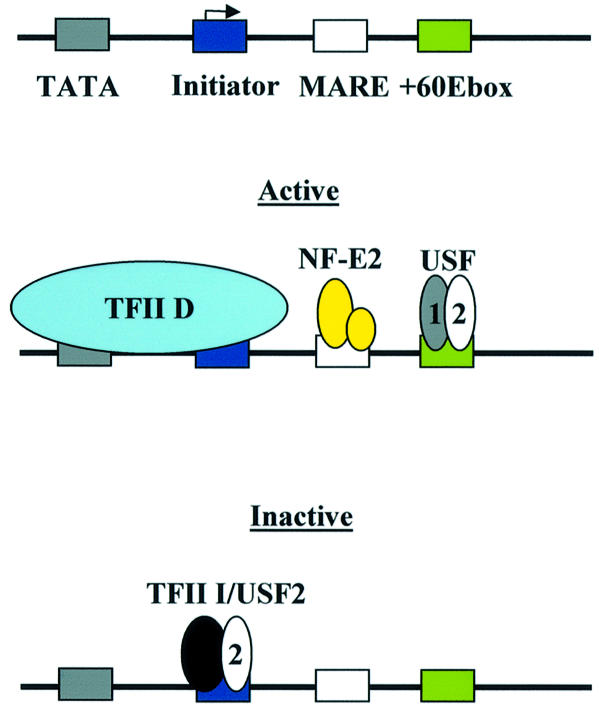
Summary and model of protein–DNA interactions at the human β-globin promoter. The human β-globin promoter consists of a TATA-box and an initiator. These sequences are bound by the TFII-D complex in cells expressing the β-globin gene (active) (13,14). Additional sequences required for the formation of active transcription complexes are MARE and E-box elements located in the downstream promoter region. These sequences are bound by NF-E2 (p45 and p18) and USF, respectively. It is proposed that in cells not expressing the β-globin gene (inactive), TFII-D is not bound and the initiator sequence is occupied by protein complexes consisting of TFII-I and USF2.
The interaction of TFII-I and USF2 with the β-globin promoter in K562 cells could contribute to the repression of this gene at the embryonic stage of erythroid development by recruiting histone deacetylase activities. Other studies also implicate USF2 as a potential transcriptional repressor (44). However, in adult cells, USF2 and USF1 interact with the E-box at +60 and stimulate transcription. This conclusion is supported by our observation that in vitro transcription of the β-globin gene is inhibited by pre-incubating the protein extracts with USF2 antibodies. Antibodies against USF1 were not as efficient in these experiments, which could be explained by the possibility that either the USF1 epitope is not recognized by the antibody under transcription conditions or that USF2 is able to act as a homodimer via the +60 E-box.
The observation that USF proteins interact with a downstream promoter element to facilitate formation of active transcription complexes of the β-globin gene is interesting in light of the fact that Kadonaga and co-workers recently demonstrated that genes containing downstream promoter elements in Drosophila require the activity of NC2 (21). NC2 was originally characterized as a component of a co-activator fraction that was shown to be involved in mediating the function of USF (USA, USF stimulating activity) (45). It is thus possible that downstream promoter elements in Drosophila and higher eukaryotes are regulated by similar mechanisms.
There are known mutations in the human population leading to mild thalassemias in the region of the NF-E2-binding site, one at +22 and another at +33, underscoring the importance of this element in vivo (46). There are also known mutations in the +60 E-box (47). However, because this element is located within the coding region of the gene, mutations of this element also affect translation, stability or function of the protein. The phenotype in individuals with single nucleotide mutations in the +60 E-box is rather mild, possibly because the mutations did not completely abolish USF binding. However, one case was characterized by a deletion of one nucleotide in codon 5 and an insertion of one nucleotide between codons 2 and 3 (changing the E-box from CACCTG to CACTCTG) (48). This mutation eliminates the E-box and led to a more severe phenotype. The authors of this study hypothesized that the problem was a severe instability of the protein due to low abundance, which could in fact be the consequence of inefficient transcription.
In summary, our data provide evidence that E-box motifs located in the downstream promoter region of the β-globin gene significantly contribute to the formation of active transcription complexes and that HLH proteins differentially interact with the initiator and downstream promoter during erythroid development.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Gail Green for expert technical assistance and our colleagues Padraic Levings, Christof Dame and Thomas Yang (University of Florida) for stimulating discussions. We thank Emery Bresnick for providing us with detailed protocols for the ChIP assay and Tom Yang and members of the Yang laboratory, particularly Christine Mione and Sarah Rodriguez, for assistance with in vivo footprinting. We are grateful to Doug Engel (University of Michigan) and Mike Kilberg (University of Florida) for critically reading the manuscript. This work was supported by a research grant from the NIH (DK 52356) and by the Howard Hughes Medical Institute Research Resources Program (University of Florida) to J.B.
REFERENCES
- 1.Bulger M. and Groudine,M. (1999) Looping versus linking: toward a model for long-distance gene activation. Genes Dev., 13, 2465–2477. [DOI] [PubMed] [Google Scholar]
- 2.Higgs D. (1998) Do LCRs open chromatin domains? Cell, 95, 299–302. [DOI] [PubMed] [Google Scholar]
- 3.Engel J.D. and Tanimoto,K. (2000) Looping, linking and chromatin activity: new insights into β-globin locus regulation. Cell, 100, 499–502. [DOI] [PubMed] [Google Scholar]
- 4.Levings P.P. and Bungert,J. (2002) The human β-globin locus control region: a center of attraction. Eur. J. Biochem., 269, 1589–1599. [DOI] [PubMed] [Google Scholar]
- 5.Reik A., Telling,A., Zitnik,G., Cimbora,D., Epner,E. and Groudine,M. (1998) The locus control region is necessary for gene expression in the human β-globin locus but not for the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol., 18, 5992–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender M.A., Bulger,M., Close,J. and Groudine,M. (2000) Globin gene switching and DNaseI sensitivity of the endogenous β-globin locus in mice does not require the locus control region. Mol. Cell, 5, 387–393. [DOI] [PubMed] [Google Scholar]
- 7.Tanimoto K., Liu,Q., Bungert,J. and Engel,J.D. (1999) Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature, 398, 344–348. [DOI] [PubMed] [Google Scholar]
- 8.Miller I.J. and Bieker,J.J. (1993) A novel, erythroid cell-specific transcription factor that binds to the CACC element related to the kruppel family of nuclear proteins. Mol. Cell. Biol., 13, 2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuez B., Michalovich,D., Bygrave,A., Ploemacher,R. and Grosveld,F. (1995) Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature, 375, 316–318. [DOI] [PubMed] [Google Scholar]
- 10.Perkins A.C., Sharpe,A.H. and Orkin,S.H. (1995) Lethal β-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature, 375, 318–322. [DOI] [PubMed] [Google Scholar]
- 11.Tanimoto K., Liu,Q., Grosveld,F., Bungert,J. and Engel,J.D. (2000) Context-dependent EKLF responsiveness defines the developmental specificity of the human ε-globin gene in erythroid cells of YAC transgenic mice. Genes Dev., 14, 2778–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armstrong J.A., Bieker,J.J. and Emerson,B.M. (1998) A Swi/SNF-related chromatin-remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell, 95, 93–104. [DOI] [PubMed] [Google Scholar]
- 13.Lewis B.A., Kim,T.K. and Orkin,S.H. (2000) A downstream element in the human β-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc. Natl Acad. Sci. USA, 97, 7122–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis B.A. and Orkin,S.H. (1995) A functional initiator in the human β-globin promoter. J. Biol. Chem., 270, 28139–28144. [DOI] [PubMed] [Google Scholar]
- 15.Smale S.T., Jain,A., Kaufmann,J., Emami,K.H., Lo,K. and Garraway,I.P. (1998) The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harbor Symp. Quant. Biol., 63, 21–31. [DOI] [PubMed] [Google Scholar]
- 16.Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- 17.Roy A.L., Meisterernst,M., Pognonec,P. and Roeder,R.G. (1991) Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature, 354, 245–248. [DOI] [PubMed] [Google Scholar]
- 18.Du H., Roy,A.L. and Roeder,R.G. (1993) Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the AdML promoters. EMBO J., 12, 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bungert J., Kober,I., During,F. and Seifart,K.H. (1992) Transcription factor eUSF is an essential component of isolated transcription complexes on the duck histone H5 gene and it mediates the interaction of TFII-D with a TATA-deficient promoter. J. Mol. Biol., 223, 885–898. [DOI] [PubMed] [Google Scholar]
- 20.Kutach A.K. and Kadonaga,J.T. (2000) The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol., 20, 4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willy P.J., Kobayashi,R. and Kadonaga,J.T. (2000) A basal transcription factor that activates or represses transcription. Science, 290, 982–985. [DOI] [PubMed] [Google Scholar]
- 22.Leach K.M., Nightingale,K., Igarashi,K., Levings,P.P., Engel,J.D., Becker,P.B. and Bungert J. (2001) Reconstitution of human β-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol. Cell. Biol., 21, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung H.L., Kim,A.Y., Hong,W., Rakowski,C. and Blobel,G.A. (2001) Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem., 276, 10715–10721. [DOI] [PubMed] [Google Scholar]
- 24.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Globin Gene Server Website (1994) http://globin.cse.psu.edu, last accessed 20 August 2001. Pennsylvania State University.
- 26.Selden R.F., Howie,K.B., Rowe,M.E., Goodman,H.M. and Moore D.D. (1986) Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol. Cell. Biol., 6, 3173–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsberg E.C., Downs,K.M., Christensen,H.M., Im,H., Nuzzi,P.A. and Bresnick,E. (2000) Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA, 97, 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornstra I.K. and Yang,T.P. (1992) Multiple in vivo footprints are specific to the active allele of the X-linked human hypoxanthine phosphoribosyltransferase gene 5′region: implications for X chromosome inactivation. Mol. Cell. Biol., 6, 4122–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 30.Hornstra I.K. and Yang,T.P. (1993) In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal. Biochem., 213, 179–193. [DOI] [PubMed] [Google Scholar]
- 31.Sawado T., Igarashi,K. and Groudine,M. (2001) Activation of β-major globin gene transcription is associated with recruitment of NF-E2 to the β-globin LCR and gene promoter. Proc. Natl Acad. Sci. USA, 98, 10226–10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee-Kang S.H., Vieira,K. and Bungert,J. (2002) Combining chromatin immunoprecipitation and DNA footprinting: a novel method to analyze protein–DNA interactions in vivo. Nucleic Acids Res., 30, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy A.L., Du,H., Gregor,P.D., Novina,C.D., Martinez,E. and Roeder,R.G. (1997). Cloning of an INR- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J., 16, 7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wobbe R. and Struhl,K. (1990) Yeast and human TATA-binding proteins have nearly identical DNA-sequence requirements for transcription in vitro. Mol. Cell. Biol., 10, 3859–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart J.J. and Stargell,L.A. (2001) The stability of the TFIIA-TBP-DNA complex is dependent on the sequence of the TATAAA element. J. Biol. Chem., 276, 30078–30084. [DOI] [PubMed] [Google Scholar]
- 36.Coulson J.M., Edgson,J.L., Marshall-Jones,Z.V., Mulgrew,R., Quinn,J.P. and Woll,P.J. (2002) Upstream stimulatory factor activates the vasopressin promoter via multiple motifs, including a non-canonical E-box. Biochem. J., online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manzano-Winkler B., Novina,C.D. and Roy,A.L. (1996) TFII-I is required for transcription of the naturally TATA-less but initiator containing Vβ promoter. J. Biol. Chem., 271, 12076–12081. [DOI] [PubMed] [Google Scholar]
- 38.Goetz T.L., Lloyd,T.L. and Griswold,M.D. (1996) Role of E-box and initiator in the expression of the rat follicle-stimulating hormone receptor. J. Biol. Chem., 271, 33317–33324. [DOI] [PubMed] [Google Scholar]
- 39.Breen G. and Jordan,E.M. (2000) Upstream stimulatory factor 2 stimulates transcription through an initiator element in the mouse cytochrome c oxidase subunit Vβ promoter. Biochim. Biophys. Acta, 1517, 119–127. [DOI] [PubMed] [Google Scholar]
- 40.Clark M.P., Chow,C.-W., Rinaldo,J.E. and Chalkley,R. (1998) Multiple domains for initiator binding proteins TFII-I and YY 1 are present in the initiator and upstream regions of the rat XDH/XO TATA-less promoter. Nucleic Acids Res., 26, 2813–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh C.-H., Yet,S.-F., Layne,M.D., Watanabe,M., Hong,A.M., Perrella,M.A. and Lee,M.-E. (1999) Genomic cloning and promoter analysis of aortic preferentially expressed gene-1. J. Biol. Chem., 274, 14344–14351. [DOI] [PubMed] [Google Scholar]
- 42.Tussie-Luna M.I., Bayarsaihan,D., Seto,E., Ruddle,F.H. and Roy,A.L. (2002) Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc. Natl Acad. Sci. USA, 99, 12807–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson K., Christensen,H., Zhao,B. and Bresnick,E.H. (2001) Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell, 8, 465–471. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y.-H., Layne,M.D., Watanabe,M., Yet,S.-F. and Perrella,M.A. (2001) Upstream stimulatory factors regulate aortic preferentially expressed gene-1 expression in vascular smooth muscle cells. J. Biol. Chem., 276, 47658–47663. [DOI] [PubMed] [Google Scholar]
- 45.Meisterernst M., Roy,A.L., Lieu,H.M. and Roeder,R.G. (1991) Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell, 66, 981–993. [DOI] [PubMed] [Google Scholar]
- 46.Ho P.J., Sloane-Stanley,J., Athanassiadou,A., Wood,W.G. and Thein,S.L. (1999) An in vitro system for expression analysis of mutations of the β-globin gene: validation and application to two mutations in the 5′ UTR. Br. J. Haematol., 106, 938–947. [DOI] [PubMed] [Google Scholar]
- 47.Higgs D.R., Thein,S.L. and Wood,W.G. (2001) The biology of the thalassaemia syndromes. In Weatherall,D.J. and Clegg,J.B. (eds), The Thalassaemia Syndromes, 4th Edn. Blackwell Science, Oxford, UK, pp. 133–191.
- 48.Keser I., Kayisli,O.G., Yesilipek,A., Ozes,O.N. and Luleci (2001) Hb Antalya [codons 3–5(Leu-Thr-Pro>Ser-Asp-Ser]: a new unstable variant leading to chronic microcytic anemia with high HbA2. Hemoglobin, 25, 369–373. [DOI] [PubMed] [Google Scholar]